Abstract
Phloretin hydrolase catalyzes the hydrolytic C-C cleavage of phloretin to phloroglucinol and 3-(4-hydroxyphenyl)propionic acid during flavonoid degradation in Eubacterium ramulus. The gene encoding the enzyme was cloned by screening a gene library for hydrolase activity. The insert of a clone conferring phloretin hydrolase activity was sequenced. Sequence analysis revealed an open reading frame of 822 bp (phy), a putative promoter region, and a terminating stem-loop structure. The deduced amino acid sequence of phy showed similarities to a putative protein of the 2,4-diacetylphloroglucinol biosynthetic operon from Pseudomonas fluorescens. The phloretin hydrolase was heterologously expressed in Escherichia coli and purified. The molecular mass of the native enzyme was approximately 55 kDa as determined by gel filtration. The results of sodium dodecyl sulfate-polyacrylamide gel electrophoresis and the deduced amino acid sequence of phy indicated molecular masses of 30 and 30.8 kDa, respectively, suggesting that the enzyme is a homodimer. The recombinant phloretin hydrolase catalyzed the hydrolysis of phloretin to equimolar amounts of phloroglucinol and 3-(4-hydroxyphenyl)propionic acid. The optimal temperature and pH of the catalyzed reaction mixture were 37°C and 7.0, respectively. The Km for phloretin was 13 ± 3 μM and the kcat was 10 ± 2 s−1. The enzyme did not transform phloretin-2′-glucoside (phloridzin), neohesperidin dihydrochalcone, 1,3-diphenyl-1,3-propandione, or trans-1,3-diphenyl-2,3-epoxy-propan-1-one. The catalytic activity of the phloretin hydrolase was reduced by N-bromosuccinimide, o-phenanthroline, N-ethylmaleimide, and CuCl2 to 3, 20, 35, and 85%, respectively. Phloroglucinol and 3-(4-hydroxyphenyl)propionic acid reduced the activity to 54 and 70%, respectively.
The dihydrochalcone phloretin (2′,4,4′,6′-tetrahydroxydihydrochalcone) (structure is shown in Fig. 1) has been described as an intermediate in the degradation of the flavone apigenin and the flavanone naringenin as catalyzed by human intestinal bacteria (23, 28). In addition, glycosides of phloretin are present in apples and derived products (15). Flavones, flavanones, and dihydrochalcones are subclasses of flavonoids, a group of secondary plant metabolites ingested in considerable amounts with food. Flavonoids are reported to have effects on different stages of cancerogenesis, the immune system, and hemostasis in animals (16). Owing to their antioxidant properties, they are able to scavenge free oxygen radicals and thereby prevent lipid peroxidation (21). Because of these interesting features of flavonoids, research has been carried out on their bioavailability in the human body (22). It has been known for several decades that the microbiota of the human gut contributes to the conversion of ingested flavonoids (8, 19).
FIG. 1.
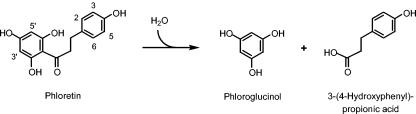
Hydrolysis of phloretin catalyzed by phloretin hydrolase.
Eubacterium ramulus, which is present in the human gut (29), is able to degrade flavonoids of various subclasses (23). The organism is a gram-positive, anaerobic, nonsporulating, chain-forming rod which was first described by Moore and Holdeman in 1974 (18) and reisolated from human feces by Schneider et al. (24). Its ability to degrade flavonoids has been extensively studied (2, 23-25). The flavone apigenin is presumably reduced to the flavanone naringenin in a first step which is followed by an isomerization to the corresponding chalcone structure. The chalcone is reduced to the dihydrochalcone phloretin, which is subsequently hydrolyzed to phloroglucinol and 3-(4-hydroxyphenyl)propionic acid (23). Although the degradation pathway of flavones by E. ramulus has successfully been elucidated, the enzymes involved have so far escaped purification and characterization.
Phloretin hydrolases (EC 3.7.1.4) catalyzing the formation of phloroglucinol and 3-(4-hydroxyphenyl)propionic acid (Fig. 1) have been purified from the saprobe Erwinia herbicola (5) and the mycelial felts of Aspergillus niger (17). The hydrolytic cleavage of a carbon-carbon bond adjacent to a carbonyl as catalyzed by this enzyme is a rare kind of reaction and is noteworthy from an enzymological point of view (7). Here we describe the cloning and sequencing of the encoding gene and the subsequent purification and characterization of the recombinant phloretin hydrolase of E. ramulus expressed in Escherichia coli.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
For cultivation of Eubacterium ramulus (wK1), which was previously isolated by Schneider et al. (24), the anoxic techniques applied were essentially those of Hungate (12) and Bryant (3). Cultures were grown under strictly anoxic conditions in 16-ml tubes fitted with butyl rubber stoppers and screw caps. The tubes contained 10 ml of standard medium for growth of E. ramulus. One liter of standard medium consisted of 9 g of tryptically digested peptone from meat, 1 g of proteose peptone, 3 g of meat extract, 4 g of yeast extract, 6 g of glucose, 3 g of NaCl, 2 g of Na2HPO4, 0.5 ml of Tween 80, 0.25 g of cystine, 0.25 g of cysteine, 0.1 g of MgSO4 · 7H2O, 5 mg of FeSO4 · 7H2O, and 3.4 mg of MnSO4 · 2H2O and was adjusted to pH 7.0. E. coli DH5α (9) was cultivated on standard I agar (Merck, Darmstadt, Germany) plates under oxic or anoxic conditions. E. coli DH5α(pPH3) was cultivated on standard I agar plates supplemented with ampicillin (100 μg ml−1). All cultures were incubated overnight at 37°C.
Preparation of cell extracts.
E. coli DH5α(pPH3) cultures were grown overnight in standard I medium supplemented with ampicillin (100 mg liter−1). The cell extracts were prepared under oxic conditions at 4°C. The cells were centrifuged (10,000 × g, 20 min, 4°C) and washed twice with 50 mM potassium phosphate buffer (pH 7.0). Each gram of cell pellet was resuspended in approximately 3 ml of TE buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA). Following the addition of lysozyme to a final concentration of 0.2 mg ml−1, the cell suspension was incubated for 20 min at 37°C with gentle shaking. Cell extracts (approximately 17 mg of protein ml−1) were obtained by centrifugation (39,200 × g, 30 min, 4°C).
Enzyme and protein assays.
Phloretin hydrolase was assayed in 50 mM potassium phosphate buffer (pH 7.0) containing 60 μM phloretin (Roth, Karlsruhe, Germany) added from a 1.2 mM stock solution in dimethyl sulfoxide (DMSO). DMSO (final concentration, 5% [vol/vol]) reduced the phloretin hydrolase activity by approximately 15%. However, 5% DMSO was necessary to completely dissolve higher concentrations of phloretin. The reaction was started by the addition of cell extract or enriched or purified protein solution (between 5 and 335 μg of protein ml−1). The hydrolysis of phloretin was monitored by measuring the decrease in absorption at 288 nm in a Cary 1 UV-visible spectrophotometer (Varian, Darmstadt, Germany). The transformation of phloretin to 3-(4-hydroxyphenyl)propionic acid and phloroglucinol was confirmed by high-performance liquid chromatography (HPLC) analysis as follows: samples were taken immediately after starting the reaction as well as after 10, 20, and 30 min and mixed with 1 volume of methanol-H2O-acetic acid (50:45:5, vol/vol/vol) to stop the reaction. The mixture was centrifuged at 12,000 ×g for 5 min, and the supernatant was subjected to HPLC analysis. The Km value of the purified enzyme for phloretin was determined at concentrations ranging from 4 to 130 μM. The influence of temperature and pH on enzyme activity was tested at temperatures between 20 and 50°C and pH values between 5.4 and 8.0. Inhibitors (1 mM) were preincubated with the purified enzyme for 10 min at 4°C. Phloroglucinol and 3-(4-hydroxyphenyl)propionic acid were tested for their possible inhibitory effect on phloretin hydrolase at concentrations of 120 μM. The substrate preference of the enzyme was determined by testing 60 μM phloretin-2′-glucoside (Roth), neohesperidin dihydrochalcone (Evesa, Cádiz, Spain), 1,3-diphenyl-1,3-propandione, or trans-1,3-diphenyl-2,3-epoxy-propan-1-one (Fluka, Deisenhofen, Germany). All compounds were added from stock solutions in DMSO. The protein concentration was determined according to the method of Bradford (1) using the Bio-Rad (Richmond, Calif.) dye reagent and bovine serum albumin as the standard.
Detection of phloroglucinol.
The phloroglucinol concentration was determined according to the method of Chatterjee and Gibbins using the vanillin-HCl reagent (4).
HPLC.
Phloretin and 3-(4-hydroxyphenyl)propionic acid were determined by HPLC analysis according to the method of Braune et al. (2). Methanol and 2% aqueous acetic acid served as the mobile phase to form a gradient as follows: from 5 to 30% methanol in 20 min, from 30 to 50% methanol in 5 min, from 50 to 65% methanol in 5 min, 65% methanol maintained for 5 min, and from 65 to 100% methanol in 7 min. The flow rate was 0.8 ml min−1.
Cloning of the phloretin hydrolase gene (phy).
Genomic DNA was isolated from E. ramulus (wK1) using the InViSorb genomic DNA kit and following the protocol of the manufacturer (InViTek, Berlin, Germany). The DNA (∼10 μg) was digested with Sau3AI (4 U) for 4 min at 37°C. The reaction was stopped by heating to 65°C for 20 min. The DNA fragments obtained were ligated into the BamHI-digested dephosphorylated plasmid pUC18 (31). E. coli DH5α was transformed with this gene library by electroporation with a Gene Pulser (Bio-Rad) according to the manual of the manufacturer. Approximately 4,000 clones were screened for phloretin hydrolase activity using the fluorescence-quenching test with 25 mM (final concentration) phloretin as quencher for 0.5 mM 1,6-diphenyl-1,3,5-hexatriene (27). The plasmid DNA from two clones that were positive in the fluorescence-quenching test, E. coli DH5α(pPH1) and E. coli DH5α(pPH2), was isolated using the Wizard Minipreps DNA purification system (Promega, Madison, Wisc.). To analyze the cloned fragments, pPH1 and pPH2 were digested with the endonucleases BamHI, DraI, EcoRI, and HindIII. The insert size of both plasmids was 4.3 kb. The size of the insert of the plasmid pPH2 was reduced by using the HindIII restriction site. The resulting DNA fragment of 2.4 kb from the plasmid pPH2 was ligated into a HindIII-digested, dephosphorylated pUC18 plasmid. After electrotransformation, the resulting clone E. coli DH5α(pPH3) was tested for its ability to hydrolyze phloretin by using the fluorescence-quenching test (27) and HPLC analysis, respectively. The purified plasmid was sequenced (double-strand sequencing; AGOWA, Berlin, Germany).
Enzyme purification.
All purification steps were carried out under oxic conditions. The cell extract of E. coli DH5α(pPH3) containing the heterologously expressed enzyme phloretin hydrolase was prepared by lysozyme treatment as described above. Ammonium sulfate was added to the cell extract to achieve 55% saturation. Following centrifugation (10,000 × g, 10 min, 4°C) the ammonium sulfate concentration of the supernatant was increased to 60% saturation. The pellet was resuspended in 50 mM potassium phosphate buffer (pH 7.0) and loaded onto a DEAE Sephacel column (2.6 by 6 cm, DEAE Sephacel resin; Amersham Biosciences, Freiburg, Germany), which was equilibrated with buffer A (50 mM potassium phosphate [pH 7.0]), using the HiLoad fast-performance liquid chromatography system (Amersham Biosciences). The column was washed with 40 ml of buffer A at a flow rate of 2 ml min−1 followed by a 100-ml linear gradient of 0 to 1 M KCl in buffer A. Fractions of 2 ml were collected, and the fractions containing high phloretin hydrolase activity were pooled. The pooled fractions were concentrated with a Centriprep 10 cartridge (Amicon, Witten, Germany), and the salt concentration of the retentate was raised to 1 M ammonium sulfate. The solution was applied to a phenyl Sepharose column (HiLoad 16/10; Amersham Biosciences) and washed at a flow rate of 2 ml min−1 with 40 ml of 1 M ammonium sulfate in buffer A. The protein was eluted with a 60-ml linear gradient of 1 to 0 M ammonium sulfate in buffer A. The active fractions of 2 ml were pooled and concentrated using a Centriprep 10 cartridge (Amicon). In order to determine the molecular mass of the enzyme, the purified enzyme was loaded onto a Superdex 200 column (1.0 by 30 cm; Amersham Biosciences), which had previously been calibrated using an LMW gel filtration calibration kit (Amersham Biosciences). The proteins were eluted with 50 mM potassium phosphate buffer (pH 7) containing 100 mM KCl at a flow rate of 0.5 ml min−1. Fractions of 0.4 ml were collected.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out according to the method of Laemmli (14). Phosphorylase b (94 kDa), albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), trypsin inhibitor (20.1 kDa), and α-lactalbumin (14.4 kDa) (Amersham Biosciences) were used as standards. Proteins were stained with the Bio-Safe Coomassie G250 stain (Bio-Rad) according to the manual of the manufacturer.
Nucleotide sequence accession number.
The sequence encompassing the phy gene and the adjacent regions from E. ramulus has been submitted to GenBank under the accession number AF548616.
RESULTS
Gene sequence of phloretin hydrolase.
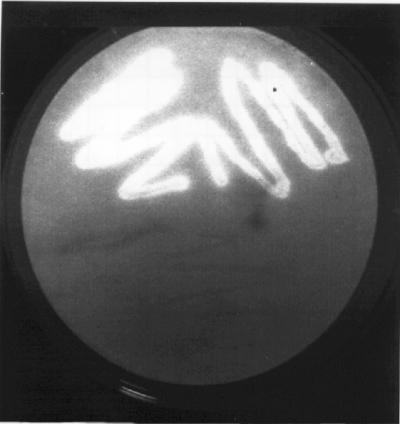
The specific activity of the phloretin hydrolase in cell extracts of E. ramulus was 0.0064 U mg of protein−1. The gene encoding this enzyme was identified by screening a gene library of E. ramulus in E. coli for phloretin hydrolase activity using the fluorescence-quenching test (27). Of the approximately 4,000 clones analyzed, two positive clones, E. coli DH5α(pPH1) and E. coli DH5α(pPH2), were detected and isolated. The ability of the two clones to hydrolyze phloretin was confirmed by the fluorescence-quenching test (Fig. 2) and HPLC analysis. The plasmids isolated from these clones were analyzed by restriction analysis (data not shown). The resulting restriction patterns were identical, indicating that both plasmids harbor the same fragment of chromosomal DNA inserted in the same orientation and, therefore, most likely the same phloretin hydrolase gene. However, the phloretin hydrolase activity of E. coli DH5α(pPH2) was higher than that of E. coli DH5α(pPH1). Therefore, E. coli DH5α(pPH2) was used for further analysis. The size of the cloned fragment in plasmid pPH2 was reduced, resulting in the subclone E. coli DH5α(pPH3). The sequence analysis of the 2,280-bp genomic insert in pPH3 revealed two open reading frames. Only open reading frames that were complete, that were >180 bp in length, and that started with ATG were taken into consideration. The smaller one, ORF1, encompasses 315 bp (nucleotides 270 to 584 of the insert). This open reading frame was able to encode a protein of 12,585 Da, whose sequence, however, showed no similarities to protein sequences deposited in databases. The second open reading frame, downstream of ORF1 and oriented in the same direction, consists of 822 bp (nucleotides 753 to 1574 of the insert) corresponding to a protein of 274 amino acids. Since the resulting molecular mass of 30,846 Da is in accordance with that observed for the purified phloretin hydrolase (30 kDa) (see below) in SDS-PAGE gels, this latter open reading frame, named phy, most probably encodes the phloretin hydrolase. A putative promoter region was identified by alignment with promoter sequences of two bacterial strains phylogenetically closely related to E. ramulus: Eubacterium sp. strain VPI 12708 and Clostridium sp. strain TO-931 (Fig. 3A). The aligned promoter regions of the bai (bile acid-inducible) operons were deduced from the transcription start sites, which had been determined by primer extension analysis (32). The alignment revealed a putative transcription start site 91 bases upstream of the translation start site of the E. ramulus phy gene. A putative terminating stem-loop structure was found 44 bases downstream of the stop codon of phy with a stem of 16 bp and a loop of 6 bases, followed by a poly(U) sequence (Fig. 3B). The promoter and terminator sequences identified suggest that the phloretin hydrolase gene is transcribed in a monocistronic form. Several databases were searched for sequences showing similarity to the deduced amino acid sequence of the phloretin hydrolase (Phy). The search revealed similarities of Phy to several putative proteins. Two of these were of interest, namely the ones derived from the open reading frame AF1466 from Archaeoglobus fulgidus (13) and phlG from Pseudomonas fluorescens (26). The alignment of Phy (amino acids 33 to 269) with the hypothetical protein derived from AF1466 (amino acids 14 to 250) showed 33% identical and 46% similar amino acids, whereas the alignment of Phy (amino acids 36 to 213) with the hypothetical protein deduced from phlG (amino acid residues 55 to 237) showed 24% identical and 41% similar amino acids. The conserved amino acids are primarily located in the central region of the aligned sequences (Fig. 4). While no function for the hypothetical protein of AF1466 has been proposed, PhlG is thought to be involved in the biosynthesis of 2,4-diacetylphloroglucinol since this gene is part of the 2,4-diacetylphloroglucinol operon (26).
FIG. 2.
Comparison of phloretin-degrading E. coli DH5α(pPH2) (top left) and E. coli DH5α(pPH1) (top right) with phloretin-nondegrading E. coli DH5α(pUC18) (bottom) subjected to the fluorescence-quenching test (27). A membrane, soaked in a mixture of 1,6-diphenyl-1,3,5-hexatriene and phloretin, on an agar plate was inoculated with the various bacteria.
FIG. 3.
(A) Alignment of the putative phy gene promoter region of E. ramulus (nucleotides 645 to 665; GenBank accession number AF548616) with the bai operon promoter regions of Eubacterium sp. strain VPI 12708 and Clostridium sp. strain TO-931 (32). Arrows denote transcription start sites, and conserved regions are shaded in grey. The underlined sequences represent the putative −10 regions. (B) Putative transcriptional terminator sequence downstream of the E. ramulus phy gene (nucleotides 1615 to 1669; GenBank accession number AF548616). The underlined bases represent the palindromic sequences probably forming a stem-loop structure followed by a poly(U) sequence.
FIG. 4.
Alignment of amino acid sequences of the phloretin hydrolase from E. ramulus (ErPhy; GenBank accession number AF548616), the conserved hypothetical protein deduced from the open reading frame AF1466 of A. fulgidus (AfChp; GenBank accession number AE001001.1 [13]), and the putative protein derived from phlG of the 2,4-diacetylphloroglucinol biosynthetic operon of P. fluorescens (PfPhlG; GenBank accession number AF207529 [26]). Amino acid residues which are identical in two of the sequences are shaded in grey, whereas amino acids identical in all aligned sequences are shaded in black. The conserved tryptophan motif is marked by asterisks.
Purification of the recombinant phloretin hydrolase.
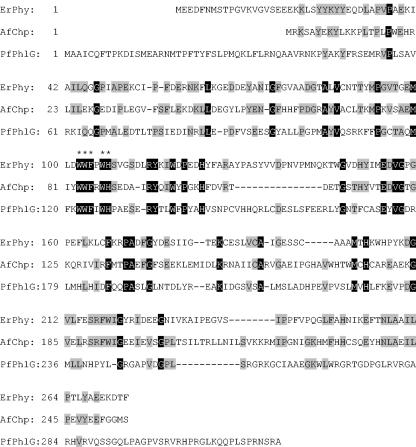
The phloretin hydrolase activity in extracts from E. coli DH5α(pPH3) was approximately 100-fold higher than that in cell extracts from E. ramulus (0.7 U mg of protein−1 versus 0.0064 U mg of protein−1). SDS-PAGE analysis of the cell extract of E. coli DH5α(pPH3) revealed a predominant protein band of approximately 30 kDa (Fig. 5), which is in close agreement with the molecular mass of 30.4 kDa deduced from the amino acid sequence of the phy gene. This predominant protein band was not detected in the cell extract from E. coli DH5α(pUC18). The heterologously expressed E. ramulus phloretin hydrolase was purified from E. coli DH5α(pPH3) by ammonium sulfate fractionation and column chromatography with an overall 13-fold enrichment and a recovery of 0.9% (Table 1). In the cell extracts, phloretin hydrolase amounted to 7.5% of total protein. Phenyl Sepharose chromatography was the most effective purification step. The resulting enzyme preparation was >95% homogeneous as judged by SDS-PAGE (Fig. 5).
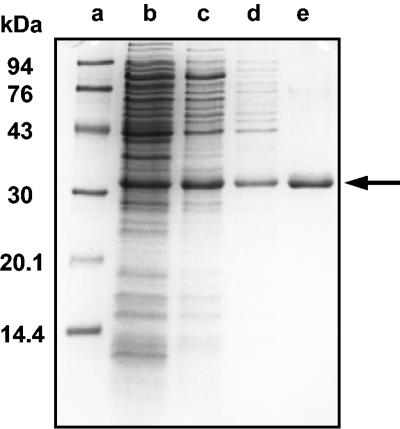
FIG. 5.
SDS-PAGE of steps in the purification of the recombinant phloretin hydrolase from E. ramulus. Lane a: molecular mass standards; lane b: cell extract (24 μg of protein); lane c: (NH4)2SO4 precipitate (55 to 60% saturation) (11 μg of protein); lane d: pooled DEAE Sephacel fractions (8 μg of protein); lane e: pooled phenyl Sepharose fractions (1.3 μg of protein). The arrow indicates phloretin hydrolase. The gel was stained with Bio-Safe Coomassie G250 stain.
TABLE 1.
Purification of phloretin hydrolase from E. coli DH5α(pPH3)
| Purification step | Total activity (U) | Total protein (mg) | Sp act, (U mg of protein−1) | Purification factor (fold) | Recovery (%) |
|---|---|---|---|---|---|
| Cell extract | 166.0 | 252 | 0.7 | 1 | 100 |
| Dissolved (NH4)2SO4 precipitate (55-60% saturation) | 25.7 | 24 | 1.1 | 1.7 | 15.5 |
| DEAE Sephacel | 3.3 | 1.6 | 2.1 | 3.2 | 2.0 |
| Phenyl Sepharose | 1.5 | 0.17 | 8.7 | 13.2 | 0.9 |
Properties of the recombinant phloretin hydrolase.
The apparent native molecular mass of the recombinant phloretin hydrolase was approximately 55 kDa, as determined by gel filtration. The subunit size of 30.8 kDa suggests that the native recombinant phloretin hydrolase consists of two identical polypeptides. The hydrophobicity plot analysis indicates that the enzyme is located in the cytoplasm, since no hydrophobic regions of notable size were found. The enzyme catalyzed the hydrolysis of phloretin to equimolar amounts of phloroglucinol and 3-(4-hydroxyphenyl)propionic acid. While phloroglucinol was determined colorimetrically, 3-(4-hydroxyphenyl)propionic acid was identified by its retention time and its UV spectrum in HPLC-UV analysis. Maximal phloretin hydrolase activity was observed at a temperature of 37°C and a pH of 7.0. The Km value for phloretin was 13 ± 3 μM, and the kcat was 10 ± 2 s−1. The phloretin hydrolase transformed neither the glycosides phloretin-2′-glucoside (phloridzin) and neohesperidin dihydrochalcone nor compounds showing a structure similar to phloretin, such as 1,3-diphenyl-1,3-propandione and trans-1,3-diphenyl-2,3-epoxy-propan-1-one. The phloretin hydrolase was found to be inhibited strongly by N-bromosuccinimide, which oxidizes tryptophan residues. At a concentration of 1 mM, the enzyme activity was reduced to 3% by this compound. The metal-ion-chelating agent o-phenanthroline (1 mM) considerably reduced the activity of the enzyme to 20% of the control level. The sulfhydryl reagent N-ethylmaleimide reduced the phloretin hydrolase activity at a concentration of 1 mM to 35%, while CuCl2 had little inhibitory effect on the enzyme, reducing its activity only to 85%. At a concentration of 120 μM, the products of phloretin hydrolysis, phloroglucinol and 3-(4-hydroxyphenyl)propionic acid, reduced the activity to 54 and 70%, respectively. MgCl2 or CaCl2 (both 1 mM) did not affect the phloretin hydrolase activity.
DISCUSSION
The phloretin hydrolase (Phy) from E. ramulus was shown to hydrolyze phloretin to 3-(4-hydroxyphenyl)propionic acid and phloroglucinol. This reaction is part of the apigenin degradation pathway (23). The gene encoding Phy was cloned to allow for the characterization of the underlying reaction and to retrieve information on the organization of the genes involved in anaerobic bacterial flavonoid degradation. The resulting recombinant enzyme was purified and initially characterized in order to gain insight into the catalytic mechanism, which is of interest because the hydrolytic cleavage of a C-C bond adjacent to a carbonyl group is a rare kind of reaction in biochemistry (7).
So far, phloretin hydrolases have been purified from the saprobe E. herbicola (5) and from the fungus A. niger (17). However, the gene sequence encoding Phy from E. ramulus is the first one to be determined for a phloretin hydrolase gene. Moreover, phy represents the first gene that encodes an enzyme involved in the degradation of flavonoids by bacteria. Database searches revealed similarity of the phloretin hydrolase from E. ramulus with several putative proteins. The amino acid sequence of one of these hypothetical proteins, PhlG, has been deduced from phlG, which is part of the 2,4-diacetylphloroglucinol biosynthetic operon of P. fluorescens (26). However, PhlG does not appear to be involved in the biosynthesis of 2,4-diacetylphloroglucinol. In view of the sequence similarity of Phy and PhlG, it may be hypothesized that PhlG plays a role in the degradation of 2,4-diacetylphloroglucinol via monoacetylphloroglucinol (26). The hydrolysis of the C-acetyl bonds to form monoacetylphloroglucinol or even phloroglucinol from 2,4-diacetylphloroglucinol represents a reaction homologous to that catalyzed by the phloretin hydrolase. In addition, the substrates and products involved in both reactions are all derivatives of phloroglucinol, which could explain the sequence similarity observed. The sequences of Phy and PhlG showed no similarities to any other protein sequences with known functions deposited in databases. No conserved domains or binding sites were found in the sequence of the phloretin hydrolase of E. ramulus. C-C-cleaving hydrolases catalyzing reactions similar to the reaction carried out by phloretin hydrolase belong to the α/β-hydrolase fold superfamily (11). These enzymes contain the serine catalytic triad serine-aspartate-histidine (20). At the serine position the conserved sequence motif GXSXG has been observed (6, 10). Since the phloretin hydrolase sequence of E. ramulus has no sequence similarity to other C-C-cleaving hydrolases and also does not contain the conserved sequence motif GXSXG, it can be assumed that this enzyme belongs to another enzyme family.
Compared to the purified phloretin hydrolases from E. herbicola (213 U mg−1) (5) and A. niger (3.6 U mg−1) (17), the specific activity of the E. ramulus enzyme purified from E. coli (8.7 U mg−1) was similar to that of the fungal enzyme. However, the affinity for phloretin of the recombinant phloretin hydrolase from E. ramulus with a Km of 13 μM was higher than that of the corresponding E. herbicola enzyme (Km, 38 μM) (5) or of the phloretin hydrolase from A. niger (Km, 0.3 to 0.4 mM) (17). The activity of the phloretin hydrolase from E. ramulus was reduced by CuCl2 as reported for the corresponding enzymes from A. niger (17) and E. herbicola (5). The sensitivity of the E. ramulus enzyme to N-ethylmaleimide suggests the involvement of cysteine in the catalysis. However, although cysteine residues occur in the sequence, these are not conserved in the sequences aligned in Fig. 4. The almost complete inhibition of the recombinant phloretin hydrolase of E. ramulus by N-bromosuccinimide indicates that tryptophan may play a role in catalysis. Three tryptophan residues were found in all three of the aligned sequences that were part of the conserved motif WWFXWH (Fig. 4) which could be involved in substrate binding. The strong inhibition of phloretin hydrolase by the metal-ion-chelating agent o-phenanthroline indicates that metal ions, such as iron, may be involved in catalysis. In contrast, the phloretin hydrolase of A. niger is hardly affected by o-phenanthroline (17). The products of phloretin hydrolysis, phloroglucinol and 3-(4-hydroxyphenyl)propionic acid, reduced the activity of the E. ramulus phloretin hydrolase, while the enzyme from E. herbicola was not inhibited by these compounds even at higher concentrations (5).
The inability of the E. ramulus enzyme to hydrolyze phloretin-2′-glucoside is in accordance with that of the enzyme from E. herbicola (5), indicating that the cleavage of the glycosidic bond is a prerequisite for further degradation by the phloretin hydrolases of these two organisms. In contrast, the phloretin hydrolase from A. niger was reported to degrade glycosides (17). The fungal phloretin hydrolase generally shows a broad substrate spectrum, and hydroxylated acetophenone derivatives and 2′,4,4′-trihydroxydihydrochalcone are even more efficiently converted than phloretin (17). In contrast, the phloretin-related substances 1,3-diphenyl-1,3-propandione and trans-1,3-diphenyl-2,3-epoxy-propan-1-one were not degraded by the recombinant phloretin hydrolase of E. ramulus. These compounds have no hydroxyl groups at all, indicating that those substituents attached to the phenyl rings play a crucial role in catalysis. This fact is not surprising since even the number and position of the hydroxyl groups were shown to have an influence on the accessibility of the compounds. Small structural changes in the substrate phloretin result in greatly reduced bacterial degradation in rat intestine (30).
Based on cloning and overexpression of the gene encoding the phloretin hydrolase from E. ramulus presented herein, further research will be carried out to unravel the mechanism underlying the hydrolysis of phloretin. The knowledge of the gene sequence of this enzyme involved in flavonoid degradation by gut bacteria allows the assignment of phloretin-cleaving activities to organisms with similar gene sequences.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (grant number INK 26/B1-1).
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Braune, A., M. Gütschow, W. Engst, and M. Blaut. 2001. Degradation of quercetin and luteolin by Eubacterium ramulus. Appl. Environ. Microbiol. 67:5558-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant, M. P. 1972. Commentary on the Hungate technique for culture of anaerobic bacteria. Am. J. Clin. Nutr. 25:1324-1328. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee, A. K., and L. N. Gibbins. 1969. Determination of phloroglucinol in the presence of phloretin using a modified vanillin reagent. Anal. Biochem. 30:436-442. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee, A. K., and L. N. Gibbins. 1969. Metabolism of phloridzin by Erwinia herbicola: nature of the degradation products, and the purification and properties of phloretin hydrolase. J. Bacteriol. 100:594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz, E., and K. N. Timmis. 1995. Identification of functional residues in a 2-hydroxymuconic semialdehyde hydrolase. A new member of the α/β hydrolase-fold family of enzymes which cleaves carbon-carbon bonds. J. Biol. Chem. 270:6403-6411. [DOI] [PubMed] [Google Scholar]
- 7.Fleming, S. M., T. A. Robertson, G. J. Langley, and T. D. Bugg. 2000. Catalytic mechanism of a C-C hydrolase enzyme: evidence for a gem-diol intermediate, not an acyl enzyme. Biochemistry 39:1522-1531. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths, L. A., and A. Barrow. 1972. Metabolism of flavonoid compounds in germ-free rats. Biochem. J. 130:1161-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 10.Hofer, B., L. D. Eltis, D. N. Dowling, and K. N. Timmis. 1993. Genetic analysis of a Pseudomonas locus encoding a pathway for biphenyl/polychlorinated biphenyl degradation. Gene 130:47-55. [DOI] [PubMed] [Google Scholar]
- 11.Hotelier, T., L. Renault, X. Cousin, V. Negre, P. Marchot, and A. Chatonnet. 2004. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins. Nucleic Acids Res. 32:D145-D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hungate, R. E. 1969. A roll tube method for cultivation of strict anaerobes, p. 117-132. In J. R. Norris and D. W. Ribbons (ed.), Methods in microbiology. Academic Press, New York, N.Y.
- 13.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, S. Peterson, C. I. Reich, L. K. McNell, J. H. Badger, A. Glodek, L. Zhou, R. Overbeek, J. D. Gocayne, J. F. Weidmen, L. McDonald, T. Utterback, M. D. Cotton, T. Spriggs, P. Artiach, B. P. Kaine, S. M. Sykes, P. W. Sadow, K. P. D'Andrea, C. Bowman, C. Fujii, S. A. Garland, T. M. Mason, G. J. Olsen, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Lu, Y., and Y. Foo. 1997. Identification and quantification of major polyphenols on apple pomace. Food Chem. 59:187-194. [Google Scholar]
- 16.Middleton, E., and C. Kandaswami. 1994. The impact of plant flavonoids on mammalian biology: implications for immunity, inflammation and cancer, p. 619-652. In J. B. Harborne (ed.), The flavonoids: advances in research since 1986. Chapman and Hall, London, United Kingdom.
- 17.Minamikawa, T., N. P. Jayasankar, B. A. Bohm, I. E. Taylor, and G. H. Towers. 1970. An inducible hydrolase from Aspergillus niger, acting on carbon-carbon bonds, for phlorrhizin and other C-acylated phenols. Biochem. J. 116:889-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore, W. E., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakagawa, Y., M. R. Shetlar, and S. H. Wender. 1965. Urinary products from quercetin in neomycin-treated rats. Biochim. Biophys. Acta 97:233-241. [DOI] [PubMed] [Google Scholar]
- 20.Ollis, D. L., E. Cheah, M. Cygler, B. Dijkstra, F. Frolow, S. M. Franken, M. Harel, S. J. Remington, I. Silman, and J. Schrag. 1992. The α/β hydrolase fold. Protein Eng. 5:197-211. [DOI] [PubMed] [Google Scholar]
- 21.Rice-Evans, C. 2001. Flavonoid antioxidants. Curr. Med. Chem. 8:797-807. [DOI] [PubMed] [Google Scholar]
- 22.Scalbert, A., and G. Williamson. 2000. Dietary intake and bioavailability of polyphenols. J. Nutr. 130:2073-2085. [DOI] [PubMed] [Google Scholar]
- 23.Schneider, H., and M. Blaut. 2000. Anaerobic degradation of flavonoids by Eubacterium ramulus. Arch. Microbiol. 173:71-75. [DOI] [PubMed] [Google Scholar]
- 24.Schneider, H., A. Schwiertz, M. D. Collins, and M. Blaut. 1999. Anaerobic transformation of quercetin-3-glucoside by bacteria from the human intestinal tract. Arch. Microbiol. 171:81-91. [DOI] [PubMed] [Google Scholar]
- 25.Schneider, H., R. Simmering, L. Hartmann, H. Pforte, and M. Blaut. 2000. Degradation of quercetin-3-glucoside in gnotobiotic rats associated with human intestinal bacteria. J. Appl. Microbiol. 89:1027-1037. [DOI] [PubMed] [Google Scholar]
- 26.Schnider-Keel, U., A. Seematter, M. Maurhofer, C. Blumer, B. Duffy, C. Gigot-Bonnefoy, C. Reimmann, R. Notz, G. Defago, D. Haas, and C. Keel. 2000. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 182:1215-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoefer, L., A. Braune, and M. Blaut. 2001. A fluorescence quenching test for the detection of flavonoid transformation. FEMS Microbiol. Lett. 204:277-280. [DOI] [PubMed] [Google Scholar]
- 28.Schoefer, L., R. Mohan, A. Schwiertz, A. Braune, and M. Blaut. 2003. Anaerobic degradation of flavonoids by Clostridium orbiscindens. Appl. Environ. Microbiol. 69:5849-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmering, R., B. Kleessen, and M. Blaut. 1999. Quantification of the flavonoid-degrading bacterium Eubacterium ramulus in human fecal samples with a species-specific oligonucleotide hybridization probe. Appl. Environ. Microbiol. 65:3705-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skjevrak, I., E. Solheim, and R. R. Scheline. 1986. Dihydrochalcone metabolism in the rat: trihydroxylated derivatives related to phloretin. Xenobiotica 16:35-45. [DOI] [PubMed] [Google Scholar]
- 31.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 32.Wells, J. E., and P. B. Hylemon. 2000. Identification and characterization of a bile acid 7α-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7α-dehydroxylating strain isolated from human feces. Appl. Environ. Microbiol. 66:1107-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]