Abstract
Thermotoga maritima, a fermentative, anaerobic, hyperthermophilic bacterium, was found to attach to bioreactor glass walls, nylon mesh, and polycarbonate filters during chemostat cultivation on maltose-based media at 80°C. A whole-genome cDNA microarray was used to examine differential expression patterns between biofilm and planktonic populations. Mixed-model statistical analysis revealed differential expression (twofold or more) of 114 open reading frames in sessile cells (6% of the genome), over a third of which were initially annotated as hypothetical proteins in the T. maritima genome. Among the previously annotated genes in the T. maritima genome, which showed expression changes during biofilm growth, were several that corresponded to biofilm formation genes identified in mesophilic bacteria (i.e., Pseudomonas species, Escherichia coli, and Staphylococcus epidermidis). Most notably, T. maritima biofilm-bound cells exhibited increased transcription of genes involved in iron and sulfur transport, as well as in biosynthesis of cysteine, thiamine, NAD, and isoprenoid side chains of quinones. These findings were all consistent with the up-regulation of iron-sulfur cluster assembly and repair functions in biofilm cells. Significant up-regulation of several β-specific glycosidases was also noted in biofilm cells, despite the fact that maltose was the primary carbon source fed to the chemostat. The reasons for increased β-glycosidase levels are unclear but are likely related to the processing of biofilm-based polysaccharides. In addition to revealing insights into the phenotype of sessile T. maritima communities, the methodology developed here can be extended to study other anaerobic biofilm formation processes as well as to examine aspects of microbial ecology in hydrothermal environments.
Mesophilic bacteria in natural and pathogenic environments are often associated with biofilms. This localization facilitates interactions and coexistence in an optimized microenvironment while at the same time limiting the adverse consequences of competition and selectivity (12). The establishment of a sessile community of cells encapsulated by a polysaccharide matrix on a surface involves a complex series of steps: initial attachment, production of exopolysaccharides, early biofilm development, mature biofilm formation, and detachment of cells, perhaps as communities (49, 96). These steps have been investigated for several mesophilic bacteria, including Pseudomonas aeruginosa (20, 105), Bacillus cereus (71), Vibrio cholerae (127), and a Streptococcus sp. (100). Biofilm formation apparently requires expression of a distinct set of genes that differentiate sessile from planktonic cells, including those related to chemotaxis, motility, exopolysaccharide biosynthesis, and stress response (88). However, this set of genes may only comprise about 1% of the total genome such that differences between planktonic and sessile cells may be subtle (28, 121). This is not surprising, since biofilm-bound populations likely include newly recruited cells that have a planktonic phenotype as well as cells that represent various stages of biofilm formation (90, 119). Furthermore, even interactions between planktonic cells and surfaces can affect gene expression. For B. cereus, planktonic cells grown in the presence of biofilm substratum (glass wool) shared common differentially expressed genes with biofilm-bound cells (71). Thus, composite planktonic and sessile communities likely contain a continuous distribution of distinct phenotypes that have temporal and spatial signatures (96).
The capacity to form biofilms is not limited to aerobic, mesophilic bacteria. Biofilms are also evident in high-temperature environments, such as terrestrial geothermal settings and hydrothermal vents (83). Several anaerobic hyperthermophiles (microorganisms with optimal growth temperatures at or above 80°C) have been shown to produce exopolysaccharides (3, 31, 67). These exopolysaccharides form the basis for biofilms, which have been observed in pure cultures of Archaeoglobus fulgidus (52), Thermotoga maritima (84), and Thermococcus litoralis (87), as well as in cocultures of T. maritima and Methanococcus jannaschii (63, 84). Biofilm formation was induced by elevated pH, decreased and increased growth temperature, high salt, and exposure to UV light, oxygen, or antibiotic in A. fulgidus (52) and by ammonium chloride in T. litoralis (87).
A key challenge that must be addressed to further explore biofilm formation processes in hyperthermophilic anaerobes is the experimental complexity associated with the growth of these organisms. This problem was addressed with a high-temperature, anaerobic chemostat that was used to generate biofilms in cultures of T. maritima that could be sampled and examined for differential gene expression patterns by whole-genome cDNA microarrays comparing planktonic to sessile cells. Transcriptional patterns related to the biofilm phenotype in this hyperthermophilic microorganism were then determined and compared to biofilm formation in less thermophilic microorganisms. Such information concerning biofilm formation mechanisms in hyperthermophiles is needed to develop a better understanding of the microbial ecology in hydrothermal habitats, particularly in regard to surface colonization.
MATERIALS AND METHODS
Microorganism and growth conditions.
T. maritima (DSM 3109) was grown anaerobically on sea salts medium (SSM) containing 40-g/liter sea salts (Sigma Chemical, St. Louis, Mo.), 1-g/liter yeast extract (Fisher Scientific, Pittsburgh, Pa.), 3.1-g/liter PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer (Sigma), 2-g/liter tryptone, 2-ml/liter 0.05% resazurin, and 10-ml/liter 10× Wolin minerals (117). Growth medium was adjusted to pH 6.8 with KOH (Fisher Scientific) and autoclaved prior to use. Batch cultures (50 ml) were inoculated under N2 (high-purity nitrogen; National Welders, Raleigh, N.C.) headspace, as previously described (77, 85), and were grown at 80°C for 8 to 10 h in oil baths. Maltose (Sigma Chemical, St. Louis) was added to SSM (final concentration, 5 g/liter) as a carbon source prior to inoculation. Continuous cultivation of T. maritima was performed in a 2-liter five-neck, round-bottom flask, as previously described (77, 85). A 50-ml batch culture was used to inoculate 1 liter of SSM supplemented with 5-g/liter maltose in the flask. This seed culture was grown at 80°C for 8.5 h under continuous nitrogen sparging, after which medium was fed at a dilution rate of 0.25 h−1. Medium for continuous cultivation was prepared in 9-liter batches at a 1.2× concentration as mentioned above, to which 1 liter of a filter-sterilized maltose solution (50 g) was added immediately after autoclaving. The pH of the culture was continuously monitored with a Chemcadet pH controller (Cole Parmer, Vernon Hills, Ill.) and adjusted by the addition of 1 M NaOH. Temperature was controlled with a Digi-Sense controller (Cole-Parmer, Vernon Hills, Ill.) such that variations were typically ±0.8°C and verified by a mercury glass thermometer inserted into the culture. Steady-state conditions were monitored by following cell counts (see below) and optical densities at 600 nm. All planktonic cell samples were collected from the outlet line into sterile pyrex bottles (see below), from which 1 ml of cells was fixed in glutaraldehyde for cell counting.
Biofilm substrata and collection.
Nylon mesh (Sefar America, Hamden, Conn.) and polycarbonate filters (Poretics 0.22-μm pore diameter; Fisher Scientific, Pittsburgh, Pa.) were used as substrata for biofilm formation. Twelve squares of mesh (13.3 by 9.8 cm) were cut, rolled tightly, and tied with polycarbonate string. Three rolled mesh squares were tied to one another at the ends. Polycarbonate filters were tied to the center of each set of mesh squares to be used for biofilm imaging, while the mesh itself provided biomass for RNA samples within the biofilm. The mesh and loose polycarbonate filters were placed in the reactor and autoclaved prior to startup. The strings were suspended in the growing culture until the sample was collected, whereby they were pulled quickly through one of the five necks of the reactor. The mesh samples and polycarbonate filters were rinsed twice in sterile medium while on ice to remove loosely adhered planktonic cells. Polycarbonate filters were removed from each tube and placed in 2.5% glutaraldehyde (Sigma) to fix the biofilm cells for examination and imaging under the microscope (see below). The mesh squares were separated from the strings and submerged in 50-ml conical tubes containing 300 mM NaCl (Fisher Scientific). The conical tubes were vortexed vigorously (4°C) to remove biofilm material from the mesh, after which the mesh was removed and the resulting suspension was centrifuged (10,000 × g, 20 min, 4°C) to pellet the biofilm cell material. Rinse media and 300 mM NaCl were chilled at 4°C prior to use. RNA was extracted as described below.
Imaging and microscopy methods.
Epifluorescent micrographs were taken with a SPOT digital camera (Southern Micro Instruments, Atlanta, Ga.) attached to a Nikon (Labophot-2) microscope (Southern Micro Instruments) with 100× oil immersion lens. Planktonic cell suspensions were fixed in 2.5% glutaraldehyde and stained with acridine orange (1 g/liter; Fisher Scientific) to determine cell densities (77, 85). Biofilm cells on polycarbonate filters were fixed as described above, stained in acridine orange (1 g/liter), and dried briefly under vacuum prior to imaging. A scanning electron microscope (JEOL JSM-35CF Microscope, North Carolina State University, Department of Veterinary Medicine) was also used to image biofilm cells on polycarbonate filters. Filters were fixed in 2.5% glutaraldehyde and critically point dried in CO2. Images of reactor walls of the continuous culture were also taken regularly with a Nikon Coolpix 950 digital camera.
RNA sample collection.
Approximately 200-ml samples of planktonic cells were withdrawn through culture outlet (77) into sterile Pyrex bottles on ice. Fifty milliliters of cells was collected prior to sampling to eliminate existing fluid in the lines. The 200-ml samples were used for RNA extractions and processed as described previously (14). Biofilm pellets were rinsed once after being extracted in 300 mM NaCl (4°C) and used immediately for RNA isolation (i.e., RNase-inhibiting buffers were added directly after rinsing step). Total RNA from planktonic cells was extracted from samples from three different time points (Fig. 1) during the steady-state operation; approximately 1 mg of RNA was obtained from each sampling time from which 100 μg was pooled. Similarly, total RNA from the three rolls of mesh from one point in the middle of the chemostat run was pooled to produce a biofilm sample. The cDNA generated from the planktonic and biofilm cells was hybridized to glass slides containing the targeted microarray, scanned, and analyzed, as described previously (15).
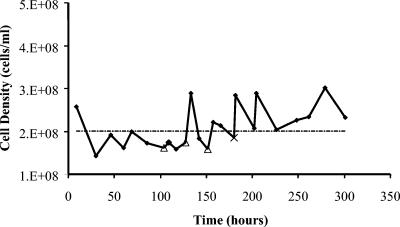
FIG. 1.
T. maritima growth in 1.5-liter continuous culture at 80°C. Planktonic samples were collected at points designated by ▵, and biofilm samples were collected at the time noted by ×.
Construction of the whole-genome cDNA microarray.
Open reading frames (ORFs) were identified from the T. maritima MSB8 genome available at http://www.tigr.org/tigrscripts/CMR2GenomePage3.spl?database=btm". DNA primers were designed with similar annealing temperatures and minimal hairpin formation with Genomax (Informax, Bethesda, Md.). Probes were PCR amplified in a PTC-100 Thermocycler (MJ Research, Inc., Waltham, Mass.) using Taq polymerase (Boehringer, Indianapolis, Ind.) and T. maritima genomic DNA, isolated as described previously (14). Purification of PCR products and microarray construction were performed by protocols described elsewhere (14). PCR products were randomized within plates before printing with a random number generator, and each gene was applied as a spot six times on each array.
Labeling and hybridization.
The whole-genome microarray was interrogated by methods previously described (14). Briefly, first-strand cDNA was prepared from T. maritima total RNA using Stratascript (Stratagene, La Jolla, Calif.) and random hexamer primers (Invitrogen Life Technologies, Carlsbad, Calif.); 5-[3-Aminoallyl]-2′-deoxyuridine-5′-triphosphate (Sigma) was used for dye incorporation, as described elsewhere (34). Each biofilm or planktonic cDNA sample was labeled with Cyanine-3 and Cyanine-5, and the samples hybridized to different arrays. The slides were scanned and processed with a Perkin-Elmer Scanarray Express Lite.
Statistical analyses and determination of differential gene expression.
Replication of treatments, arrays, dyes, and cDNA spots allowed the use of analysis of variance (ANOVA) models for data analysis (122). Cyanine-3-labeled biofilm cDNA and Cyanine-5-labeled planktonic cDNA were hybridized on one chip, and Cyanine-3-labeled planktonic cDNA and Cyanine-5-labeled biofilm cDNA were hybridized on another chip. The data import code and statistical analysis procedures reported previously (14) were used to analyze spot intensities obtained from Quantarray. Briefly, a linear normalization ANOVA model (122) was used to estimate global variation in the form of fixed (dye, D; treatment, T) and random [array, A; spot, A(BS); block, A(B)] effects and random error using the model log2(yijklmn) = μ + Ai + Dj + Tk + Ai(Bl) +Ai(Sm · Bl) + ɛijklm. A gene-specific ANOVA model was then used to partition the remaining variation into gene-specific effects using the model rijklm = μ + Ai + Dj + Tk + Ai(Bl) +Ai(Sm · Bl) + ɛijklm. Volcano plots were used to visualize interesting contrasts or comparisons between two treatments (122). A statistical test with the null hypothesis of no differential expression was performed for each of the 1,880 ORFs included on the array. A Bonferroni correction was used to adjust α for the expected increase in false positives due to multiple tests (122). The Bonferroni correction, calculated by dividing 0.05 by 1,880, yielded a corrected α of 0.00003, equivalent to a −log10 (P value) of 4.5. Genes meeting this significance criterion and showing fold changes of ±2.0 or greater were selected for further examination.
For complete information on significance of expression changes and fold changes, see our website (to be included on our microarray data page at http://www.che.ncsu.edu/extremophiles/microarray/).
RT-PCR confirmation of gene expression.
Real-time reverse trasnscription-PCR (RT-PCR) was used to confirm the microarray results of four up-regulated genes (TM0851, 2.3-fold; TM1645, 8.1-fold; TM1848, 6.9-fold; TM1867, 5.1-fold) and one unchanged, control gene (TM0403, 1.2-fold). Primers were designed with Genomax software: TM0851 (GCATAACCGTCAGGATAGGAAG and TTCGACGTGAAGAGGTACACAC), TM1645 (TGTCATGCTGGACAATCTCTCT and ACTTCCACGATCACGTTAGGAT), TM1848 (ATGGAAGCACTTACCACCAGTT and CCAGTCACCTGTCTCTTTGATG), TM1867 (GGAGAACATGGAGATTCAGAGG and ATCGCACTTCTGACAAATCTGA), and TM0403 (AGGTGATGCTTCTCATAGCGGT and ATCCTAATGCAATCCAGCAGATCCA). RT of RNA to cDNA was performed as described above. RT-PCR was performed with the SYBRGREEN kit and iCycler iQ real-time PCR detection system (Bio-Rad Laboratories, Hercules, Calif.) according to the manufacturer's protocols. Briefly, reactions for 10 ng of samples were carried out for the five genes at three different temperatures to determine the optimum S-curves. Optimization indicated that all reactions could be performed at 55°C. Standard curves (20, 4, 0.8, and 0.16 ng) for biofilm samples were run along with 10 ng of planktonic and biofilm samples for each gene. Quantitative results were calculated with vendor-provided software (Bio-Rad Laboratories). In all cases, RT-PCR results exhibited the same patterns as those obtained from cDNA microarray analysis. Fold changes calculated from real time PCR were as follows: TM0851, 6.7-fold; TM1645, 10.0-fold; TM1867, 51.1-fold; TM1848, 17.4-fold; and TM0403, 1.3-fold. In all cases of differentially expressed genes, the microarray tended to underestimate the fold changes calculated by real-time PCR, which is not surprising given the smaller dynamic range of microarray scanners when compared to real time PCR.
RESULTS AND DISCUSSION
T. maritima growth in continuous culture and biofilm formation.
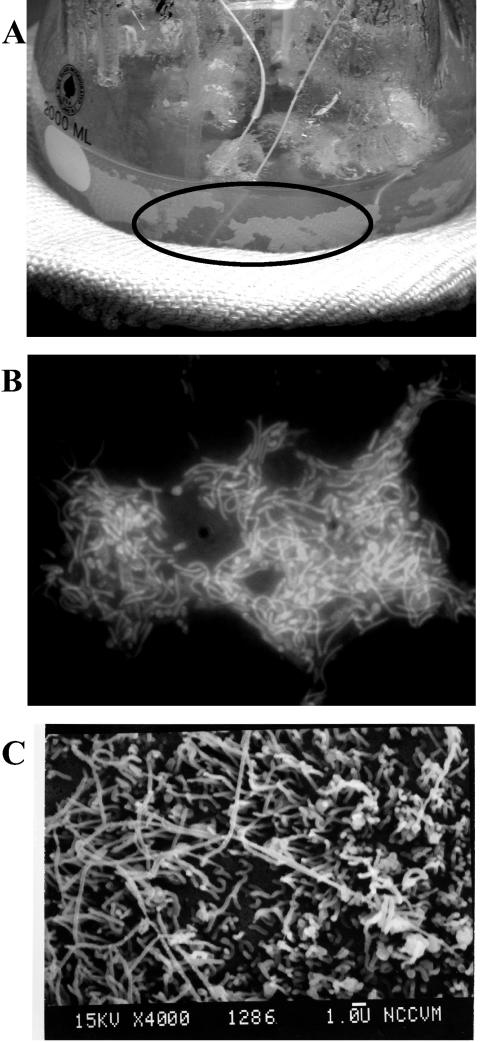
Because efforts with batch culture were unsuccessful in generating sufficient attached cellular material for transcriptional analysis, a high-temperature anaerobic chemostat was operated to collect T. maritima biofilm formed on removable nylon mesh. The mesh was used to create a compact, high-surface-area substratum for biofilm attachment; materials like this have been used successfully to study P. aeruginosa biofilms (19). T. maritima (Topt 80°C) (37) was grown in continuous culture (dilution rate, D = 0.25 h−1) for over 300 h (Fig. 1). Figure 2A shows the formation of substantial wall growth in the 80°C reactor (86). Epifluorescent micrographs of polycarbonate filters placed in the chemostat showed significant cell attachment at 80°C (Fig. 2B); this was supported by scanning electron microscopy (SEM) analysis of biofilm cells on the filters which showed cells associated with rope-like structures, consistent with SEM analysis of mesophilic biofilms (Fig. 2C) (22, 24, 40).
FIG. 2.
T. maritima biofilm formation on (A) nylon mesh and reactor walls during continuous cultivation. Polycarbonate filters inserted into the culture showed microcolony formation by (B) epifluorescent microscopy and (C) scanning electron microscopy.
Whole-genome cDNA microarray analysis of differential gene expression of sessile and planktonic T. maritima.
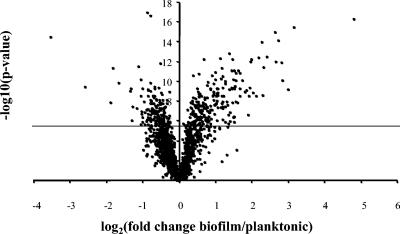
Despite the inherent heterogeneity of the biofilm state, planktonic and sessile T. maritima cells could be differentiated by transcriptional response patterns as determined by cDNA microarray analysis (Fig. 3). Table 1 lists genes exhibiting significant expression changes (twofold or higher, −log10 P value of ≥4.5) for biofilm cells as compared to planktonic cells. The cDNA microarray results were confirmed by real-time RT-PCR (see Materials and Methods). For T. maritima, approximately 114 genes of the entire genome were differentially expressed twofold or higher at this significance level; 43 of these 114 genes were originally annotated as hypothetical proteins (65). T. maritima gene expression patterns were further analyzed according to function, genomic location, and in comparison to biofilm gene expression profiles in mesophilic bacteria. A complete list of expression changes for predicted operons responding significantly between biofilm and planktonic cells is shown in Table 2. Where appropriate, gene annotations are updated with information from comparative genomics and functional studies subsequent to the publication of the T. maritima genome sequence (65).
FIG. 3.
Volcano plot showing differential gene expression in planktonic and biofilm T. maritima cells grown in chemostat culture at 80°C. A horizontal line indicates Bonferroni correction.
TABLE 1.
T. maritima ORFs differentially regulated in biofilma
| Gene description | Source of gene description (reference) | Gene ID | Fold change | −Log10P value |
|---|---|---|---|---|
| Upregulated in biofilmb | ||||
| Aspartate dehydrogenase | (124) | TM1643 | 27.9 | 16.3 |
| ThiH protein, putative | 28% identity, 347 aa with E. coli ThiH (53, 104) conserved CX3CX2C [4Fe-4S] cluster binding motif | TM1267 | 8.9 | 15.4 |
| Nicotinate-nucleotide pyrophosphorylase (NadC) | Crystallized (GI:34811257; 1O4U_A, 1O4U_B) | TM1645 | 8.1 | 9.1 |
| Hypothetical protein | (65) | TM1266 | 7.2 | 10.0 |
| Quinolinate synthetase A (NadA) | 54% identity, 298 aa with P. abyssi (PAB2345) and P. horikoshii (PH0013), 3 conserved cysteines | TM1644 | 7.1 | 11.9 |
| Cellobiose phosphorylase | (79) | TM1848 | 6.9 | 12.9 |
| Cation-transporting ATPase, P-type | COG2217, 30% identity, 724 aa with E. coli ZntA (93) | TM0317 | 6.6 | 14.1 |
| SufD homolog, similar to ABC permease components | COG0719 | TM1370 | 6.3 | 12.0 |
| SufB homolog, similar to ABC ATP-binding components | (80) | TM1369 | 6.3 | 15.0 |
| Iron(II) transport protein B (FeoB) | 33% identity, 718 aa with E. coli FeoB (43, 60) | TM0051 | 5.4 | 12.4 |
| l-Lactate dehydrogenase | (72) | TM1867 | 5.1 | 11.0 |
| Ubiquinone/menaquinone methyltransferase | COG0500, SAM-dependent methyltransferases, 53% identity, 452 aa with PF0738 | TM0318 | 5.0 | 11.4 |
| Biotin synthetase, putative (BioB homolog) | 22% identity, 281 aa with E. coli BioB (108) conserved CX3CX2C motif, CX2C motif | TM1269 | 5.0 | 8.6 |
| IscS/SufS homolog, cysteine desulfurase | (59) | TM1371 | 4.8 | 13.9 |
| Sms/RadA homolog, DNA recombination/repair protein | 39% identity, 452 aa with E. coli Sms/RadA (6), conserved CX2CX10CX2C motif is a zinc finger in E. coli Sms/RadA | TM0199 | 4.6 | 12.4 |
| FTR1, predicted high-affinity Fe2+/Pb2+ permease | 25% identity 228 aa with S. cerevisiae Ftr1p; two conserved REXXE iron-binding motifs (95) | TM0417 | 4.4 | 8.5 |
| Esterase, putative | (65) | TM0053 | 4.1 | 8.8 |
| Iron(II) transport protein A (FeoA) | 42% identity 57 aa with E. coli FeoA (43) | TM0050 | 4.0 | 12.2 |
| Cysteine synthase (CysK) | Crystal structure (GI:34810052, IO58A-IO8D) | TM0665 | 3.9 | 12.0 |
| Serine acetyltransferase (CysE) | 43% identity, 195 aa with S. xylosus CysE (26) | TM0666 | 3.9 | 9.3 |
| FpaA family protein, contains flavodoxin domain and β-metallo-lactamase domain | COG0426, 28% identity, 366 aa with D. gigas ROO (27) | TM0755 | 3.8 | 8.5 |
| Hypothetical protein | (65), 32% identity, 78 aa with TTE1569 | TM1268 | 3.7 | 6.6 |
| Uncharacterized homolog of γ-carboxymuconolactone decarboxylase | COG0599 | TM0316 | 3.6 | 9.1 |
| NADH oxidase | 46% identity, 440 aa with T. neapolitana NADH: polysulfide oxidoreductase | TM0395 | 3.5 | 9.0 |
| Ubiquinone/menaquinone biosynthesis methyltransferase, putative (UbiE homolog) | COG2226 | TM0753 | 3.3 | 10.1 |
| HycB domain containing protein, related to hydrogenase components | COG1142, 2 CX2CX2CX3C motifs, 1 CX2CX7CX2C motif, 1 CX2CX4CX3C motif | TM0396 | 3.2 | 10.8 |
| Heavy metal binding protein | (65) | TM0320 | 3.2 | 9.2 |
| Uncharacterized conserved protein | COG3862, CX2CX3C motif | TM1434 | 3.2 | 8.5 |
| Predicted zinc-dependent protease | COG2738 | TM1511 | 3.1 | 11.0 |
| NAD+-dependent α-glucuronidase | (99) | TM0752 | 3.1 | 7.9 |
| Protein containing Bcp domain and nitroreductase domain | COG1225 (Bcp domain), COG0778 (nitroreductase domain) | TM0386 | 3.0 | 3.1 |
| Hypothetical protein | (65) | TM0338 | 3.0 | 7.9 |
| Hypothetical protein | (65) | TM0052 | 3.0 | 10.1 |
| Predicted GTPase | COG1160 | TM0445 | 2.9 | 10.8 |
| β-d-Galactosidase | 40% identity, 670 aa with B. stearothermophilus β-galactosidase (36) | TM0310 | 2.9 | 9.5 |
| IscU/SufA homolog, iron-sulfur cluster assembly scaffold | 3 conserved cysteines bind a [2Fe-2S] cluster (8, 58) | TM1372 | 2.9 | 5.9 |
| Uncharacterized conserved protein | COG0011, pfam01910, localized with TauABC (4 species) | TM0486 | 2.9 | 9.4 |
| ABC transporter, permease protein, TauC family | COG0600 | TM0485 | 2.9 | 11.1 |
| ATP-dependent Clp protease, ATPase subunit, ClpA homolog | COG0542 | TM0198 | 2.8 | 9.2 |
| Glutamate synthase domain 3 (GltB) | COG0070 | TM0394 | 2.8 | 6.5 |
| Possible endonuclease or sugar phosphate isomerase | COG1082, pfam01261 | TM0422 | 2.8 | 6.3 |
| Hypothetical protein | (65) | TM0054 | 2.8 | 10.2 |
| Predicted membrane protein | COG3462 | TM0315 | 2.8 | 12.2 |
| Uncharacterized conserved protein | COG0718 | TM0687 | 2.8 | 7.9 |
| Cystathionine γ-synthase/β-lyase | COG0626 | TM1270 | 2.7 | 9.3 |
| Predicted transcriptional regulator, ACR/TetR related | pfam00440 | TM0823 | 2.7 | 9.2 |
| Sugar ABC transporter, permease protein, UpgA family | COG1175 | TM0419 | 2.7 | 11.0 |
| Sugar ABC transporter, periplasmic sugar-binding protein, MalE-related | COG2182 | TM0418 | 2.7 | 6.9 |
| ABC transporter, ATP-binding subunit, TauA family | COG0715 | TM0484 | 2.6 | 5.9 |
| Hypothetical protein | (65) | TM0714 | 2.6 | 9.2 |
| Thioredoxin peroxidase | COG1225, 59% identity, 157 aa with T. acidophilum (Tal1368M) | TM0780 | 2.6 | 12.8 |
| Hypothetical protein | (65) | TM0319 | 2.6 | 6.6 |
| Predicted HHH nucleic acid binding protein | COG1623, pfam02457 | TM0200 | 2.6 | 5.7 |
| Glycerol uptake facilitator protein | COG0580 | TM1429 | 2.5 | 7.6 |
| FixX homolog, putative ferredoxin | COG2440, CX3CX4CX3CC motif, CX2CX2CX3C motif | TM1533 | 2.5 | 5.3 |
| FixC homolog | 42% identity, 438 aa with A. caulinodans FixC (4) | TM1532 | 2.5 | 7.9 |
| PotD, spermidine/putrescine ABC transporter, PBP | 39% identity, 320 aa with E. coli potD (45) | TM1375 | 2.5 | 2.6 |
| Transcriptional regulator, biotin repressor family | Crystallized (GI:22218828, 1J5YA) | TM1602 | 2.4 | 9.7 |
| Predicted CAAX amino-terminal protease | pfam0257 | TM1529 | 2.4 | 10.0 |
| RAD55-related, RecA superfamily ATPase | COG0467, 78% identity, 235 aa with PH0824, PAB2180, PH1931 | TM0370 | 2.4 | 7.4 |
| Pyridine nucleotide-disulfide oxidoreductase, putative | pfam00070 | TM0754 | 2.4 | 5.9 |
| Oxidoreductase, aldo/keto reductase family | COG0467 | TM1743 | 2.4 | 7.2 |
| NADH:polysulfide oxidoreductase | 86% identity, 440 aa with T. neapolitana (GI:21702687) | TM0379 | 2.3 | 8.5 |
| Hypothetical protein | (65) | TM0002 | 2.3 | 9.4 |
| Heat shock operon repressor HrcA | (65, 78) | TM0851 | 2.3 | 7.3 |
| Conserved hypothetical protein, GGDEF domain | COG2199, pfam00990 | TM1588 | 2.3 | 7.5 |
| Glutamate synthase domain 2, GltB | COG0069, pfam01645, CX2CX2C motif | TM0397 | 2.3 | 7.9 |
| UDP-sugar disphosphatase precursor | 38% identity, 504 aa with E. coli UshA (11) | TM1878 | 2.3 | 7.9 |
| Putative endonuclease | COG1833, pfam01986 | TM0664 | 2.3 | 10.5 |
| Uncharacterized conserved protein | COG4198, pfam06245 | TM1510 | 2.3 | 6.1 |
| Membrane bound protein LytR, putative transcriptional regulator | COG1316, pfam03816 | TM1866 | 2.3 | 9.8 |
| FixA homolog, electron transfer flavoprotein | 45% identity, 241 aa with A. caulinodans FixA (4) | TM1530 | 2.3 | 1.9 |
| Sugar ABC transporter, ATP-binding protein, MalK homolog | COG3839 | TM0421 | 2.3 | 11.3 |
| Heat shock protein, class I | (62) | TM0374 | 2.3 | 8.5 |
| Conserved hypothetical protein | (65) | TM0387 | 2.3 | 7.8 |
| Predicted coenzyme A-binding protein | COG1832 | TM1435 | 2.2 | 7.3 |
| Conserved hypothetical protein | pfam03706 | TM1390 | 2.2 | 10.2 |
| Hypothetical protein | (65) | TM1534 | 2.2 | 12.3 |
| Frameshift | (65) | TM0621 | 2.2 | 5.2 |
| Glutamate synthase domain 1, GltB | COG0067 | TM0398 | 2.2 | 5.2 |
| Putative sensor histidine kinase | (65) | TM0187 | 2.2 | 5.3 |
| Conserved hypothetical protein | pfam01139 | TM1357 | 2.1 | 8.8 |
| ABC transporter, ATP-binding subunit, SalX domain | COG1136 | TM0352 | 2.1 | 9.2 |
| Endoglucanase (extracellular) | (15, 57) | TM1525 | 2.1 | 8.6 |
| Iron(III) ABC transporter, ATP-binding protein, putative | COG1120, 50% identity, 242 aa with P. furiosis (PF0909) | TM0191 | 2.1 | 7.3 |
| Putative transcriptional regulator | COG1318, 58% identity, 150 aa with P. horikoshii (PH0283) | TM0369 | 2.1 | 8.0 |
| Iron-sulfur cluster-binding protein, putative | COG2768, 2 CX2CX2CX3C motifs (65) | TM0034 | 2.1 | 6.0 |
| Heat shock serine protease, periplasmic | (46, 47) | TM0571 | 2.1 | 9.3 |
| Hypothetical protein | (65) | TM0003 | 2.0 | 8.7 |
| Permease, putative | COG0477, 36% identity, 409 aa with B. subtilis YceI (50) | TM1603 | 2.0 | 7.7 |
| Octaprenyl pyrophosphate synthase | (32, 51) | TM1535 | 2.0 | 7.6 |
| Membrane protein, putative | COG3374 | TM1536 | 2.0 | 8.5 |
| ABC transporter, ATP-binding protein, TauB family | COG1116 | TM0483 | 2.0 | 10.2 |
| RNA polymerase σA factor | (13, 65) | TM1451 | 2.0 | 10.9 |
| Putative Holliday junction resolvase | COG0816 | TM1545 | 2.0 | 10.9 |
| ABC transporter, permease subunit, SalY family | COG0577 | TM0351 | 2.0 | 8.9 |
| Down-regulated in biofilmc | ||||
| K+ channel, beta subunit | COG0667, pfam00248 | TM0313 | −2.0 | 9.3 |
| Cyclomaltodextrinase | (54) | TM1835 | −2.0 | 9.6 |
| Putative regulator, XRE family HTH | COG1917, pfam1381 | TM0656 | −2.0 | 6.7 |
| Hypothetical protein | (65) | TM0794 | −2.1 | 6.9 |
| Predicted dehydrogenase | COG0673, 67% identity, 325 aa with P. furiosus (PF0554) | TM0312 | −2.1 | 5.6 |
| (3R)-Hydroxymyristoyl-(acyl carrier protein) dehydratase | COG0764 (65) | TM0801 | −2.1 | 10.1 |
| Uncharacterized conserved protein | COG3906 | TM0606 | −2.1 | 8.5 |
| Bacteriocin | 33% identity, 251 aa to B. linens linocin M18 (109) | TM0785 | −2.2 | 11.4 |
| Maltose ABC transporter, permease protein | (118) | TM1836 | −2.2 | 2.0 |
| Predicted glycosyltransferase | COG0438, pfam00534 | TM0392 | −2.2 | 7.3 |
| Hypothetical protein | (65) | TM1241 | −2.4 | 7.4 |
| Uncharacterized conserved protein | COG3471 | TM0786 | −2.4 | 6.0 |
| Ribosomal protein L7/L12 | (65) | TM0457 | −2.5 | 9.3 |
| Ribosomal protein L10 | (65) | TM0456 | −2.6 | 9.0 |
| Cold shock protein | (76, 120) | TM1874 | −2.8 | 9.6 |
| Rubredoxin | 68% identity, 51 aa with P. furiosis rubredoxin PF1282 (9, 21), 2 conserved CXXC motifs | TM0659 | −3.1 | 9.9 |
| Superoxide reductase (neelaredoxin) | 57% identity, 128 aa with P. furiosis SOR (PF1281) (38, 125) | TM0658 | −3.5 | 11.4 |
| Rubrerythrin | 58% identity, I65 aa with A. fulgidus rubrerythrin (AF1640) (115) | TM0657 | −3.7 | 7.8 |
| NADPH-dependent alkyl hydroperoxide reductase | 69% identity, 214 aa with P. horikoshii (PH1217) (44) | TM0807 | −6.0 | 9.4 |
| Protein distantly related to bacterial ferritins | COG2406, 82% identity, 183 aa with M. acetivorans strain CZA (MA2882) | TM0560 | −11.6 | 14.5 |
Locus description based on conserved domain searches (CDD, NCBI) and similarity to characterized proteins. Species used: Archaeoglobus fulgidus, Azorhizobium caulinodans, Brevibacterium linens, Bacillus stearothermophilus, Bacillus subtilis, Desulfovibrio gigas, Escherichia coli K-12, Methanosarcina acetivorans strain C2A, Pyrococcus furiosus, Pyrococcus horikoshii, Pyrococcus abyssi, Thermoplasma acidophilum, Thermotoga neapolitana, Saccharomyces cerevisiae, and Staphylococcus xylosus. Bcp, bacterioferritin comigratory protein.
Genes up-regulated 2.0-fold or greater in biofilm. Significance based on Bonferroni-corrected significance criterion with −log10 P value of >4.6.
Genes down-regulated 2.0-fold or greater in biofilm. Significance based on Bonferroni-corrected significance criterion with −log10 P value of >4.6.
TABLE 2.
Differential expression of genes in biofilm-bound cells as related to predicted T. maritima operons
| Putative function | Gene ID | Gene description | Fold change | −Log10P valuec |
|---|---|---|---|---|
| Iron transportb | TM0050 | Iron(II) transport protein A (FeoA) | 4.0 | 12.2 |
| TM0051 | Iron(II) transport protein B (FeoB) | 5.4 | 12.4 | |
| TM0052 | Hypothetical protein | 3.0 | 10.1 | |
| TM0053 | Esterase, putative | 4.1 | 8.8 | |
| TM0054 | Hypothetical protein | 2.8 | 10.2 | |
| DNA processingb | TM0198 | ATP-dependent Clp protease, ATPase subunit, ClpA family | 2.8 | 9.2 |
| TM0199 | Sms/RadA homolog, DNA recombination/repair protein | 4.6 | 12.4 | |
| TM0200 | Predicted HHH nucleic acid binding protein | 2.6 | 5.7 | |
| TM0201 | NADP-reducing hydrogenase, subunit D, putative | 1.5 | 2.7 | |
| TM0202 | TauA homolog | 1.7 | 5.5 | |
| Cation transport systemb | TM0315 | Predicted membrane protein | 2.8 | 12.2 |
| TM0316 | Uncharacterized homolog of γ-carboxymuconolactone decarboxylase subunit | 3.6 | 9.1 | |
| TM0317 | Cation-transporting ATPase, P-type | 6.6 | 14.1 | |
| TM0318 | Ubiquinone/menaquinone methyltransferase | 5.0 | 11.4 | |
| TM0319 | Hypothetical protein | 2.6 | 6.6 | |
| TM0320 | Heavy metal binding protein | 3.2 | 9.2 | |
| ABC transporta | TM0351 | ABC transporter, permease subunit, SalY family | 2.0 | 8.9 |
| TM0352 | ABC transporter, ATP-binding subunit, SalX domain | 2.1 | 9.2 | |
| TM0353 | Predicted membrane fusion protein (COG0845) | 1.4 | 6.4 | |
| DNA repaira | TM0369 | Predicted transcriptional regulator | 2.1 | 8.0 |
| TM0370 | RAD55-related, RecA superfamily ATPase (COG0467) | 2.4 | 7.4 | |
| Thermal stress | TM0373 | DnaK protein (62) | 1.9 | 8.5 |
| TM0374 | Heat shock protein, class I | 2.3 | 8.5 | |
| Glutamate synthesisa | TM0394 | Glutamate synthase domain 3 (GltB) | 2.8 | 6.5 |
| TM0395 | NADH oxidase | 3.5 | 9.0 | |
| TM0396 | HycB domain containing protein, related to hydrogenase components | 3.2 | 10.8 | |
| TM0397 | Glutamate synthase domain 2, GltB | 2.3 | 7.9 | |
| TM0398 | Glutamate synthase domain 1, GltB | 2.2 | 5.2 | |
| Sugar and iron transportb | TM0417 | FTR1, predicted high affinity Fe2+/Pb2+ permease | 4.4 | 8.5 |
| TM0418 | Sugar ABC transporter, periplasmic sugar-binding protein, MalE-related | 2.7 | 6.9 | |
| TM0419 | Sugar ABC transporter, permease protein, UpgA family | 2.7 | 11.0 | |
| TM0420 | Sugar ABC transporter, permease protein | 1.4 | 7.6 | |
| TM0421 | Sugar ABC transporter, ATP-binding protein, MalK homolog | 2.3 | 11.3 | |
| TM0422 | Possible endonuclease or sugar phosphate isomerase | 2.8 | 6.3 | |
| Sulfur compound transport systemsa | TM0483 | ABC transporter, ATP-binding protein, TauB family | 2.0 | 10.2 |
| TM0484 | ABC transporter, ATP-binding subunit, TauA family | 2.6 | 5.9 | |
| TM0485 | ABC transporter, permease protein, TauC family | 2.9 | 11.1 | |
| TM0486 | Uncharacterized conserved protein | 2.9 | 9.4 | |
| Oxygen detoxificationa | TM0657 | Rubrerythrin | −3.7 | 7.8 |
| TM0658 | Superoxide reductase (neelaredoxin) | −3.5 | 11.4 | |
| TM0659 | Rubredoxin | −3.1 | 9.9 | |
| Amino acid metabolismb | TM0664 | Putative endonuclease | 2.3 | 10.5 |
| TM0665 | Cysteine synthase | 3.9 | 12.0 | |
| TM0666 | Serine acetyltransferase (CysE) | 3.9 | 9.3 | |
| Oxygen detoxification/electron transferb | TM0752 | NAD+-dependent α-glucuronidase | 3.1 | 7.9 |
| TM0753 | Ubiquinone/menaquinone biosynthesis methyltransferase, putative (UbiE homolog) | 3.3 | 10.1 | |
| TM0754 | Oxidoreductase | 2.4 | 5.9 | |
| TM0755 | FpaA family protein, contains flavodoxin domain and β-metallolactamase domain (COG0426) | 3.8 | 8.5 | |
| Biotin/thiamine-synthesisb | TM1266 | Hypothetical protein | 7.2 | 10.0 |
| TM1267 | ThiH protein, putative | 8.9 | 15.4 | |
| TM1268 | Hypothetical protein | 3.7 | 6.6 | |
| TM1269 | Biotin synthetase, putative (BioB homolog) | 5.0 | 8.6 | |
| TM1270 | Cystathionine γ-synthase/β-lyase | 2.7 | 9.3 | |
| Iron-sulfur cluster assemblya | TM1368 | SufC homolog, similar to ABC ATP-binding components | 1.1 | 2.0 |
| TM1369 | SufB homolog, similar to ABC permease components | 6.3 | 15.0 | |
| TM1370 | SufD homolog, similar to ABC permease components | 6.3 | 12.0 | |
| TM1371 | SufS/IscS homolog, cysteine desulfurase | 4.8 | 13.9 | |
| TM1372 | SufA/IscU homolog, iron-sulfur cluster assembly scaffold | 2.9 | 5.9 | |
| Sugar/electron transfer cascadeb | TM1524 | Endoglucanase (intracellular) | 1.8 | 7.1 |
| TM1525 | Endoglucanase (extracellular) | 2.1 | 8.6 | |
| TM1526 | Hypothetical protein (COG1633) | 1.7 | 6.3 | |
| TM1527 | IscR homolog, putative | 1.8 | 6.0 | |
| TM1528 | 1,4-Dihydroxy-2-naphthoate octaprenyltransferase, MenA homolog | 1.7 | 6.0 | |
| TM1529 | Predicted CAAX amino terminal protease | 2.4 | 10.0 | |
| TM1530 | FixA homolog, electron transfer flavoprotein | 2.3 | 1.9 | |
| TM1531 | FixB homolog, electron transfer flavoprotein | 2.0 | 4.9 | |
| TM1532 | FixC homolog | 2.5 | 7.9 | |
| TM1533 | FixX homolog | 2.5 | 5.3 | |
| TM1534 | Hypothetical protein | 2.2 | 12.3 | |
| TM1535 | Octaprenyl pyrophosphate synthase (OPP) | 2.0 | 7.6 | |
| TM1536 | Putative membrane protein | 2.0 | 8.5 | |
| Nicotinate synthesisb | TM1643 | Aspartate dehydrogenase | 27.9 | 16.3 |
| TM1644 | Quinolinate synthetase A (NadA) | 7.1 | 11.9 | |
| TM1645 | Nicotinate-nucleotide pyrophosphorylase (NadC) | 8.1 | 9.1 | |
| Maltose utilization and transportb | TM1834 | α-glucosidase | −1.5 | 5.1 |
| TM1835 | Cyclomaltodextrinase | −2.0 | 9.6 | |
| TM1836 | Maltose ABC transporter, permease protein | −2.2 | 2.0 | |
| TM1837 | Maltose transport system permease protein | −1.9 | 8.1 | |
| TM1838 | Hypothetical protein | −1.4 | 5.7 | |
| TM1839 | Maltose ABC transporter, periplasmic maltose-binding protein | 1.0 | 0.4 |
Complete operon predicted by www.tigr.org. Partial operon predicted by www.tigr.org. Note that some −log10 P values are below the significance criterion Bonferroni factor of 4.6.
Oxidative and thermal stress response.
Biofilm formation has been observed as a response to oxidative stress in the hyperthermophilic archaeon A. fulgidus (52), and certain aerobic mesophilic biofilms showed increased expression of oxidative stress genes (29, 91). Increased protein levels of superoxide dismutase and alkyl hydroperoxide reductase in aerobic mesophile biofilms have also been reported (90). Here, the observed down-regulation of predicted operon members rubrerythin (TM0657, −3.7-fold), superoxide reductase (SOR, TM0658, −3.4-fold), and rubredoxin (TM0659, −3.1-fold) in biofilm cells, along with an ahpC-related alkylhydroperoxide reductase (TM0807, −6.0-fold), was somewhat unexpected. All four proteins share high identity (57 to 67%) to homologs in Pyrococcus species (9, 16, 38, 44). The Pyrococcus homologs of rubredoxin and SOR are known to be involved in the NADPH-dependent detoxification of dioxygen to H2O2 (38), while a Pyrococcus horikoshii AhpC homolog (69% identity, 214 amino acids [aa] with TM0807) participates in a pathway responsible for the reduction of H2O2 to alcohol (44). However, an AhpC-related gene (TM0780) encoding a putative thioredoxin peroxidase/bacterioferritin comigratory protein (Bcp) was up-regulated 2.6-fold in biofilm cells (39). A second related gene (TM0386, 3.0-fold), containing an apparently unique combination of a Bcp thioredoxin peroxidase domain and a nitroreductase domain, was also up-regulated.
Several possible explanations exist for the down-regulation of the SOR gene cluster. Lower expression of these genes has been observed during the stationary phase (M. R. Johnson and R. M. Kelly, unpublished data); therefore, decreases in expression here may reflect similarities between stationary-phase and biofilm cells. Alternatively, down-regulation of these genes may suggest lower residual oxygen exposure for cells trapped within a biofilm matrix, although both T. maritima planktonic and biofilm cells were cultured in the same anaerobic chemostat. Finally, the down-regulation may reduce exposure of biofilm cells to hydrogen peroxide during oxygen detoxification. Work in E. coli K-12 has implicated cysteine as the reductant responsible for rapid recycling of iron(II) to iron(III), allowing reactions between hydrogen peroxide and iron(III) to drive the formation of DNA-damaging hydroxyl radicals (75). Indications of DNA damage in T. maritima biofilm cells were observed in the up-regulation of genes encoding a homolog of Sms/RadA (TM0199, 4.6-fold) involved in recombination and repair in E. coli K-12 (6), a putative endonuclease specific to archaea (TM0664, 2.3-fold), and a predicted endonuclease (TM1545, 2.0-fold) related to proteins involved in recombination events. DNA protection and repair proteins were also induced in Listeria monocytogenes biofilms (107).
Gene expression analysis of biofilm-bound T. maritima cells revealed the induction of genes implicated in thermal stress response. Here, biofilm cells displayed 2.3-fold-higher expression of the CIRCE (controlling inverted repeat of chaperone expression)-binding HrcA repressor (TM0851), which controls expression of major heat shock operons in a number of species. We have previously noted the conservation of the CIRCE element upstream of the T. maritima hrcA-dnaJ-grpE and groESL operons (TM0849 to TM0851) but not upstream of dnaK-smHSP (TM0373 to TM0374) (78). Thermal stress genes have been shown to be up-regulated in biofilms of P. aeruginosa (groES and dnaK) (121) and S. mutans (grpE and dnaK) (100). T. maritima grpE (TM0850, 1.7-fold), dnaJ (TM0849, 1.4-fold), dnaK (TM0373, 1.9-fold), heat shock protein class I gene (TM0374, 2.3-fold), and groES (TM0373, 1.4-fold) were all up-regulated in biofilm cells. It was interesting that a cold shock protein (TM1874, −2.8 fold) recently shown to act as a functional homolog of product of the RNA-binding E. coli K-12 cspA gene was down-regulated (76). Schembri et al. (91) also observed down-regulation of the cold shock protein encoded by cspA in a microarray experiment comparing E. coli K-12 biofilm and exponential-phase planktonic cells.
Exopolysaccharide biosynthesis and degradation.
A 2.0-fold upregulation of TM1535 (octaprenyl pyrophosphate synthase) involved in isoprenoid chain synthesis was observed in biofilm cells. Isoprenoid chains serve as scaffolds or lipid carriers for the assembly of monosaccharides into linear and/or branched polysaccharide chains via glycosyl transferases (112). Despite the presence in the T. maritima genome of a large cluster of glycosyltransferases, very few were differentially regulated between biofilm and planktonic cells. In fact, a number were expressed 1.5- to 1.7-fold higher in planktonic cells, including TM0630, an NDP-sugar epimerase related to UDP-glucose-4-epimerases; TM0627, a putative NDP-linked sugar glycosyltransferase; and TM0818, a techoic acid biosynthesis protein related to GumM in Xanthomonas campestris pv. gum (data not shown). A glycosyltransferase (TM0392, −2.2) predicted to be involved in the synthesis of NDP-linked sugars was also down-regulated in biofilm cells, while a homolog of E. coli K-12 ushA which encodes a periplasmic protein with UDP-sugar hydrolase activity (11) was up-regulated (TM1878, 2.3 fold). It is possible that exopolysaccharide synthesis occurs both in biofilm and planktonic cells, since commonalities in expression patterns have been observed between biofilm cells and planktonic cells in the presence of biofilm substratum (71).
Glycosyl hydrolases may also be involved in exopolysacchride synthesis and/or degradation. Induction of an NAD+-dependent family 4 α-glucuronidase (TM0752, 3.1-fold) (99) and a β-galactosidase (TM0310, 2.9-fold) was observed in the apparent absence of growth substrates related to these enzymes. Both proteins have been observed to be up-regulated during early stationary phase in T. maritima-M. jannaschii coculture experiments when the formation of biofilm material is observed (M. R. Johnson and R. M. Kelly, unpublished). Cellobiose phosphorylase (CepA) (TM1848) (65) exhibited a 6.9-fold expression increase in biofilm cells compared to planktonic cells at 80°C. CepA from Thermotoga neopolitana has sole substrate specificity for cellobiose (126), which it converts to d-glucose and glucose-1-phosphate (68). Characterization of the T. maritima homolog revealed substrate specificity for cellobiose in the hydrolysis reaction but relaxed synthetic specificity for the reverse reaction, allowing mannose, xylose, glucosamine, 2- and 6-deoxy-d-glucose, and β-d-glucoside to act as glucosyl acceptors for glucose-1-P (79). The strong up-regulation of this gene was unexpected, since maltose (α-1,6) and not cellobiose (β-1,4) was used as the primary carbon source in the growth medium. The up-regulation of the operon (TM1524 to TM1536) containing β-endoglucanases Cel12B (TM1524) and Cel12A (TM1525) (18, 56), previously shown to be up-regulated on carboxymethylcellulose, barley, and konjac glucomannan (14), was also noted. Further work will be necessary to determine whether the induction of glycoside hydrolases in biofilm cells is related to the synthesis or breakdown of exopolysaccharide-based biofilm material or the sloughing of biofilm.
ABC transporters.
Several ABC transporter genes were differentially expressed in biofilm cells. Despite the fact that maltose was the primary carbohydrate in the growth medium, genes within a maltose utilization and transport operon (TM1834 to TM1839) were down-regulated in biofilm cells. On the contrary, genes predicted to encode an uncharacterized multiple-sugar transport system (TM0418 to TM0421) downstream of the FTR1-related iron transporter (TM0417) were up-regulated in biofilm cells along with a gene sharing domain similarity with sugar phosphate isomerases (TM0422, 2.8-fold).
Additional homologs to ABC transporters were up-regulated during biofilm growth (Table 2). It was particularly intriguing to note the up-regulation of two genes which bear similarity to genes encoding antimicrobial peptide exporters. TM0352 (2.1-fold) is predicted to encode an ATP-binding ABC subunit (COG1136), while TM0351 (2.0-fold) possibly encodes an ABC-associated permease component (COG0577). Three additional upstream genes encode a putative membrane fusion protein (TM0353), outer membrane protein (TM0354), and TolC protein (TM0355). These genes (TM0353 to TM0355) were not expressed differentially between biofilm and planktonic conditions. A distantly related, though not well conserved, multiprotein system is essential for biofilm adhesion in Pseudomonas fluorescens WCS365, consisting of an ABC ATPase, ABC permease, outer membrane protein, and large adhesion protein with repetitive domains which is secreted via the transporter (35). A glycerol uptake facilitator protein (TM1429) (2.5-fold) was also up-regulated in T. maritima biofilm cells; ferric iron and glycerol may be required for antimicrobial peptide release as shown during biofilm growth of bacilli (123).
Response of iron/sulfur uptake and utilization genes in biofilm cells.
Biofilm cells showed increased expression of iron and sulfur uptake systems, consistent with up-regulation of genes encoding iron-sulfur cluster-containing proteins and components of a chaperone system involved in iron-sulfur cluster formation and repair. Predicted operons containing these genes are present in a number of distinct regions of the T. maritima genome. Known Fe-S clusters or cysteine-rich sequence motifs in the corresponding proteins are noted in Table 2.
Up-regulation of iron uptake is important in mesophile biofilm formation processes (10), which likely relates to the observed induction of genes encoding iron acquisition proteins in T. maritima biofilm cells. Increased expression was noted for genes encoding homologs of FeoB (TM0051, 5.4-fold), which is a G protein-like iron(II) transport system characterized in several species (2), and FeoA (TM0050, 4.4-fold), also presumed to be involved in iron transport (33). A second putative transporter gene (TM0417, 4.4-fold) related to yeast FTR1 high-affinity Fe2+ permeases (95), and the ATP-binding subunit of a putative iron(III) ABC transporter, FepC (TM0191, 2.1-fold), were also induced in biofilm cells. A protein distantly related to bacterial ferritins (TM0560 and COG2406) was the most highly downregulated gene in biofilm cells (−11.6-fold), presumably reflecting a decreased need for iron sequestration (5). Iron uptake regulation mechanisms have not been determined experimentally for T. maritima, but a small, statistically significant increase in the expression of a ferric uptake regulator (fur) homolog was noted (TM0122, 1.5-fold). Sequences resembling Fur binding sites are found upstream of the predicted iron transporter TM0417 and also upstream of TM0122, which precedes a similarly regulated set of ABC transporter components related to metal uptake systems (data not shown).
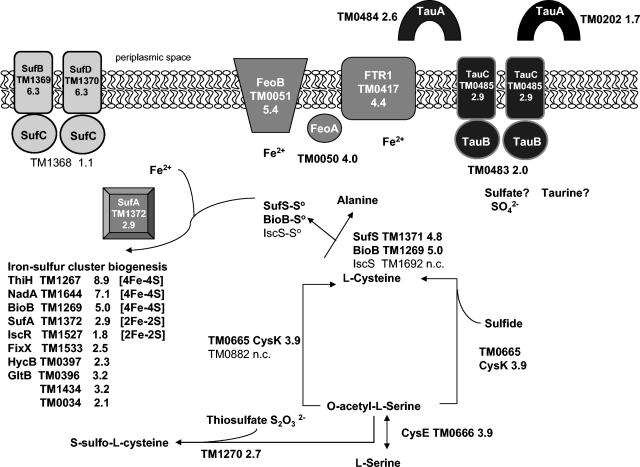
Genes (TM0483 to TM0485) homologous to two E. coli K-12 ABC transporter systems for sulfonates (25, 111) were preferentially induced in biofilm cells. E. coli K-12 and Rhodobacter capsulatus (61) tauABC encode taurine uptake ABC transporters, while the ssuABC operon encodes an alkane sulfonate transport system in E. coli K-12 and B. subtilis (25, 110). Although the natural substrates of the two tauABC-related systems in T. maritima have not been determined, sulfates and cysteine are present in the growth medium. Imported taurine and sulfates are typically incorporated via the cysteine biosynthesis pathway, but no recognizable homolog to the E. coli TauD desulfonation enzyme is apparent in the T. maritima genome. However, homologs to the uncharacterized conserved ORF (TM0486) are found upstream of tauABC homologs in two Streptococcus pneumoniae strains, Clostridium acetobutylicum, and Corynebacterium glutamicum (94). Crystal structures of two proteins related to TM0486 (pfam01910) suggest a ferredoxin-like fold and a possible role in protein-protein interaction regulated by the binding of sulfate ions (103). Several genes encoding predicted serine and cysteine biosynthesis enzymes were up-regulated here, including cysteine synthase (TM0665, 3.9-fold), serine acetyltransferase (TM0666, 3.9-fold), and a cystathione β-lyase/cystathione γ-synthase homolog predicted to be involved in cysteine degradation (TM1270, 2.7-fold) (Fig. 4).
FIG. 4.
Predicted pathway for iron-sulfur cluster biogenesis in T. maritima biofilm cells. Expression data suggest a number of known iron-sulfur cluster binding proteins and proteins with conserved cysteine-rich motifs as plausible targets for the SufABCDS iron-sulfur cluster assembly chaperone complex. Note that fold changes are listed after gene identification numbers. n.c., no change in expression.
The up-regulation of genes encoding members of a predicted iron-sulfur cluster chaperone complex offers insight into the apparent need of biofilm cells to acquire iron and sulfur from the environment and increase synthesis of cysteine. Iron-sulfur cluster synthesis and repair in biofilms may be a more general phenomenon, as a recent report indicates the upregulation of the iron-sulfur chaperones nifSU in mature biofilms of E. coli K-12 (7). Three paralogous cysteine desulfurases—IscS, NifS, and SufS—have been characterized in E. coli K-12 (101). While TM1371 and TM1372 have been referred to as iscS and iscU in characterization efforts, the lack of other isc genes in this genomic region has been noted (8, 58). Recent characterization of the SufABCDES iron-sulfur cluster assembly complex in E. coli K-12 (101) suggests a more appropriate designation of sufS (TM1371) and sufA (TM1372), given the colocalization with sufBCD homologs and the known role of SufABCDS in iron-sulfur cluster assembly under conditions of iron limitation and oxidative stress in E. coli K-12 (97) and Erwinia chrysanthemi (64). A homolog to SufE, which stimulates the cysteine desulfurase activity of SufS in E. coli K-12 (70), is not identifiable in T. maritima. The proteins encoded by sufC (TM1368) and sufB (TM1369) have been shown to interact in T. maritima cells (80); despite the lack of differential expression of the sufC homolog, the distantly related sufB and sufD (TM1370) are both expressed 6.3-fold higher in biofilm. Structural characterization of SufS/IscS (TM1371) has revealed conformational flexibility consistent with a role in iron-sulfur cluster donation to a variety of proteins, while SufA (TM1372) may act as a scaffold for iron-sulfur cluster assembly (59). A second SufS/IscS homolog in T. maritima, previously designated NifS (TM1692) (41), was not differentially expressed here.
Two putative regulators found in the T. maritima genome (TM0567 and TM1527) bear sequence similarity to IscR, a negative regulator of the Isc “housekeeping” iron-sulfur cluster assembly complex in E. coli K-12 (92). Three cysteine residues in E. coli IscR coordinate a [2Fe-2S] cluster which, when destabilized, disrupts DNA binding to IscR and allows transcription of the Isc operon (92). All three conserved cysteine residues are present in TM1527, located within a biofilm up-regulated gene string encoding FixABCX homologs and a hypothetical protein (TM1534, 2.2-fold) with a conserved CXXCX12CXXC motif. While the FixABCX proteins of T. maritima have not yet been characterized, homologous proteins function in electron transfer chains in other bacteria, including Rhizobium meliloti (23), Rhizobium leguminosarum (30), Azorhizobium caulinodans (42), and E. coli K-12 (116).
Additional plausible targets for Fe-S cluster assembly complexes are suggested by differential expression data. TM0034 (2.1-fold) contains two cysteine-rich sequence motifs, which are predicted to bind iron-sulfur clusters (Fig. 4). Up-regulated genes in a glutamate synthesis operon (TM0394 to TM0398) encode a putative NADH oxidase (TM0395) and three domains of glutamate synthase, a multiple iron-sulfur cluster binding complex (82). Also up-regulated is an iron-sulfur cluster binding protein (TM0396, 3.2-fold) that shares identity (44% identity, 143 amino acids) with a carbon monoxide dehydrogenase from A. fulgidus. O'Toole and Kolter (73) have shown that glutamate- and/or iron-containing medium can restore the ability of some biofilm-defective P. fluorescens strains to form biofilm.
Two separate predicted operons encoding a number of cofactor biosynthesis enzymes (TM1266 to TM1270 and TM0787 to TM0789) were overexpressed in T. maritima biofilm cells. Expression changes for the putative thi1-thiC homologs (TM0787 and TM0788), which are most closely related to archaeal thiamine biosynthesis enzymes, but largely absent from other eubacteria, were considerably lower (<2.0-fold) than those of thiH-bioB-metC (TM1267, TM1269, and TM1270). E. coli K-12 iscS mutants have been shown to be deficient in thiamine biosynthesis (53), likely as a result of degradation of an iron-sulfur cluster in the ThiH protein (56). E. coli K-12 ThiH is involved in biosynthesis of the thiamine thiazole ring, a process which requires sulfur donation from cysteine to ThiS via IscS (53, 104). The iron-sulfur cluster binding motif found in E. coli K-12 ThiH is conserved in the T. maritima ThiH homolog (TM1267, 8.9-fold).
A connection to iron-sulfur cluster assembly is also apparent in the up-regulation of genes encoding enzymes involved in nicotinate biosynthesis (TM1643 to TM1645). E. coli iscS mutants have been shown to require NAD as well as thiamine, presumably due to defects in assembly of the iron-sulfur cluster of quinolinate synthetase, NadA (53). A NifS cysteine desulfurase homolog has also been shown to be required for NAD biosynthesis in B. subtilis (98). Increases in expression were observed here for genes encoding NadA (TM1644, 7.1-fold), NadC (TM1645, 8.1-fold), and an NADP+-dependent l-aspartate dehydrogenase (TM1643, 27.9-fold) recently shown to convert l-aspartate to iminoaspartate (124) as an alternative to an NadB-type l-aspartate oxidase in the T. maritima NAD biosynthesis pathway. Increased NAD and/or NADH pools in sessile T. maritima may relate to the up-regulation of genes encoding l-lactate dehydrogenase (TM1867, 5.1-fold) (72), putative NADH oxidases (TM0379, 2.3-fold; TM0395, 3.5-fold), and the predicted dihydrolipoamide dehydrogenase (TM0381, 1.7-fold). l-Lactate dehydrogenase induction has also been observed in B. cereus biofilms (71) and may be involved in regenerating NAD+ (66) in conjunction with NADH oxidases (38, 117).
Regulation of biofilm formation and maintenance.
The genome sequence of T. maritima reveals the apparent lack of orthologs to a number of known biofilm-induced regulators, including RpoS, BmrAB, and CpsR. However, putative transcriptional regulators induced in sessile T. maritima cells included the sensor histidine kinase TM0187 (2.2-fold) and the response regulator TM1360 (1.6-fold). While the roles of these proteins have not yet been determined, the importance of a variety of related proteins in signaling processes during mesophilic biofilm formation has been well established (55). Small but statistically significant expression changes (1.8-fold) were also observed for TM0842, a CheY-related response regulator, and TM0841, a similarly regulated S-layer-like array protein sharing 35% identity (460 aa) with Thermus thermophilus SlpM (69), an activator of bacterial cell surface-layer protein synthesis.
Regulation of sigma factor expression influences biofilm formation in a number of species (1, 48). Little functional information is available for T. maritima sigma factors (13); however, the up-regulation of homologs to sigA (TM1451, 2.0-fold) and sigE (TM1598, 1.6-fold) in biofilm cells hints at a possible role for these proteins as global regulators during T. maritima biofilm formation. In contrast, the only two other T. maritima sigma factor homologs, sigH (TM0534) and fliA (TM0902), showed little fluctuation in expression levels for the two conditions compared here. Putative regulatory proteins induced in biofilm cells included members of the LytR (TM1866, 2.3-fold), biotin repressor (TM1602, 2.4-fold), and AcrR/TetR (TM0823, 2.7-fold) families. None of these proteins has been characterized in T. maritima, although regulators with TetR DNA binding domains have previously been shown to be important in biofilm formation in mesophiles (17, 48).
No function is known for the predicted transcriptional regulator TM1602 (2.4-fold); however, the major facilitator superfamily permease (TM1603) bears sequence similarity to a B. subtilis transporter conferring resistance to the toxic oxyanion tellurite, TeO3(2−) (50) (Table 1). The IscS cysteine desulfurase and the CysK cysteine synthase of Geobacillus stearothermophilis also confer tellurite resistance on E. coli K-12, presumably protecting cells from superoxide-mediated iron-sulfur cluster degradation (102, 113). Complementation of a tellurite-hypersensitive E. coli K-12 iscS mutant with G. stearothermophilus iscS confers tellurite resistance and relieves a growth requirement for thiamine but not nicotinic acid (102). The isolation of a number of tellurite- and selenite-resistant strains of bacteria from hydrothermal vents near sulfide rocks and bacterial biofilms suggests that the expression changes observed here may indicate an adaptive response to an iron-sulfur cluster degradation stimulus in the natural environment of T. maritima (81).
Summary.
The complex nature of biofilm formation processes, the dynamic physical and chemical characteristics of these microenvironments, and the likely heterogeneity of cellular states comprising biofilm populations make assigning a definitive biofilm phenotype difficult for T. maritima. Nonetheless, clear transcriptional differences were ascertained here that relate to cells involved in surface colonization. There is still much to be understood about biofilm formation and dynamics for T. maritima, but this work provides evidence for biofilm formation by T. maritima, a methodology for generating sufficient biofilm populations on nylon mesh in a high-temperature anaerobic chemostat for subsequent investigation of transcriptional response comparing planktonic and sessile cells, as well as a list of candidate genes whose expression patterns suggest a role in this process.
Acknowledgments
This work was supported in part by grants from the National Science Foundation, NASA Exobiology Program, and the Department of Energy (Energy Biosciences Program). S.B.C. acknowledges support from an NIEHS Traineeship in Bioinformatics.
We thank M. Dykstra at the Electron Microscopy Center, NCSU School of Veterinary Medicine, for assistance with electron microscopy; and Stephanie Bridger, Ubie Sullivan, Jennifer Strayhorn, and Leon Kluskens for their assistance in generation of the PCR products used to construct the array. We also thank R. Wolfinger and K. Scott, SAS Institute, Cary, N.C., for help with implementing the mixed-model analysis and the NCSU Genome Research Laboratory for assistance with microarray development and use.
REFERENCES
- 1.Adams, J. L., and R. J. C. McLean. 1999. Impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ. Microbiol. 65:4285-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 3.Antón, J., I. Meseguer, and F. Rodriguez-Valera. 1988. Production of an extracellular polysaccharide by Haloferax mediterranei. Appl. Environ. Microbiol. 54:2381-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arigoni, F., P. A. Kaminski, H. Hennecke, and C. Elmerich. 1991. Nucleotide sequence of the fixABC region of Azorhizobium caulinodans ORS571: similarity of the fixB product with eukaryotic flavoproteins, characterization of fixX, and identification of nifW. Mol. Gen. Genet. 225:514-520. [DOI] [PubMed] [Google Scholar]
- 5.Baaghil, S., A. Lewin, G. R. Moore, and N. E. Le Brun. 2003. Core formation in Escherichia coli bacterioferritin requires a functional ferroxidase center. Biochemistry 42:14047-14056. [DOI] [PubMed] [Google Scholar]
- 6.Beam, C. E., C. J. Saveson, and S. T. Lovett. 2002. Role for radA/sms in recombination intermediate processing in Escherichia coli. J. Bacteriol. 184:6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. [DOI] [PubMed] [Google Scholar]
- 8.Bertini, I., J. A. Cowan, C. Del Bianco, C. Luchinat, and S. S. Mansy. 2003. Thermotoga maritima IscU. Structural characterization and dynamics of a new class of metallochaperone. J. Mol. Biol. 331:907-924. [DOI] [PubMed] [Google Scholar]
- 9.Blake, P. R., J. B. Park, F. O. Bryant, S. Aono, J. K. Magnuson, E. Eccleston, J. B. Howard, M. F. Summers, and M. W. Adams. 1991. Determinants of protein hyperthermostability: purification and amino acid sequence of rubredoxin from the hyperthermophilic archaebacterium Pyrococcus furiosus and secondary structure of the zinc adduct by NMR. Biochemistry 30:10885-10895. [DOI] [PubMed] [Google Scholar]
- 10.Bollinger, N., D. J. Hassett, B. H. Iglewski, J. W. Costerton, and T. R. McDermott. 2001. Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J. Bacteriol. 183:1990-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns, D. M., and I. R. Beacham. 1986. Nucleotide sequence and transcriptional analysis of the E. coli ushA gene, encoding periplasmic UDP-sugar hydrolase (5′-nucleotidase): regulation of the ushA gene, and the signal sequence of its encoded protein product. Nucleic Acids Res. 14:4325-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caldwell, D. E., and J. W. Costerton. 1996. Are bacterial biofilms constrained to Darwin's concept of evolution through natural selection? Microbiol. SEM 12:347-358. [PubMed] [Google Scholar]
- 13.Camarero, J. A., A. Shekhtman, E. A. Campbell, M. Chlenov, T. M. Gruber, D. A. Bryant, S. A. Darst, D. Cowburn, and T. W. Muir. 2002. Autoregulation of a bacterial sigma factor explored by using segmental isotopic labeling and NMR. Proc. Natl. Acad. Sci. USA 99:8536-8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chhabra, S. R., K. R. Shockley, S. B. Conners, K. L. Scott, R. D. Wolfinger, and R. M. Kelly. 2003. Mixed model analysis of carbohydrate-induced differential expression patterns in the hyperthermophilic bacterium Thermotoga maritima. J. Biol. Chem. 278:7540-7552. [DOI] [PubMed] [Google Scholar]
- 15.Chhabra, S. R., K. R. Shockley, D. E. Ward, and R. M. Kelly. 2002. Regulation of endo-acting glycosyl hydrolases in the hyperthermophilic bacterium Thermotoga maritima grown on glucan- and mannan-based polysaccharides. Appl. Environ. Microbiol. 68:545-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clay, M. D., C. A. Cosper, F. E. Jenney, Jr., M. W. Adams, and M. K. Johnson. 2003. Nitric oxide binding at the mononuclear active site of reduced Pyrococcus furiosus superoxide reductase. Proc. Natl. Acad. Sci. USA 100:3796-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dakhova, O. N., N. E. Kurepina, V. V. Zverlov, V. A. Svetlichnyi, and G. A. Velikodvorskaya. 1993. Cloning and expression in Escherichia coli of Thermotoga neapolitana genes coding for enzymes of carbohydrate substrate degradation. Biochem. Biophys. Res. Commun. 194:1359-1364. [DOI] [PubMed] [Google Scholar]
- 19.Davies, D. G., A. M. Chakrabarty, and G. G. Geesey. 1993. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 59:1181-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies, D. G., and G. G. Geesey. 1995. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl. Environ. Microbiol. 61:860-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Day, M. W., B. T. Hsu, L. Joshua-Tor, J. B. Park, Z. H. Zhou, M. W. Adams, and D. C. Rees. 1992. X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus. Protein Sci. 1:1494-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Domingues, M. R., J. C. Araujo, M. B. A. Varesche, and R. F. Vazoller. 2002. Evaluation of thermophilic anaerobic microbial consortia using fluorescence in situ hybridization (FISH). Water Sci. Technol. 45:27-33. [PubMed] [Google Scholar]
- 23.Donald, R. G. K., D. W. Nees, C. K. Raymond, A. I. Loroch, and R. A. Ludwig. 1986. Characterization of three genomic loci encoding Rhizobium sp. strain ORS571 N2 fixation genes. J. Bacteriol. 165:72-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donlan, R. M. 2000. Biofilm control in industrial water systems: approaching an old problem in new ways, p. 333-360. In L. V. Evans (ed.), Biofilms: recent advances in their study and control. Harwood Academic Publishers, Singapore.
- 25.Eichhorn, E., J. R. van der Ploeg, and T. Leisinger. 2000. Deletion analysis of the Escherichia coli taurine and alkanesulfonate transport systems. J. Bacteriol. 182:2687-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiegler, H., and R. Bruckner. 1997. Identification of the serine acetyltransferase gene of Staphylococcus xylosus. FEMS Microbiol. Lett. 148:181-187. [DOI] [PubMed] [Google Scholar]
- 27.Frazao, C., G. Silva, C. M. Gomes, P. Matias, R. Coelho, L. Sieker, S. Macedo, M. Y. Liu, S. Oliveira, M. Teixeira, A. V. Xavier, C. Rodrigues-Pousada, M. A. Carrondo, and J. Le Gall. 2000. Structure of a dioxygen reduction enzyme from Desulfovibrio gigas. Nat. Struct. Biol. 7:1041-1045. [DOI] [PubMed] [Google Scholar]
- 28.Ghigo, J. M. 2003. Are there biofilm-specific physiological pathways beyond a reasonable doubt? Res. Microbiol. 154:1-8. [DOI] [PubMed] [Google Scholar]
- 29.Golovlev, E. L. 2002. The mechanism of formation of Pseudomonas aeruginosa biofilm, a type of structured population. Microbiology 71:249-254. [PubMed] [Google Scholar]
- 30.Gronger, P., S. S. Manian, H. Reilander, M. O'Connell, U. B. Priefer, and A. Puhler. 1987. Organization and partial sequence of a DNA region of the Rhizobium leguminosarum symbiotic plasmid pRL6JI containing the genes fixABC, nifA, nifB and a novel open reading frame. Nucleic Acids Res. 15:31-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guezennec, J. 1999. Microbial exopolysaccharides from extreme environments. Agro Food Ind. Hi-Tech 10:34-35. [Google Scholar]
- 32.Guo, R. T., C. J. Kuo, C. C. Chou, T. P. Ko, H. L. Shr, P. H. Liang, and A. H. Wang. 2004. Crystal structure of octaprenyl pyrophosphate synthase from hyperthermophilic Thermotoga maritima and mechanism of product chain length determination. J. Biol. Chem. 279:4903-4912. [DOI] [PubMed] [Google Scholar]
- 33.Hantke, K. 2003. Is the bacterial ferrous iron transporter FeoB a living fossil? Trends Microbiol. 11:192-195. [DOI] [PubMed] [Google Scholar]
- 34.Hasseman, J. 2001. TIGR microarray protocols. [Online.] http://www.tigr.org/tdb/microarray/protocolsTIGR.shtml.
- 35.Hinsa, S. M., M. Espinosa-Urgel, J. L. Ramos, and G. A. O'Toole. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49:905-918. [DOI] [PubMed] [Google Scholar]
- 36.Hirata, H., T. Fukazawa, S. Negoro, and H. Okada. 1986. Structure of a β-galactosidase gene of Bacillus stearothermophilus. J. Bacteriol. 166:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber, R., T. A. Langworthy, H. Konig, M. Thomm, C. R. Woese, U. B. Sleytr, and K. O. Stetter. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 144:324-333. [Google Scholar]
- 38.Jenney, F., M. Verhage, X. Cui, and M. W. W. Adams. 1999. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science 286:306-309. [DOI] [PubMed] [Google Scholar]
- 39.Jeong, W., M. K. Cha, and I. H. Kim. 2000. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J. Biol. Chem. 275:2924-2930. [DOI] [PubMed] [Google Scholar]
- 40.Kacklany, S. C., S. B. Levery, J. S. Kim, B. L. Reuhs, L. W. Lion, and W. C. Ghiorse. 2001. Structure and carbohydrate analysis of the exopolysaccharide capsule of Pseudomonas putida G7. Environ. Microbiol. 3:774-784. [DOI] [PubMed] [Google Scholar]
- 41.Kaiser, J. T., T. Clausen, G. P. Bourenkow, H. D. Bartunik, S. Steinbacher, and R. Huber. 2000. Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly. J. Mol. Biol. 297:451-464. [DOI] [PubMed] [Google Scholar]
- 42.Kaminski, P. A., F. Norel, N. Desnoues, A. Kush, G. Salzano, and C. Elmerich. 1988. Characterization of the fixABC region of Azorhizobium caulinodans ORS571 and identification of a new nitrogen fixation gene. Mol. Gen. Genet. 214:496-502. [DOI] [PubMed] [Google Scholar]
- 43.Kammler, M., C. Schön, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kashima, Y., and K. Ishikawa. 2003. Alkyl hydroperoxide reductase dependent on thioredoxin-like protein from Pyrococcus horikoshii. J. Biochem. (Tokyo) 134:25-29. [DOI] [PubMed] [Google Scholar]
- 45.Kashiwagi, K., S. Miyamoto, E. Nukui, H. Kobayashi, and K. Igarashi. 1993. Functions of PotA and PotD proteins in spermidine-preferential uptake system in Escherichia coli. J. Biol. Chem. 268:19358-19363. [PubMed] [Google Scholar]
- 46.Kim, D. Y., D. R. Kim, S. C. Ha, N. K. Lokanath, C. J. Lee, H. Y. Hwang, and K. K. Kim. 2003. Crystal structure of the protease domain of a heat-shock protein HtrA from Thermotoga maritima. J. Biol. Chem. 278:6543-6551. [DOI] [PubMed] [Google Scholar]
- 47.Kim, D. Y., and K. K. Kim. 2002. Crystallization and preliminary X-ray studies of the protease domain of the heat-shock protein HtrA from Thermotoga maritima. Acta Crystallogr. D Biol. Crystallogr. 58:170-172. [DOI] [PubMed] [Google Scholar]
- 48.Kojic, M., and V. Venturi. 2001. Regulation of rpoS gene expression in Pseudomonas: involvement of a TetR family regulator. J. Bacteriol. 183:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolter, R., and R. Losick. 1998. One for all and all for one. Science 280:226-227. [DOI] [PubMed] [Google Scholar]
- 50.Kumano, M., A. Tamakoshi, and K. Yamane. 1997. A 32 kb nucleotide sequence from the region of the lincomycin-resistance gene (22°-25°) of the Bacillus subtilis chromosome and identification of the site of the lin-2 mutation. Microbiology 143:2775-2782. [DOI] [PubMed] [Google Scholar]
- 51.Kuo, T. H., and P. H. Liang. 2002. Reaction kinetic pathway of the recombinant octaprenyl pyrophosphate synthase from Thermotoga maritima: how is it different from that of the mesophilic enzyme. Biochim. Biophys. Acta 1599:125-133. [DOI] [PubMed] [Google Scholar]
- 52.LaPaglia, C., and P. L. Hartzell. 1997. Stress-induced production of biofilm in the hyperthermophile Archaeoglobus fulgidus. Appl. Environ. Microbiol. 63:3158-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lauhon, C. T., and R. Kambampati. 2000. The iscS gene in Escherichia coli is required for the biosynthesis of 4-thiouridine, thiamin, and NAD. J. Biol. Chem. 275:20096-20103. [DOI] [PubMed] [Google Scholar]
- 54.Lee, M. H., Y. W. Kim, T. J. Kim, C. S. Park, J. W. Kim, T. W. Moon, and K. H. Park. 2002. A novel amylolytic enzyme from Thermotoga maritima, resembling cyclodextrinase and alpha-glucosidase, that liberates glucose from the reducing end of the substrates. Biochem. Biophys. Res. Commun. 295:818-825. [DOI] [PubMed] [Google Scholar]
- 55.Lejeune, P. 2003. Contamination of abiotic surfaces: what a colonizing bacterium sees and how to blur it. Trends Microbiol. 11:179-184. [DOI] [PubMed] [Google Scholar]
- 56.Leonardi, R., S. A. Fairhurst, M. Kriek, D. J. Lowe, and P. L. Roach. 2003. Thiamine biosynthesis in Escherichia coli: isolation and initial characterisation of the ThiGH complex. FEBS Lett. 539:95-99. [DOI] [PubMed] [Google Scholar]
- 57.Liebl, W., P. Ruile, K. Bronnenmeier, K. Riedel, F. Lottspeich, and I. Greif. 1996. Analysis of a Thermotoga maritima DNA fragment encoding two similar thermostable cellulases, CelA and CelB, and characterization of the recombinant enzymes. Microbiology 142:2533-2542. [DOI] [PubMed] [Google Scholar]
- 58.Mansy, S. S., G. Wu, K. K. Surerus, and J. A. Cowan. 2002. Iron-sulfur cluster biosynthesis. Thermotoga maritima IscU is a structured iron-sulfur cluster assembly protein. J. Biol. Chem. 277:21397-21404. [DOI] [PubMed] [Google Scholar]
- 59.Mansy, S. S., S. P. Wu, and J. A. Cowan. 2004. Iron-sulfur cluster biosynthesis: biochemical characterization of the conformational dynamics of Thermotoga maritima IscU and the relevance for cellular cluster assembly. J. Biol. Chem. 279:10469-10475. [DOI] [PubMed] [Google Scholar]
- 60.Marlovits, T. C., W. Haase, C. Herrmann, S. G. Aller, and V. M. Unger. 2002. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. USA 99:16243-16248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masepohl, B., F. Fuhrer, and W. Klipp. 2001. Genetic analysis of a Rhodobacter capsulatus gene region involved in utilization of taurine as a sulfur source. FEMS Microbiol. Lett. 205:105-111. [DOI] [PubMed] [Google Scholar]
- 62.Michelini, E. T., and G. C. Flynn. 1999. The unique chaperone operon of Thermotoga maritima: cloning and initial characterization of a functional Hsp70 and small heat shock protein. J. Bacteriol. 181:4237-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muralidharan, V., K. D. Rinker, I. S. Hirsh, E. J. Bouwer, and R. M. Kelly. 1997. Hydrogen transfer between methanogens and fermentative heterotrophs in hyperthermophilic cocultures. Biotechnol. Bioeng. 56:268-278. [DOI] [PubMed] [Google Scholar]
- 64.Nachin, L., L. Loiseau, D. Expert, and F. Barras. 2003. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 22:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, L. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 66.Neves, A. R., R. Ventura, N. Mansour, C. Shearman, M. J. Gasson, C. Maycock, A. Ramos, and H. Santos. 2002. Is the glycolytic flux in Lactococcus lactis primarily controlled by the redox charge? Kinetics of NAD+ and NADH pools determined by 13C NMR. J. Biol. Chem. 277:28088-28098. [DOI] [PubMed] [Google Scholar]
- 67.Nicolaus, B., M. C. Manca, I. Romano, and L. Lama. 1993. Production of an exopolysaccharide from two thermophilic archaea belonging to the genus Sulfolobus. FEMS Microbiol. Lett. 109:203-206. [Google Scholar]
- 68.Nidetzky, B., C. Eis, and M. Albert. 2000. Role of non-covalent enzyme-substrate interactions in the reaction catalysed by cellobiose phosphorylase from Cellulomonas uda. Biochem. J. 351:649-659. [PMC free article] [PubMed] [Google Scholar]
- 69.Olabarria, G., L. A. Fernandez-Herrero, J. L. Carrascosa, and J. Berenguer. 1996. slpM, a gene coding for an “S-layer-like array” overexpressed in S-layer mutants of Thermus thermophilus HB8. J. Bacteriol. 178:357-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ollagnier-de-Choudens, S., D. Lascoux, L. Loiseau, F. Barras, E. Forest, and M. Fontecave. 2003. Mechanistic studies of the SufS-SufE cysteine desulfurase: evidence for sulfur transfer from SufS to SufE. FEBS Lett. 555:263-267. [DOI] [PubMed] [Google Scholar]
- 71.Oosthuizen, M. C., B. Steyn, J. Theron, P. Cosette, D. Lindsay, A. von Holy, and V. S. Brözel. 2002. Proteomic analysis reveals differential protein expression by Bacillus cereus during biofilm formation. Appl. Environ. Microbiol. 68:2770-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ostendorp, R., W. Liebl, H. Schurig, and R. Jaenicke. 1993. The L-lactate dehydrogenase gene of the hyperthermophilic bacterium Thermotoga maritima cloned by complementation in Escherichia coli. Eur. J. Biochem. 216:709-715. [DOI] [PubMed] [Google Scholar]
- 73.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 74.Pan, G., A. L. Menon, and M. W. Adams. 2003. Characterization of a [2Fe-2S] protein encoded in the iron-hydrogenase operon of Thermotoga maritima. J. Biol. Inorg. Chem. 8:469-474. [DOI] [PubMed] [Google Scholar]
- 75.Park, S., and J. A. Imlay. 2003. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J. Bacteriol. 185:1942-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Phadtare, S., J. Hwang, K. Severinov, and M. Inouye. 2003. CspB and CspL, thermostable cold-shock proteins from Thermotoga maritima. Genes Cells 8:801-810. [DOI] [PubMed] [Google Scholar]
- 77.Pysz, M. A., K. D. Rinker, K. R. Shockley, and R. M. Kelly. 2001. Continuous cultivation of hyperthermophiles. Hyperthermophilic Methods Enzymol. 330:31-40. [DOI] [PubMed] [Google Scholar]
- 78.Pysz, M. A., D. E. Ward, K. R. Shockley, C. I. Montero, S. B. Conners, M. R. Johnson, and R. M. Kelly. 2004. Transcriptional analysis of dynamic heat-shock response by the hyperthermophilic bacterium Thermotoga maritima. Extremophiles 8:209-217. [DOI] [PubMed] [Google Scholar]
- 79.Rajashekhara, E., M. Kitaoka, Y. K. Kim, and K. Hayashi. 2002. Characterization of a cellobiose phosphorylase from a hyperthermophilic eubacterium, Thermotoga maritima MSB8. Biosci. Biotechnol. Biochem. 66:2578-2586. [DOI] [PubMed] [Google Scholar]
- 80.Rangachari, K., C. T. Davis, J. F. Eccleston, E. M. Hirst, J. W. Saldanha, M. Strath, and R. J. Wilson. 2002. SufC hydrolyzes ATP and interacts with SufB from Thermotoga maritima. FEBS Lett. 514:225-228. [DOI] [PubMed] [Google Scholar]
- 81.Rathgeber, C., N. Yurkova, E. Stackebrandt, J. T. Beatty, and V. Yurkov. 2002. Isolation of tellurite- and selenite-resistant bacteria from hydrothermal vents of the Juan de Fuca Ridge in the Pacific Ocean. Appl. Environ. Microbiol. 68:4613-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ravasio, S., B. Curti, and M. A. Vanoni. 2001. Determination of the midpoint potential of the FAD and FMN flavin cofactors and of the 3Fe-4S cluster of glutamate synthase. Biochemistry 40:5533-5541. [DOI] [PubMed] [Google Scholar]
- 83.Reysenbach, A. L., and E. Shock. 2002. Merging genomes with geochemistry in hydrothermal ecosystems. Science 296:1077-1082. [DOI] [PubMed] [Google Scholar]
- 84.Rinker, K. D. 1998. Growth physiology and bioenergetics of the hyperthermophilic archaeon Thermococcus litoralis and bacterium Thermotoga maritima. Ph.D. thesis. North Carolina State University, Raleigh.
- 85.Rinker, K. D., C. J. Han, and R. M. Kelly. 1999. Continuous culture as a tool for investigating the growth physiology of heterotrophic hyperthermophiles and extreme thermoacidophiles. J. Appl. Microbiol. 85:118-127. [DOI] [PubMed] [Google Scholar]
- 86.Rinker, K. D., and R. M. Kelly. 2000. Effect of carbon and nitrogen sources on growth dynamics and exopolysaccharide production for the hyperthermophilic archaeon Thermococcus litoralis and bacterium Thermotoga maritima. Biotechnol. Bioeng. 69:537-547. [DOI] [PubMed] [Google Scholar]
- 87.Rinker, K. D., and R. M. Kelly. 1996. Growth physiology of the hyperthermophilic archaeon Thermococcus litoralis: development of a sulfur-free defined medium, characterization of an exopolysaccharide, and evidence of biofilm formation. Appl. Environ. Microbiol. 62:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sauer, K. 27 May 2003, posting date. The genomics and proteomics of biofilm formation. Genome Biol. 4:219. [Online.] http://genomebiology.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schembri, M. A., K. Kjaergaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 92.Schwartz, C. J., J. L. Giel, T. Patschkowski, C. Luther, F. J. Ruzicka, H. Beinert, and P. J. Kiley. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 98:14895-14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sharma, R., C. Rensing, B. P. Rosen, and B. Mitra. 2000. The ATP hydrolytic activity of purified ZntA, a Pb(II)/Cd(II)/Zn(II)-translocating ATPase from Escherichia coli. J. Biol. Chem. 275:3873-3878. [DOI] [PubMed] [Google Scholar]
- 94.Snel, B., G. Lehmann, P. Bork, and M. A. Huynen. 2000. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 28:3442-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stearman, R., D. S. Yuan, Y. Yamaguchi-Iwai, R. D. Klausner, and A. Dancis. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552-1557. [DOI] [PubMed] [Google Scholar]
- 96.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 97.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 98.Sun, D., and P. Setlow. 1993. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis nadB gene and a nifS-like gene, both of which are essential for NAD biosynthesis. J. Bacteriol. 175:1423-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Suresh, C., M. Kitaoka, and K. Hayashi. 2003. A thermostable non-xylanolytic alpha-glucuronidase of Thermotoga maritima MSB8. Biosci. Biotechnol. Biochem. 67:2359-2364. [DOI] [PubMed] [Google Scholar]
- 100.Svensater, G., J. Welin, J. C. Wilkins, D. Beighton, and I. R. Hamilton. 2001. Protein expression by planktonic and biofilm cells of Streptococcus mutans. FEMS Microbiol. Lett. 205:139-146. [DOI] [PubMed] [Google Scholar]
- 101.Takahashi, Y., and U. Tokumoto. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277:28380-28383. [DOI] [PubMed] [Google Scholar]
- 102.Tantaleán, J. C., M. A. Araya, C. P. Saavedra, D. E. Fuentes, J. M. Pérez, I. L. Calderón, P. Youderian, and C. C. Vásquez. 2003. The Geobacillus stearothermophilus V iscS gene, encoding cysteine desulfurase, confers resistance to potassium tellurite in Escherichia coli K-12. J. Bacteriol. 185:5831-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tao, X., R. Khayat, D. Christendat, A. Savchenko, X. Xu, S. Goldsmith-Fischman, B. Honig, A. Edwards, C. H. Arrowsmith, and L. Tong. 2003. Crystal structures of MTH1187 and its yeast ortholog YBL001c. Proteins 52:478-480. [DOI] [PubMed] [Google Scholar]
- 104.Taylor, S. V., N. L. Kelleher, C. Kinsland, H. J. Chiu, C. A. Costello, A. D. Backstrom, F. W. McLafferty, and T. P. Begley. 1998. Thiamin biosynthesis in Escherichia coli. Identification of this thiocarboxylate as the immediate sulfur donor in the thiazole formation. J. Biol. Chem. 273:16555-16560. [DOI] [PubMed] [Google Scholar]
- 105.Tolker-Nielsen, T., U. C. Brinch, P. C. Ragas, J. B. Andersen, C. S. Jacobsen, and S. Molin. 2000. Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 182:6482-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tremoulet, F., O. Duche, A. Namane, B. Martinie, and J. C. Labadie. 2002. A proteomic study of Escherichia coli O157:H7 NCTC 12900 cultivated in biofilm or in planktonic growth mode. FEMS Microbiol. Lett. 215:7-14. [DOI] [PubMed] [Google Scholar]
- 107.Tremoulet, F., O. Duche, A. Namane, B. Martinie, The European Listeria Genome Consortium, and J. C. Labadie. 2002. Comparison of protein patterns of Listeria monocytogenes grown in biofilm or in planktonic mode by proteome analysis. FEMS Microbiol. Lett. 210:25-31. [DOI] [PubMed] [Google Scholar]
- 108.Ugulava, N. B., B. R. Gibney, and J. T. Jarrett. 2001. Biotin synthase contains two distinct iron-sulfur cluster binding sites: chemical and spectroelectrochemical analysis of iron-sulfur cluster interconversions. Biochemistry 40:8343-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Valdes-Stauber, N., and S. Scherer. 1996. Nucleotide sequence and taxonomical distribution of the bacteriocin gene lin cloned from Brevibacterium linens M18. Appl. Environ. Microbiol. 62:1283-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.van der Ploeg, J. R., R. Iwanicka-Nowicka, T. Bykowski, M. M. Hryniewicz, and T. Leisinger. 1999. The Escherichia coli ssuEADCB gene cluster is required for the utilization of sulfur from aliphatic sulfonates and is regulated by the transcriptional activator Cbl. J. Biol. Chem. 274:29358-29365. [DOI] [PubMed] [Google Scholar]
- 111.van der Ploeg, J. R., M. A. Weiss, E. Saller, H. Nashimoto, N. Saito, M. A. Kertesz, and T. Leisinger. 1996. Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J. Bacteriol. 178:5438-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.van Kranenburg, R., H. R. Vos, I. I. van Swam, M. Kleerebezem, and W. M. de Vos. 1999. Functional analysis of glycosyltransferase genes from Lactococcus lactis and other gram-positive cocci: complementation, expression, and diversity. J. Bacteriol. 181:6347-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vasquez, C. C., C. P. Saavedra, C. A. Loyola, M. A. Araya, and S. Pichuantes. 2001. The product of the cysK gene of Bacillus stearothermophilus V mediates potassium tellurite resistance in Escherichia coli. Curr. Microbiol. 43:418-423. [DOI] [PubMed] [Google Scholar]
- 114.Verhagen, M. F., T. O'Rourke, and M. W. Adams. 1999. The hyperthermophilic bacterium, Thermotoga maritima, contains an unusually complex iron-hydrogenase: amino acid sequence analyses versus biochemical characterization. Biochim. Biophys. Acta 1412:212-229. [DOI] [PubMed] [Google Scholar]
- 115.Wakagi, T. 2003. Sulerythrin, the smallest member of the rubrerythrin family, from a strictly aerobic and thermoacidophilic archaeon, Sulfolobus tokodaii strain 7. FEMS Microbiol. Lett. 222:33-37. [DOI] [PubMed] [Google Scholar]
- 116.Walt, A., and M. L. Kahn. 2002. The fixA and fixB genes are necessary for anaerobic carnitine reduction in Escherichia coli. J. Bacteriol. 184:4044-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ward, D. E., C. J. Donnelly, M. E. Mullendore, J. van der Oost, W. M. de Vos, and E. J. Crane. 2001. The NADH oxidase from Pyrococcus furiosus. Implications for the protection of anaerobic hyperthermophiles against oxidative stress. Eur. J. Biochem. 268:5816-5823. [DOI] [PubMed] [Google Scholar]
- 118.Wassenberg, D., W. Liebl, and R. Jaenicke. 2000. Maltose-binding protein from the hyperthermophilic bacterium Thermotoga maritima: stability and binding properties. J. Mol. Biol. 295:279-288. [DOI] [PubMed] [Google Scholar]
- 119.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Welker, C., G. Bohm, H. Schurig, and R. Jaenicke. 1999. Cloning, overexpression, purification, and physicochemical characterization of a cold shock protein homolog from the hyperthermophilic bacterium Thermotoga maritima. Protein Sci. 8:394-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 122.Wolfinger, R. D., G. Gibson, E. D. Wolfinger, L. Bennett, H. Hamadeh, P. Bushel, C. Afshari, and R. S. Paules. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8:625-637. [DOI] [PubMed] [Google Scholar]
- 123.Yan, L., K. G. Boyd, D. R. Adams, and J. G. Burgess. 2003. Biofilm-specific cross-species induction of antimicrobial compounds in bacilli. Appl. Environ. Microbiol. 69:3719-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang, Z., A. Savchenko, A. Yakunin, R. Zhang, A. Edwards, C. Arrowsmith, and L. Tong. 2003. Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643. J. Biol. Chem. 278:8804-8808. [DOI] [PubMed] [Google Scholar]
- 125.Yeh, A. P., Y. Hu, F. E. Jenney, Jr., M. W. Adams, and D. C. Rees. 2000. Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states. Biochemistry 39:2499-2508. [DOI] [PubMed] [Google Scholar]
- 126.Yernool, D. A., J. K. McCarthy, D. E. Eveleigh, and J. D. Bok. 2000. Cloning and characterization of the glucooligosaccharide catabolic pathway β-glucan glucohydrolase and cellobiose phosphorylase in the marine hyperthermophile Thermotoga neapolitana. J. Bacteriol. 182:5172-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yu, H., M. J. Schurr, and V. Deretic. 1995. Functional equivalence of Escherichia coli σE and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J. Bacteriol. 177:3259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]