Abstract
Large amounts of tylosin, zinc-bacitracin, and avilamycin are currently used as prophylactics in New Zealand broiler production. Avoparcin was also used from 1977 to 2000. A total of 382 enterococci were isolated from 213 fecal samples (147 individual poultry farms) using enrichment broths plated on m-Enterococcus agar lacking antimicrobials. These isolates were then examined to determine the prevalence of antimicrobial resistance. Of the 382 isolates, 5.8% (22 isolates) were resistant to vancomycin, and 64.7% were resistant to erythromycin. The bacitracin MIC was ≥256 μg/ml for 98.7% of isolates, and the avilamycin MIC was ≥8 μg/ml for 14.9% of isolates. No resistance to ampicillin or gentamicin was detected. Of the 22 vancomycin-resistant enterococci (VRE) isolates, 18 (81.8%) were Enterococcus faecalis, 3 were Enterococcus faecium, and 1 was Enterococcus durans. However, when the 213 fecal enrichment broths were plated on m-Enterococcus agar containing vancomycin, 86 VRE were recovered; 66% of these isolates were E. faecium and the remainder were E. faecalis. Vancomycin-resistant E. faecium isolates were found to have heterogenous pulsed-field gel electrophoresis (PFGE) patterns of SmaI-digested DNA, whereas the PFGE patterns of vancomycin-resistant E. faecalis isolates were identical or closely related, suggesting that this VRE clone is widespread throughout New Zealand. These data demonstrate that vancomycin-resistant E. faecalis persists in the absence and presence of vancomycin-selective pressure, thus explaining the dominance of this VRE clone even in the absence of avoparcin.
In the last 2 decades, enterococci have emerged as an important cause of nosocomial infection; this has resulted in an increased interest in identifying reservoirs of both antimicrobial-resistant strains of enterococci and genes encoding resistance determinants. Food animals have been previously implicated as reservoirs for antibiotic-resistant enterococci, following the use of antimicrobial growth promotants and prophylactics (9). Use of avoparcin in food animals has been linked to an increase in vancomycin-resistant enterococci (VRE) in broilers and pigs (8). Because of the potential for spread of resistance through the food chain, a European Union-wide ban was imposed in 1997 on avoparcin use in animal husbandry. Since this discontinuation, a decrease in the prevalence of VRE in Danish poultry has been observed (5). This trend has not been seen in Norway (10, 11); however, in both countries it has been reported that VRE are still present in broilers 5 years after avoparcin was discontinued (16). In Denmark, broiler sheds have been identified as potential sources of recolonization and persistence of VRE strains (17).
In New Zealand, the use of avoparcin was discontinued in June 2000, after being used as a prophylactic in poultry production since 1977. A previous study in New Zealand found that VRE were only isolated from farms that were using, or had used, avoparcin as a growth promotant (24). Interestingly, 82% of VRE isolated from New Zealand broilers were found to be VanA-type Enterococcus faecalis isolates (24), a situation not seen in Europe where VanA-type Enterococcus faecium isolates make up the majority of VRE isolated from human, animal, or environmental sources (1, 9, 10, 12, 13, 19, 26). A study of vancomycin susceptibility in humans and animals in Denmark found that <1% of VRE isolated were E. faecalis (1). All New Zealand poultry vancomycin-resistant E. faecalis (VREF) recovered to date have closely related pulsed-field gel electrophoresis (PFGE) patterns, indicating a high degree of clonality (24).
In New Zealand, the nationwide prevalence of VRE in poultry farms is unknown, previous work having surveyed samples from only eight poultry farms (24). In this communication, we carried out a comprehensive survey of 147 individual poultry farms from all three major New Zealand poultry suppliers. Large amounts of tylosin, zinc-bacitracin, and the oligosaccharide antimicrobial avilamycin are used as prophylactics in New Zealand broiler production. The prevalence of resistance to these compounds in New Zealand broiler-derived enterococci is unknown. We report here on both the level of resistance to these antimicrobial compounds and the prevalence and species distribution of VRE in New Zealand poultry farms 2 years after the discontinuation of avoparcin.
MATERIALS AND METHODS
Bacterial isolates.
Poultry-derived enterococci were isolated from broilers from three poultry suppliers as described below. E. faecalis ATCC 29212 and E. faecium ATCC 19434 were used to generate intragenic probes for species identification. E. faecalis AR01/DG (23) was used to generate intragenic probes for vanA and ermB.
Sampling and isolation of VRE from poultry.
From February 2002 to February 2003, broiler fecal samples were obtained from 147 broiler farms (one individual broiler fecal sample per farm). These farms were run by three suppliers: A (97 farms), B (30 farms), and C (20 farms). Sixty-six fecal samples were also obtained from individual broilers from five antimicrobial-free trial farms (supplier A). For each fecal sample, a pea-sized scoop (approximately 0.5 g) was emulsified in 10 ml of Streptococcus faecalis (enrichment) medium (Bacto SF Medium; Difco Laboratories, Detroit, Mich.). After incubation for 48 h at 35°C, dilutions were made to 10−3, and 100 μl was spread onto m-Enterococcus agar plates (Becton Dickinson and Co., Sparks, Md.) that either were antimicrobial free or contained 32 μg of vancomycin/ml. All m-Enterococcus agar plates were incubated for 48 h at 37°C. From each fecal sample, two enterococci were chosen from the antimicrobial-free m-Enterococcus plates to be representative of the enterococcal population for that sample. If colonies were present on the medium containing vancomycin, one colony was chosen for further antimicrobial susceptibility testing. Chosen colonies were plated onto bile esculin azide agar plates (Becton Dickinson and Co.) to further select and purify single colonies of enterococci.
Antimicrobial susceptibility testing.
MICs for the isolates of vancomycin (American Pharmaceutical Partners, Inc., Los Angeles, Calif.), erythromycin (Sigma-Aldrich Chemicals, St. Louis, Mo.), gentamicin (Sigma), ampicillin (Roche Molecular Biochemicals, Mannheim, Germany), bacitracin (50,000 IU/g; Aldrich Chemical Co., Milwaukee, Wis.), and avilamycin (Eli Lilly and Co., Indianapolis, Ind.) were determined by microdilution following National Committee for Clinical Laboratory Standards (NCCLS) guidelines (25). As no NCCLS breakpoints exist for avilamycin and bacitracin, the resistant breakpoint concentrations used were as follows: avilamycin, MIC ≥ 8 μg/ml; bacitracin, MIC ≥ 256 μg/ml. E. faecalis ATCC 29212 and E. faecium ATCC 19434 were used as quality control organisms (i.e., fully susceptible).
PCR amplification.
For species identification, intragenic probes for efaA and aac(6′)-Ii were generated by PCR from E. faecalis ATCC 29212 and E. faecium ATCC 19434, respectively, as described previously (24). An intragenic probe for vanA and ermB (24) was generated from E. faecalis AR01/DG (23). All PCR products were sequenced to ensure homology with the published sequence for these genes. PCRs were performed with 100-μl volumes with 1 U of Taq DNA Polymerase (Roche Molecular Biochemicals), in accordance with the manufacturer's instructions and the PCR program described previously (21). The DNA template was prepared by dissolving a bacterial colony in 50 μl of water and freeze-thawing at −20°C. PCR products were purified with a PCR purification kit (Roche).
Southern hybridization.
The DNA from enterococcal colonies was bound to nitrocellulose filters as previously described (24). Radiolabeled PCR products were prepared by incorporation of [α-32P]dCTP-labeled deoxynucleotides (Amersham Pharmacia Biotech, Buckinghamshire, England) using Ready-To-Go DNA labeling beads (Amersham). Southern transfer and hybridization were performed as previously described (24).
PFGE.
Genomic DNA embedded in agarose was prepared essentially as described by Keis et al. (21) for the preparation of clostridial genomic DNA, except that all steps were carried out aerobically and bacteria were grown to an optical density at 650 nm of 0.6 in 10 ml of brain heart infusion broth. DNA plugs were digested with SmaI as previously described (24). PFGE was performed by contour-clamped homogeneous electric field electrophoresis with the CHEF-DRIII system (Bio-Rad Laboratories, Richmond, Calif.). Gels were routinely run at 6 V/cm at 14°C at an included angle of 120° on a 1.2% agarose gel (Amersham) with pulse times of 5 to 25 s for 22 h. The Low Range PFG marker (New England Biolabs, Inc., Beverley, Mass.) containing lambda concatemers and lambda-digested HindIII fragments was used as a size standard.
DNA sequencing and analysis.
PCR products were sequenced directly. Sequencing reactions were carried out by using a PRISM Ready Reaction DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems, Inc., Warrington, United Kingdom) and a model ABI377 automated DNA sequencer (Applied Biosystems).
RESULTS
Isolation and characterization of VRE isolates from poultry.
From February 2002 to February 2003, individual broiler fecal samples were obtained from three different poultry suppliers (A, B, and C) from a total of 147 individual farms. A further 66 broiler fecal samples were obtained from five antimicrobial-free trial farms (supplier A). A total of 382 enterococci were isolated from 213 fecal samples (147 individual poultry farms) using enrichment broths plated on m-Enterococcus agar lacking antimicrobials (Table 1). With species-specific intragenic probes, 152 (40%) isolates were identified as E. faecalis, 181 (47%) were E. faecium, and 49 (13%) consisted of other unidentified Enterococcus species. Of the 382 isolates, 5.8% were vancomycin resistant and 64.7% were erythromycin resistant. The bacitracin MIC for 98.7% of isolates was ≥256 μg/ml, and the avilamycin MIC for 14.9% of isolates was ≥8 μg/ml (Table 1). No resistance to ampicillin or gentamicin was detected. All vancomycin-resistant isolates contained the vanA gene, while 85% (210) of the 247 erythromycin-resistant isolates contained the ermB gene. Of the 22 vancomycin-resistant isolates, 18 (81.8%) were E. faecalis, 3 (13.6%) were E. faecium, and 1 was Enterococcus durans. Twenty of the 22 VRE were also erythromycin resistant and carried the ermB gene. The avilamycin MIC for 4 isolates was ≥256 μg/ml, and the bacitracin MIC for 22 isolates was ≥256 μg/ml.
TABLE 1.
Antimicrobial resistance profiles in enterococci isolated from broiler fecal samples
| Suppliera | No. of isolates testeda | Number (%) resistant
|
|||
|---|---|---|---|---|---|
| Vancomycin | Avilamycinc | Erythromycin | Bacitracinc | ||
| A | 185 | 12 (6.5) | 34 (18.4) | 145 (78.4) | 183 (98.9) |
| Ab | 104 | 4 (3.9) | 19 (18.3) | 77 (74) | 103 (99) |
| B | 56 | 1 (1.8) | 2 (3.6) | 10 (17.9) | 55 (98.2) |
| C | 37 | 5 (13.5) | 2 (5.4) | 15 (40.5) | 36 (97.3) |
| Total | 382 | 22 (5.8) | 57 (14.9) | 247 (64.7) | 377 (98.7) |
Enterococci were isolated using m-Enterococcus agar lacking antimicrobials.
Samples taken from supplier A, with broilers currently on an antimicrobial-free feeding regime.
See Materials and Methods for breakpoints used.
When the 213 fecal enrichment broths were plated on m-Enterococcus agar containing vancomycin, a total of 86 broths were positive for VRE. The prevalence of VRE in broiler fecal samples from different suppliers varied from 27 to 52%. Importantly, we noted a change in the species distribution of VRE isolates when vancomycin was included in the isolation medium. With this procedure, 57 (66%) of these VRE isolates were E. faecium isolates containing the vanA gene, and 29 (34%) were E. faecalis isolates containing the vanA gene. Of the 86 VRE, 79 were resistant to erythromycin, and all of these contained the ermB gene. Avilamycin MICs were ≥8 μg/ml for 11 VRE, and bacitracin MICs were ≥256 μg/ml for 84 VRE.
DNA fingerprinting of VRE from poultry.
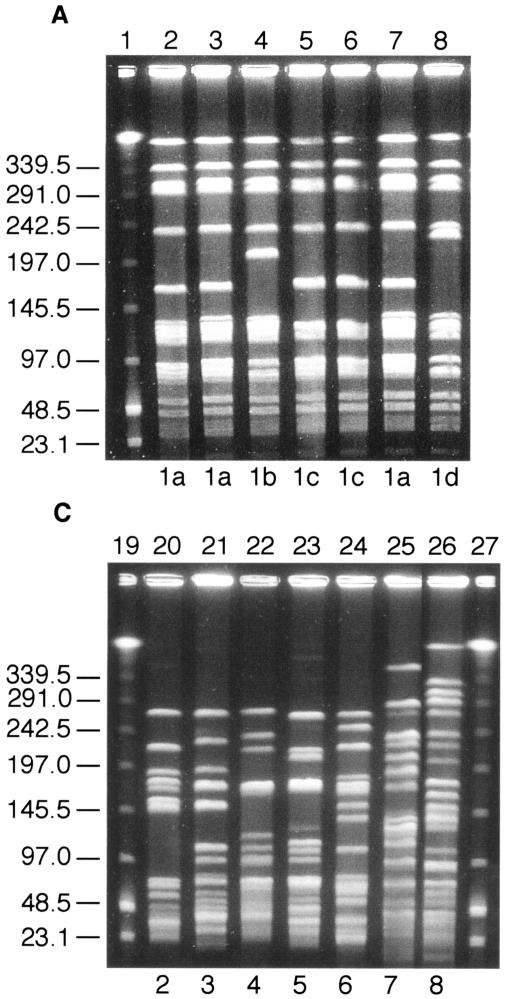
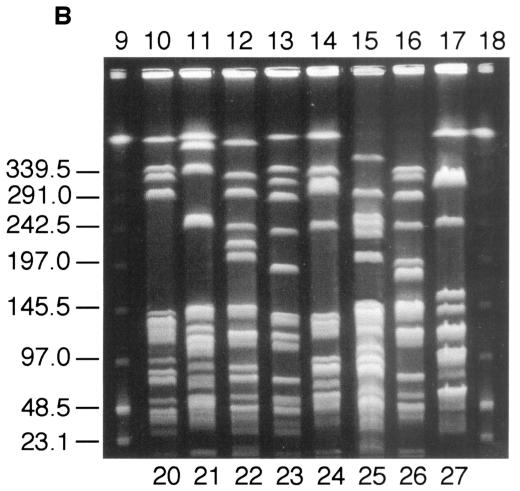
DNA fingerprinting of the poultry VRE was carried out by comparing the SmaI digestion patterns obtained after PFGE, using the criteria described by Tenover et al. (27). Thirty-two VREF isolates were chosen at random from all three suppliers representing different geographical areas. Of these, all had PFGE pattern 1a (identical to the previously described PFGE pattern of the VREF poultry clone) (24) or the closely related patterns 1b, 1c, or 1d (Fig. 1A). In comparison, eight randomly chosen vancomycin-sensitive E. faecalis isolates showed no relatedness in their PFGE patterns (Fig. 1B). Twenty vancomycin-resistant E. faecium isolates were also examined, and 18 unrelated PFGE fingerprints were present, 7 of which are shown in Fig. 1C.
FIG. 1.
Representative SmaI PFGE patterns among VRE isolated from broilers. (A) VREF isolates. Lane 1, lambda DNA ladder standard; lanes 2 to 3, PFGE pattern 1a; lane 4, PFGE pattern 1b; lanes 5 to 6, PFGE pattern 1c; lane 7, PFGE pattern 1a; lane 8, PFGE pattern 1d. (B) Vancomycin-sensitive E. faecalis isolates. Lane 9, lambda DNA ladder standard; lane 10, PFGE pattern 20; lane 11, PFGE pattern 21; lane 12, PFGE pattern 22; lane 13, PFGE pattern 23; lane 14, PFGE pattern 24; lane 15, PFGE pattern 25; lane 16, PFGE pattern 26; lane 17, PFGE pattern 27; lane 18, lambda DNA ladder standard. (C) Vancomycin-resistant E. faecium isolates. Lane 19, lambda DNA ladder standard; lane 20, PFGE pattern 2; lane 21, PFGE pattern 3; lane 22, PFGE pattern 4; lane 23, PFGE pattern 5; lane 24, PFGE pattern 6; lane 25, PFGE pattern 7; lane 26, PFGE pattern 8; lane 27, lambda DNA ladder standard. Numbers to the left of all gels indicate molecular mass markers, in kilodaltons.
DISCUSSION
This is the first report of the nationwide prevalence of antimicrobial resistance in broiler chickens in New Zealand. Across 147 poultry farms, the prevalence of VRE in the enterococcal population was approximately 5.8%, while 27 to 52% of broiler fecal samples contained VRE. Three years after the discontinuation of avoparcin in Norway, it was found that in 99% of broiler fecal samples from previously avoparcin-exposed farms VRE were still present (10). In Denmark, it was reported that the prevalence of VRE in broilers dropped from over 80% in 1995 to less than 5% in 1998 (5); however, a recent study conducted 5 years after the discontinuation of avoparcin found VRE in 74.5% of broiler flocks (16). The data from these two Danish studies were obtained by different isolation procedures, and therefore it is difficult to make valid comparisons. What we can see from all three countries is the persistence of VRE in broilers, even in the absence of the selective pressure imposed by avoparcin use.
Our survey of antimicrobial resistance in broiler animals was based on resistance-monitoring protocols utilized by the Danish Integrated Antimicrobial Resistance Monitoring and Research Programme (DANMAP). The determination of antimicrobial resistance among food animals in Denmark uses randomly selected bacterial isolates (e.g., one isolate per flock or herd) with a target number of 200 E. faecalis and/or E. faecium isolates per year (6). While this sampling strategy will detect only the most prevalent microflora, changes in resistance can be compared over time and used to reveal trends in the incidence of antimicrobial resistance (2-5, 7, 20). Kühn et al. (22) has reported that the diversity of the enterococcal population obtained in such analyses is comparable, irrespective of whether many samples (one isolate analyzed per sample) are collected or only a few samples (several isolates analyzed per sample) are analyzed. In the DANMAP surveillance program, broiler cloacal swabs were routinely enriched overnight in Enterococcosel broth before being plated onto solid medium (5, 7, 14, 20). The use of a broth enrichment step has been noted in several studies as effective at increasing the detection level of VRE in fecal samples (15, 18), and we therefore used this procedure in the present study.
Although the use of avoparcin has been discontinued in New Zealand, three other antimicrobial compounds are currently used in broiler production. Zinc-bacitracin use is extensive, and the resistance levels to this compound reflect this. The zinc-bacitracin MIC for nearly 99% of poultry-derived enterococci is ≥256 μg/ml. The use of avilamycin is not as prevalent, and the percentage of isolates for which the MIC is ≥8 μg/ml varied from 3.6 to 18.4%. Tylosin is the other major antimicrobial used in broiler production, and resistance to this compound provides cross-resistance to the related macrolide erythromycin. The levels of resistance to erythromycin varied markedly in samples from different suppliers (17.9 to 78.4%). There was a notable difference in resistance levels to erythromycin (tylosin) in supplier A isolates (78.4%), in comparison to isolates from supplier B (17.9%) and supplier C (40.5%). Interestingly, no real difference in antimicrobial resistance levels was seen between isolates obtained from antimicrobial-free and standard poultry farms owned by supplier A. The reason for this is unknown but may be due to the short length of time that these poultry farms have been antimicrobial free; therefore, future monitoring is warranted. Furthermore, the impact of contaminated broiler sheds, animal feed, or animals on antimicrobial-resistant bacteria cannot be ruled out.
A high percentage of the VRE isolated (91.6%) were erythromycin resistant and contained the ermB gene. A previous study of VRE isolated from broilers in New Zealand showed that vancomycin resistance was genetically linked to resistance to macrolides (24). This suggests that use of tylosin could select for the persistence of VRE in New Zealand poultry. Levels of erythromycin resistance are also high in vancomycin-susceptible isolates, but these levels varied from 76.8% in supplier A isolates to 16% in supplier C isolates. The percentage of isolates for which MICs of zinc-bacitracin and avilamycin were raised was similar to levels found for vancomycin-susceptible isolates, suggesting that these antimicrobials are not selecting for vancomycin resistance.
The most striking difference in antimicrobial sensitivity patterns in New Zealand broilers, compared to resistance patterns in other countries (1, 9, 10, 12, 13, 19, 26), is the predominance of VanA-type E. faecalis (VREF) (24). PFGE of both vancomycin-sensitive E. faecalis and vancomycin-resistant E. faecium isolates showed a heterogenous population. PFGE of VREF isolates showed that all were closely related and of the same clonal lineage that had previously been described in New Zealand poultry and humans (24). This VREF clone was found in samples from all three suppliers and in samples from all geographical regions of New Zealand. The fact that vancomycin-susceptible E. faecalis isolates showed no relatedness to the predominant VREF clone further supports the idea of clonal dissemination rather than horizontal transfer of vancomycin resistance. Consistent with this hypothesis is the low frequency of vancomycin resistance transfer noted with this VREF clone (24).
Differences in species distribution were also noted when comparing data using the two different isolation methodologies employed. In VRE that were isolated from medium lacking vancomycin, 81% of isolates were found to carry vanA-positive E. faecalis. This was not the case when enrichments were plated directly on medium containing vancomycin, where only 35% of the VRE were E. faecalis. The reason for the difference in species distribution may be due to the retention of vancomycin resistance in the VREF clone in a nonselective vancomycin-free environment. It has previously been shown that vancomycin resistance in several VREF strains was rapidly lost upon transfer to medium lacking vancomycin (24). In contrast, subculturing of the common poultry VREF clone continuously for 28 days in medium lacking vancomycin revealed no loss of resistance (24). This may explain the dominance of VREF when vancomycin resistance is not selected for, especially if the VREF isolates belong to the common poultry clonal lineage.
In New Zealand, a clonal lineage of VREF is present nationwide in broiler animals from all major suppliers. The dominance of this clone suggests either an ecological fitness advantage over other VRE isolates or a higher stability of vanA resistance determinants in the absence of selection pressure. Further work is needed to ascertain what novel traits this clone has that lead to its predominance in VRE isolated from broilers in New Zealand.
Acknowledgments
This work was funded by an Otago Medical Research Foundation grant.
We thank the poultry industry of New Zealand for their cooperation in this study.
REFERENCES
- 1.Aarestrup, F. M., P. Ahrens, M. Madsen, L. V. Pallesen, R. L. Poulsen, and H. Westh. 1996. Glycopeptide susceptibility among Danish Enterococcus faecium and Enterococcus faecalis isolates of animal and human origin and PCR identification of genes within the VanA cluster. Antimicrob. Agents Chemother. 40:1938-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aarestrup, F. M., F. Bager, N. E. Jensen, M. Madsen, A. Meyling, and H. C. Wegener. 1998. Surveillance of antimicrobial resistance in bacteria isolated from food animals to antimicrobial growth promoters and related therapeutic agents in Denmark. APMIS 106:606-622. [DOI] [PubMed] [Google Scholar]
- 3.Aarestrup, F. M., H. Kruse, E. Tast, A. M. Hammerum, and L. B. Jensen. 2000. Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microb. Drug Resist. 6:63-70. [DOI] [PubMed] [Google Scholar]
- 4.Aarestrup, F. M., and P. M. McNicholas. 2002. Incidence of high-level evernimicin resistance in Enterococcus faecium among food animals and humans. Antimicrob. Agents Chemother. 46:3088-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aarestrup, F. M., A. M. Seyfarth, H. D. Emborg, K. Pedersen, R. S. Hendriksen, and F. Bager. 2001. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bager, F. 2000. DANMAP: monitoring antimicrobial resistance in Denmark. Int. J. Antimicrob. Agents 14:271-274. [DOI] [PubMed] [Google Scholar]
- 7.Bager, F., F. M. Aarestrup, M. Madsen, and H. C. Wegener. 1999. Glycopeptide resistance in Enterococcus faecium from broilers and pigs following discontinued use of avoparcin. Microb. Drug Resist. 5:53-56. [DOI] [PubMed] [Google Scholar]
- 8.Bager, F., M. Madsen, J. Christensen, and F. M. Aarestrup. 1997. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev. Vet. Med. 31:95-112. [DOI] [PubMed] [Google Scholar]
- 9.Bates, J., Z. Jordens, and J. B. Selkon. 1993. Evidence for an animal origin of vancomycin-resistant enterococci. Lancet 342:490-491. [DOI] [PubMed] [Google Scholar]
- 10.Borgen, K., G. S. Simonsen, A. Sundsfjord, Y. Wasteson, Ø. Olsvik, and H. Kruse. 2000. Continuing high prevalence of VanA-type vancomycin-resistant enterococci on Norwegian poultry farms three years after avoparcin was banned. J. Appl. Microbiol. 89:478-485. [DOI] [PubMed] [Google Scholar]
- 11.Borgen, K., M. Sørum, H. Kruse, and Y. Wasteson. 2000. Persistence of vancomycin-resistant enterococci (VRE) on Norwegian broiler farms. FEMS Microbiol. Lett. 191:255-258. [DOI] [PubMed] [Google Scholar]
- 12.Butaye, P., L. A. Devriese, and F. Haesebrouck. 2001. Differences in antibiotic resistance patterns of Enterococcus faecalis and Enterococcus faecium strains isolated from farm and pet animals. Antimicrob. Agents Chemother. 45:1374-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devriese, L. A., M. Ieven, H. Goossens, P. Vandamme, B. Pot, J. Hommez, and F. Haesebrouck. 1996. Presence of vancomycin-resistant enterococci in farm and pet animals. Antimicrob. Agents Chemother. 40:2285-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emborg, H.-D., J. S. Andersen, A. M. Seyfarth, S. R. Andersen, J. Boel, and H. C. Wegener. 2002. Correlation between the occurrence of resistance to growth promoters among Enterococcus faecium isolated from broilers and broiler meat. Int. J. Food Microbiol. 84:273-284. [DOI] [PubMed] [Google Scholar]
- 15.Gambarotto, K., M. C. Ploy, P. Turlure, C. Grélaud, C. Martin, D. Bordessoule, and F. Denis. 2000. Prevalence of vancomycin-resistant enterococci in fecal samples from hospitalized patients and nonhospitalized controls in a cattle-rearing area of France. J. Clin. Microbiol. 38:620-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heuer, O. E., K. Pedersen, J. S. Andersen, and M. Madsen. 2002. Vancomycin-resistant enterococci (VRE) in broiler flocks 5 years after the avoparcin ban. Microb. Drug Resist. 8:133-138. [DOI] [PubMed] [Google Scholar]
- 17.Heuer, O. E., K. Pedersen, L. B. Jensen, M. Madsen, and J. E. Olsen. 2002. Persistence of vancomycin-resistant enterococci (VRE) in broiler houses after the avoparcin ban. Microb. Drug Resist. 8:355-361. [DOI] [PubMed] [Google Scholar]
- 18.Ieven, M., E. Vercauteren, P. Descheemaeker, F. van Laer, and H. Goossens. 1999. Comparison of direct plating and broth enrichment culture for the detection of intestinal colonization by glycopeptide-resistant enterococci among hospitalized patients. J. Clin. Microbiol. 37:1436-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iversen, A., I. Kühn, A. Franklin, and R. Möllby. 2002. High prevalence of vancomycin-resistant enterococci in Swedish sewage. Appl. Environ. Microbiol. 68:2838-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen, L. B., A. M. Hammerum, F. Bager, and F. M. Aarestrup. 2002. Streptogramin resistance among Enterococcus faecium isolated from production animals in Denmark in 1997. Microb. Drug Resist. 8:369-374. [DOI] [PubMed] [Google Scholar]
- 21.Keis, S., C. F. Bennett, V. K. Ward, and D. T. Jones. 1995. Taxonomy and phylogeny of industrial solvent-producing clostridia. Int. J. Syst. Bacteriol. 45:693-705. [DOI] [PubMed] [Google Scholar]
- 22.Kühn, I., A. Iversen, L. G. Burman, B. Olsson-Liljequist, A. Franklin, M. Finn, F. Aarestrup, A. M. Seyfarth, A. R. Blanch, X. Vilanova, H. Taylor, J. Caplin, M. A. Moreno, L. Dominguez, I. A. Herrero, and R. Möllby. 2003. Comparison of enterococcal populations in animals, humans, and the environment—a European study. Int. J. Food Microbiol. 88:133-145. [DOI] [PubMed] [Google Scholar]
- 23.Manson, J. M., S. Keis, J. M. B. Smith, and G. M. Cook. 2003. Characterization of a vancomycin-resistant Enterococcus faecalis (VREF) isolate from a dog with mastitis: further evidence of a clonal lineage of VREF in New Zealand. J. Clin. Microbiol. 41:3331-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manson, J. M., S. Keis, J. M. B. Smith, and G. M. Cook. 2003. A clonal lineage of VanA-type Enterococcus faecalis predominates in vancomycin-resistant enterococci isolated in New Zealand. Antimicrob. Agents Chemother. 47:204-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial susceptibility testing; eleventh informational supplement. Document M100-S11. National Commitee for Clinical Laboratory Standards, Wayne, Pa.
- 26.Schouten, M. A., J. A. Hoogkamp-Korstanje, J. F. Meis, and A. Voss. 2000. Prevalence of vancomycin-resistant enterococci in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 19:816-822. [DOI] [PubMed] [Google Scholar]
- 27.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]