Abstract
Ephemeral blooms of filamentous bacteria are a common phenomenon in the water column of oligo- to mesotrophic lakes. It is assumed that the appearance of such morphotypes is favored by selective predation of bacterivorous protists and that filter-feeding zooplankton plays a major role in suppressing these bacteria. The phylogenetic affiliation of the important bloom-forming filamentous bacteria in freshwaters is presently unknown. Here we report the identification of dominant members of a filamentous bacterial assemblage during a bloom of such morphotypes in a mesotrophic lake. By molecular cloning and fluorescence in situ hybridization with specific oligonucleotide probes, up to 98% of filamentous cells in lake water could be assigned to a clade of almost identical (99% similarity) 16S rRNA gene sequence types, the cosmopolitan freshwater LD2 cluster. For a period of less than 1 week, members of the LD2 clade constituted >40% of the total bacterial biomass, potentially favored by high grazing of planktivorous protists. This is probably the most pronounced case of dominance by a single bacterioplankton species ever observed in natural freshwaters. In enclosures artificially stocked with the metazoan filter feeder Daphnia, bacteria related to the LD2 clade formed a significantly larger fraction of filaments than in enclosures where Daphnia had been removed. However, in the presence of higher numbers of Daphnia individuals, the LD2 bacteria, like other filaments, were eventually eliminated both in enclosures and in the lake. This points at the potential importance of filter-feeding zooplankton in controlling the occurrence and species composition of filamentous bacterial morphotypes in freshwater plankton.
Filamentous bacteria are a conspicuous and ecologically distinct component of many freshwater systems. They form permanent planktonic populations in hypertrophic lakes (47) and may cause undesired foam formations of activated sludge (51). Yet the occurrence of such morphotypes is not limited to environments with extreme substrate or nutrient loadings. High abundances and activities of filamentous morphotypes have also been reported from an oligotrophic high mountain lake and from anthropogenically acidified Czech lakes (35, 38, 49).
In oligo- to mesotrophic lakes, filament-forming bacteria appear to occupy a particular niche that is clearly distinct from the optimal growth environment of small unicellular morphotypes. For one, they are nonmotile, so they are probably disadvantaged in an environment in which the organic substrates are discontinuously distributed in particulate “hot spots,” e.g., lake snow (12, 13). On the other hand, filamentous pelagic bacteria are distinctly favored if assemblages are exposed to high grazing by hetero- or mixotrophic nanoflagellates (17, 21-23, 45, 46) or bacterivorous ciliates (37, 42). Many of these small protists are highly size-selective feeders, preferably ingesting prey with a cell length range of around 1 to 3 μm (15, 43). Filamentous forms with a cell length of more than 10 μm are either completely avoided by nanoflagellates or ingested at substantially lower rates in the presence of alternative prey (17, 52). As a consequence, blooms of grazing-resistant bacterial morphotypes may follow blooms of heterotrophic flagellates in the spring plankton of lakes (22, 25, 36). Such shifts can also be artificially induced by specific uncoupling of the planktonic food web in size fractionation experiments (24, 26, 45). In contrast to small unicellular morphotypes, filamentous bacteria are believed to be sensitive to predation by filter-feeding metazooplankton, in particular, by cladocerans (14, 19, 20).
It is, however, unclear if the occurrence of filamentous morphotypes in lakes is only controlled by the above-described direct and indirect top-down mechanisms (30). Filamentous bacteria in lakes might also differ from small unicellular morphotypes in their specific substrate requirements or affinity for limiting nutrients. In addition, filament formation might be due to phenotypic plasticity of small cells but might also be a permanent morphological feature of some bacteria. Such questions cannot be studied comparatively in lakes at the community level only, and they cannot be resolved from the behavior of model organisms in laboratory studies (17). Obviously, filaments that cause foaming in activated sludge (51) and filaments that form more than 50% of the microbial biomass in an ultraoligotrophic high mountain lake (35) are physiologically and ecologically very different organisms. Therefore, it is necessary to develop identification systems and to distinguish between key populations of filamentous bacteria in different freshwater systems.
Molecular biological techniques focusing on comparative sequence analysis of rRNA genes have become an invaluable element for the study of diversity and community composition of microbial assemblages. One popular combination of molecular tools has been termed the “full-cycle rRNA approach” (1). In this procedure, the 16S rRNA gene (rDNA) diversity in a particular environmental sample is first studied by PCR amplification, cloning, and sequencing. Phylogenetic analysis of environmental sequence types is followed by the design of specific rRNA-targeted DNA probes. Whole-cell fluorescence in situ hybridization (FISH) with these probes is then applied for the staining and microscopic visualization of specific microbial populations in the environment studied. The FISH technique allows quantification of population sizes, as well as distinction of bacterial morphotypes (33, 35). It thus conserves potentially relevant information about environmental microbes that is lost by other techniques, e.g., molecular fingerprinting (27) or reverse line blotting (55).
In the present study, we limited the analysis of environmental 16S rDNA sequence types to one particular phylogenetic group of interest. During one of several large-scale mesocosm field experiments on cascading trophic interactions (53), a conspicuous bloom of filamentous bacteria was observed in the water column of a mesotrophic lake. A preliminary FISH analysis revealed that the majority of these filaments were members of the Cytophaga-Flavobacterium cluster of the phylum Bacteroidetes. We thus attempted to identify the dominant filamentous phylotypes within this lineage and to follow their in situ population dynamics. To obtain information about the regulating factors, we studied the development of these filaments in the context of the planktonic food webs both in the lake and in zooplankton-manipulated mesocosms.
MATERIALS AND METHODS
Experimental design, sampling, and fixation.
Samples were collected in May 2001 from the surface waters of Schöhsee, a mesotrophic dimictic lake in northern Germany (area, 82 ha; maximum depth, 30 m). In parallel, a mesocosm experiment was performed during that period. Twelve transparent polyethylene bags (2.5-m length, 1.7-m3 volume) were filled with lake water that had been prefiltered through 50-μm nylon gauze to remove the larger zooplankton. After 2 days, cladocerans (Daphnia hyalina × galeata) from stock cultures of the Max Planck Institute for Limnology (Plön, Germany) were added to replicate mesocosms at seeding concentrations of 1.25, 2.5, 5, 10, and 20 individuals liter−1. Two mesocosms without added zooplankton served as controls.
Samples (10 liters) were obtained approximately twice weekly from the lake water surface and from the mesocosms after gentle mixing of the top 2 m. Chlorophyll a was measured in situ with a submersible fluorometer (Fluoroprobe; BBE Moldaenke, Kiel, Germany). Subsamples (2 ml) for total counts of bacteria were fixed with 1% paraformaldehyde and 0.05% glutaraldehyde (final concentration), frozen in liquid nitrogen 10 min later, and stored at −80°C until flow cytometric analysis (7, 10). Subsamples (100 ml) for total counts of filamentous bacteria and heterotrophic flagellates were fixed with a formaldehyde solution (final concentration, 2% [vol/vol]) and stored at 4°C until evaluation (usually within 24 h after sampling). For FISH, portions of 10 to 30 ml were fixed with a formaldehyde solution (final concentration, 2%) for <24 h at room temperature, filtered onto white membrane filters (type GTTP; diameter, 47 mm; pore size, 0.2 μm; Millipore, Bedford, Mass.), and rinsed with sterile Millipore water. These preparations were stored at −20°C until further processing. Samples were fixed with acid Lugol's solution (final concentration, 1%) and stored at 4°C for the quantification of ciliates.
Total counts, biovolumes, and bacterial production.
Subsamples for filament and heterotrophic flagellate abundances (1 to 2 and 5 ml, respectively) were filtered onto black membrane filters (diameter, 25 mm; pore size, 0.2 μm; Millipore) and stained with 4′,6′-diamidino-2-phenylindole (DAPI; final concentration, 1 μg ml−1). Filters were stored at −20°C until microscopic evaluation. At least 500 cells per sample were counted by epifluorescence microscopy with UV excitation (Axioplan, filter set Zeiss 01; Carl Zeiss, Jena, Germany) as described previously (53). Determination of total and bacterial abundance by flow cytometry was based on the methods published by del Giorgio et al. (7) and Gasol et al. (10). Samples were stained with SYTO 13 (diluted in dimethyl sulfoxide) at 5 μM (Molecular Probes, Leiden, The Netherlands) and subsequently run through a FACScalibur flow cytometer (Becton Dickinson, San Jose, Calif.) with an excitation wavelength of 488 nm. A solution (106 ml−1) of yellow-green 0.92-μm latex beads (Polyscience, Chicago, Ill.) was compared with Trucount controls (Becton Dickinson) before each measurement series and then used as an internal standard. Acquisition and analysis of flow cytometric data was conducted with Cell-Quest software (Becton Dickinson). Abundances of randomly chosen samples were cross-checked with manual counts from DAPI-stained slides. Ciliates were counted in sedimentation chambers with an inverted microscope (Zeiss Axiovert 100; Carl Zeiss). Zooplankton was fixed with a formaldehyde solution (final concentration, 4%).
Mean cell sizes of nonfilamentous bacteria were determined for selected samples with a semiautomated image analysis system (analySIS 3.0; Soft Imaging Systems, Münster, Germany) linked to the epifluorescence microscope. Images were captured with a cooled slow-scan charge-coupled device camera (resolution, 1,276 by 1,008 pixels; 1 pixel = 0.05 μm in the microscopic image) at a magnification of ×1,250, and cell sizes were determined from binarized images as described by Massana et al. (29). Total bacterial biovolume was calculated by multiplying abundances by mean cell volume. Filamentous bacteria were operationally defined as cells exceeding 5 μm in length because cells above this size are typically inedible for unicellular planktonic protists (17). Abundances of filamentous bacteria in three size classes (5 to 10, 10 to 20, and >20 μm) were determined with an ocular micrometer. Total filament biovolumes were calculated by multiplying the respective cell numbers by the mean cell volume of each size class, obtained by measuring at least 30 cells of each size class with the image analysis system at a magnification of ×1,250 and by assuming cylindrical geometry and a mean cell width of 0.65 μm. Zooplankton was counted under a dissecting microscope.
Heterotrophic bacterial production was determined as incorporation of [3H]thymidine in accordance with standard protocols (2). All samples were incubated for 1 h at in situ temperature and then treated and measured as described by Zöllner et al. (53). Background absorption of radioactivity was accounted for by formalin-fixed controls. On 17 May, size-fractionated [3H]thymidine incorporation into the mesocosms was determined by filtration of the samples after incubation with radiotracers through polycarbonate filters (pore size, 3 μm; Millipore) (53).
16S rDNA clone libraries.
For the construction of a 16S rDNA clone library, 100 ml of formalin-fixed, unfiltered lake water obtained on 19 May was filtered onto Durapore membrane filters (pore size, 0.2 μm; Millipore) and stored at −80°C until further processing. DNA was extracted in accordance with previously published protocols (40). Briefly, samples were pretreated with sodium dodecyl sulfate, lysozyme, and proteinase K. Extraction was performed by phenol-chloroform-isoamyl alcohol (25:24:1), followed by chloroform-isoamyl alcohol (24:1), and subsequent concentration and rinsing. The appropriate DNA concentration for subsequent PCR was determined by agarose gel electrophoresis. Two nanograms of extracted DNA was used as the template for the amplification of environmental 16S rDNAs by PCR. For PCR, 5 μl of bovine serum albumin (stock concentration, 3 mg ml−1), 5 μl of 10× PCR buffer, 2 μl of deoxynucleoside triphosphates (stock concentration, 2.5 mM), 0.5 μl of the general bacterial primers GM3F and GM4R (31) (stock concentration, 15 μM), and 0.25 μl of TaKaRa-Taq DNA polymerase (stock concentration, 5 U per reaction mixture; TaKaRa BIO Inc., Shiga, Japan) were adjusted to a final volume of 48 μl with sterile water. The PCR was run on an Eppendorf Mastercycler (Eppendorf, Hamburg, Germany) under the following conditions: 1 cycle of 94°C for 5 min; 35 cycles of 94°C for 30 s, 47°C for 45 s, and 74°C for 1 min 30 s; and 1 cycle of 74°C for 10 min. The amplified rDNAs were purified with the QIAquick PCR purification kit (QIAGEN, Hilden, Germany), inserted into the TOPO vector (TOPO TA cloning kit; Invitrogen, Karlsruhe, Germany), and cloned into competent cells of Escherichia coli as described by the manufacturer. The transformed cells were plated on Luria-Bertani agar plates containing 50 μg of ampicillin ml−1 and incubated overnight at 37°C. The clones were screened for the right-sized inserts, and plasmid preparations were done with the QIAprep Spin Miniprep Kit (QIAGEN). For a first screening, the plasmid DNAs were sequenced with the vector primer M13F (5′-GTA AAA CGA CGG CCA G-3′). The partial sequences obtained were submitted to the BLAST queuing system (http://www.ncbi.nlm.nih.gov/BLAST/BLAST.cgi) for tentative phylogenetic positioning. Nearly full-length sequences were obtained from those inserts that were related to members of the phylum Bacteroidetes by additional sequencing with primers GM1F (31) and M13R (5′-CAG GAA ACA GCT ATG AC-3′).
Phylogenetic analysis and probe design.
Partial sequences were assembled manually with the Sequencher software (Gene Codes Corp., Ann Arbor, Mich.) and tested for chimeras through the Ribosomal Database Project CHIMERA_CHECK program. All sequences were again analyzed via BLAST to identify their closest relatives. Phylogenetic analyses were performed with the ARB software package (www.arb-home.de). The ARB database (June 2002 release) was complemented with sequences from the GenBank database that were related to freshwater lineages of the Cytophaga-Flavobacteria cluster (11, 54). For the reconstruction of a phylogenetic tree, only nearly complete (i.e., longer than 1,300 nucleotides) 16S rDNA sequences affiliated with this subphylum were considered. A 50% base frequency filter was applied to these sequences to exclude highly variable positions. The respective ARB tools were used to perform maximum-parsimony, neighbor-joining, and maximum-likelihood analyses. Downloaded partial sequences of closely related sequence types from freshwater samples were subsequently added to the consensus tree in accordance with maximum-parsimony criteria, without introducing changes in the topology based on the complete sequences.
Specific oligonucleotide probes for new and available sequences within the freshwater LD2 and CL500-6 clusters as defined by Zwart et al. (54), as well as for two other groups of newly obtained sequences within the Cytophaga-Flavobacteria cluster, were designed with the ARB software package and the 16S rRNA accessibility information provided by Fuchs et al. (9). Stringent conditions for FISH were established by analysis of fluorescence intensities of the target cells from hybridizations at increasing concentrations of formamide in the hybridization buffer (34).
FISH.
Whole-cell in situ hybridizations on sections from the polycarbonate filters were performed with 16S rRNA-targeted oligonucleotide probes EUB I-III (5), CF319a (many groups of the Cytophaga-Flavobacterium cluster of the phylum Bacteroidetes) (28), and R-FL615 for the freshwater cfIII (Flectobacillus sp.) lineage (45) and newly designed specific probes. All probes were obtained from ThermoHybaid (Interactiva Division, Ulm, Germany). Hybridizations with directly Cy3 monolabeled probes were performed as described with 35 to 40% formamide in the hybridization buffer (see Results for hybridization conditions of newly designed probes) (34). After FISH, the filter sections were counterstained with DAPI (final concentration, 1 μg ml−1) and evaluated on a Zeiss Axioplan II epifluorescence microscope equipped with 40× and 63× Plan Neofluar oil objective lenses (Carl Zeiss). The filter sets were Chroma HQ 41007 (Chroma Tech. Corp., Brattleboro, Vt.) for probe fluorescence and Zeiss 01 for DAPI. The relative abundance of cells hybridized with the specific probe was determined as a fraction of all DAPI-stained filaments (i.e., cells of >5 μm). At least 200 cells per preparation were counted. From lake samples, the length of 40 to 50 hybridized cells was measured and mean cell volumes were estimated as described above. Images of Cy3 and DAPI fluorescence were captured with a SPOT color charge-coupled device camera (Diagnostic Instruments, Sterling Heights, Mich.) mounted on the Axioplan II microscope.
Statistical analyses.
Testing for differences between pairs of treatments with and without Daphnia was performed by the nonparametric Mann-Whitney rank sum test. For this, the individual data sets from the two mesocosms without Daphnia were pooled and compared to the respective pooled data sets of the mesocosms with Daphnia (nominal density, 5 individuals liter−1). This analysis was performed for total bacterial counts, for counts of total filamentous bacteria, for the relative contribution of bacteria from the LD2 clade to all filamentous cells, and for thymidine incorporation. For the comparison of cell lengths of bacteria of the LD2 clade at different sampling dates, Kruskal-Wallis analysis of variance on ranks followed by Dunn's post-hoc comparisons was applied. All analyses were carried out with SigmaStat (SPSS Inc., Chicago, Ill.).
Nucleotide sequence accession numbers.
Newly generated sequences that were included in the phylogenetic analysis have been submitted to GenBank under accession numbers AJ697697 to AJ697708.
RESULTS
Development of the planktonic community in the lake.
Surface water temperatures in Schöhsee increased from 11.9 to >18°C on May 15 and declined thereafter, to 14.7°C. The lake was thermally stratified during the study period. Pronounced P depletion and an increasing ratio of total N to total P were observed (data not shown). Chlorophyll a concentrations in the lake ranged around 3 μg liter−1; inside the mesocosms they declined from 5.3 ± 1.5 to 0.7 ± 0.6 μg liter−1. Phytoplankton was dominated by flagellates, particularly by members of the genus Chrysochromulina.
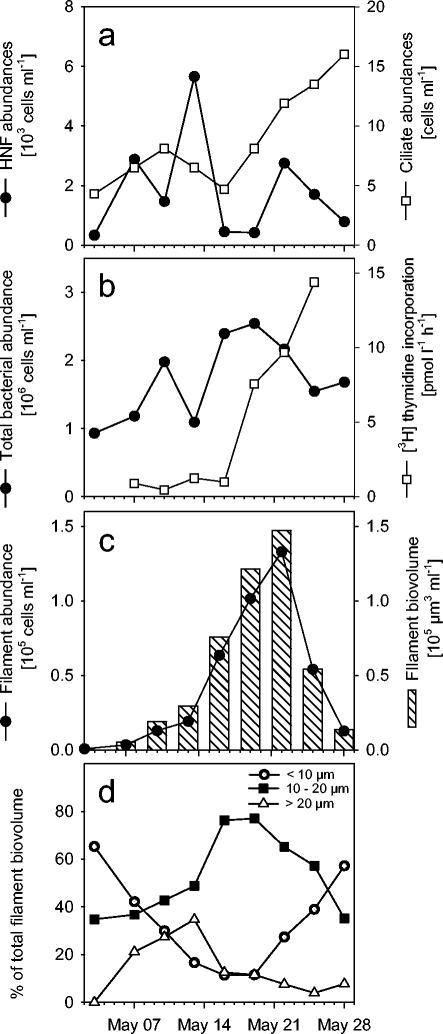
The Daphnia densities in lake water increased from <1 individual liter−1 at the beginning of the investigation to 3 individuals liter−1 on 18 May, and to 18 individuals liter−1 on 26 May. Heterotrophic flagellates contributed from 3 to 59% of the total flagellate community; they varied between approximately 300 and 3,000 cells liter−1 and developed a distinct maximum during the first half of the study period (Fig. 1a). In contrast, ciliate densities tripled during the second half, from <5 to 15 cells ml−1 (Fig. 1a), mainly because of a bloom of nanociliates of <20 μm.
FIG. 1.
Development of the microbial assemblages in Schöhsee during the study period. Panels: a, abundances of heterotrophic nanoflagellates (HNF) and ciliates; b, total bacterial numbers and [3H]thymidine uptake rate; c, abundances and biovolume of filamentous bacterial morphotypes; d, relative contributions of three size fractions to the biovolume of filamentous bacteria.
Total bacterial abundances in lake water increased during the study period from 1 to 2.5 × 106 cells ml−1 (Fig. 1b). The uptake rate of tritiated thymidine increased by more than 10-fold during the second half of the investigation period, in parallel with a bloom of filamentous cells (Fig. 1b and c). Threadlike bacterial morphotypes increased in abundance and biovolume increased by more than 2 orders of magnitude within 3 weeks, from 800 to 1.3 × 105 cells ml−1 and from 900 to 1.5 × 105 μm3 ml−1, respectively. Both the abundances and biovolumes of filamentous bacteria peaked on 22 May and declined precipitously during the following week (Fig. 1c). The biovolume size distribution of filamentous bacteria shifted from an initial dominance of cells <10 μm long to distinctively longer morphotypes and back to shorter cells when filament abundances were declining (Fig. 1d).
Community dynamics in mesocosms with different Daphnia densities.
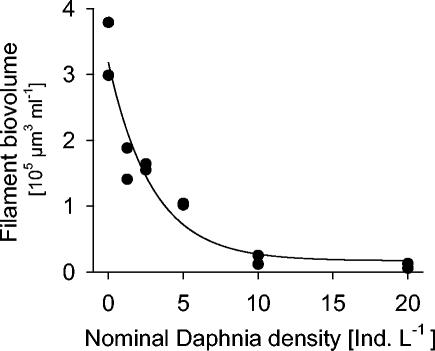
In samples from 19 May, the total biovolume of filamentous bacteria in mesocosms steeply declined with increasing Daphnia seeding densities, to less than 5% of the amount that was present without cladocerans (Fig. 2). In parallel, the fraction of [3H]thymidine that was incorporated into bacterial cells of >3 μm decreased from >30% in the Daphnia-free controls to around 10% in the mesocosms with the highest cladoceran concentrations.
FIG. 2.
Biovolumes of filamentous bacteria that developed in replicate lake water mesocosms that had been stocked with different Daphnia densities 2 weeks earlier. Ind. L−1, individuals per liter.
At a nominal Daphnia density of 5 individuals liter−1, the filament biomass was reduced by more than 70% compared to that of controls where zooplankton had been removed. These two sets of mesocosms were therefore selected for a detailed analysis of the microbial assemblages over the study period. Total bacterial abundances in the enclosures ranged between <1 × 106 and 4 × 106 cells ml−1 (Table 1). In contrast to the development in the lake, cell numbers showed a trend to decline, yet this was masked by differences between replicates. A conspicuous bloom of total bacterial abundances in one of the control enclosures was also reflected in increased bacterial uptake of tritiated thymidine (Table 1). Overall, there was no statistically significant difference between the two treatments with respect to total bacterial numbers or bacterial thymidine uptake.
TABLE 1.
Development of total bacterial abundances and thymidine incorporation rates in replicate mesocosmsa
| Date | Bacterial abundance (106 cells ml−1)
|
Thymidine uptake rate (pmol liter−1 h−1)
|
||||||
|---|---|---|---|---|---|---|---|---|
| + Daphnia
|
Control
|
+ Daphnia
|
Control
|
|||||
| A | B | A | B | A | B | A | B | |
| 4 May 2001 | 2.79 | 4.00 | 3.19 | 3.14 | ||||
| 7 May 2001 | 1.37 | 1.52 | 1.80 | 1.43 | 6.10 | 9.44 | 7.52 | 3.09 |
| 10 May 2001 | 1.84 | 1.68 | 1.52 | 1.43 | 1.13 | 2.37 | 1.18 | 1.46 |
| 13 May 2001 | 1.38 | 1.21 | 1.12 | 0.63 | 7.43 | 4.88 | 4.81 | 3.46 |
| 16 May 2001 | 1.37 | 2.55 | 1.07 | 1.06 | 3.98 | 8.86 | 5.76 | 4.03 |
| 19 May 2001 | 1.34 | 1.35 | 1.03 | 1.72 | 7.07 | 2.49 | 7.13 | 21.04 |
| 22 May 2001 | 0.85 | 0.94 | 0.93 | 2.97 | 3.02 | 2.10 | 4.72 | 15.32 |
| 25 May 2001 | 1.12 | 0.88 | 0.80 | 0.81 | 4.43 | 4.32 | 5.72 | 17.15 |
| 28 May 2001 | 0.99 | 1.83 | 0.83 | 1.17 | ||||
Mesocosms A and B were stocked with 5 Daphnia individuals liter−1 or contained no zooplankton (control).
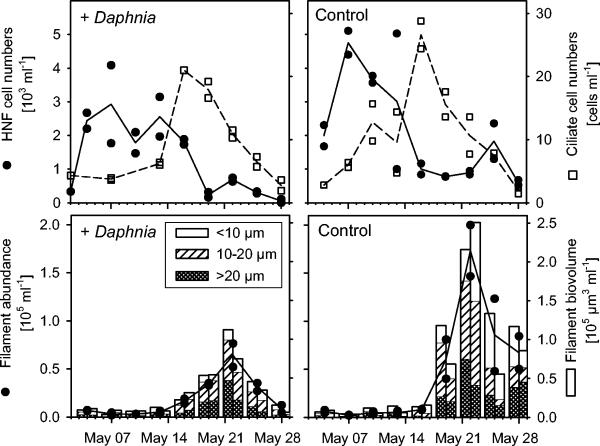
Heterotrophic flagellates and nanociliates formed successive blooms in the mesocosms both in the presence and in the absence of Daphnia (Fig. 3). The total abundances and biovolumes of filamentous bacteria were significantly higher in the control mesocosms without cladocerans (Fig. 3). In these treatments, a considerable density of threadlike bacteria (>0.5 × 105 cells ml−1) was maintained until the end of the investigation, whereas filaments almost completely disappeared from mesocosms with Daphnia.
FIG. 3.
(Upper panels) Dynamics of heterotrophic nanoflagellates (HNF) and ciliates in experimental mesocosms either seeded with 5 Daphnia individuals liter−1 (+Daphnia) or without zooplankton (control). (Lower panels) Abundances and biovolumes of filamentous bacterial morphotypes.
Phylogenetic analysis.
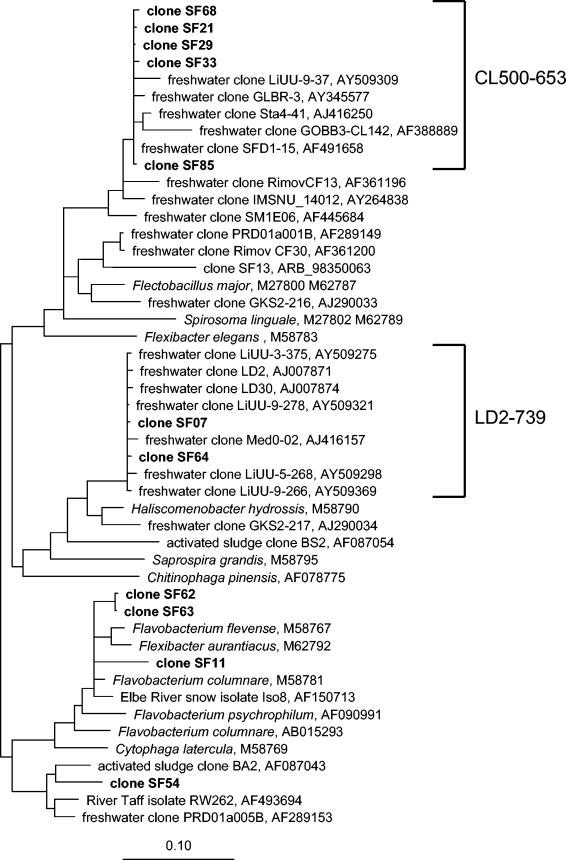
Only 21 of 96 sequenced clones were affiliated with the Cytophaga-Flavobacterium lineage of the phylum Bacteroidetes, whereas the majority were related to plastids of algae and were not considered further. The clone sequences analyzed were affiliated with three known groups of freshwater bacteria, the CL500-6, LD2, and cfIII (Flectobacillus) lineages (11, 54) (Fig. 4). In addition, a group of sequence types fell outside the currently defined typical freshwater clades. These sequences were related to Flavobacterium spp., Flexibacter spp., and a bacterium isolated from aggregates of the river Elbe (clones SF62, SG63, and SF11). One sequence type (clone SF54) was related to a river isolate and to clone sequences from activated sludge.
FIG. 4.
Phylogenetic positions of 16S rDNA sequences from Schöhsee affiliated with the Cytophaga-Flavobacterium lineage of the phylum Bacteroidetes. Only some of the nearly identical clones are depicted. The brackets indicate sequences targeted by newly designed specific FISH probes LD2-739 and CL500-653. Bar, 10% estimated sequence divergence.
FISH with specific oligonucleotide probes.
Between 33 and 100% of all hybridized filament-forming bacteria in lake water and in the mesocosms could be detected by probe CF319a (data not shown). Filament-forming bacterial morphotypes were detected with specific probes R-FL615, LD2-739 (5′-GCG TCA ATA CAG ATC CAG-3′; 40% formamide in hybridization buffer), and CL500-653 (5′-GTA CCC CTC CAA CAC TCA-3′; 35% formamide in hybridization buffer) (Fig. 5). Bacteria affiliated with Flectobacillus sp. and the CL500 cluster were rare cells <10 μm in length. A substantial amount of the longer morphotypes was hybridized by probe LD2-739 (Fig. 5). FISH with probes designed for other phylogenetic lineages harboring clone sequences did not stain any filamentous cells in selected samples (data not shown).
FIG. 5.
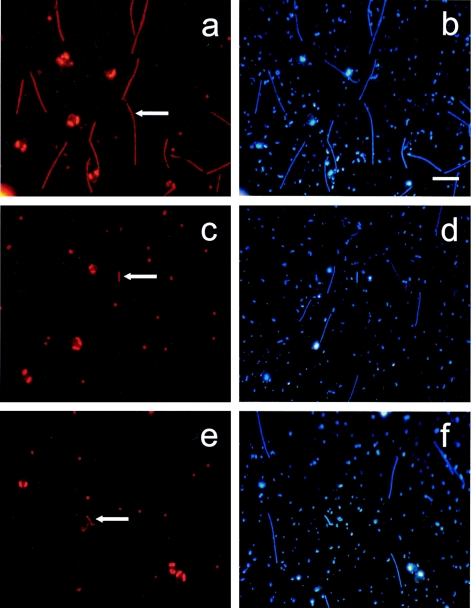
Photomicrographs of common and rare filamentous bacteria hybridized with probes for different lineages of the Cytophaga-Flavobacteria cluster. Left panels, hybridized cells (arrows); right panels, same microscopic field, all cells (DAPI staining). Panels: a and b, probe LD2-739; c and d, probe CL500-653; e and f, probe R-FL615. Bar, 10 μm. Other orange fluorescent objects visible in the panels are cyanobacteria and algal plastids.
Population dynamics and morphology of members of the LD2 clade.
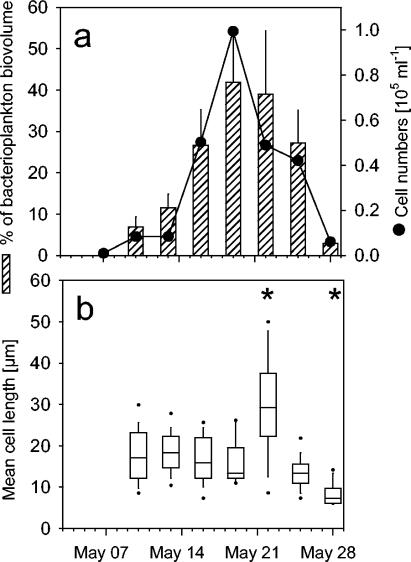
FISH with probe LD2-739 revealed that members of the LD2 clade accounted for most of the filament bloom observed in Schöhsee (Fig. 1c, 5a, and 6a). Within 2 weeks, the abundances of these bacteria increased by 2 orders of magnitude, from <1,000 to almost 105 cells ml−1. During this period, their highest apparent growth rate ranged around 0.6 day−1. From cell size measurements of all bacteria and of filaments of the LD2 clade, it was estimated that this group formed up to 42% of the total microbial biovolume on 19 May (Fig. 6a).
FIG. 6.
(a) Cell numbers of filaments hybridized with probe LD2-739 in Schöhsee and their mean fraction of the total bacterial biovolume (+1 standard deviation). (b) Cell length distributions of filaments related to the LD2 clade at different sampling dates. Box plots depict medians and 50th, 75th, and 90th percentiles. Asterisks indicate dates with significant differences from the first sampling date.
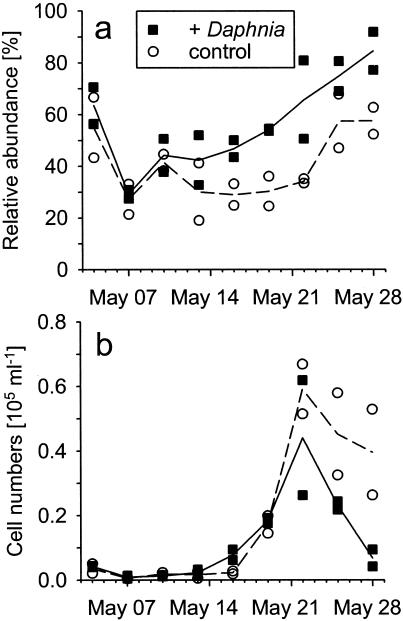
The mean cell length of these bacteria initially ranged around 20 μm (Fig. 6b). It significantly increased to >30 μm when their abundances started to decline and significantly decreased below the original cell length in parallel with their disappearance in the plankton (P < 0.05) (Fig. 6b). Bacteria of the LD2 clade formed a significantly higher fraction of the total filaments in the mesocosms that had been seeded with 5 individuals of Daphnia liter−1 than in the Daphnia-free controls (P < 0.05) (Fig. 7a). A bloom and subsequent decline of LD2 filaments as observed in lake water were also found in mesocosms containing Daphnia (Fig. 7b). In contrast, LD2 filaments showed only a slight population decrease in mesocosms without cladocerans (Fig. 7b) at the end of the study period.
FIG. 7.
Development of relative (a) and absolute (b) abundances of filaments related to the LD2 clade in mesocosms with and without Daphnia. The treatment definitions are given in the legend to Fig. 3.
DISCUSSION
Development of filamentous bacteria in lake plankton.
In oligo- to mesotrophic lakes with plankton successions comparable to that of Schöhsee, the development of filamentous bacteria is most pronounced in late spring and early summer, a period that is characterized by high primary and bacterial productivity, high abundances of phagotrophic protists, and the absence of filter-feeding cladocerans (48). During the subsequent clear-water phase, Daphnia reaches maximal population densities, thereby suppressing filamentous bacteria and inducing a shift toward smaller cells. This succession has been described for Lake Constance (14), and also in Schöhsee maximal filament numbers were found during the period before the clear-water phase (20). Other types of water bodies, e.g., shallow eutrophic lakes, may show a different pattern, such as a dominance of filaments during midsummer or throughout the year (47). Since the blooms of filamentous bacteria may be short-lived (Fig. 1), they are probably often overlooked by routine monthly samplings. However, their appearance can be reproducibly induced in food web manipulation experiments that trigger increased concentrations of phagotrophic protists (24, 45).
Dominant freshwater filaments within the uncultured LD2 lineage.
So far, filamentous lake bacteria have been considered either as a single compartment (22, 47, 49) or as model isolates (17, 52) or they have been divided into rather large phylogenetic categories (24, 35, 45). In contrast, this study determined the precise phylogenetic affiliation and population dynamics of a key group of filamentous bacteria during a bloom of such morphotypes in a mesotrophic lake (Fig. 4, 5, and 6a). Members of the uncultured LD2 clade are regarded as one among many groups of 16S rDNA sequence types that occur in various freshwaters (54). We show that these bacteria dominated the spring assemblage of filament-forming microbes in Schöhsee and transiently formed more than 40% of the total microbial biovolume (Fig. 6a). Bacterial lineages harboring typical model isolates of filamentous bacteria (17) were rare in the study system (Fig. 5e).
This remarkable dominance of a single phylogenetic group of bacteria illustrates the potential of microbes to accumulate biomass in the water column even at modest growth rates of <1 day−1 if they escape from control by grazing (or viral lysis). A comparable enrichment of particular groups of planktonic bacteria has never been observed before during field studies, only after experimental manipulations. For example, presumably grazing-resistant filamentous bacteria related to Flectobacillus sp. could be enriched to high cell densities after an artificially induced bloom of heterotrophic flagellates in a eutrophic reservoir (45).
A cosmopolitan group of filamentous bacteria?
So far, sequence types within the LD2 clade have been collected from various meso- to eutrophic freshwater systems. Originally, the lineage has been defined by Zwart et al. on the basis of 16S rRNA sequences from the shallow eutrophic lakes Loosdrecht and Ijssel (54), but similar sequence types were also obtained from the eutrophic Swedish lakes Erken and Limmaren during cyanobacterial blooms (A. Eiler and S. Bertilson, GenBank submission). Unfortunately, Zwart and coworkers did not include this group in a recent survey on the occurrence of various phylogenetic lineages of freshwater bacteria across a large range of European lakes (55). Preliminary data (J. Pernthaler, unpublished observations) indicate that the LD2 filaments were also present in a mountain lake in Austria and a hypertrophic shallow lagoon in Uruguay.
16S rDNA sequence types of such high similarity (Fig. 4) are commonly regarded as representatives of a single bacterial species (39). This limited range of phylogenetic diversity may reflect the well-defined niche that is occupied by filament-forming bacteria in meso- to eutrophic lakes harboring cladoceran zooplankton. So far, sequence types related to LD2 have been only retrieved from such habitats (Fig. 4); it is thus conceivable that the whole clade represents a single, ecologically coherent type of microorganisms. However, there might be considerable genotypic and ecophysiological diversity beneath the level of 16S rDNA diversity (8, 16).
Factors controlling filament abundances.
Doubtlessly, the higher resistance of filamentous bacterial morphotypes to size-selective mortality is an important factor favoring their occurrence in the water column during periods of high protistan grazing. This has been illustrated numerous times in laboratory systems and in experimental and correlative studies in the field (6, 17, 20, 21, 36, 45, 46). Yet the dynamics of pelagic protists in our study do not unequivocally support such an interpretation, as there was a conspicuous time lag between the blooms of heterotrophic flagellates and of filamentous bacterial morphotypes in the lake, as well as in the enclosures (Fig. 1a and 3) Other protistan bacterivores, e.g., particular oligotrichous species, such as Halteria sp., sometimes might also contribute to a scenario of overall high bacterial grazing pressure (44). Ciliates showed distinct blooms in parallel with the rise of filamentous bacteria both in the lake and in the mesocosms (Fig. 1a and 2). In addition, some members of the phytoplankton genus Chrysochromulina, which was common during the study period, are known mixotrophs (32). Chrysochromulina cells from Schöhsee with ingested fluorescently labeled bacteria were repeatedly observed during the study period (E. Zöllner, personal observation).
We can only speculate which other factors might have favored the occurrence of bacteria of the LD2 clade in Schöhsee, e.g., a potential to accumulate and store limiting nutrients during their grazing-induced release (41). Recent experiments with mixed bacterial assemblages in chemostat cultures revealed that protist grazing favors filamentous bacteria only in combination with phosphorus limitation but not with carbon limitation (30). Dissolved P was below detection in Schöhsee surface waters during most of the study period, suggesting P limitation of bacterial growth.
In contrast, there is much clearer evidence of top-down, i.e., predation, control of filamentous bacterial morphotypes by cladocerans. Threadlike bacteria were virtually absent in mesocosms with nominal Daphnia seeding densities of 10 or more individuals liter−1 (Fig. 2). These results not only confirm earlier findings from Schöhsee and other lakes (19, 20) but also introduce a quantitative aspect to the relationship between Daphnia grazing and the survival of filamentous morphotypes. At nominal Daphnia densities of 5 individuals liter−1, the filament biomass was reduced to <30% of the amount found in mesocosms without cladocerans (Fig. 2 and 3). This provided the opportunity to specifically study the sensitivity of bacteria of the LD2 clade to Daphnia predation. Since the relative abundance of these filaments was significantly higher in mesocosms containing Daphnia (Fig. 7a), we suggest that members of the LD2 clade were more resistant to Daphnia grazing than were the other filamentous bacteria. Therefore, these bacteria may experience a selective advantage in lake bacterioplankton if Daphnia is present at low densities. Increased Daphnia populations in the mesocosms and in the lake, however, also resulted in the elimination of LD2 filaments (Fig. 2).
Other factors than zooplankton grazing might potentially also contribute to the control of the abundance of freshwater filaments. In a high mountain lake that does not feature cladocerans, threadlike bacteria were visibly infected by a morphologically conspicuous filamentous type of virus-like particles (18). The abundances of filaments and their likely parasites significantly covaried over a seasonal cycle. On the other hand, Weinbauer and Höfle (50) reported that bacteria of >2.4 μm in the oxic layer of the eutrophic Pluβsee were never visibly infected by viruses. In view of our experimental evidence (Fig. 2) and of the absence of such filamentous virus-like particles in most lakes (18), this factor appears to play a secondary role in the present investigation.
Phenotypic plasticity or permanently filamentous morphotypes.
Currently two modes of filament formation in planktonic assemblages are discussed. On the one hand, laboratory studies have demonstrated that some bacteria may exhibit high phenotypic plasticity and that the filamentous morphotype may specifically be triggered in the presence of a size-selective protistan predator (17, 46). Such induced filamentation has been explained as a consequence of an enhanced growth rate (17), but it might potentially also be related to other causes, e.g., specific signaling compounds (22).
On the other hand, some bacterial species may always resemble filamentous or other complex morphotypes that are protected against protistan grazing, e.g., Ancalomicrobium sp. (3). Their apparent presence or absence in the water column may be a simple consequence of their relative abundances. Our data indicate that this might be the case for filaments of the LD2 cluster. During the whole study period, we never observed cells hybridizing with probe LD2-739 with a cell length of <5 μm (Fig. 6b). However, the biomass of all filaments (Fig. 1) and of members of the LD2 clade in lake samples was significantly shifted toward the smallest cell length class (<10 μm), in parallel with their decline in total abundances (Fig. 6b). This might indicate that the mortality inflicted by Daphnia upon filaments is also size selective, probably owing to the mesh size of the cladoceran filtration apparatus (4). Thus, a limited phenotypic plasticity outside the feeding range of protistan predators may also help threadlike bacteria to survive grazing by Daphnia.
Linking the genotype with the phenotype.
We were able to assign a specific morphotype to a group of freshwater bacteria that were previously known only from 16S rDNA sequences, thereby better defining its functional ecological niche. Our study thus illustrates the potential of combining molecular identification and specific FISH staining to throw light on the ecology of single bacterial populations in the plankton. Yet it can only provide preliminary information about this apparently widespread group of lake bacteria. It would be interesting to learn more about the controlling factors, habitat range, and in situ substrate requirements of filamentous bacteria of the LD2 lineage in freshwater environments.
Acknowledgments
We thank Silke Wetzel, Heike Brockmöller, and Elke Allers for technical assistance. We furthermore thank the mesocosm team and technical staff of the Max Planck Institute for Limnology and the Leibniz Institute for Marine Sciences for help during mesocosm field work.
This study was supported by grants from the German Science Foundation (JU367/4-1 to K.J.), by the German Ministry of Education and Research (BMBF01 LC0021/TP4 to J.P.), and by the Max Planck Society.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, R. T. 1993. Estimating production of heterotrophic bacterioplankton via incorporation of tritiated thymidine, p. 495-503. In P. Kemp, B. F. Sherr, E. B. Sherr, and J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis, Boca Raton, Fla.
- 3.Bianchi, M. 1989. Unusual bloom of star-like prosthecate bacteria and filaments as a consequence of grazing pressure. Microb. Ecol. 17:137-141. [DOI] [PubMed] [Google Scholar]
- 4.Brendelberger, H. 1991. Filter mesh size of cladocerans predicts retention efficiency for bacteria. Limnol. Oceanogr. 36:884-894. [Google Scholar]
- 5.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 6.Degans, H., E. Zöllner, K. Van der Gucht, L. De Meester, and K. Jürgens. 2002. Rapid Daphnia-mediated changes in microbial community structure: an experimental study. FEMS Microbiol. Ecol. 42:137-149. [DOI] [PubMed] [Google Scholar]
- 7.Del Giorgio, P. A., D. F. Bird, Y. T. Prairie, and D. Planas. 1996. Flow cytometric determination of bacterial abundance in lake plankton with the green nucleic acid stain SYTO 13. Limnol. Oceanogr. 41:783-789. [Google Scholar]
- 8.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk, Jr. 1992. How close is close? 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in-situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasol, J. M., U. L. Zweifel, F. Peters, J. A. Fuhrman, and Hagström. 1999. Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl. Environ. Microbiol. 65:4475-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glöckner, F.-O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossart, H. P., L. Riemann, and F. Azam. 2001. Bacterial motility in the sea and its ecological implications. Aquat. Microb. Ecol. 25:247-258. [Google Scholar]
- 13.Grossart, H.-P., and M. Simon. 1997. Formation of macroscopic organic aggregates (lake snow) in a large lake: the significance of transparent exopolymer particles, phytoplankton, and zooplankton. Limnol. Oceanogr. 42:1651-1659. [Google Scholar]
- 14.Güde, H. 1988. Direct and indirect influences of crustacean zooplankton on bacterioplankton of Lake Constance. Hydrobiologia 159:63-73. [Google Scholar]
- 15.Hahn, M. W., and M. G. Höfle. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35:113-121. [DOI] [PubMed] [Google Scholar]
- 16.Hahn, M. W., H. Lünsdorf, Q. Wu, M. Schauer, M. G. Höfle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahn, M. W., E. R. B. Moore, and M. G. Höfle. 1999. Bacterial filament formation, a defense mechanism against flagellate grazing, is growth rate controlled in bacteria of different phyla. Appl. Environ. Microbiol. 65:25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofer, J. S., and R. Sommaruga. 2001. Seasonal dynamics of viruses in an alpine lake: importance of filamentous forms. Aquat. Microb. Ecol. 26:1-11. [Google Scholar]
- 19.Jürgens, K. 1994. Impact of Daphnia on planktonic microbial food webs: a review. Mar. Microb. Food Webs 8:295-324. [Google Scholar]
- 20.Jürgens, K., H. Arndt, and K. O. Rothhaupt. 1994. Zooplankton-mediated change of bacterial community structure. Microb. Ecol. 27:27-42. [DOI] [PubMed] [Google Scholar]
- 21.Jürgens, K., H. Arndt, and H. Zimmermann. 1997. Impact of metazoan and protozoan grazers on bacterial biomass distribution in microcosm experiments. Aquat. Microb. Ecol. 12:131-138. [Google Scholar]
- 22.Jürgens, K., and H. Güde. 1994. The potential importance of grazing-resistant bacteria in planktonic systems. Mar. Ecol. Prog. Ser. 112:169-188. [Google Scholar]
- 23.Jürgens, K., and C. Matz. 2002. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 81:413-434. [DOI] [PubMed] [Google Scholar]
- 24.Jürgens, K., J. Pernthaler, S. Schalla, and R. Amann. 1999. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl. Environ. Microbiol. 65:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jürgens, K., and G. Stolpe. 1995. Seasonal dynamics of crustacean zooplankton, heterotrophic nanoflagellates and bacteria in a shallow, eutrophic lake. Freshw. Biol. 33:27-38. [Google Scholar]
- 26.Langenheder, S., and K. Jurgens. 2001. Regulation of bacterial biomass and community structure by metazoan and protozoan predation. Limnol. Oceanogr. 46:121-134. [Google Scholar]
- 27.Lindström, E. S. 1998. Bacterioplankton community composition in a boreal forest lake. FEMS Microbiol. Ecol. 27:163-174. [Google Scholar]
- 28.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 29.Massana, R., J. M. Gasol, P. K. Bjornsen, N. Blackburn, A. Hagstrom, S. Hietanen, B. H. Hygum, J. Kuparinen, and C. PedrosAlio. 1997. Measurement of bacterial size via image analysis of epifluorescence preparations: description of an inexpensive system and solutions to some of the most common problems. Sci. Mar. 61:397-407. [Google Scholar]
- 30.Matz, C., and K. Jurgens. 2003. Interaction of nutrient limitation and protozoan grazing determines the phenotypic structure of a bacterial community. Microb. Ecol. 45:384-398. [DOI] [PubMed] [Google Scholar]
- 31.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nygaard, K., and A. Tobiesen. 1993. Bacterivory in algae—a survival strategy during nutrient limitation. Limnol. Oceanogr. 38:273-279. [Google Scholar]
- 33.Pernthaler, J., A. Alfreider, T. Posch, S. Andreatta, and R. Psenner. 1997. In situ classification and image cytometry of pelagic bacteria from a high mountain lake (Gossenköllesee, Austria). Appl. Environ. Microbiol. 63:4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 35.Pernthaler, J., F.-O. Glöckner, S. Unterholzner, A. Alfreider, R. Psenner, and R. Amann. 1998. Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Appl. Environ. Microbiol. 64:4299-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pernthaler, J., B. Sattler, K. Šimek, A. Schwarzenbacher, and R. Psenner. 1996. Top-down effects on the size-biomass distribution of a freshwater bacterioplankton community. Aquat. Microb. Ecol. 10:255-263. [Google Scholar]
- 37.Posch, T., J. Jezbera, J. Vrba, K. Šimek, J. Pernthaler, S. Andreatta, and B. Sonntag. 2004. Size selective feeding in Cyclidium glaucoma (Ciliophora, Scuticociliatida) and its effects on bacterial community structure: a study from a continuous cultivation system. Microb. Ecol. 42:217-227. [DOI] [PubMed]
- 38.Posch, T., J. Pernthaler, A. Alfreider, and R. Psenner. 1997. Cell-specific respiratory activity of aquatic bacteria studied with the tetrazolium reduction method, Cyto-Clear slides, and image analysis. Appl. Environ. Microbiol. 63:867-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossello-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 40.Schauer, M., R. Massana, and C. Pedros-Alio. 2000. Spatial differences in bacterioplankton composition along the Catalan coast (NW Mediterranean) assessed by molecular fingerprinting. FEMS Microbiol. Ecol. 33:51-59. [DOI] [PubMed] [Google Scholar]
- 41.Sherr, B. F., E. B. Sherr, and T. Berman. 1983. Grazing, growth, and ammonium excretion rates of a heterotrophic microflagellate fed with 4 species of bacteria. Appl. Environ. Microbiol. 45:1196-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shikano, S., L. S. Luckinbill, and Y. Kurihara. 1990. Changes of traits in a bacterial population associated with protozoal predation. Microb. Ecol. 20:75-84. [DOI] [PubMed] [Google Scholar]
- 43.Šimek, K., and T. H. Chrzanowski. 1992. Direct and indirect evidence of size-selective grazing on pelagic bacteria by freshwater nanoflagellates. Appl. Environ. Microbiol. 58:3715-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Šimek, K., K. Jurgens, J. Nedoma, M. Comerma, and J. Armengol. 2000. Ecological role and bacterial grazing of Halteria spp.: small freshwater oligotrichs as dominant pelagic ciliate bacterivores. Aquat. Microb. Ecol. 22:43-56. [Google Scholar]
- 45.Šimek, K., J. Pernthaler, M. G. Weinbauer, K. Hornák, J. R. Dolan, J. Nedoma, M. Mašín, and R. Amann. 2001. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67:2723-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Šimek, K., J. Vrba, J. Pernthaler, T. Posch, P. Hartman, J. Nedoma, and R. Psenner. 1997. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl. Environ. Microbiol. 63:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sommaruga, R., and R. Psenner. 1995. Permanent presence of grazing-resistant bacteria in a hypertrophic lake. Appl. Environ. Microbiol. 61:3457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sommer, U., Z. M. Gliwicz, W. Lampert, and A. Duncan. 1986. The PEG-model of seasonal succession of planktonic events in fresh waters. Arch. Hydrobiol. 106:433-471. [Google Scholar]
- 49.Vrba, J., J. Nedoma, L. Kohout, J. Kopacek, L. Nedbalova, P. Rackova, and K. Šimek. 2003. Massive occurrence of heterotrophic filaments in acidified lakes: seasonal dynamics and composition. FEMS Microbiol. Ecol. 46:281-294. [DOI] [PubMed] [Google Scholar]
- 50.Weinbauer, M. G., and M. G. Höfle. 1998. Size-specific mortality of lake bacterioplankton by natural virus communities. Aquat. Microb. Ecol. 15:103-113. [Google Scholar]
- 51.Westlund, A. D., E. Hagland, and M. Rothman. 1998. Operational aspects on foaming in digesters caused by Micothrix parvicella. Water Sci. Technol. 38:29-34. [Google Scholar]
- 52.Wu, O. L. L., J. Boenigk, and M. W. Hahn. 2004. Successful predation of filamentous bacteria by a nanoflagellate challenges current models of flagellate bacterivory. Appl. Environ. Microbiol. 70:332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zöllner, E., B. Santer, M. Boersma, H. G. Hoppe, and K. Jürgens. 2003. Cascading predation effects of Daphnia and copepods on microbial food web components. Freshw. Biol. 48:2174-2193. [Google Scholar]
- 54.Zwart, G., B. C. Crump, M. Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]
- 55.Zwart, G., E. J. van Hannen, M. P. Kamst-van Agterveld, K. Van der Gucht, E. S. Lindstrom, J. Van Wichelen, T. Lauridsen, B. C. Crump, S. K. Han, and S. Declerck. 2003. Rapid screening for freshwater bacterial groups by using reverse line blot hybridization. Appl. Environ. Microbiol. 69:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]