Abstract
The bacterium Ornithobacterium rhinotracheale has been recognized as an emerging pathogen in poultry since about 10 years ago. Knowledge of this bacterium and its mechanisms of virulence is still very limited. Here we report the development of a transformation system that enables genetic modification of O. rhinotracheale. The system is based on a cryptic plasmid, pOR1, that was derived from an O. rhinotracheale strain of serotype K. Sequencing indicated that the plasmid consisted of 14,787 nucleotides. Sequence analysis revealed one replication origin and several rep genes that control plasmid replication and copy number, respectively. In addition, pOR1 contains genes with similarity to a heavy-metal-transporting ATPase, a TonB-linked siderophore receptor, and a laccase. Reverse transcription-PCR demonstrated that these genes were transcribed. Other putative open reading frames exhibited similarities with a virulence-associated protein in Actinobacillus actinomycetemcomitans and a number of genes coding for proteins with unknown function. An Escherichia coli-O. rhinotracheale shuttle plasmid (pOREC1) was constructed by cloning the replication origin and rep genes from pOR1 and the cfxA gene from Bacteroides vulgatus, which codes for resistance to the antibiotic cefoxitin, into plasmid pGEM7 by using E. coli as a host. pOREC1 was electroporated into O. rhinotracheale and yielded cefoxitin-resistant transformants. The pOREC1 isolated from these transformants was reintroduced into E. coli, demonstrating that pOREC1 acts as an independent replicon in both E. coli and O. rhinotracheale, fulfilling the criteria for a shuttle plasmid that can be used for transformation, targeted mutagenesis, and the construction of defined attenuated vaccine strains.
The gram-negative bacterium Ornithobacterium rhinotracheale was identified and recognized as a pathogen for birds in 1991 by van Beek et al. (9). The bacterium normally causes infection of the respiratory tract but may disseminate to other body sites, resulting in osteitis, meningitis, and joint infections (12). O. rhinotracheale infections in flocks occur worldwide. They are increasingly recognized as a problem in chicken and turkey farming. The major economic losses due to O. rhinotracheale infection result from the rejection of carcasses for consumption, growth retardation, and mortality. O. rhinotracheale is a member of the Flavobacteriaceae that belong to the Cytophaga-Flavobacterium-Bacteroides phylum (10). Related pathogens are the bird pathogens Riemerella anatipestifer and Coenonia anatina (10).
Infections with O. rhinotracheale can be successfully treated with antibiotics, but the bacterium rapidly develops antibiotic resistances (5). Infections of O. rhinotracheale can spread both horizontally by aerosols and vertically via infected eggs. Vaccination against O. rhinotracheale infections with a bacterin has been found to be suitable for the prevention of infections by the vertical route, but to be effective the bacterin has to be administered with an oily adjuvant (13). To prevent horizontal transfer of the bacterium, vaccination with a live avirulent strain seems to be effective (14). The construction of genetically well-defined and attenuated strains, however, is seriously hampered by the lack of tools to genetically manipulate the pathogen. For the related Flavobacteriaeceae a few tools have been developed (7), but these plasmids have been found not to be suitable for transformation of O. rhinotracheale (unpublished data).
An attractive approach for the development of a plasmid that can be exploited for genetic manipulation of O. rhinotracheale isolates is to modify an endogenous O. rhinotracheale plasmid. One strain of O. rhinotracheale has been reported to carry such a plasmid (A. Back, S. Sprenger, G. Rajashekara, D. A. Halvorson, and K. V. Nagaraja, Abstr. 48th North Central Avian Disease Conference, Des Moines, Iowa, p. 15-18, 1997). Here the isolation and characterization of the native plasmid pOR1 from O. rhinotracheale and the construction of a shuttle vector that can replicate both in E. coli and O. rhinotracheale are reported. This shuttle vector paves the way for genetic modification and characterization of the O. rhinotracheale genome and for the development of genetically defined vaccine strains.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains of O. rhinotracheale used in this study (Table 1) were kindly provided by P. van Empel (Intervet International, Boxmeer, The Netherlands). The characteristics of the strains have previously been described (11). O. rhinotracheale strains were grown at 37°C in an atmosphere of 5% CO2 on Colombia agar plates (Oxoid, Basingstoke, England) or in Todd-Hewitt broth (Oxoid). The broth cultures were incubated on an orbital shaker (120 rpm) at 37°C in a tightly sealed 50-ml Falcon tube.
TABLE 1.
Characteristics and origins of the O. rhinotracheale strains used in this study
| Strain | Serotype | Origin | Country (state) |
|---|---|---|---|
| 7 | A | Guinea fowl | France |
| 11 | B | Turkey | Germany |
| 12 | B | Turkey | The Netherlands |
| 18 | B | Turkey | Germany |
| 35 | D | Turkey | France |
| 45 | E | Turkey | Germany |
| 99-0118 | K | Turkey | United States (Minnesota) |
| 215 | K | Turkey | South Africa |
| 97-0321 | K | Turkey | United States (Iowa) |
DNA isolation and hybridization.
Total genomic DNA from bacterial strains was isolated by using the High Pure PCR template kit (Roche Diagnostics Nederland B.V., Almere, The Netherlands). Southern blot hybridization was done with Hybond N membranes (Amersham Biosciences, Roosendaal, The Netherlands). The probes were labeled with [32P]dATP by using a random primer labeling kit (Boehringer, Mannheim, Germany). The hybridizations were done under conditions of high stringency with a final wash of 0.5× SSPE (1× SSC is 0.18 M NaCl, 10 mM NaH2PO4, and 1mM EDTA [pH 7.7]) at 65°C.
Cloning and sequencing of pOR1.
The plasmid pOR1 was isolated from total genomic DNA from strain 99-0118 by CsCl gradient centrifugation (8) in a TL-100 ultracentrifuge (Beckman, Palo Alto, Calif.). The full-length pOR1 was cloned with its unique BamHI site into lambda EMBL3 phage (Promega Corp., Madison, Wis.) with E. coli JM109 as a host. The pOR1 DNA was subcloned as HindIII fragments into the pGEM7Zf(+) cloning vector (Promega Corp.) with E. coli DH5α as a host. Serial deletion clones for sequencing purposes were made by using the Erase-a-Base system (Promega Corp.). The DNA sequence was determined by using the Big-Dye terminator system on a 310 Genetic Analyzer (Perkin-Elmer Applied Biosystems, Foster City, Calif.).
Expression studies on pOR1 genes.
Total RNA was isolated from O. rhinotracheale cultured for 2 days on a blood agar plates. RNA was isolated by using RNasol B (Tel-Test, Friendswood, Tex.) under the manufacturer's conditions. Residual DNA was removed from the RNA preparation by using RQ1 RNase-free DNase (Promega). The reverse transcription (RT) of the RNA was done by using the Access RT-PCR system (Promega). The RT reaction was done at 37°C during 60 min with the downstream primer 2, 4, or 6 (Table 2). An aliquot of the RT reaction mixture was used in the PCR. The 35 PCR cycles were run under the following conditions: annealing at 55°C for 30 s, extension at 74°C for 1 min, and melting at 94°C for 30 s. The primers that were used in the RT reactions are listed in Table 2.
TABLE 2.
PCR and RT-PCR primers used in this study
| Primer | Position in POR1 | Sequence | Target gene |
|---|---|---|---|
| PCR | |||
| 16S-F | GAGAATTAATTTACGGATTAAG | 16S RNA | |
| 16S-R | TTCGCTTGGTCTCCGAAGAT | 16S RNA | |
| rep | GAAAGGATTTAAGCGGAC | rep2 | |
| M13U | CACGACGTTGTAAAACGACGGCCAG | ||
| RT | |||
| 1 | 4599-4615 | CGTGATAGATTACGGAG | Siderophore receptor |
| 2 | 4832-4849 | ATGATTGGACATTTAGCA | |
| 3 | 6759-6776 | CCAATGTAAGAGGGAAAG | Cd resistance |
| 4 | 7164-7181 | GTATAGCAATGGGTGGAT | |
| 5 | 10575-10592 | CAACCGAAGACAGGACAC | Laccase |
| 6 | 11210-11227 | GAGGAAGGTGATTGGTTC | |
| 7 | EGFPup | ACCCTCGTGACCACCCTGACCTAC | egfP |
| 8 | EGFPdown | GACCATGTGATCGCGCTTCTCGTT |
Heavy metal resistance.
Resistance to heavy metal ions was determined on Columbia agar plates with the heavy metal salts CdCl2, CoSO4, CuSO4, and HgCl2 (Merck, Darmstadt, Germany) at concentrations of 0.01, 0.1, 1, or 10 mM. The plates were inoculated with the O. rhinotracheale strains, and after 2 days of incubation the colonies were scored on a scale from 0 to 3 (0, no colonies; 1, visible colonies; 2, intermediate-size colonies; 3, colonies similar to those of the control). The MIC was defined as the lowest concentration at which colonies with a score of 2 were obtained.
Construction of pOREC1.
The shuttle vector pOREC1 consists of pGEM7 with the replication origin of pOR1 and the cfxA gene of Bacteroides vulgatus (EMBL/GenBank accession number U38243). Construction of pOREC1 was carried out in two steps. First, the replication origin of pOR1 was cloned as a BamHI/NdeI fragment of 2,681 bp into the BamHI and EcoRI sites of pGEM7Zf(+). Prior to the ligation, the NdeI and EcoRI sites were made blunt with the Klenow fragment of DNA polymerase. In the second step, the cfxA gene was cut from pCP29 as a BamHI/PstI fragment of 2.1 kbp (kindly provided by Sarika Agarwal, University of Wisconsin) and ligated into the BamHI/NsiI sites of the former construct.
Cloning of the gene encoding enhanced green fluorescent protein (EGFP) on pOREC1.
The gene encoding EGFP was excised from plasmid pEGFP (Clontech Laboratories Inc., Palo Alto, Calif.) as a 962-bp PvuII-EcoRI fragment. This fragment was cloned in the NdeI and EcoRI sites of pORECI, yielding a construct with the EGFP gene under the control of the lac promoter. This construct was introduced in O. rhinotracheale by electroporation.
Electroporation of O. rhinotracheale.
Transformation of O. rhinotracheale was done by electroporation with a GenePulser (Bio-Rad, Uppsala, Sweden). Electrocompetent cells were prepared from bacteria grown for 2 days in 10 ml of Todd-Hewitt broth in 50-ml Falcon tubes on an orbital shaker (120 rpm, 37°C). The optical density at 600 nm of the cultures was between 0.3 and 0.4. The cultures were washed two times with 25 ml of sterile distilled water and finally washed with 10% glycerol. The cell pellet was resuspended in approximately 100 μl of 10% glycerol, resulting in a suspension of 109 to 1010 CFU per ml. Electroporation was done in disposable cuvettes with an electrode distance of 2 mm, using 40 μl of cell suspension and 5 μl of DNA solution containing approximately 1 mg of DNA per ml. All manipulations of the bacteria were done at room temperature. The settings of the GenePulser were as follows: 25-μF capacity, 2,500 V, and 800 Ω parallel resistor. Directly after the pulse, 960 μl of Todd-Hewitt broth was added, and the bacteria were recovered for at least 90 min at 37°C. After recovery, the bacteria were plated on selective plates (cefoxitin, 5 μg/ml) and incubated for at least 3 days at 37°C with 5% CO2.
The electroporation survival rate was measured by comparing the CFU of serial dilutions of the cell suspension before and after the electric pulse. It was observed that the survival rate was close to 100%, which implies that the O. rhinotracheale cell wall is resistant to an electric pulse and probably difficult to be made permeable.
PCR and RT primers.
The primers used in a PCR to identify O. rhinotracheale from its 16S rRNA genes were primers 16S-F and 16S-R. These primers amplify a 784-bp fragment of the 16S rRNA gene of O. rhinotracheale (accession number L19156). The primers used to identify the presence of pOREC1 in transformants were primer rep, which is located in the rep2 gene of pOR1, and primer M13U, which is located in pGEM7Zf(+). The sequences of these primers and those of the primers used in the RT-PCR to demonstrate expression of pOR1 genes and egfp are listed in Table 2.
Assembly and annotation of pOR1 sequence.
The nucleotide sequence of pOR1 was determined by using a combination of subcloning and primer walking. The full-length sequence of pOR1 was assembled from the single-read sequences by using the SeqMan program of the Lasergene package (DNAStar Inc., Madison, Wis.). The whole pOR1 sequence was covered at least three times by single reads, and each base was identified with a reliability of at least 95% (Q value larger than 20).
The annotation of potential coding sequences was done by GeneMark.hmm at http://dixie.biology.gatech.edu/GeneMark/heuristic.cgi (2) and with BlastX at http://www.ncbi.nlm.nih.gov/BLAST/BLAST.cgi of the National Institutes of Health. To predict the potential function of the genes, the encoded proteins were compared with the proteins of the EMBL/GenBank database by using BlastP. Repetitive sequences and AT contents were determined by using the GeneQuest program of Lasergene.
Nucleotide sequence accession number.
The nucleotide sequence of the pOR1 plasmid has been deposited in the EMBL/GenBank database under accession number AY513488.
RESULTS
Cloning of pOR1 and prevalence among O. rhinotracheale isolates.
Plasmid pOR1 was isolated by CsCl density gradient centrifugation from O. rhinotracheale strain 99-0118. The plasmid banded at a higher density than genomic DNA, indicating that pOR1 is circular and supercoiled. The entire plasmid pOR1 was cloned into lambda phage EMBL3, using its unique BamHI site.
The occurrence of pOR1 among O. rhinotracheale isolates was investigated by screening total DNA of a panel of O. rhinotracheale isolates by dot blot DNA hybridization with the full-length pOR1 clone in lambda EMBL3 phage as a probe. The O. rhinotracheale strain 99-0118, which carries pOR1, originates from Minnesota and has the rare serotype K. The panel consisted of two other isolates that have serotype K, 34 non-K serotype isolates that originated in Minnesota, and 20 isolates that were isolated from different bird species from Europe and Africa. In one of the other serotype K strains (97-0321) that originated in Minnesota, pOR1 sequences were detected. All other isolates, including the serotype K isolate (strain 215) from South Africa, yielded no positive signals under the conditions employed. Southern blot analysis of the HindIII-digested DNAs from isolates 99-0118 and 97-0321 with pOR1 as a probe yielded identical patterns for both strains (data not shown). This indicates that both strains carry pOR1.
Determination and annotation of pOR1 sequences.
The nucleotide sequence of pOR1 comprised 14,787 bp. The plasmid has a GC content of 35.7%, which is similar to the 37 to 39% reported for the whole genome of O. rhinotracheale (10). The AT and GC nucleotides are unevenly distributed on the plasmid. Part of the plasmid, encompassing open reading frames (ORFs) 4 to 9 (see below), has a higher GC content (38.8%) than the remainder of the plasmid (31.2%), which might indicate that pOR1 has a mosaic composition.
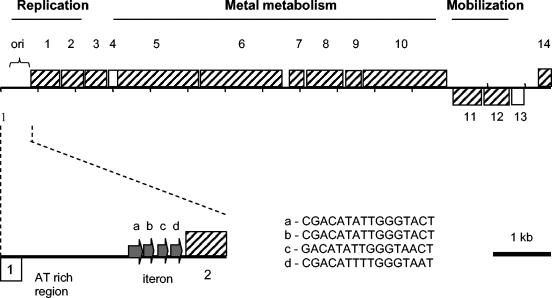
The ORFs on the plasmid were identified by using the GeneMark.hmm program (2). Thirteen regions that start with an initiation codon with the potential to encode a protein of more than 100 amino acids were found on pOR1. A map of the ORFs of pOR1 is presented in Fig. 1. ORFs encoding proteins of less than the arbitrarily set length of 100 amino acids were included only when they had significant homology to a protein sequence in the database. The vapD gene (17) is therefore included in Fig. 1 and Table 3, whereas two other genes encoding proteins of less than 100 amino acids (approximately 80 amino acids) are excluded, one in the origin region of pOR1 and one in front of ORF 14. To attribute a function to the putative genes, the encoded proteins were compared with the EMBL/GenBank database by using BlastP. The genes from the database with the best match to the putative genes are listed in Table 3.
FIG. 1.
Linear presentation of pOR1. In the upper panel, the pOR1 sequence of 14,787 bp is indicated by the solid line. The ORFs of pOR1 are indicated by boxes. The ORFs on the direct strand are positioned above the sequence line, and those on the reverse strand are positioned below the sequence line. Dashed ORFs have orthologs in the EMBL/GenBank database. In the lower panel, the replication region of pOR1 is enlarged. Indicated are the four direct repeats of the iteron and their nucleotide sequences (a to d).
TABLE 3.
Characteristics of putative genes of pOR1
| ORF | Gene | Size (amino acids) | Similar gene product | Probable function | e value | Gen info identifier no. | Species |
|---|---|---|---|---|---|---|---|
| 1 | repA | 325 | Replication protein A | Plasmid replication | 3e-72 | 1075495 | Bacteroides vulgatus |
| 2 | rep2 | 143 | Replication protein 2 | 3e-24 | 10956802 | Riemerella anatipestifer | |
| 3 | trans | 201 | Transposase | Transposition | le-41 | 16082872 | Yersinia pestis |
| 4 | orf4 | 109 | None | ||||
| 5 | orf5 | 741 | TonB-linked outer membrane receptor | Iron uptake | 2e-33 | 29345912 | Bacteroides thetaiotamicron |
| 6 | yvgW | 675 | Heavy metal ion-transporting ATPase | Cadmium efflux | 1e-159 | 21398552 | Cytophaga hutchinsonii |
| 7 | orf7 | 103 | None | ||||
| 8 | merP | 291 | Periplasmic transport protein | Mercury efflux | 6e-04 | 2935549 | Pseudomonas alcaligenes |
| 9 | mlpB | 111 | Putative lipoprotein | Unknown | 2e-19 | 4467968 | Myxococcus xanthus |
| 10 | pcoA | 768 | Copper resistance protein | Copper binding | 1e-72 | 13632794 | Escherichia coli |
| 11 | orf11 | 187 | Chromosome assembly protein homolog | Chromosome assembly | 0.002 | 15606061 | Aquifex aeolicus |
| 12 | mbpB | 268 | Mobilization protein | Plasmid mobilization | 2e-10 | 915338 | Bacteroides fragilis |
| 13 | orf13 | 135 | None | ||||
| 14 | vapD | 93 | Virulence associated protein | Unknown | 2e-24 | 6136169 | Actinobacillus actinomycetemcomitans |
Three functional regions on pOR1 can be distinguished. These regions are involved in plasmid replication (ORFs 1 and 2), heavy metal metabolism (ORFs 5 to 10), and mobilization (ORFs 11 and 12). ORFs 1 and 2 are homologous to the replication genes repA and rep2 of B. vulgatus and R. anatipestifer plasmids, respectively. These Rep proteins are usually found in plasmids with a theta-form replication mode (3). In front of the repA gene four imperfect repeats that may function as iterons that typically regulate plasmid replication can be recognized (Fig. 1). No other iteron sequences are present elsewhere on the plasmid. The replication origin of the plasmid is most likely located in front of the iteron (4). The region of 262 bp before the iteron has a high AT content (76%), which is characteristic of plasmid replication origins.
ORF 3 encodes a protein of 201 amino acid residues. This protein is similar to the proximal halves of a large number of bacterial transposases that are about 400 amino acid residues in size. Therefore, ORF 3 is most likely the truncated transposase of an insertion sequence element.
ORF 5 encodes a protein that is highly similar to TonB-linked siderophore receptors. Siderophore receptors allow the bacteria to recruit iron from the environment.
The proteins encoded by ORFs 6 to 10 show similarities to proteins that are involved in resistance to heavy metal ions. ORF 6 encodes a protein that is highly similar to numerous bacterial heavy metal ion-transporting ATPases of the CPx-type ATPase family. These proteins facilitate efflux of heavy metal ions such as cadmium, cobalt, and copper across the bacterial cell envelope (16). The protein encoded by the small ORF 7 has no significant similarity to any proteins in the databases. The 291 amino acid residues encoded by ORF 8 have similarity to the MerP protein, which is involved in mercury resistance. However, this is unlikely, since MerP proteins are much smaller (approximately 90 amino acid residues) than the ORF 8 protein. In addition, the merP genes are part of an operon encoding proteins that are involved in sensing, transport, and reduction of mercury ions, which is not the case for pOR1. The ORF 9 protein is similar to a single protein, MlpB of Myxococcus xanthus. No function has been attributed to MlpB, and the function of ORF 9 is unknown.
The protein encoded by ORF 10 has a high similarity to two groups of proteins. The highest similarity (e-72) was found with bacterial proteins involved in copper resistance, such as PcoA of E. coli and CopA of Xanthomonas campestris and Pseudomonas species. A lower but very significant similarity (4e-37) was found with the large family of the laccases. Laccases are enzymes (EC 1.10.3.2) that are widely distributed among bacteria and fungi (6). The laccases require copper binding for activity. The ORF 10 protein has the four copper binding domains that are found in all laccases (see annotation at EMBL/GenBank AY513488 and at www.llu.edu/llu/medicine/micro/laccase/www.llu.edu/llu/medicine/micro/laccase/), which strongly suggests that the ORF 10 protein is a laccase.
The proteins encoded by ORFs 11 and 12 may form a functional unit that is involved in plasmid mobilization. The functional entity of these genes is also reflected by their reverse orientation with respect to the other genes of pOR1. The protein encoded by ORF 11 has a weak similarity to a putative chromosome assembly protein of Aquifex aeolicus, but that encoded by ORF 12 shows a clear similarity to the plasmid mobilization protein MbpB of Bacteroides fragilis.
The protein encoded by ORF 14 is similar to the virulence-associated protein D (VapD) that is present in at least 13 bacterial species, among which are some known bird pathogens such as R. anatipestifer. Although the name of these proteins suggests involvement in bacterial virulence, no function has been attributed to these proteins (17).
Expression of heavy metal resistance-encoding genes of pOR1.
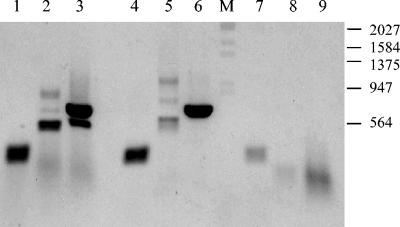
RT-PCR was used to test the expression of three of the functionally related genes of pOR1. The three genes are ORF 5, encoding the putative TonB-linked siderophore receptor; ORF 6, encoding the putative heavy metal-transporting ATPase; and ORF 10, encoding the putative copper resistance or the putative laccase. RT-PCR was carried out on total RNA from strain 99-0118 grown on agar on plates for 2 days. RT-PCR products were obtained for all three genes, which is indicative of gene transcription (Fig. 2).
FIG. 2.
Transcription of pOR1 genes. An ethidium bromide-stained agarose gel of DNA fragments amplified by RT-PCR of genes located on pOR1 in O. rhinotracheale strain 99-0118 is shown. Lanes 1, 2, and 3: RT-PCR products of the putative siderophore receptor, the Cd resistance gene, and the laccase, respectively. Lanes 4, 5, and 6: PCR on the cloned pOR1 sequences. Lanes 7, 8, and 9: RT-PCR of the RNase-treated RNA preparation. The molecular weight marker (M) was EcoRI- and HindIII-digested phage lambda DNA. The fragment sizes of the marker (in base pairs) are indicated.
The functionality of the putative heavy metal resistance protein encoded by ORF 6 was tested by its ability to protect O. rhinotracheale from toxic heavy metal ions. Strains 99-0118 and 97-0321, which carry pOR1, were exposed to various concentrations of the heavy metal ions cobalt, chromium, cadmium, mercury, and copper. Strain 215 of serotype K, strain 7 of serotype A, and strains 11 and 18 of serotype B, which all lack pOR1, served as controls in these experiments. The growth of the six strains was fully inhibited by mercury (0.05 mM), while cobalt and chromium were not inhibitory to the O. rhinotracheale strains at a concentration of 1 mM. Cadmium inhibited growth of the three serotype K strains at 0.05 mM, while three non-serotype K strains were inhibited at the lower concentration of 0.01 mM cadmium. Copper inhibited the growth of the four strains that do not carry pOR1 at a concentration of 0.5 mM, while the two pOR1-carrying strains were inhibited at concentrations 10 times higher (5 mM). These data indicate that the putative heavy metal ATPase of pOR1 hardly protects the bacteria from heavy metal toxicity. Only a slightly higher copper resistance is correlated with the presence of pOR1. The higher cadmium resistance of the plasmid-carrying stains is unlikely to be correlated with the presence of the plasmid, since strain 215, which has the same serotype K but not pOR1, exhibited similar levels of resistance to cadmium.
Transformation of O. rhinotracheale and construction of a shuttle vector.
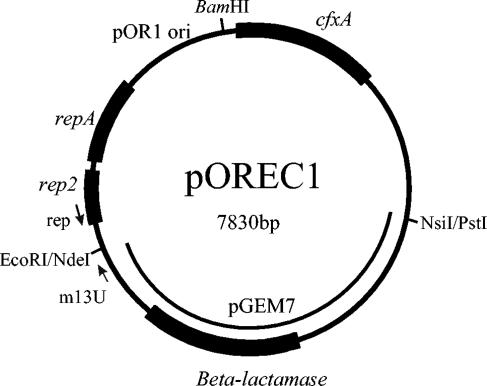
To establish a transformation system for O. rhinotracheale, an E. coli-O. rhinotracheale shuttle vector was constructed. This shuttle vector, pOREC1, was made by cloning the putative replication origin, the iteron, and the repA and rep2 genes (ORFs 2 and 3) needed for replication in O. rhinotracheale into pGEM7. As a selection marker, the cefoxitin resistance gene cfxA from B. vulgatus was included. A map of the shuttle vector pOREC1 is presented in Fig. 3.
FIG. 3.
Organization of the shuttle plasmid pOREC1. Boxes represent the genes that are involved in plasmid replication and selection in O. rhinotracheale (repA, rep2, and cfxA) and E. coli (beta-lactamase). The restriction sites and the part of the plasmid derived from pGEM7 that were used in the construction of pOREC1 are indicated.
Transformation of O. rhinotracheale was achieved by electroporation with 0.5 μg of pOREC1 that was isolated from E. coli. Several O. rhinotracheale strains were subjected to electroporation with pOREC1, and cefoxitin-resistant colonies were found after 4 days of incubation. The efficiency of the transformation was approximately 400 recombinants per μg of plasmid DNA. No strain differences in the efficiency of transformation were found. The presence of pOREC1 in the recombinant O. rhinotracheale was confirmed by PCR with primers M13U and rep (Fig. 3). In addition, recombinants were verified as O. rhinotracheale by a species-specific PCR on the 16S rRNA gene with primers OR16S-F and OR16S-R. Transformation of E. coli with total DNA of the O. rhinotracheale recombinants again resulted in transformed E. coli with pOREC1. This indicates that pOREC1 was present in O. rhinotracheale as an independent replicon. The transformation of both O. rhinotracheale and E. coli with pOREC1 demonstrates the functionality of the replication origin and rep genes of pOR1.
Expression of EGFP on pOREC1.
The expression of EGFP by O. rhinotracheale can be a useful tool to monitor the tissue localization of the bacteria after experimental infection of cultured cells and animals. The gene encoding the EGFP protein was cloned into pOREC1 between the replication origin of pOR1 and the pGEM7 part of the shuttle vector (Fig. 3). The EGFP gene was placed under the control of the lac promoter. It was demonstrated by RT-PCR with EGFP-specific primers that transcripts of the EGFP gene were present in O. rhinotracheale transformed with the pOREC1-EGFP construct, demonstrating that the lac promoter is active in O. rhinotracheale. However, the bacteria that transcribed the EGFP gene did not show any fluorescence.
DISCUSSION
In this report, the successful transformation of O. rhinotracheale by using a newly constructed plasmid shuttle vector, pOREC1, is described. The transformation of O. rhinotracheale for the first time paves the way to genetic manipulation of this bacterium and will aid in a better understanding of the virulence of this bird pathogen. The genetic manipulation of O. rhinotracheale also may aid in the development, by directed mutagenesis, of a defined attenuated vaccine strain of O. rhinotracheale.
The bacterium O. rhinotracheale has been subjected to numerous fruitless attempts at transformation with shuttle vectors from other Flavobacteriaceae and wide-host-range plasmids (R. Jansen, unpublished data). Most likely this lack of success can be attributed to the failure of the introduced plasmids to replicate within O. rhinotracheale, since for these other vectors the same transformation protocol and the same selection marker, the cfxA gene, as for pOREC1 were employed.
In the construction of the functional shuttle plasmid pOREC1,the replication origin, the iteron region, and the two putative replication genes repA and rep2 of pOR1 were used. The presence of two replication genes, repA and rep2, is uncommon for plasmids with theta-like replication. Most of these plasmids have a single repA gene. A second replication gene similar to rep2 is also present on pCFC1 of R. anatipestifer and on pUCL287 of Tetragenococcus halophilus (1, 15). In T. halophilus the Rep2 protein is not essential for replication but is involved in copy number control and segregational stability (1). It is possible that rep2 of O. rhinotracheale has a similar function.
The plasmid pOR1 was detected in only two isolates of the numerous O. rhinotracheale strains that were analyzed. These two isolates might represent the same strain, since both isolates originate from the same geographical region and have the rare serotype K. The apparent rare occurrence of pOR1 suggests that the plasmid was introduced into O. rhinotracheale on a single recent occasion and has not spread in the population. The selective advantage for the bacterium of carrying pOR1 is unclear. Apart from the replication genes, one can speculate on the advantage to the bacterium of three of the other genes. First, the putative laccase might be a virulence factor for O. rhinotracheale, as was suggested for the pathogenic fungus Cryptococcus neoformans (6). The laccase of C. neoformans possibly is involved in protection of the pathogen against killing by alveolar macrophages. The laccase prevents the production of hydroxyl radicals by oxidizing Fe(II) to Fe(III) in the macrophage. However, attempts to demonstrate laccase activity in O. rhinotracheale by using the polymerization activity of the laccase in pigment formation failed (R. Jansen, unpublished data). The activity of other bacterial laccases also is enigmatic, and the assays that were developed for plant and fungal laccases are not suitable for the bacterial family of laccases.
The second gene that may be beneficial for O. rhinotracheale is that for the heavy metal-transporting ATPase. This enzyme may aid the bacterium in scavenging for copper, an essential component of the laccase. Our experiments did not reveal such a function, as inferred from the equal sensitivity of the various strains to the heavy metals cadmium and mercury. The slightly higher resistance to copper that was observed is unlikely to have physiological significance, considering the extremely high concentrations (0.5 mM) at which the effect became apparent.
The third gene that might be advantageous to the bacterium is the gene encoding the TonB-linked siderophore receptor. The corresponding protein might aid the bacterium in scavenging for iron, which is essential for bacterial growth, or, alternatively, for copper to aid the laccase activity.
Clearly, the most important conclusions from the present work are that transformation of O. rhinotracheale is feasible and that the developed transformation system can be exploited to genetically modify the bacterium. The efficiency of transformation with the shuttle vector pOREC1 is still rather low and needs improvement for efficient use in, for instance, transposon mutagenesis or DNA displacement by homologous recombination. The low efficiency of transformation is likely caused by the rather poor permeability of the O. rhinotracheale membrane during electroporation and/or by restriction of the entering plasmid DNA. Optimal electroporation may be achieved by growing the bacteria under conditions that weaken the cell wall. The absence of fluorescence in O. rhinotracheale transformed with the pOREC 1 shuttle vector containing the EGFP gene likely resulted from inefficient translation of the mRNA, possibly because the codon usage of EGFP is not suitable for O. rhinotracheale. The codon usage of the genes from plasmid pOR1 differs strongly from the codon usage of EGFP. Most notable is that only 3.6% of the EGFP codons have an A or T residue at the first position, while the codons of the native O. rhinotracheale genes have 58% A or T at the first position.
Acknowledgments
We gratefully acknowledge L. Heijmen-van Dijk and C. Zwaagstra for DNA sequencing.
R.J. is a recipient of the Paul K. Storm Memorial Grant awarded by Intervet International B.V., Boxmeer, The Netherlands.
REFERENCES
- 1.Benachour, A., J. Frere, S. Flahaut, G. Novel, and Y. Auffray. 1997. Molecular analysis of the replication region of the theta-replicating plasmid pUCL287 from Tetragenococcus (Pediococcus) halophilus ATCC33315. Mol. Gen. Genet. 255:504-513. [DOI] [PubMed] [Google Scholar]
- 2.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chattoraj, D. K. 2000. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 37:467-476. [DOI] [PubMed] [Google Scholar]
- 4.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devriese, L. A., J. Hommez, P. Vandamme, K. Kersters, and F. Haesebrouck. 1995. In vitro antibiotic sensitivity of Ornithobacterium rhinotracheale strains from poultry and wild birds. Vet. Rec. 137:435-436. [DOI] [PubMed] [Google Scholar]
- 6.Liu, L., R. P. Tewari, and P. R. Williamson. 1999. Laccase protects Cryptococcus neoformans from antifungal activity of alveolar macrophages. Infect. Immun. 67:6034-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McBride, M. J., and S. A. Baker. 1996. Development of techniques to genetically manipulate members of the genera Cytophaga, Flavobacterium, Flexibacter, and Sporocytophaga. Appl. Environ. Microbiol. 62:3017-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., p. 1.42-1.52. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.van Beek, P., P. van Empel, G. van den Bosch, P. Storm, J. Bongers, and J. du Preez. 1994. Respiratory problems, growth retardation and arthritis in turkeys and broilers caused by a Pasteurella-like organism: Ornithobacterium rhinotracheale or ′Taxon 28′. Tijdsch. Diergeneeskunde 119:99-101. [PubMed] [Google Scholar]
- 10.Vandamme, P., P. Segers, M. Vancaneyt, K. van Hover, R. Mutters, J. Hommez, F. Dewirst, B. Paster, K. Kersters, E. Falsen, L. Devrieze, M. Bisgaard, K-H. Hinz, and W. Mannheim. 1994. Ornithobacterium rhinotracheale gen. nov., sp. nov., isolated from the avian respiratory tract. Int. J. Syst. Bacteriol. 44:24-37. [DOI] [PubMed] [Google Scholar]
- 11.van Empel, P. 1998. Ornithobacterium rhinotracheale. Ph.D. thesis. Utrecht University, Utrecht, The Netherlands.
- 12.van Empel, P., M. Vrijenhoek, D. Goovaerts, and H. van den Bosch. 1999. Immuno-histochemical and serological investigation of experimental Ornithobacterium rhinotracheale infection in chickens. Avian Pathol. 28:187-193. [DOI] [PubMed] [Google Scholar]
- 13.van Empel, P., and H. van den Bosch. 1998. Vaccination of chickens against Ornithobacterium rhinotracheale infection. Avian Dis. 42:572-578. [PubMed] [Google Scholar]
- 14.van Empel, P., and H. Hafez. 1999. Ornithobacterium rhinotracheale. Avian Pathol. 28:217-227. [DOI] [PubMed] [Google Scholar]
- 15.Weng, S., W. Lin, Y. Chang, and C. Chang. 1999. Identification of a virulence-associated protein homolog gene and ISRa1 in a plasmid of Riemerella anatipestifer. FEMS Microbiol. Lett. 179:11-19. [DOI] [PubMed] [Google Scholar]
- 16.Williams, L. E., J. K. Pittman, and J. L. Hall. 2000. Emerging mechanisms for heavy metal transport in plants. Biochim. Biophys. Acta 1465:104-126. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida, Y., Y. Nakano, Y. Yamashita, and T. Koga. 1997. The gnd gene encoding a novel 6-phosphoglyconate dehydrogenase and its adjacent region of Actinobacillus actinomycetemcomitans chromosomal DNA. Biochem. Biophys. Res. Commun. 230:220-225. [DOI] [PubMed] [Google Scholar]