Abstract
The aim of this work was to identify genes responsible for host recognition in the lactococcal phages sk1 and bIL170 belonging to species 936. These phages have a high level of DNA identity but different host ranges. Bioinformatic analysis indicated that homologous genes, orf18 in sk1 and orf20 in bIL170, could be the receptor-binding protein (RBP) genes, since the resulting proteins were unrelated in the C-terminal part and showed homology to different groups of proteins hypothetically involved in host recognition. Consequently, chimeric bIL170 phages carrying orf18 from sk1 were generated. The recombinant phages were able to form plaques on the sk1 host Lactococcus lactis MG1614, and recombination was verified by PCR analysis directly with the plaques. A polyclonal antiserum raised against the C-terminal part of phage sk1 ORF18 was used in immunogold electron microscopy to demonstrate that ORF18 is located at the tip of the tail. Sequence analysis of corresponding proteins from other lactococcal phages belonging to species 936 showed that the N-terminal parts of the RBPs were very similar, while the C-terminal parts varied, suggesting that the C-terminal part plays a role in receptor binding. The phages investigated could be grouped into sk1-like phages (p2, fd13, jj50, and φ7) and bIL170-like phages (P008, P113G, P272, and bIL66) on the basis of the homology of their RBPs to the C-terminal part of ORF18 in sk1 and ORF20 in bIL170, respectively. Interestingly, sk1-like phages bind to and infect a defined group of L. lactis subsp. cremoris strains, while bIL170-like phages bind to and infect a defined group of L. lactis subsp. lactis strains.
Lactococcus lactis is the most important bacterium used for starter cultures by the dairy industry. An important problem in industrial milk fermentation is infection of the starter bacteria by bacteriophages, which leads to bacterial lysis. The consequences of phage infection are fermentation delay, alteration of product quality, and in severe cases loss of the product. All these outcomes result in considerable economic loss to dairies (5, 22). Industrial phage ecology is dominated by three phage species: the predominant species 936 and species c2 and P335, which also have considerable importance (5, 22).
The first step in phage infection is adsorption of the phage to the host cell. Despite the fact that little information concerning the adsorption process of lactococcal phages is available, it seems to be a two-step process that starts with reversible binding to specific carbohydrates exposed on the surface of the cell wall (30, 37, 39), which is followed by, at least in species c2, irreversible binding to a protein in the cell membrane (14, 30, 38). Information on receptor-binding proteins (RBPs) in phages infecting gram-positive bacteria is sparse compared to the information available for phages infecting gram-negative bacteria. However, recently, Duplessis and Moineau identified the first RBP genes for the Streptococcus thermophilus phages DT1 and MD4 (11). These authors reported that a variable region in the C-terminal part of the protein encoded by orf18 of phage DT1 was responsible for host recognition. Subsequently, the RBP genes were identified in three prolate-headed phages belonging to species c2 infecting L. lactis (35). For these phages the homologous genes orf2 in CHL92, orf35 in bIL67, and orfl15 in c2 were identified as the RBP genes. The central part of the RBPs was found to be responsible for host recognition.
In this work we identified the RBP genes in the lytic L. lactis phages sk1 and bIL170 belonging to the important species 936. The genomes of both of these phages have been completely sequenced (7, 10). The sk1 genome has 28,451 nucleotides and contains 54 open reading frames (7). The phage bIL170 genome is larger, with 31,754 nucleotides and 64 open reading frames (10). sk1 and bIL170 are closely related and have a high level of DNA identity (84% nucleotide identity for 80% of the shortest genome) and the same overall gene organization (10). Especially the late-transcribed regions encoding the structural genes are very similar in the two phages. Despite the similarities between sk1 and bIL170, they have different host ranges. Consequently, they constitute a useful model for identifying and studying genes that are important for host recognition.
MATERIALS AND METHODS
Bacteria, bacteriophages, and growth conditions.
The bacterial strains and bacteriophages used in this study are listed in Table 1.
TABLE 1.
Bacterial strains, phages, and plasmids used in this study
| Bacterial strain, phage, or plasmid | Relevant feature(s) | Source and/or reference |
|---|---|---|
| E. coli strains | ||
| XL1-Blue MRF′ | Transformation host, Tetr | Stratagene, La Jolla, Calif. |
| BL21D | T7 expression strain | Invitrogen, Carlsbad, Calif. |
| L. lactis strains | ||
| Bu2 | L. lactis subsp. lactis biovar diacetylactis | 19 |
| F7/2 | L. lactis subsp. lactis biovar diacetylactis | 28 |
| FD13 | L. lactis subsp. cremoris | E. W. Nielsenb |
| IL1403a | L. lactis subsp. lactis, plasmid free (AE005176) | 3 |
| KH | L. lactis subsp. cremoris | 34 |
| MG1614 | L. lactis subsp. cremoris, plasmid free | 13 |
| V32.2 | L. lactis subsp. cremoris | E. W. Nielsen |
| Phagesc | ||
| bIL66 | Propagated on IL1403 | 2 |
| bIL170a | Propagated on IL1403 (AF009630) | 10 |
| P008 | Propagated on F7/2 | 28 |
| P113G | Propagated on IL1403 | 27 |
| P272 | Propagated on IL1403 | 27 |
| fd13 | Propagated on FD13 | E. W. Nielsen |
| jj50 | Propagated on KH | 23 |
| p2 | Propagated on KH | 17 |
| sk1a | Propagated on MG1614 (AF011378) | 6 |
| φ7 | Propagated on KH | E. W. Nielsen |
| Plasmids | ||
| pJDC9-6 | E. coli-L. lactis low-copy-number shuttle vector, Ermr derivative of pJDC9 with a replicon from pJW566 | 8; J. Josephsen |
| pKDU1 | pJDC9-6 with an insert of nucleotides 13117 to 15469 from phage sk1, Emr | This study |
| pRSET-B | T7 expression vector, Ampr | Invitrogen |
| pKDU2 | pRSET-B with an insert of nucleotides 14197 to 14672 from phage sk1, Ampr | This study |
Bacterial strain or phage with known genome sequence. The GenBank accession number is indicated in parentheses.
Department of Food Science, The Royal Veterinary and Agricultural University, Frederiksberg, Denmark.
Phages belonging to species 936.
Escherichia coli XL1-Blue MRF′ was grown in Luria-Bertani medium (33) at 37°C with 12.5 μg of tetracycline per ml. Erythromycin (200 μg/ml) was added when appropriate. L. lactis strains were cultured at 30°C in M17 broth (Oxoid Ltd., Basingstoke, Hampshire, England) supplemented with 0.5% (wt/vol) glucose (GM17). Erythromycin (5 μg/ml) was added when appropriate. Phages were propagated on their hosts growing in GM17 containing 5 mM CaCl2 (GM17-Ca2+). Media for plaque assays containing agarose instead of agar were prepared essentially as described by Lillehaug (26).
Preparation of plasmid and phage DNA.
E. coli and L. lactis plasmid DNA was isolated by using a QIAprep spin plasmid miniprep kit (QIAGEN, Inc., Chatsworth, Calif.) as recommended by the manufacturer, except that L. lactis initially was incubated with lysozyme (20 mg/ml) for 15 min at 37°C. Phage DNA was isolated as described by Josephsen et al. (21).
Molecular DNA techniques.
Molecular DNA techniques were performed essentially as described by Sambrook and Russell (33). Primers (Table 2) used for PCR amplification of DNA fragments were obtained from MWG Biotech AG (Ebersberg, Germany). orf18 in sk1 was amplified by using primers sk1orf16F and sk1orf20R, and the PCR product obtained was digested with XbaI and PstI. The PCR fragments were cloned in pJDC9-6, and the resulting plasmid was designated pKDU1 (Table 1). The 3′ fragment of orf18 in sk1, covering nucleotides 14197 to 14672, was amplified with primers sk1orf18IHF and sk1orf18IHR. After digestion with BamHI and EcoRI, PCR fragments were cloned in the T7 expression vector pRSET-B (Invitrogen, Carlsbad, Calif.). The resulting plasmid was designated pKDU2 (Table 1). Genes analogous to orf18 and orf20 from sk1 and bIL170 were PCR amplified in phages fd13, φ7, jj50, p2, P008, P113G, P272, and bIL66 by using primers bIL170orf19F and bIL170orf22R covering nucleotides 15522 to 16866 in bIL170.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)a | Position | Restriction site |
|---|---|---|---|
| sk1orf16F | GCTCTAGAATAGAAAAAGAATCACGACAA | 13117 | XbaI |
| sk1orf20R | AACTGCAGAAAGTATGAGCGACAACTCTC | 15469 | PstI |
| sk1orf18F | GGGTAGATAGACCATTTCGTC | 14496 | |
| bIL170orf22R | CTAACATGCGTGTAGATCGTA | 17409 | |
| sk1orf18IHF | CGGGATCCTAACGGTGTTGATATAAACAA | 14197 | BamHI |
| sk1orf18IHR | CGGAATTCTTTGTTTCGCTTTCTATTTTG | 14672 | EcoRI |
| bIL170orf19F | GCTCTAGAGTATCAATTGGTGGTTATCC | 15522 | XbaI |
| bIL170orf22R | AACTGCAGTGAGCCTGCGTCATT | 16866 | PstI |
Underlined sequences are restriction sites.
Electrotransformation of plasmid DNA into E. coli was performed essentially as described by Sambrook and Russell (33), while electroporation of L. lactis was performed as described by Holo and Nes (18).
DNA sequence analysis.
DNA sequence analysis of the PCR products was performed with a CEQ 2000 DNA analysis system (Beckman Coulter, Inc., Fullerton, Calif.). Sequences were assembled by using the ContigExpress program of the Vector NTI Suite 7 software package (Invitrogen). Sequences were aligned by using the ClustalX (version 1.81) program (36). BLAST searches (1) were conducted by using the National Center for Biotechnology Information homepage (http://www.ncbi.nlm.nih.gov/BLAST/).
Production and identification of recombinant phages with altered host specificity.
Recombinant phages with altered host specificity were obtained as previously described (11, 35). Plasmid pKDU1 was transformed into L. lactis IL1403 (host strain for bIL170). The transformed strain was grown to an optical density at 600 nm of 0.3 and infected with phage bIL170 at a multiplicity of infection of 0.01. After overnight incubation, 200-μl samples of the phage lysate were tested in plaque assays against L. lactis MG1614 (host strain for phage sk1). Production of recombinant phages was confirmed by a PCR assay performed directly with single plaques. The PCR assay included four primers, sk1orf16F, sk1orf20R, sk1orf18F, and bIL170orf22R (Table 2).
Host range determination.
Host ranges of the phages listed in Table 1 were determined by using the spot test method. Ten-microliter drops of suitable phage dilutions were deposited on top of a soft layer containing 200 μl of a fresh overnight culture. Clearing zones were recorded after incubation overnight.
Phage adsorption assay with SYBR Gold-stained phages.
Phages were SYBR Gold stained essentially as described by Noble and Fuhrman (31). A phage lysate with a titer of at least 1010 PFU/ml was mixed 20:1 (vol/vol) with a 1,000-fold diluted SYBR Gold stock solution (Molecular Probes, Inc., Eugene, Oreg.) and incubated overnight in the dark at 4°C. Phages were mixed 1:1 with host cells grown to an optical density at 600 nm of 0.6, and phage adsorption was examined with a fluorescence microscope (Axioplan 2; Zeiss, Inc., Jena, Germany) at a magnification of ×1,000 with blue light (450- to 490-nm) excitation. If the phage titer was low (<1010 PFU/ml), a 10-min preincubation step was included before inspection by microscopy. Adsorption of phages to the host cells was indicated by a fluorescent halo around the cells.
Expression and purification of proteins.
The 3′-terminal part of orf18 in sk1, cloned in pRSET-B (pKDU2), was expressed by using the T7 expression system (Invitrogen). The resulting proteins were subsequently purified by using the Probond purification system (Invitrogen). Both systems were used as recommended by the manufacturer.
Preparation of polyclonal antisera.
A rabbit was immunized by subcutaneous injection of 200 mg of purified protein consisting of the C-terminal part of ORF18 (158 amino acids) of phage sk1. Before injection the protein solution was mixed 1:1 with Freund's incomplete adjuvant (Sigma-Aldrich Chemie GmbH, Steinheim, Germany). The rabbit was immunized four times at 3-week intervals. Antisera were obtained 14 days after the last immunization.
SDS-PAGE, Western blotting, and dot blotting.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), Western blotting, and dot blotting were carried out essentially as described by Harlow and Lane (16). The NuPage precast gel system (Invitrogen) was used for SDS-PAGE and Western blotting. For dot blotting, 109 phage were collected in each dot on a nitrocellulose membrane (BA85; Schleicher and Schüll, Dassel, Germany) by using a Minifold I SRC 96 dot blotter (Schleicher and Schüll). The polyclonal antisera were diluted 1:20,000 and used for overnight incubation of the blots. Alkaline phosphatase-conjugated anti-rabbit immunoglobulin G (Sigma-Aldrich) was used at a 1:30,000 dilution during a 2-h incubation. As substrate for the alkaline phosphatase, 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium (Sigma-Aldrich) was used according to the instructions of the manufacturer.
Immunogold labeling and electron microscopy.
Phages were incubated overnight at room temperature with polyclonal antisera that were diluted 10-fold. Subsequently, the phage-polyclonal antibody mixture was incubated for 2 h with anti-rabbit immunoglobulin G-5-nm gold conjugate (BioCell, Oxford, England) diluted 1:10 to 1:40. After the phage-antibody complexes were fixed with 0.1% (vol/vol) glutaraldehyde for 30 min, samples were attached to 300-mesh carbon-coated grids (Electron Microscopy Sciences, Fort Washington, Pa.) for 5 min. Liquid was removed from the grids, and the preparations were negatively stained in 2% (wt/vol) uranyl acetate for 5 min. Air-dried grids were examined with a transmission electron microscope (CM100; Philips, Eindhoven, The Netherlands) at 80 kV and at an instrument magnification of ×52,000.
Nucleotide sequence accession numbers.
Sequence data for phage gene homologues to orf18 and orf20 in sk1 and bIL170 have been deposited in the GenBank database under accession numbers AF539441 (p2), AF539444 (fd13), AF539448 (jj50), AF539450 (φ7), AF539449 (P008), AF539451 (P113G), AF539443 (P272), and AF539445 (bIL66).
RESULTS AND DISCUSSION
Bioinformatic analysis of phages sk1 and bIL170.
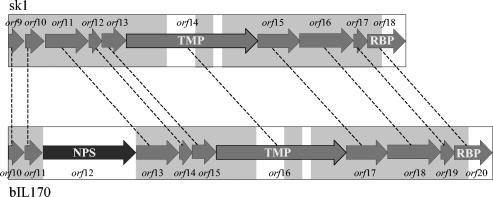
Due to the completely different host ranges of phages sk1 and bIL170 (Table 3), putative RBP genes were assumed to show no or only partial homology in the two phages. The first step in identifying the RBP genes was to demarcate the DNA region in both phages encoding the tail proteins. Previously, Chandry et al. (7) suggested that the region encoding tail proteins in phage sk1 included orf9 to orf18 on the basis of a comparison of the gene sizes and predicted isoelectric points of the resulting proteins in the phage lambda tail region with corresponding information for sk1. A homologous tail region in bIL170 consisting of orf10 to orf20 was identified (10). This region of the bIL170 genome included a gene (orf12) with no homologous gene in phage sk1, as well as two regions of nonhomology to sk1 in the center of orf16 (orf14 in sk1) and a region with no homology in the 3′ end of orf20 (orf18 in sk1) (Fig. 1).
TABLE 3.
Infection and binding abilities of lytic lactococcal bacteriophagesa
| Phageb |
L. lactis subsp. lactis strains
|
L. lactis subsp. cremoris strains
|
|||||
|---|---|---|---|---|---|---|---|
| IL1403 | F7/2 | Bu2 | KH | MG1614 | FD13 | V32.2 | |
| bIL170 | + | + | + | − | − | − | − |
| P008 | + | + | + | − | − | − | − |
| bIL66 | + | + | + | − | − | − | − |
| P113G | + | + | + | − | − | − | − |
| P272 | + | + | + | − | − | − | − |
| sk1 | − | − | − | + | + | + | + |
| fd13 | − | − | − | + | + | + | + |
| φ7 | − | − | − | + | + | + | + |
| jj50 | − | − | − | + | + | + | + |
| p2 | − | − | − | + | + | + | + |
Infection was determined by a spot test, and binding ability was determined by using SYBR Gold-stained phages.
Phages showing homology in the C-terminal part of the RBPs (Fig. 5) are grouped together.
FIG. 1.
Gene organization of the genome regions in sk1 and bIL170 encoding tail proteins. Homologous genes in phages sk1 and bIL170 are connected by dashed lines. The grey boxes indicate DNA sequences with homology in sk1 and bIL170. The open boxes indicate DNA sequences with no homology in sk1 and bIL170. NPS, putative neck passage structure; TMP, putative tape measure protein.
BLAST searches were conducted with the predicted protein sequences encoded by the genes in phages sk1 and bIL170 which exhibited no or only partial homology. ORF12 in bIL170 showed 68% amino acid identity over the whole sequence (653 amino acids) with ORF51, a putative neck passage structure in phage TP901-1 (4, 20). This is consistent with electron micrographs of sk1 and bIL170 showing that bIL170 contains a collar that is absent in sk1 (data not shown) and that deletion of orf12 did not impair propagation of bIL170 (10).
National Center for Biotechnology Information BLAST analyses of OFR14 (sk1) and ORF16 (bIL170) revealed that the proteins showed approximately 18% amino acid identity to ORF45, an experimentally determined tape measure protein in phage TP901-1 (32). In addition, the 3D-PSSM fold program (25) suggested that both proteins fold in an α-helix-like structure that also has been found for the tape measure protein gpH in phage lambda (24). Hence, it appears likely that orf14 and orf16 encode tape measure proteins in phages sk1 and bIL170, respectively.
orf18 in sk1 and orf20 in bIL170 showed 89% identity in the 5′ 340 nucleotides (Fig. 1), while the remaining 460 nucleotides of the genes showed no homology. At the protein level, the unique C-terminal part of ORF18 in sk1 (approximately 140 amino acids) showed 33% amino acid identity to ORF49, an experimentally determined base plate protein in TP901-1, and 27% amino acid identity to ORF18, a putative RBP, in phage BK5-T (29). For comparison, the C-terminal part of ORF20 in bIL170 (approximately 140 amino acids) showed 43% identity to the C-terminal part of proteins hypothetically involved in host recognition in two L. lactis IL1403 prophages, bIL309 (ORF53) and bIL286 (ORF57) (9). Hence, the variable C-terminal regions of the two proteins and their homologies to different hypothetical host recognition proteins suggest that ORF18 in sk1 and ORF20 in bIL170 are the RBPs with binding specificity found in the C-terminal region. By analogy, the C-terminal part of the RBP in phage lambda, specified by gene J, was also found to be essential for host recognition (40). We speculate that the homologous N-terminal parts of ORF18 and ORF20 interact with conserved proteins at the distant part of the tail, while the variable C-terminal regions are exposed to the environment and recognize specific bacterial receptors.
Identification of the RBP gene in sk1 and bIL170 by homologous recombination.
The information derived from the bioinformatic analysis prompted us to generate recombinant bIL170 phages carrying the putative RBP gene from sk1. orf18 in sk1, including approximately 800 bp of flanking DNA on each side, was cloned into pJDC9-6 (pKDU1) (Table 1) and used to transform L. lactis IL1403. The DNA flanking orf18 in sk1 was very similar (approximately 90%) to DNA flanking orf20 in bIL170 (data not shown), which favored homologous recombination. L. lactis IL1403(pKDU1) was infected with bIL170, and the phage progeny were tested for recombinant phages with altered host specificities by screening for plaques on the sk1 host strain L. lactis MG1614.
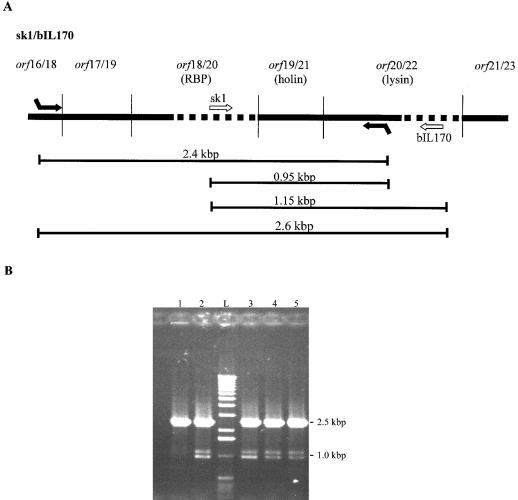
Twelve plaques presumed to contain chimeric bIL170 phageswere obtained. They were all pinpoint and turbid, and the recombinant phages could not be propagated further. Consequently, recombination was confirmed by a PCR assay performed directly on the plaques. The assay mixture included a pair of primers (sk1orf16F and sk1orf20R [Table 2]) annealing in regions homologous in sk1 and bIL170 to verify the presence of phages, recombinant or not, by the formation of a 2.4-kbp PCR product (Fig. 2A). To detect recombinant phages, two additional primers (sk1orf18F and bIL170orf22R [Table 2], annealing to nonhomologous areas in sk1 and bIL170, respectively) were included in the assay mixture (Fig. 2A), which generated two additional DNA fragments (1.15 and 0.95 kbp). Finally, primers sk1orf16F and bIL170orf22R were expected to generate a 2.6-kbp product from all bIL170 phages, recombinant or not.
FIG. 2.
PCR assay for confirmation of homologous recombination between orf18 from sk1 and the bIL170 genome. (A) Combined gene map for phages sk1 and bIL170, showing the putative RBP gene and surrounding genes. Areas of homology and nonhomology are indicated by solid and dotted lines, respectively. The arrows represent primers. Primers sk1orf16F and sk1orf20R (Table 2) bind to both sk1 and bIL170 DNA. Primers sk1orf18F and bIL170orf22R (Table 2) bind exclusively to sk1 and bIL170 DNA, respectively. The lines below the gene map indicate the DNA distances between the primers. (B) PCR analysis with all four primers used in the reaction. Lane 1, control PCR performed directly with a lysate from L. lactis IL1403(pKDU1) infected with phage bIL170; lanes 2 to 5, PCR performed directly with single plaques; lane L, 1-kb DNA ladder (Life Technologies Inc., Rockville, Md.).
All plaques containing putative chimeric phages were analyzed by PCR assays, and recombination was confirmed by fragments that were approximately 1.1 and 0.9 kbp long (Fig. 2B, lanes 2 to 5). As a control, the assay mixture was applied directly onto progeny phages from L. lactis IL1403(pKDU1) infected with bIL170, which resulted in a single approximately 2.5-kbp PCR product (Fig. 2B, lane 1). This PCR product was probably a mixture of 2.4- and 2.6-kbp PCR products. In addition, no PCR products were formed when the assay was performed with L. lactis MG1614, L. lactis IL1403, and samples of the bacterial lawn outside plaques in the recombination plaque assay (data not shown). Hence, the 1.1- and 0.9-kbp fragments indicated that there was homologous recombination between sk1 and bIL170 and did not arise from unexpected PCR with remnant DNA fragments in the lysate originating from the host strain or the cloned sk1 DNA. These results suggest that recombinant bIL170 phages revealing the host specificity of sk1 were obtained in the plaques and indicate that orf18 and orf20 in sk1 and bIL170 are involved in host recognition.
We were not able to propagate the bIL170 recombinant phages containing orf18 from phage sk1, although a number of different experimental conditions were tried. Duplessis and Moineau (M. Duplessis and S. Moineau, Lactic acid bacteria: genetics, metabolism and applications, book of abstracts, abstr. no. F19, 2002) recently emphasized that host specificity is determined not only by the phage RBP and the bacterial receptor but also by internal host factors. They found that some S. thermophilus phages that adsorbed efficiently to specific strains could not produce progeny phages, as determined by plaque assays. Such incompatibility between phage bIL170 and L. lactis MG1614 in DNA replication, gene expression, and/or phage assembly may explain our finding that recombinant bIL170 phages formed pinpoint plaques and could not be propagated further. This was also suggested by the low frequency at which chimeric bIL170 phages appeared (less than 10−9). Although this frequency is comparable to the frequency found for construction of chimeric S. thermophilus DT1 phages with the host specificity of phage MD4 (11), Stuer-Lauridsen et al. (35) obtained chimeric L. lactis bIL67 phages with the host specificity of phage CHL92 at a 100- to 1,000-fold-higher frequency. This may be explained by the overlapping host ranges of bIL67 and CHL92 (35), suggesting a closer relationship of the two phages. This possibly permits more successful propagation of phage bIL67 in a host for phage CHL92.
Immunogold labeling and electron microscopy.
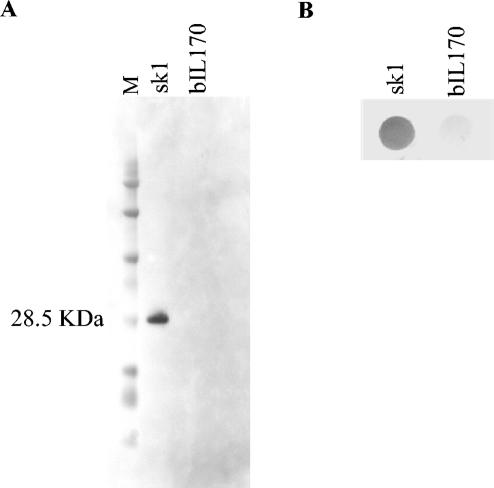
The 3′ part of orf18, encompassing 474 nucleotides, was cloned in a T7 expression vector (pKDU2 [Table 1]). The protein (158 amino acids) was expressed and purified from E. coli. Subsequently, polyclonal antibodies were raised against the purified protein. The specificity of the antiserum was tested with phage sk1 in a Western blot (Fig. 3A). The results demonstrated that there was specific binding to a 28.5-kDa protein from phage sk1, which corresponded to the calculated size of ORF18. As a control, no reaction was seen with phage bIL170. A concomitant dot blot analysis showed that the antibodies were also able to bind to native sk1 phages but not to native bIL170 phages (Fig. 3B).
FIG. 3.
Western (A) and dot blot (B) analyses of sk1 and bIL170 phages performed with antibodies raised against the C-terminal part of ORF18 from sk1. Lane M contained a protein marker (BioWhittaker Molecular Applications, Rockland, Maine).
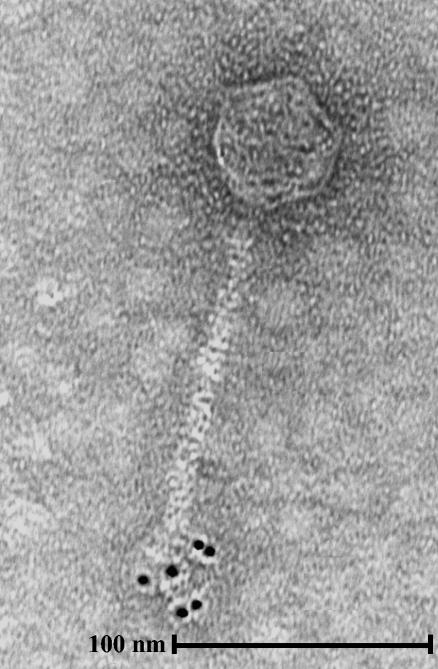
Subsequently, the antiserum was used in immunogold labeling of phage sk1 and analyzed by electron microscopy. The polyclonal antibodies specifically bound to the tip of the tail (Fig. 4), which supported the results of the recombination experiments showing that orf18 in phage sk1 encodes the RBP. Binding of the polyclonal antibodies to the tip of the tail was seen in several independent experiments.
FIG. 4.
Immunogold electron micrograph of phage sk1. The primary antibodies were those described in the legend to Fig. 3. Secondary antibodies conjugated to gold particles that were 5 nm in diameter were used.
RBP gene sequences in phages with host ranges similar to those of sk1 and bIL170.
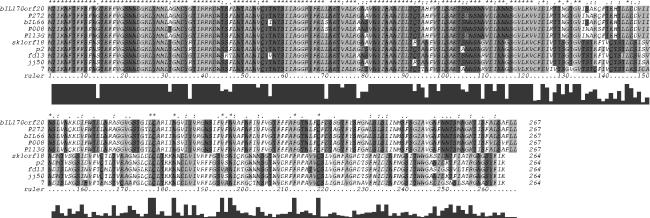
The presumed RBP genes in phages with host ranges and host binding abilities similar to those of sk1 and bIL170 (Table 3) were PCR amplified and sequenced. Alignment of the deduced amino acid sequences from all phages (sk1, p2, fd13, jj50, 7, bIL170, P008, P113G, P272, and bIL66) showed that the N-terminal parts (approximately 135 amino acids) were about 79% identical (Fig. 5). Based on homologies, the C-terminal parts (approximately 130 amino acids) of the RBPs fell into two groups: the RBPs of sk1-like phages (sk1, p2, fd13, jj50 and 7), which showed 64% identity, and the RBPs of bIL170-like phages (bIL170, P008, P113G, P272, and bIL66), which showed 90% identity (Fig. 5). A third type of RBPs has recently been identified in the 936-species phages φ645 and φ712 (12).
FIG. 5.
Alignment of the deduced amino acid sequences of the RBPs in lytic lactococcal phages. The stars above the sequences indicate positions at which there is a single, fully conserved residue. The alignment scores are shown below the ruler.
Table 3 shows that phages with homology in the C-terminal region of RBP bind to and infect the same lactococcal strains. These results are supported by the observation that phages with a RBP similar to that of bIL170 (P008, P113G, P272, and bIL66) were unable to bind to a receptor-negative mutant of L. lactis IL1403 selected for its inability to adsorb phage bIL170 (12). In conclusion, these results further verify that orf18 in phage sk1 and orf20 in phage bIL170 encode the RBPs, which allows the establishment of two host specificity groups within 936-species phages based on the binding specificity of the C-terminal region of the RBPs.
Interestingly, it was observed that phages belonging to the sk1 host specificity group infected only L. lactis subsp. cremoris strains, whereas members of the bIL170 host specificity group infected only L. lactis subsp. lactis strains. The same pattern, with only a few exceptions, has been determined previously for a group of lytic phages tested on different strains of L. lactis (15).
Conclusion.
The results presented in this paper strongly suggest that orf18 in phage sk1 and orf20 in phage bIL170 encode RBPs. Moreover, two groups of lytic lactococcal phages belonging to species 936 have been identified based on host specificity, host binding ability, and the amino acid sequence of the RBP. The two groups are represented by phages sk1 and bIL170.
Acknowledgments
This work was supported by the Danish Government Food Research Programme (FØTEK 3) and by the Danish Dairy Research Foundation through the Center for Advanced Food Studies.
Thomas Janzen (Chr. Hansen A/S, Hørsholm, Denmark) is thanked for help with fluorescence microscopy.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bidnenko, E., D. Ehrlich, and M.-C. Chopin. 1995. Phage operon involved in sensitivity to the Lactococcus lactis abortive infection mechanism AbiD1. J. Bacteriol. 177:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brøndsted, L., S. Østergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 5.Brüssow, H. 2001. Phages of dairy bacteria. Annu. Rev. Microbiol. 55:283-303. [DOI] [PubMed] [Google Scholar]
- 6.Chandry, P. S., B. E. Davidson, and A. J. Hillier. 1994. Temporal transcription map of the Lactococcus lactis bacteriophage sk1. Microbiology 140:2251-2261. [DOI] [PubMed] [Google Scholar]
- 7.Chandry, P. S., S. C. Moore, J. D. Boyce, B. E. Davidson, and A. J. Hillier. 1997. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol. Microbiol. 26:49-64. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. D., and D. A. Morrison. 1988. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene 64:155-164. [DOI] [PubMed] [Google Scholar]
- 9.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M.-C. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crutz-Le Coq, A. M., B. Cesselin, J. Commissaire, and J. Anba. 2002. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148:985-1001. [DOI] [PubMed] [Google Scholar]
- 11.Duplessis, M., and S. Moineau. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41:325-336. [DOI] [PubMed] [Google Scholar]
- 12.Dupont, K., T. Janzen, F. K. Vogensen, J. Josephsen, and B. Stuer-Lauridsen. 2004. Identification of Lactococcus lactis genes required for bacteriophage adsorption. Appl. Environ. Microbiol. 70:5808-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geller, B. L., R. G. Ivey, J. E. Trempy, and B. Hettinger-Smith. 1993. Cloning of a chromosomal gene required for phage infection of Lactococcus lactis subsp. lactis C2. J. Bacteriol. 175:5510-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godon, J., D. Delorme, S. D. Ehrlich, and P. Renault. 1992. Divergence of genomic sequences between Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. cremoris. Appl. Environ. Microbiol. 58:4045-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Higgins, D. L., R. B. Sanozky-Dawes, and T. R. Klaenhammer. 1988. Restriction and modification activities from Streptococcus lactis ME2 are encoded by a self-transmissible plasmid, pTN20, that forms cointegrates during mobilization of lactose-fermenting ability. J. Bacteriol. 170:3435-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jahns, A., A. Schäfer, A. Geis, and M. Teuber. 1991. Identification, cloning and sequencing of the replication region of Lactococcus lactis ssp. lactis biovar diacetylactis Bu2 citrate plasmid pSL2. FEMS Microbiol. Lett. 64:253-258. [DOI] [PubMed] [Google Scholar]
- 20.Johnsen, M. G., H. Neve, F. K. Vogensen, and K. Hammer. 1995. Virion positions and relationships of lactococcal temperate bacteriophage TP901-1 proteins. Virology 212:595-606. [DOI] [PubMed] [Google Scholar]
- 21.Josephsen, J., N. Andersen, H. Behrndt, E. Brandsborg, G. Christiansen, M. B. Hansen, S. Hansen, E. W. Nielsen, and F. K. Vogensen. 1994. An ecological study of lytic bacteriophages of Lactococcus lactis subsp. cremoris isolated in a cheese plant over a five year period. Int. Dairy J. 4:123-140. [Google Scholar]
- 22.Josephsen, J., and H. Neve. 1998. Bacteriophages and lactic acid bacteria, p. 385-436. In S. Salminen and A. von Wright (ed.), Lactic acid bacteria. Microbiology and functional aspects. Marcel Dekker, Inc., New York, N.Y.
- 23.Josephsen, J., and F. K. Vogensen. 1989. Identification of three different plasmid-encoded restriction/modification systems in Streptococcus lactis subsp. cremoris W56. FEMS Microbiol. Lett. 59:161-166. [Google Scholar]
- 24.Katsura, I., and R. W. Hendrix. 1984. Length determination in bacteriophage lambda tails. Cell 39:691-698. [DOI] [PubMed] [Google Scholar]
- 25.Kelley, L. A., R. M. MacCallum, and M. J. E. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 26.Lillehaug, D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J. Appl. Microbiol. 83:85-90. [DOI] [PubMed] [Google Scholar]
- 27.Loof, M., and M. Teuber. 1986. Heteroduplex analysis of the genomes of Streptococcus lactis “subsp. diacetylactis” bacteriophages of the P008-type isolated from German cheese factories. Syst. Appl. Microbiol. 8:226-229. [DOI] [PubMed] [Google Scholar]
- 28.Loof, M., J. Lembke, and M. Teuber. 1983. Characterization of the genome of the Streptococcus lactis “subsp. diacetylactis” bacteriophage P008 wide-spread in German cheese factories. Syst. Appl. Microbiol. 4:413-423. [DOI] [PubMed] [Google Scholar]
- 29.Mahanivong, C., J. D. Boyce, B. E. Davidson, and A. J. Hillier. 2001. Sequence analysis and molecular characterization of the Lactococcus lactis temperate bacteriophage BK5-T. Appl. Environ. Microbiol. 67:3564-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monteville, M. R., B. Ardestani, and B. L. Geller. 1994. Lactococcal bacteriophages require a host cell wall carbohydrate and a plasma membrane protein for adsorption and ejection of DNA. Appl. Environ. Microbiol. 60:3204-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 32.Pedersen, M., S. Østergaard, J. Bresciani, and F. K. Vogensen. 2000. Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901 -1 involved in tail length determination and baseplate assembly. Virology 276:315-328. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sanders, M. E., and T. R. Klaenhammer. 1981. Evidence for plasmid linkage of restriction and modification in Streptococcus cremoris KH. Appl. Environ. Microbiol. 42:944-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuer-Lauridsen, B., T. Janzen, J. Schnabl, and E. Johansen. 2003. Identification of the host determinant of two prolate-headed phages infecting Lactococcus lactis. Virology 309:10-17. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valyasevi, R., W. E. Sandine, and B. L. Geller. 1990. The bacteriophage kh receptor of Lactococcus lactis subsp. cremoris KH is the rhamnose of the extracellular wall polysaccharide. Appl. Environ. Microbiol. 56:1882-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valyasevi, R., W. E. Sandine, and B. L. Geller. 1991. A membrane protein is required for bacteriophage c2 infection of Lactococcus lactis subsp. lactis C2. J. Bacteriol. 173:6095-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valyasevi, R., W. E. Sandine, and B. L. Geller. 1994. Lactococcus lactis ssp. lactis C2 bacteriophage sk1 receptor involving rhamnose and glucose moieties in the cell wall. J. Dairy Sci. 77:1-6.7962868 [Google Scholar]
- 40.Wang, J., M. Hofnung, and A. Charbit. 2000. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J. Bacteriol. 182:508-512. [DOI] [PMC free article] [PubMed] [Google Scholar]