Abstract
The aim of this work was to identify genes in Lactococcus lactis subsp. lactis IL1403 and Lactococcus lactis subsp. cremoris Wg2 important for adsorption of the 936-species phages bIL170 and φ645, respectively. Random insertional mutagenesis of the two L. lactis strains was carried out with the vector pGh9:ISS1, and integrants that were resistant to phage infection and showed reduced phage adsorption were selected. In L. lactis IL1403 integration was obtained in the ycaG and rgpE genes, whereas in L. lactis Wg2 integration was obtained in two genes homologous to ycbC and ycbB of L. lactis IL1403. rgpE and ycbB encode putative glycosyltransferases, whereas ycaG and ycbC encode putative membrane-spanning proteins with unknown functions. Interestingly, ycaG, rgpE, ycbC, and ycbB are all part of the same operon in L. lactis IL1403. This operon is probably involved in biosynthesis and transport of cell wall polysaccharides (WPS). Binding and infection studies showed that φ645 binds to and infects L. lactis Wg2, L. lactis IL1403, and L. lactis IL1403 strains with pGh9:ISS1 integration in ycaG and rgpE, whereas bIL170 binds to and infects only L. lactis IL1403 and cannot infect Wg2. These results indicate that φ645 binds to a WPS structure present in both L. lactis IL1403 and L. lactis Wg2, whereas bIL170 binds to another WPS structure not present in L. lactis Wg2. Binding of bIL170 and φ645 to different WPS structures was supported by alignment of the receptor-binding proteins of bIL170 and φ645 that showed no homology in the C-terminal part.
Lactococcus lactis is widely used in starter cultures for cheese production. Bacteriophage contamination during the fermentation process is a major problem, causing lysis of the starter bacteria and consequently slow or failed fermentation of the milk. Bacteriophage infection requires specific recognition between the phage receptor-binding protein (RBP) and the host cell receptor. A better understanding of this recognition mechanism should increase the possibility of preventing phage infection.
Bacterial receptors have been well studied in gram-negative bacteria, especially Escherichia coli. Phages attacking E. coli recognize either lipopolysaccharides or specific proteins of the outer membrane. For example, phage lambda initially interacts reversibly and then interacts irreversibly with the outer membrane protein LamB (32, 35), which facilitates the diffusion of maltose into the cell (10). A slightly more complex mechanism is utilized by phage T5, which initially binds reversibly to polymannose O antigens in lipopolysaccharides at the E. coli surface (13, 14) and then binds irreversibly to the ferrichrome transporter FhuA (5, 19). Finally, phage T4 binds reversibly with its long tail fibers to B-type lipopolysaccharides or to the outer membrane porin OmpC (15, 16, 28), whereupon the additional short tail fibers bind irreversibly to the lipopolysaccharide core region (27).
For gram-positive bacteria the information on phage receptors is sparser. However, phages attacking L. lactis seem to bind initially to specific carbohydrate receptors exposed to the surface of the cell wall (29, 34, 40, 42). For many phages this binding step is reversible (29). Rhamnose, glucose, and galactose are often involved in this initial phage binding, as shown by the ability of these monosaccharides to competitively inhibit adsorption of several phages to cell surfaces (29, 40, 42). Binding to other carbohydrates, such as glucosamine and galactosamine, has been demonstrated for phage eb7 (20). For phages belonging to species c2, a secondary irreversible binding step requires binding to the membrane phage infection protein (PIP) (2, 12, 29, 41), while most phages of species 936, P335, and 949 do not use PIP as a secondary receptor (21); the only exception is 936-species phage kh (2, 29). Some of these phages may use other protein receptors in the membrane, as determined for 936-species phage sk1, which is able to bind to cell membranes deficient in PIP (2).
The aim of this work was to identify genes in L. lactis subsp. lactis IL1403 and L. lactis subsp. cremoris Wg2 that are important for adsorption of the 936-species phages bIL170 and φ645, respectively. Phage bIL170 and its host, L. lactis IL1403, are an excellent model system because the complete genome sequences are known (4, 7). In L. lactis IL1403 a biochemical or biological role has been assigned to 64.2% of the genes (4), and genes assumed to be involved in biosynthesis of surface polysaccharides and the PIP gene have been identified (2, 4). In bIL170 the RBP gene has recently been identified to be orf20 (9). In contrast, the genome sequences of L. lactis Wg2 and phage φ645 are not available. φ645 was selected since it infects both L. lactis Wg2 and L. lactis IL1403, whereas bIL170 infects only L. lactis IL1403.
Here we report that the genes identified, which are putatively involved in biosynthesis of cell wall polysaccharides in L. lactis IL1403 and L. lactis Wg2, are important for adsorption of phages bIL170 and φ645, respectively.
MATERIALS AND METHODS
Bacteria, bacteriophages, and growth conditions.
The bacterial strains and bacteriophages used in this study are listed in Table 1.
TABLE 1.
Phages, bacterial strains, and plasmid
| Phage, bacterial strain, or plasmid | Relevant features | Reference or source |
|---|---|---|
| Phages | ||
| bIL170a | 936-type lytic phage, propagated on IL1403 (AF009630) | 7 |
| bIL66 | 936-type lytic phage, propagated on IL1403 | 3 |
| P008 | 936-type lytic phage, propagated on IL1403 | 24 |
| P113G | 936-type lytic phage, propagated on IL1403 | 23 |
| P272 | 936-type lytic phage, propagated on IL1403 | 23 |
| φ645 | 936-type lytic phage, propagated on IL1403 and Wg2 | Chr. Hansen A/S |
| E. coli TG1 | Transformation host | Stratagene, La Jolla, Calif. |
| L. lactis strains | ||
| IL1403a | L. lactis subsp. lactis, plasmid free (AE005176) | 4 |
| Wg2 | L. lactis subsp. cremoris | 30 |
| MG1614 | Str Rifr derivative of L. lactis subsp. cremoris MG1363, plasmid free | 11 |
| Plasmid | ||
| pGh9:ISS1 | Used in insertional mutagenesis, Emr | 25 |
Phage or bacterial strain with known genome sequence. The GenBank accession number is indicated in parentheses.
E. coli TG1 was grown in Luria-Bertani medium (33) at 37°C with 200 μg of erythromycin per ml when appropriate. L. lactis strains were cultured at selected temperatures between 28 and 37.5°C in M17 broth (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) containing 0.5% glucose (L. lactis subsp. lactis IL1403 and L. lactis subsp. cremoris MG1614) or 0.5% lactose (L. lactis subsp. cremoris Wg2). Erythromycin (2 or 5 μg/ml) was added when appropriate. Phages were propagated on their hosts cultured in media supplemented with 5 mM CaCl2.
Electroporation procedure.
Electrotransformation of plasmid DNA into E. coli was performed essentially as described by Sambrook and Russell (33), while electroporation of L. lactis was performed as described by Holo and Nes (17).
Insertional mutagenesis and selection for phage-resistant L. lactis strains.
Random insertional mutagenesis in L. lactis IL1403 and L. lactis Wg2 was carried out with the vector pGh9:ISS1 essentially as described by Maguin et al. (26). The vector system combined the insertion sequence ISS1 with a thermosensitive replicon, which allowed optimal replication of plasmids in the host at 28 to 30°C and integration of the entire plasmid between duplicated ISS1 sequences at temperatures at or above 37°C.
L. lactis strains containing the vector pGh9:ISS1 were grown overnight at 30°C with 5 μg of erythromycin per ml. After this, the cultures were diluted 100-fold in M17 broth with the appropriate carbohydrate but without erythromycin and incubated for 2.5 h at 30°C; this was followed by a temperature shift to 37°C for 2.5 h. Aliquots (200 μl) of the cultures were mixed with approximately 109 bIL170 or φ645 phages and imbedded in a soft agar layer as described for standard plaque assays. Erythromycin (2 μg/ml) was added to the agar media used for this assay. Colonies appearing after 2 days of incubation at 37°C were isolated and tested for phage sensitivity by cross-streaking on M17-Ca2+ agar containing 0.5% glucose with the phage used for selection (18).
Excision of the pGh9:ISS1 vector from the chromosome of phage-resistant integrants, leaving a solitary ISS1 sequence in the chromosome, was carried out as described by Maguin et al. (26).
Phage binding assay with SYBR Gold-stained phages.
Phages were SYBR Gold stained essentially as described by Noble and Fuhrman (30). A phage lysate having a titer of at least 1010 PFU/ml was mixed with 20:1 (vol/vol) of a 1,000-fold-diluted SYBR Gold stock solution (Molecular Probes, Inc., Eugene, Oreg.) and incubated overnight in the dark at 4°C. Phages were mixed 1:1 with host cells grown to an optical density at 600 nm (OD600) of 0.6, and phage binding was examined by using a fluorescence microscope (Axioplan 2; Zeiss, Inc., Jena, Germany) and a magnification of ×1,000 with blue light (450- to 490-nm) excitation. Phage binding to the host cells was indicated by a fluorescent halo around the cells.
Phage adsorption assay.
Stationary-phase cultures of phage-resistant integrants of L. lactis IL-403 and L. lactis Wg2 grown at 37°C were diluted 100-fold in M17-Ca2+ broth, grown to an OD600 of 0.5, and infected with homologous phages at a multiplicity of infection of 0.001. The mixtures were incubated for 10 min at 30°C to allow binding of the phages to cell surfaces. After centrifugation (3 min, 15,000 × g, 4°C) the phage titer in the supernatant was determined by a standard plaque assay with the wild-type host strain. The percentage of adsorption was calculated as follows: [1 − (phage titer of supernatant after cells were removed/phage titer of a control reaction mixture without cells)] × 100.
Southern analysis of integrants with reduced phage adsorption.
Southern analyses were carried out as described by Maguin et al. (26). Chromosomal DNA was purified with a DNeasy QIAGEN tissue kit (QIAGEN, Inc., Chatsworth, Calif.) as recommended by the manufacturer and was digested with EcoRI (New England Biolabs, Inc., Beverly, Mass.). Southern blotting was performed with a chemiluminescence detection kit (ECL; Amersham Pharmacia Biotech UK Limited, Little Chalfont, Buckinghamshire, England) by using pGh9:ISS1 as the probe.
Rescue cloning and sequencing of pGh9:ISS1 flanking regions.
Chromosomal DNA from integrants digested with EcoRI (New England Biolabs) was treated with T4 DNA ligase (Roche Applied Science, Basel, Switzerland) as described by Maguin et al. (26) and was transformed into E. coli TG1 or L. lactis MG1614. Each transformation mixture was grown at 30°C with erythromycin selection. Rescued plasmids (pGh9:ISS1 including flanking chromosomal DNA) were purified from the transformants with a QIAprep spin plasmid miniprep kit (QIAGEN, Inc.) used as recommended by the manufacturer, except that L. lactis MG1614 initially was incubated with lysozyme (20 mg/ml) for 15 min at 37°C. Chromosomal DNA flanking pGh9:ISS1 was sequenced by using the reverse primer Gh9ISEcoR (Table 2), which annealed upstream in the ISS1 element.
TABLE 2.
Primers used for PCR analyses in this study
| Primer | Sequence (5′-3′) | Position | Restriction site |
|---|---|---|---|
| Gh9ISEcoR | GAAGAAATGGAACGCTC | Near EcoRI sitea | |
| ISS1F | TGTGATTATTGTCGCTGTTGG | Upstream in ISS1a | |
| sk1orf16F | GCTCTAGAATAGAAAAAGAATCACGACAAb | 13117 in sk1c | XbaI |
| sk1orf20R | AACTGCAGAAAGTATGAGCGACAACTCTCb | 15469 in sk1 | PstI |
In the vector pGh9:ISS1.
The underlined sequences are restriction sites.
The GenBank accession number for sk1 is AF011378.
Purification of total RNA from integrants of L. lactis.
Integrants of L. lactis IL1403 were grown to an OD600 of 0.6. Aliquots (1.5 ml) of the cultures were harvested by centrifugation at 20,000 × g for 5 min at 4°C and kept at −80°C. The harvested cells were mixed with 300 mg of glass beads (diameter, 106 μm; Sigma-Aldrich Chemie GmbH, Schnelldorf, Germany) and 100 μl of lysis buffer supplemented with 0.7 μl of β-mercaptoethanol from an Absolutely RNA RT-PCR miniprep kit (Stratagene, La Jolla, Calif.). Cells were disrupted by shaking them for 40 s at 6.0 m/s with a FastPrep FP120 bead beater (Bio 101 Savant, Savant Instruments, Inc., Holbrook, N.Y.). After chilling on ice, the samples were centrifuged at 20,000 × g for 2 min at 4°C. Each supernatant was mixed with 500 μl of lysis buffer containing 3.5 μl of β-mercaptoethanol, and total RNA was isolated with the Absolutely RNA RT-PCR miniprep kit (Stratagene) used as recommended by the manufacturer.
RT-PCR of total RNA.
Reverse transcription (RT)-PCR was carried out with total RNA by using the ProSTAR HF single-tube RT-PCR system (Stratagene) according to the manufacturer's instructions. As a control, RT-PCR was carried out without reverse transcriptase to ensure that the purified RNA was free of contaminating DNA. The positions of primers used in RT-PCR to determine the size of the operon are shown in Table 3. To investigate transcription from the ISS1 sequence, a forward primer (ISS1F [Table 2]) that annealed inside this sequence but downstream from the ISS1 promoter was used.
TABLE 3.
RT-PCR analysis of the L. lactis IL-1403 region containing ycaG, rgpE, ycbB, and ycbC
| Positions of primer pairs in IL-1403a | GenBank accession no. | Genes | PCR productb | Size of PCR product (kbp)c |
|---|---|---|---|---|
| 421 and 4838 | AE006257 | ybjB and rmlC | − | |
| 1343 and 3008 | AE006257 | rmlA and ybjF | + | 1.7 |
| 2822 and 4838 | AE006257 | ybjF and rmlC | + | 2.0 |
| 3563 and 1260 | AE006257/58 | rmlB and rgpA | + | 2.9 |
| 212 and 2657 | AE006258 | rgpA and rgpC | + | 2.5 |
| 2314 and 5575 | AE006258 | rgpC and ycaG | + | 3.2 |
| 2314 and 5753 | AE006258 | rgpC and rgpE | + | 3.4 |
| 6481 and 7855 | AE006258 | rgpE and rgpF | + | 1.3 |
| 6637 and 9866d | AE006258 | rgpF and ycbA | + | 3.2 |
| 8476 and 1875 | AE006258/59 | ycbA and ycbD | + | 3.8 |
| 883 and 5344 | AE006259 | ycbC and ycbG | + | 4.4 |
| 4973 and 9013 | AE006259 | ycbG and tagD1 | + | 4.0 |
| 7811 and 9318 | AE006259 | ycbJ and yccB | + | 1.5 |
| 7811 and 992 | AE006259/60 | ycbJ and guaB | − |
The numbers are the positions of the first nucleotides in the 5′ ends of the primers used with DNA sequences of L. lactis IL-1403. The number system is the system for the previously published sections of the L. lactis IL-1403 genome. The primers were 20 bp long.
−, no PCR product; +, PCR product evident.
The results were obtained by performing overlapping RT-PCR with total RNA from the integrants of L. lactis IL-1403.
The combination was tested with wild-type IL-1403 total RNA, and negative results were obtained.
Monosaccharide inhibition of phage infection.
Rhamnose (0.5 M) or glucose (0.5 M) was added to the growth medium in infection experiments that included L. lactis IL1403 and phage bIL170 as described by Monteville et al. (29). The infection was monitored by spectrophotometric measurement at 600 nm.
PCR amplification of the RBP gene in phage φ645.
The RBP gene in phage φ645 was PCR amplified by using primers sk1orf16F and sk1orf20R (Table 2) originally designed for PCR amplifying the RBP gene (orf18) in phage sk1 (9).
DNA sequence analysis.
A DNA sequence analysis was performed with an ABI PRISM 310 genetic analyzer (Perkin-Elmer, Wellesley, Mass.). Sequences were assembled by using the ContigExpress program of the Vector NTI Suite 7 software package (Invitrogen, Carlsbad, Calif.). Sequences were aligned by using the ClustalX (version 1.81) program (37). BLAST searches (1) were conducted by using the National Center for Biotechnology Information homepage (http://www.ncbi.nlm.nih.gov/BLAST/). Transmembrane helices in proteins (22) were predicted by using the Center for Biological Sequence Analysis homepage (http://www.cbs.dtu.dk/services/TMHMM/).
Nucleotide sequence accession number.
Sequence data for the RBP gene in phage φ645 have been deposited in the GenBank database under accession no. AY515015.
RESULTS
Isolation of integrants of L. lactis IL1403 and L. lactis Wg2 with reduced phage adsorption.
To identify genes involved in phage binding in L. lactis strains IL1403 and Wg2, a panel of mutants carrying random chromosomal integrations of plasmid pGh9:ISS1 was generated. Subsequently, phage-resistant mutants of L. lactis IL1403 and L. lactis Wg2 were selected on the basis of the ability to grow in the presence of the bIL170 and φ645 phages, respectively. This selection resulted in several hundred phage-resistant mutants of L. lactis Wg2 and 10 phage-resistant mutants of L. lactis IL1403. Phage-resistant integrants of L. lactis Wg2 appeared at a rate of 3 × 10−6, whereas control experiments showed that spontaneous phage-resistant mutants were obtained at a rate of 2 × 10−7. In L. lactis IL1403 phage-resistant integrants were obtained at a rate of 5 × 10−6, whereas no spontaneous mutants were obtained at a frequency of >10−7.
To screen for phage-resistant integrants with a reduced ability to adsorb phages, adsorption assays were carried out. Fifteen randomly selected phage-resistant integrants of L. lactis Wg2 were examined, and 10 adsorption-deficient integrants were retained due to low adsorption (0 to 23%) of added φ645 phage (Table 4). All 10 phage-resistant integrants of L. lactis IL1403 showed a reduced ability to adsorb bIL170 phage (0 to 27% adsorption) (Table 4). In contrast, the levels of adsorption of bIL170 and φ645 to their indicator strains, L. lactis IL1403 and L. lactis Wg2, respectively, were approximately 97%.
TABLE 4.
Phage adsorption assay for bIL170 and φ645 with pGh9:ISS1 integrants of L. lactis IL1403 and L. lactis Wg2, respectively
| Strain | Adsorption of bIL170 (%) | Adsorption of φ645 (%) |
|---|---|---|
| L. lactis IL1403 | 97 | |
| Integrant 1 | 20 | |
| Integrant 2 | 20 | |
| Integrant 3 | 10 | |
| Integrant 4 | 27 | |
| Integrant 5a | 20 | |
| Integrant 6 | 13 | |
| Integrant 7a | 10 | |
| Integrant 8a | 0 | |
| Integrant 9a | 26 | |
| Integrant 10 | 9 | |
| L. lactis Wg2 | 97 | |
| Integrant 1a | 0 | |
| Integrant 2 | 22 | |
| Integrant 3 | 23 | |
| Integrant 4 | 0 | |
| Integrant 5 | 5 | |
| Integrant 6 | 19 | |
| Integrant 7 | 0 | |
| Integrant 8a | 17 | |
| Integrant 9 | 0 | |
| Integrant 10 | 10 |
Integrant with identified integration site of pGh9:ISS1 (Fig. 2).
The reduced abilities of the phage-resistant integrants of L. lactis IL1403 and L. lactis Wg2 to adsorb phages bIL170 and φ645, respectively, were confirmed by a binding assay in which SYBR Gold-stained phages were used. The cells were subsequently observed by fluorescence microscopy to detect phage binding. None of the integrants showed visible phage binding, whereas binding to the wild-type strains was documented by the appearance of bright fluorescent halos.
Southern blot analysis of pGh9:ISS1 integrants with reduced phage adsorption.
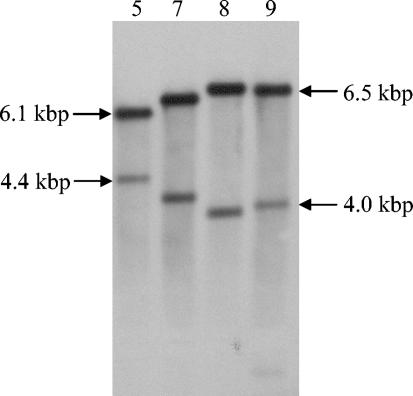
The 10 adsorption-deficient integrants of L. lactis IL1403 were examined by Southern blot analysis by using pGh9:ISS1 as the probe to identify different integrant types. Four different integrant types of L. lactis IL1403 were identified on the basis of the band patterns appearing in the Southern blot (Fig. 1). Likewise, two different integrant types of L. lactis Wg2 were identified (data not shown). The appearance of several integrants with similar band patterns in the Southern blots could have indicated that they were daughter cells of the same original integrant.
FIG. 1.
Southern analyses of chromosomal DNA from selected integrants of L. lactis IL1403 with reduced binding of phage bIL170. Chromosomal DNA was digested with EcoRI, and pGh9:ISS1 was used as a probe. The two bands in each lane represent chromosomal DNA with a flanking pGh9:ISS1 sequence and a flanking ISS1 sequence (25). The numbers at the top indicate the integrants used (see Table 4).
Analysis of pGh9:ISS1 insertion sites.
The chromosomal integration sites of pGh9:ISS1 were determined in the four integrant types of L. lactis IL1403 and in the two integrant types of L. lactis Wg2 by rescue cloning of pGh9:ISS1 and flanking chromosomal DNA. Sequence analysis showed that insertion in L. lactis IL1403 had occurred at three different sites in the rgpE gene and at one site in an open reading frame upstream from rgpE, designated ycaG (Fig. 2). rgpE encodes a putative glycosyltransferase (4), whereas the function of ycaG is unknown.
FIG. 2.
Integration sites of pGh9:ISS1 in integrants of L. lactis IL1403 (A) and L. lactis Wg2 (B) resistant to phages bIL170 and φ645, respectively. The integration sites in the resulting proteins are indicated by open triangles and numbers that correspond to the integrants shown in Table 4 and in Fig. 1. For L. lactis Wg2 the approximate integration sites are indicated in the homologous L. lactis IL1403 proteins.
For integrants of L. lactis Wg2 the derived amino acid sequence of the rescued flanking sequence from integrant 1 showed 77% identity to YcbB of L. lactis IL1403 (Fig. 2), a putative glycosyltransferase. The amino acid sequence derived from the flanking region of integrant 8 showed 62% identity to YcbC of L. lactis IL1403 (Fig. 2), a protein with no known function.
Transcription analysis.
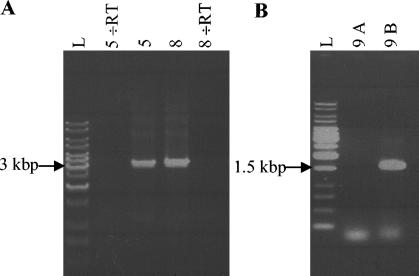
In the genome of L. lactis IL1403 the rgpE, ycaG, ycbB, and ycbC genes are located within a 6.4-kbp region (4), indicating that they could be part of an operon. Therefore, integrants of L. lactis IL1403 were examined for polar effects on downstream transcribed genes by RT-PCR. When pGh9:ISS1 was inserted into the chromosome, no polar effects on downstream transcription were observed (Fig. 3A).
FIG. 3.
RT-PCR of phage-resistant integrants of L. lactis IL1403. (A) Integrants 5 and 8 (see Table 4 and Fig. 2) with pGh9:ISS1 integrated into the ycaG and rgpE genes, respectively. Primers annealed downstream from the integrated pGh9:ISS1 in rgpF and ycbA (nucleotides 6637 and 9866; accession no. AE006258) (Table 3). (B) Integrant 9 (see Table 4 and Fig. 2) with a solitary ISS1 sequence integrated into the rgpE gene. Lane 9A, primers annealing on each side of the ISS1 sequence in the rgpC and rgpF genes (nucleotides 2314 and 7835; accession no. AE006258) (Table 3); lane 9B, primers annealing in the ISS1 sequence (ISS1F [Table 2]) and in the rgpF gene (nucleotide 7855; accession no. AE006258) (Table 3); lane L, 1-kb DNA ladder. RT, reverse transcriptase.
To examine the effects of a solitary ISS1 element integrated into the chromosome on the downstream transcription, pGh9:ISS1 was excised from the chromosome. When a primer pair annealing to chromosomal sequences flanking the solitary ISS1 sequence was used in RT-PCR, no PCR product was produced (Fig. 3B). In contrast, when the RT-PCR analysis was carried out with a primer pair annealing inside the ISS1 sequence and downstream of the ISS1 insertion, a PCR product was generated (Fig. 3B). Hence, transcription was knocked out by integration of the ISS1 sequence but was restarted from inside the ISS1 sequence, indicating that a promoter reading into the region downstream of the insertion was present in the ISS1 sequence.
As control, RT-PCR was carried out without reverse transcriptase in the reaction mixture. The results showed that PCR products were not obtained without reverse transcriptase in the reaction mixture (Fig. 3A).
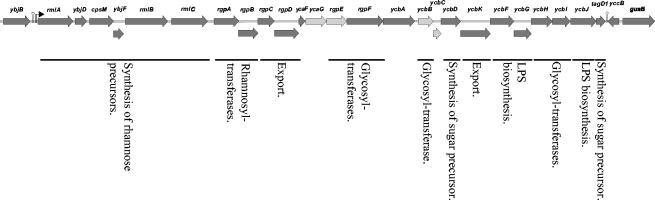
To determine if rgpE, ycaG, ycbB, and ycbC in fact were part of an operon, overlapping RT-PCR was carried out with total RNA isolated from the integrants of L. lactis IL1403 (Table 3). Interestingly, rgpE, ycaG, ycbB, and ycbC were found to be part of the same operon. The operon was determined to be approximately 25 kbp long and contained 24 genes, starting with rmlA and ending with tagD1 (Fig. 4). Attempts to conduct the same experiments with wild-type IL1403 RNA were unsuccessful as RNA containing the complete operon could not be purified. The demarcation of the operon was confirmed by identification of a hairpin-loop structure followed by a promoter-like sequence in the intergenic region upstream from rmlA. At the distant part of the operon an additional hairpin-loop structure was located in the intergenic region between tagD1 and the subsequent gene, yccB. In spite of the hairpin-loop structure, RT-PCR analysis showed that the transcript continued into yccB but that the downstream gene, guaB, was not part of the operon (Table 3). However, due to the opposite orientation of yccB we do not expect that this part of the transcript is translated into a protein. As the size of the operon was obtained by RT-PCR, the possibility that there are additional transcription start sites within the operon cannot be excluded.
FIG. 4.
Operon in L. lactis IL1403 containing the ycaG, rgpE, ycbB, and ycbC genes. The light grey arrows represent genes interrupted in integrants of L. lactis IL1403 (ycaG and rgpE) and L. lactis Wg2 (ycbB and ycbC) resistant to phages bIL170 and φ645, respectively. The displaced arrows indicate overlapping open reading frames. The bent arrow indicates the promoter, and the hairpin symbols indicate the terminators. LPS, lipopolysaccharide.
Monosaccharide inhibition of phage infection.
To clarify if glucose or rhamnose was part of the receptor for bIL170 in L. lactis IL1403, infection experiments were conducted with 0.5 M glucose or 0.5 M rhamnose in the growth medium. Rhamnose delayed the lysis of the culture for approximately 1 h, whereas glucose did not inhibit the lysis.
Infection and binding abilities of phages bIL170 and φ645.
The abilities of bIL170 and φ645 to infect and bind to L. lactis IL1403, L. lactis Wg2, and integrants with reduced phage adsorption were determined by plaque assays and binding assays performed with SYBR Gold-stained phages, respectively, to clarify if the two phages use different binding sites. φ645 was able to bind to and infect both L. lactis IL1403 and the pGh9:ISS1-integrants of L. lactis IL1403 with reduced adsorption of bIL170 (Table 5). In contrast, bIL170 did not bind to and infect L. lactis Wg2 or the integrants of this strain with reduced adsorption of φ645. Hence, the binding site for φ645 is present in both L. lactis IL1403 and L. lactis Wg2 and is different from the binding site used by bIL170, which is present only in L. lactis IL1403. A puzzling observation was that the plaques of φ645 infecting IL1403 integrants were more than twice as large as those of φ645 infecting wild-type strain L. lactis IL1403 (Table 5).
TABLE 5.
Infection and binding abilities of phages bIL170 and φ645a
| Phage | L. lactis IL1403 | L. lactis IL1403 integrant in rgpEb | L. lactis IL1403 integrant in ycaGb | L. lactis Wg2 | L. lactis Wg2 integrant in ycbBb | L. lactis Wg2 integrant in ycbCb |
|---|---|---|---|---|---|---|
| bIL170 | + (2)c | − | − | − | − | − |
| 645 | + (2) | + (5) | + (5) | + (1) | − | − |
Infection was determined by plaque assays, and binding ability was determined by using SYBR Gold-stained phages. +, positive; −, negative.
Integrants of L. lactis IL-1403 and L. lactis Wg2 with reduced adsorption of bIL170 and φ645, respectively.
The numbers in parentheses are the diameters (in millimeters) of the plaques appearing in the plaque assays.
Infection and binding abilities of phages bIL66, P008, p272, and P113G.
Phages bIL66, P008, p272, and P113G have an RBP and a host range similar to those of bIL170 (9). The infection and binding abilities of the phages with L. lactis IL1403 and integrants of L. lactis IL1403 with reduced adsorption of bIL170 were determined by plaque assays and binding assays performed with SYBR Gold-stained phages, respectively. The results showed that all the phages infected L. lactis IL1403, whereas they neither infected nor bound to integrants of L. lactis IL1403.
Alignment of the RBP in bIL170 and φ645.
The RBP genes have recently been identified in bIL170 and some other phages of species 936 (9). The primers used for PCR amplification of the RBP genes in these phages were used with phage φ645. The PCR product was sequenced, and the derived amino acid sequence was compared to the amino acid sequence of the RBP from bIL170 to determine if φ645 has an RBP that is different from that of bIL170. In Fig. 5 the deduced amino acid sequences of the RBPs in φ645 and bIL170 are aligned. In the approximately 150-amino-acid N-terminal part of the RBPs 80% identity was found between φ645 and bIL170, whereas in the approximately 100-amino-acid C-terminal part of the RBPs no homology was found. Hence, the RBPs from φ645 and bIL170 had an N-terminal homologous part and a C-terminal nonhomologous part.
FIG. 5.
Alignment of amino acid sequences of the RBPs in φ645 and bIL170. The stars above the sequences indicate positions which have a single, fully conserved residue. The alignment scores are shown below the ruler.
DISCUSSION
In this study we showed that the ycaG and rgpE genes in L. lactis IL1403 are important for adsorption of phage bIL170 to the cell surface. Likewise, we identified two genes in L. lactis Wg2 that are important for adsorption of phage φ645, which show high levels of similarity to ycbB and ycbC of L. lactis IL1403. Since φ645 infects L. lactis IL1403, we believe that it is likely that ycbB and ycbC are important for adsorption of φ645 in this strain.
Transcriptional analyses of the integrants revealed no polar effects on downstream transcribed genes due to insertion of pGh9:ISS1 into the chromosome (Fig. 3A), confirming that ycaG, rgpE, ycbB, and ycbC are involved in phage binding. In all six integrants investigated, the ISS1 sequences were orientated in the same direction as the interrupted genes, suggesting that the opposite direction results in polar effects that are lethal to the cells. pGh9:ISS1 therefore turned out to be an excellent mutagenesis tool that did not have polar effects on downstream transcribed genes. Transcription analyses of an integrant with a solitary ISS1 sequence in the chromosome (Fig. 3B) indicated that transcription restarts from a promoter inside the ISS1 sequence. This is consistent with a functional transcription promoter in the ISS1 sequence, which is localized just upstream from an open reading frame that probably encodes a transposase (6).
The two interrupted genes in L. lactis IL1403 (ycaG and rgpE), as well as ycbB and ycbC, which have high levels of similarity to the genes interrupted in phage-resistant integrants of L. lactis Wg2, turned out to be part of the same operon (Fig. 4). The operon contains genes assumed to be involved in biosynthesis and export of cell wall polysaccharides (WPSs) (4). This is in accordance with adsorption of lactococcal phages to specific carbohydrates in the cell wall (20, 29, 40, 42). Previously, genes involved in biosynthesis and export of WPSs have been identified in Streptococcus mutans (38, 39, 44), but from the investigations it is not clear if all the genes are located close to each other on the S. mutans genome or even are part of the same operon.
The function of YcaG is unknown, but predictions of transmembrane helices (see Material and Methods) in the protein suggest that it could be a membrane protein with seven transmembrane helices. We speculate that YcaG assists in transport of specific WPSs or parts of WPSs.
RgpE showed homology to several glycosyltransferases, but the specificity of the enzyme is not clear. The N-terminal two-thirds of RgpE have 24% identity and 46% similarity with RgpE of S. mutans. The S. mutans gene encodes a glycosyltransferase involved in glucose side chain formation in alternating α-1,2- and α-1,3-linked l-rhamnosyl polymers in WPSs (44). We do not believe that it is likely that RgpE in L. lactis IL1403 is involved in glucose side chain formation, as 0.5 M glucose did not competitively inhibit infection of L. lactis IL1403 by bIL170. Furthermore, no homology to RgpE from S. mutans was found in the C-terminal part of the protein. In contrast, 0.5 M rhamnose delayed the infection slightly. This is consistent with the putative functions of several upstream genes in the operon (Fig. 4) involved in synthesis of rhamnose precursors and rhamnosyl transferases (4) and with the occurrence of rhamnose in the cell wall of an L. lactis strain (40). It appears likely that the WPSs in L. lactis IL1403 have a rhamnosyl backbone, as described previously for S. mutans (44), and that bIL170 interacts partially with this backbone. However, carbohydrate side chains added to the putative rhamnosyl backbone by RgpE activity are probably crucial for phage binding.
YcbB in L. lactis IL1403 showed homology to several glycosyltransferases, which were different from the glycosyltransferases with homology to RgpE. In addition, National Center for Biotechnology Information pairwise BLAST analysis (36) of RgpE and YcbB showed no homology, suggesting that the glycosyltransferase encoded by ycbB adds different carbohydrate side chains to the putative rhamnosyl backbone than the glycosyltransferase encoded by rgpE adds. Based on these indications, at least two different types of WPS structures are present in the cell wall of L. lactis IL1403. Coexistence of different WPS structures in the same strain has previously been described by Delcour et al. (8).
Finally, the function of the small protein YcbC is also unknown. Prediction of transmembrane helices (see Materials and Methods) suggested that YcbC could be a membrane protein with three transmembrane helices. As suggested for YcaG, YcbC may assist in biosynthesis or transport of specific WPSs or parts of WPSs.
It was observed that all of the phage-resistant integrants grew slightly slower than the wild-type strains and that some of the cells were elongated and deformed. In addition, cultures of L. lactis Wg2 integrants formed aggregates at the bottoms of the tubes during growth, which was not observed for the wild type. These observations indicate that WPS affects the structure of the cell wall and the cell surface characteristics. It was observed that the plaques after infection of integrants of L. lactis IL1403 with φ645 were more than twice as big as the plaques when the wild type was infected (Table 5). We speculated that the cell wall in integrants of L. lactis IL1403 is more fragile, which facilitates lysis and liberation of phages by the φ645-produced lysin. Larger plaques were also seen when glycine was incorporated into the growth media (23). Glycine has previously been shown to weaken the bacterial cell wall (17). The functionality of WPS is far from understood, but Yamashita et al. (43) found that knockout of rmlB in S. mutans, which prevents synthesis of WPSs, resulted in increased sensitivity to acidic pH, an elevated temperature, and high osmolarity, suggesting that WPSs stabilize the cell wall under extreme environmental conditions.
As φ645 and bIL170 seem to recognize and adsorb to different structures in the cell wall, it was expected that they differ in their RBPs. The phages showed homology in the N-terminal part of the RBPs but no homology in the C-terminal part. The homologous N-terminal part of the RBPs in bIL170 and φ645 was found to be homologous to the N-terminal part of RBPs from the following other phages of species 936: φ7 (GenBank accession number AF539450), p272 (AF539443), P113G (AF539451), sk1 (orf18) (AF011378), jj50 (AF539448), p2 (AF539441), jw30 (AF539452), jw31 (AY145884), jw32 (AY145885), P008 (AF539449), fd13 (AF539444), φ936 (AF539446), φ712 (AF539442), and P475 (AF539447). This large group of phages has different host ranges despite the homologous N-terminal part of their RBPs (Dupont, unpublished results). We therefore speculated that the N-terminal part of RBPs could be involved in binding to a conserved protein at the distal part of the phage tail and probably is not essential for host recognition. It has previously been shown that lactococcal phages that differ in the C-terminal part of the RBPs bind to different host strains (9). In that study two types of RBPs were identified: the sk1-type of RBPs, which were found in phages p2, fd13, jj50, φ7, and sk1, and the bIL170 type of RBPs, which were found in phages P008, P113G, p272, bIL66, and bIL170. As determined by BLAST searches (1), the nonhomologous C-terminal part of the φ645 RBP showed 72% identity to the RBP from phage φ712, indicating that there is a third type of RBPs in 936-species phages. Interestingly, it was found that like bIL170, the phages bIL66, P008, p272, and P113G infect L. lactis IL1403, whereas they neither bind to nor infect the integrants of L. lactis IL1403 exhibiting reduced adsorption of bIL170. These results confirm that ycaG and rgpE in L. lactis IL1403 are important for adsorption of phages with an RBP similar to the RBP of bIL170.
In conclusion, this study indicated that phages bIL170 and φ645 recognize and bind to different WPS structures in the cell wall. Loss of biosynthesis or export of WPS structures or parts of these structures is apparently sufficient for total inhibition of phage infection. We speculated about why integration of pGh9:ISS1 was not obtained in genes encoding membrane proteins with a possible function as a secondary receptor, like PIP (29). The method used cannot explain this observation, since PIP mutants previously have been obtained for L. lactis MG1614 by the same method (Stuer-Lauridsen, unpublished results). For L. lactis IL1403 and L. lactis Wg2 it is therefore possible that knockout of the genes encoding the secondary membrane receptors for bIL170 and φ645 is lethal for the cell or that a secondary membrane receptor does not exist. The latter proposal is supported by investigations with phage P008, which binds irreversibly to a carbohydrate component embedded in the peptidoglycan matrix of L. lactis F7/2, resulting in DNA release (34).
Acknowledgments
This work was partially supported by the Danish Government Food Research Programme (FØTEK 3) and by the Danish Dairy Research Foundation through the Center for Advanced Food Studies.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babu, K. S., W. S. Spence, M. R. Monteville, and B. L. Geller. 1995. Characterization of a cloned gene (pip) from Lactococcus lactis required for phage infection. Dev. Biol. Stand. 85:569-575. [PubMed] [Google Scholar]
- 3.Bidnenko, E., D. Ehrlich, and M.-C. Chopin. 1995. Phage operon involved in sensitivity to the Lactococcus lactis abortive infection mechanism AbiD1. J. Bacteriol. 177:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, V., K. Schaller, and H. Wolff. 1973. A common receptor protein for phage T5 and colicin M in the outer membrane of Escherichia coli B. Biochim. Biophys. Acta 323:87-97. [DOI] [PubMed] [Google Scholar]
- 6.Cluzel P.-J., A. Chopin, S. D. Ehrlich, and M. C. Chopin. 1991. Phage abortive infection mechanism from Lactococcus lactis subsp. lactis, expression of which is mediated by an Iso-ISS1 element. Appl. Environ. Microbiol. 57:7-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crutz-Le Coq, A.-M., B. Cesselin, J. Commissaire, and J. Anba. 2002. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148:985-1001. [DOI] [PubMed] [Google Scholar]
- 8.Delcour, J., T. Ferain, M. Deghorain, E. Palumbo, and P. Hols. 1999. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie Leeuwenhoek 76:159-184. [PubMed] [Google Scholar]
- 9.Dupont, K., F. K. Vogensen, H. Neve, J. Bresciani, and J. Josephsen. 2004. Identification of the receptor-binding protein in 936-species lactococcal bacteriophages. Appl. Environ. Microbiol. 70:5801-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferenci, T., and W. Boos. 1980. The role of the Escherichia coli lambda receptor in the transport of maltose and maltodextrins. J. Supramol. Struct. 13:101-116. [DOI] [PubMed] [Google Scholar]
- 11.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geller, B. L., R. G. Ivey, J. E. Trempy, and B. Hettinger-Smith. 1993. Cloning of a chromosomal gene required for phage infection of Lactococcus lactis subsp. lactis C2. J. Bacteriol. 175:5510-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heller, K., and V. Braun. 1979. Accelerated adsorption of bacteriophage T5 to Escherichia coli F, resulting from reversible tail fiber-lipopolysaccharide binding. J. Bacteriol. 139:32-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heller, K., and V. Braun. 1982. Polymannose O-antigens of Escherichia coli, the binding sites for the reversible adsorption of bacteriophage T5+ via the L-shaped tail fibers. J. Virol. 41:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heller, K. J. 1992. Molecular interaction between bacteriophage and the gram-negative cell envelope. Arch. Microbiol. 158:235-248. [DOI] [PubMed] [Google Scholar]
- 16.Henning, U., and S. Hashemolhosseini. 1994. Receptor recognition by T-even-type coliphages, p. 291-298. In J. D. Karam (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 17.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Josephsen, J., and F. K. Vogensen. 1989. Identification of three different plasmid-encoded restriction/modification systems in Streptococcus lactis subsp. cremoris W56. FEMS Microbiol. Lett. 59:161-166. [Google Scholar]
- 19.Kadner, R. J., K. Heller, J. W. Coulton, and V. Braun. 1980. Genetic control of hydroxamate-mediated iron uptake in Escherichia coli. J. Bacteriol. 143:256-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keogh, B. P., and G. Pettingill. 1983. Adsorption of bacteriophage eb7 on Streptococcus cremoris EB7. Appl. Environ. Microbiol. 45:1946-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraus, J., and B. L. Geller. 1998. Membrane receptor for prolate phages is not required for infection of Lactococcus lactis small or large isometric phages. J. Dairy Sci. 81:2329-2335. [Google Scholar]
- 22.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 23.Lillehaug, D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J. Appl. Microbiol. 83:85-90. [DOI] [PubMed] [Google Scholar]
- 24.Loof, M., and M. Teuber. 1986. Heteroduplex analysis of the genomes of Streptococcus lactis “subsp. diacetylactis” bacteriophages of the P008-type isolated from German cheese factories. Syst. Appl. Microbiol. 8:226-229. [DOI] [PubMed] [Google Scholar]
- 25.Loof, M., J. Lembke, and M. Teuber. 1983. Characterization of the genome of the Streptococcus lactis “subsp. diacetylactis” bacteriophage P008 wide-spread in German cheese factories. Syst. Appl. Microbiol. 4:413-423. [DOI] [PubMed] [Google Scholar]
- 26.Maguin, E., H. Prévost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makhov, A. M., B. L. Trus, J. F. Conway, M. N. Simon, T. G. Zurabishvili, V. V. Mesyanzhinov, and A. C. Steven. 1993. The short tail-fiber of bacteriophage T4: molecular structure and a mechanism for its conformational transition. Virology 194:117-127. [DOI] [PubMed] [Google Scholar]
- 28.Montag, D., S. Hashemolhosseini, and U. Henning. 1990. Receptor-recognizing proteins of T-even type bacteriophages. The receptor-recognizing area of proteins 37 of phages T4 TuIa and TuIb. J. Mol. Biol. 216:327-334. [DOI] [PubMed] [Google Scholar]
- 29.Monteville, M. R., B. Ardestani, and B. L. Geller. 1994. Lactococcal bacteriophages require host cell wall carbohydrate and a plasma membrane protein for adsorption and ejection of DNA. Appl. Environ. Microbiol. 60:3204-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 31.Otto, R., W. M. de Vos, and J. Gavrieli. 1982. Plasmid DNA in Streptococcus cremoris Wg2: influence of pH on selection in chemostats of a variant lacking a protease plasmid. Appl. Environ. Microbiol. 43:1272-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randall-Hazelbauer, L., and M. Schwartz. 1973. Isolation of the bacteriophage lambda receptor from Escherichia coli. J. Bacteriol. 116:1436-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, N.Y.
- 34.Schäfer, A., A. Geis, Neve, H., and M. Teuber. 1991. Bacteriophage receptors of Lactococcus lactis subsp. diacetylactis F7/2 and Lactococcus lactis subsp. cremoris Wg2-1. FEMS Microbiol. Lett. 78:69-74. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz, M. 1975. Reversible interaction between coliphage lambda and its receptor protein. J. Mol. Biol. 99:185-201. [DOI] [PubMed] [Google Scholar]
- 36.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsukioka, Y., Y. Yamashita, Y. Nakano, T. Oho, and T. Koga. 1997. Identification of a fourth gene involved in dTDP-rhamnose synthesis in Streptococcus mutans. J. Bacteriol. 179:4411-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsukioka, Y., Y. Yamashita, T. Oho, Y. Nakano, and T. Koga. 1997. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J. Bacteriol. 179:1126-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valyasevi, R., W. E. Sandine, and B. L. Geller. 1990. The bacteriophage kh receptor of Lactococcus lactis subsp. cremoris KH is the rhamnose of the extracellular wall polysaccharide. Appl. Environ. Microbiol. 56:1882-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valyasevi, R., W. E. Sandine, and B. L. Geller. 1991. A membrane protein is required for bacteriophage c2 infection of Lactococcus lactis subsp. lactis C2. J. Bacteriol. 173:6095-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valyasevi, R., W. E. Sandine, and B. L. Geller. 1994. Lactococcus lactis ssp. lactis C2 bacteriophage sk1 receptor involving rhamnose and glucose moieties in the cell wall. J. Dairy Sci. 77:1-6.7962868 [Google Scholar]
- 43.Yamashita, Y., Y. Tsukioka, Y. Nakano, K. Tomihisa, T. Oho, and T. Koga. 1998. Biological functions of UDP-glucose synthesis in Streptococcus mutans. Microbiology 144:1235-1245. [DOI] [PubMed] [Google Scholar]
- 44.Yamashita, Y., Y. Tsukioka, K. Tomihisa, Y. Nakano, and T. Koga. 1998. Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J. Bacteriol. 180:5803-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]