Abstract
A cattle trial using artificially inoculated calves was conducted to determine the effect of the addition of colicinogenic Escherichia coli strains capable of producing colicin E7 (a 61-kDa DNase) to feed on the fecal shedding of serotype O157:H7. The experiment was divided into three periods. In period 1, which lasted 24 days, six calves were used as controls, and eight calves received 107 CFU of E. coli (a mixture of eight colicinogenic E. coli strains) per g of feed. Both groups were orally inoculated with nalidixic acid-resistant E. coli O157:H7 strains 7 days after the treatment started. In periods 2 and 3, the treatment and control groups were switched, and the colicinogenic E. coli dose was increased 10-fold. During period 3, which lasted as long as period 1, both groups were reinoculated with E. coli O157:H7. The numbers of E. coli O157:H7 were consistently greater in the control groups during the three periods, but comparisons within each time period determined a statistically significant (P < 0.05) difference only at day 21 of period 1. However, when the daily average counts were compared between the period 1 control group and the period 3 treatment group that included the same six animals, an overall reduction of 1.1 log10 CFU/g was observed, with a maximum decrease of 1.8 log10 CFU/g at day 21 (overall statistical significance, P = 0.001). Serotype O157:H7 was detected in 44% of the treatment group's intestinal tissue samples and in 64% of those from the control group (P < 0.04). These results indicated that the daily addition of 108 CFU of colicin E7-producing E. coli per gram of feed could reduce the fecal shedding of serotype O157:H7.
Escherichia coli O157:H7 is a food-borne pathogen that has been frequently linked to outbreaks attributed to the consumption of meat, produce, and water that had been contaminated with cattle manure. Multiple studies have reported that at least 20% of cattle asymptomatically carry and shed serotype O157:H7 in their feces, and it is now well established that these animals are the source of contamination (1, 8, 27). Due to the widespread distribution of this pathogen in cattle, its control will be dependent on interventions intended to reduce bovine fecal shedding (5). Several strategies are currently being explored to reduce the prevalence of E. coli O157 in cattle. Some of the proposed interventions include the use of vaccination, feed additives, diet shifts, antagonistic bacteria, and bacteriophages (28).
One of the most promising methods to reduce pathogenic microorganisms in livestock is one in which antagonistic bacteria, often referred to as probiotic, competitive exclusion (CE), or direct-fed microbial products are used. PreEmpt is a proprietary bacterial mix and commercial CE product that effectively reduces Salmonella in chickens (21). Schoeni and Doyle (26) investigated the inhibition of Campylobacter jejuni in chicks with probiotic bacteria selected from hens. CE cultures have also been able to reduce Salmonella and enterotoxigenic E. coli in swine (9, 12, 20). Lema et al. (17) fed a mix of lactic acid bacteria to lambs and observed a decrease in E. coli O157:H7 levels.
Several authors have reported the identification of bacteria with the potential to inhibit or exclude E. coli O157:H7 in the gastrointestinal tracts of cattle. In an initial report, Zhao et al. (31) identified several E. coli strains with the ability to inhibit E. coli O157:H7 in cattle. The same probiotic E. coli strains were able to significantly reduce numbers of O157:H7 in weaned calves but not in neonatal calves (30, 32). Brashears et al. (2) observed a 60% reduction in prevalence of E. coli O157:H7 in cattle after treatment with lactobacilli. Previously, Schamberger and Diez-Gonzalez (24) reported the selection of 24 E. coli strains with the ability to inhibit O157:H7. This set of anti-O157 E. coli strains was identified from a collection of 540 E. coli strains isolated from humans and nine different animal species. Further investigation characterized these strains for antibiotic resistance, virulence factors, and types of antimicrobial proteins (colicins) produced (25).
Despite initial reports showing that probiotic bacteria could reduce the levels of E. coli O157 in cattle, little is known about the ecological variables affecting their performance. In the relatively few cattle feeding studies, the influence of variables such as diet, probiotic dose frequency or level, and diversity of probiotic organisms on O157:H7 reduction has not been investigated. These factors could all influence the ability of probiotic bacteria to successfully reduce a targeted pathogen. The objective of this research project was to evaluate the ability of colicinogenic E. coli that produces colicin E7 to reduce the prevalence of E. coli O157:H7 in cattle. This research also examined the influence of two different levels of probiotic bacteria and the effect of treatment order.
MATERIALS AND METHODS
Bacterial strains.
The following eight previously characterized E. coli strains capable of producing colicin E7 were used as the probiotic organisms: bovine isolates B15, B18, and B34; human isolates H30 and H31; sheep isolates S12 and S34; and duck isolate U24 (25). The following six E. coli O157:H7 strains were also used: ATCC 43895, 86-24, MN31, 2026, 2029, and 6058 (24). Nalidixic acid-resistant (Nalr) mutants of these O157:H7 strains were selected by serial transfer into Luria-Bertani (LB) broth (Neogen, Inc., Baltimore, Md.) containing increasing concentrations of nalidixic acid (from 0.2 to 50 mg/liter, twofold daily increases; Sigma-Aldrich, St. Louis, Mo.). Nalidixic acid resistance contributed to the selective recovery of E. coli O157:H7 from cattle feces and tissues. The growth rate of Nalr mutants was not different from that of their corresponding parent strain when grown in LB broth, and only the Nalr strain MN31 became sensitive to nalidixic acid after more than 90 generations of growth in LB broth lacking this antibiotic.
Inoculum preparation.
The eight colicinogenic E. coli strains were grown individually overnight at 37°C in shaking flasks containing tryptic soy broth supplemented with 1 g of glucose (Neogen, Inc., Baltimore, Md.)/liter. Equal culture volumes of the eight strains were mixed and centrifuged at 4°C for 10 min at 10,000 × g and resuspended in distilled water. The cell suspension was blended with the feed mix described below to obtain an estimated count of 107 CFU/g for period 1 and 108 CFU/g for periods 2 and 3. Bacterial cultures were monitored with optical density measurements at 600 nm, and bacterial numbers in the feed were confirmed by spread plating samples onto sorbitol-MacConkey (SMAC; Neogen, Inc.) agar plates. The six Nalr E. coli O157:H7 strains were grown individually overnight in shaking flasks containing tryptic soy broth supplemented with 1 g of glucose/liter. Equal volumes of the strains were mixed, centrifuged at 4°C (10 min at 10,000 × g), and resuspended in distilled water at a 180-fold concentration. The O157:H7 strain suspension was mixed with 2 parts (vol/wt) starch on a total weight basis. Gelatin capsules (Torpac Inc., Fairfield, N.J.) were filled with the E. coli O157:H7-starch mix, calculated to deliver 1011 CFU to each calf. Cell numbers were confirmed by using the most-probable-number method in triplicate using serially diluted samples in 96-well plates containing 200 μl of LB broth. After overnight incubation of the 96-well plates, turbid wells were scored as positive tubes, and standard tables were used for most-probable-number determination (11). The gelatin capsules were administered orally to the calves with a metallic bolus gun.
Cattle management.
The animal experiment was conducted at the isolation barns (an animal biosafety level 2 facility) of the University of Minnesota, St. Paul, Minnesota. Sixteen castrated, weaned Holstein calves were dewormed and vaccinated against viral and bacterial diseases (infectious bovine rhinotracheitis, bovine viral diarrhea virus, parainfluenza 3 virus, bovine respiratory syncytial virus, seven Clostridium spp., and Haemophilus somnus). The calves initially weighed between 90 and 115 kg and were housed in groups of two (total of eight rooms). The calves had free access to water and were fed a corn-based diet that was incrementally increased to provide approximately 95% of the National Research Council recommendations during the course of the study (18a). The diet consisted of the following components (per 100 kg of feed): 72.6 kg of cracked corn, 20.5 kg of alfalfa pellets, 5 kg of soybean meal, 0.5 kg of limestone, 0.5 kg of salt, 0.5 kg of urea, 0.2 kg of vitamin premix (containing 2,040,000 IU of vitamin A, 200,000 IU of vitamin D, and 700 IU of vitamin E), 0.15 kg of Dynamate (IMC Feed Ingredients, Inc., Lake Forest, Ill.), and 0.05 kg of sodium selenite. All calves were initially given this feed mix for 10 days. The cattle rooms were cleaned twice daily.
Experimental design.
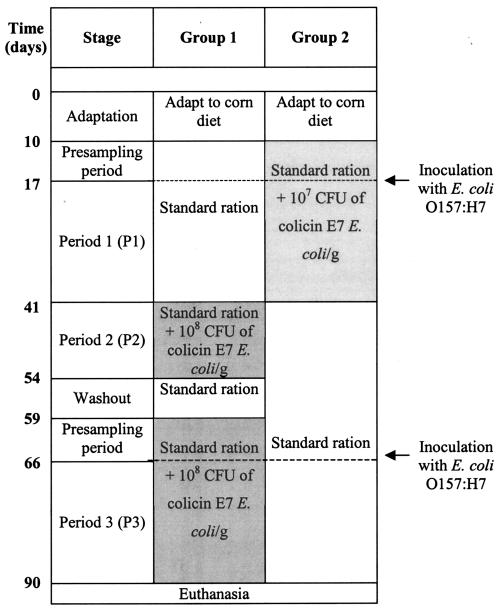
The calf rooms were randomly assigned to two groups (group 1 and group 2) with four pens per group (Fig. 1). After the calves were fed the diet described above for 10 days, group 2 was fed the same diet supplemented with 107 CFU of E. coli (the eight colicinogenic E. coli strains) per g, and group 1 continued to receive that diet. Seven days after group 2 had been fed the supplemented diet (presampling stage), both groups were orally inoculated with 1011 CFU of the mixture containing the six Nalr E. coli O157:H7 strains, and this was considered day 0 of period 1 (P1). Both groups were fed the same diet used in the presampling stage for an additional 24 days, which was the total duration of P1. During the second week of P1, two animals in group 1 died in separate rooms due to pneumonia, and their data are shown only for days 1, 3, and 6, which were not used for data analysis. Period 2 (P2) began immediately after P1 when group 1 was switched to receive feed containing 108 CFU of the mix of colicinogenic E. coli per g and group 2 was fed the diet without the supplemented E. coli. P2 lasted a total of 15 days. After this time, neither group received the colicinogenic E. coli for 5 days (washing stage). A second presampling stage started when group 1 was again fed the diet containing 108 CFU of colicinogenic E. coli per g and group 2 was given only the uninoculated feed. After 7 additional days, all calves were reinoculated with 1011 CFU of the Nalr E. coli O157:H7 strain mixture, and period 3 (P3) began. P3 lasted for a total of 24 days, and the calves were euthanized at the end of this period.
FIG. 1.
Diagram of the experimental design that describes the sequence of events that the groups of calves were subjected to during the test period.
Analysis of E. coli O157:H7 and sample collection.
During the adaptation period, the presence of natural E. coli O157:H7 was detected as follows: 10 g of cattle feces was homogenized for 2 min with 90 ml of gram-negative (GN) Hajna broth (Neogen, Inc.) supplemented with 8 mg of vancomycin (ICN Biomedicals, Inc., Irvine, Calif.), 0.05 mg of cefixime (Dynal, Inc., Lake Success, N.Y.), and 10 mg of cefsuludin (ICN Biomedicals, Inc.) per liter and incubated for 6 h at 37°C. One milliliter of enrichment samples was then mixed with 20 μl of anti-O157 immunomagnetic beads (Dynal, Inc.) in centrifuge tubes and mixed at room temperature for 30 min. The samples were placed into a magnetic rack and washed three times with a washing solution (sterile water with 5 g of bovine albumin [Sigma-Aldrich] and 0.5 ml of Tween 20 [ICN Biomedicals, Inc.] per liter). The immunomagnetic beads were suspended in 100 μl of the washing solution and spread plated onto Rainbow agar (Biolog, Inc., Hayward, Calif.) supplemented with 20 mg of novobiocin and 0.8 mg of potassium tellurite per liter or onto SMAC agar containing 0.05 mg of cefixime and 2.5 mg of potassium tellurite (CT-SMAC; Dynal, Inc.) per liter. The plates were then incubated overnight at 37°C, and black and colorless colonies on Rainbow and CT-SMAC plates, respectively, were tested by using an O157 latex agglutination test (Oxoid, Inc., Ogdensburg, N.Y.).
During the experiment, individual fecal samples were collected from the rectum of each animal on every third day for all three feeding periods. The count of Nalr E. coli O157:H7 isolates was determined as follows: fecal samples (0.5 g) were serially diluted 10-fold from 10−1 to 10−6 in phosphate-buffered saline (PBS; ICN Biomedicals, Inc.), and portions (0.1 ml) of selected dilutions were spread plated onto CT-SMAC plates containing 50 mg of nalidixic acid per liter (CTNA-SMAC). During the last 12 days of P1 and P3 and during P2, portions (1.0 ml) of the 10−1 dilutions were also spread plated, and the excess liquid on the agar surface was allowed to dry. Plates were incubated at 37°C for 24 h, and sorbitol-negative colonies were counted as presumptive E. coli O157:H7. The presence of O157:H7 in each plate was confirmed by testing three individual colonies using a latex immunoassay specific for O157 (Oxoid, Inc.). When E. coli O157:H7 colonies could not be recovered with direct plating, a selective enrichment procedure was used. One-gram fecal samples stored at 4°C for less than 24 h were mixed with 9 ml of GN broth supplemented with 50 mg of nalidixic acid/liter and incubated for 6 h at 37°C. Samples were then plated onto CTNA-SMAC plates and incubated for 24 h at 37°C. To recover colicinogenic E. coli from feces, PBS dilutions of samples were spread plated onto SMAC plates, incubated for 24 h at 37°C, and enumerated, and plates containing sorbitol-positive colonies were stored for further characterization.
At the end of the experiment, the calves were euthanized following a protocol approved by the Institutional Animal Care and Use Committee by injection of an intravenous solution of sodium pentobarbital (100 mg/kg). After the abdomens were cut and opened, 20-cm sections of ileum, cecum, spiral colon, and descending colon were cut and placed separately into sterile sampling bags. Similarly, the whole rectum and a 20-cm-diameter piece of rumen wall were cut and placed separately into sterile sampling bags. Samples were immediately transported and subjected to microbiological analysis to evaluate the colonization of the O157:H7 and colicinogenic strains. Tissue samples were rinsed with sterile distilled water, and representative 1-g samples of the mucosal layer were homogenized in tubes with 9 ml of PBS or GN broth with 50 mg of nalidixic acid/liter and analyzed quantitatively and qualitatively for E. coli O157:H7 and sorbitol-positive colonies as described above.
Identification of colicinogenic E. coli.
Sorbitol-positive colonies obtained from the highest dilutions identified in SMAC plates during the experiment were streaked onto additional SMAC plates and incubated at 37°C for 24 h. Representative colonies (suspected colicinogenic E. coli) were grown overnight in LB medium, and 25 μl was spot inoculated onto an LA agar (LB medium with 1.5% agar) plate supplemented with 0.25 mg of mitomycin C (Sigma-Aldrich)/ml. After overnight growth, the agar layer was flipped over, resulting in the putative colicin-producing colony on the bottom of petri plate. Plates were overlaid with 5 ml of LA soft agar (0.75% agar) containing 106 CFU of E. coli O157:H7 strain ATCC 43895 as the indicator organism and incubated overnight at 37°C. The plates were then observed and scored for zones of inhibition against the O157:H7 indicator lawn, and those isolates that produced a significant inhibition were selected for further confirmation as colicin producers.
DNA fingerprinting of isolated anti-O157 colicinogenic E. coli was achieved by using a modified protocol of the repetitive DNA sequences (rep-PCR) technique described by Dombek et al. (6). Bacterial cells used to derive the rep-PCR template were grown in LB broth overnight at 37°C. One milliliter of culture was placed into microcentrifuge tubes and centrifuged at 20,000 × g for 1 min. The supernatants were removed, and the cell pellets were resuspended in 1 ml of sterile water. Two microliters of cell templates was mixed with 23 μl of PCR mixture containing 45 pmol of BOX A1R primer (5′-CTACGGCAAGGCGACGCTGACG-3′), 1.25 mM each deoxynucleoside triphosphate (Fisher Scientific, Pittsburgh, Pa.), 1 U of Taq DNA polymerase (Fisher Scientific), 5 μl of Gitschier buffer (16), 2.5 μl of dimethyl sulfoxide, and 160 μg of bovine serum albumin (Fisher Scientific)/ml. The PCR was performed with a Robocycler Gradient 96 Thermocycler (Stratagene, Inc., La Jolla, Calif.) for 2 min at 95°C, followed by 30 cycles of 93°C for 30 s, 50°C for 1 min, and 65°C for 8 min. The reaction mixtures were electrophoresed on a 1.5% agarose gel. The gels were stained with ethidium bromide, and patterns of individual isolates were compared and identified by using a gel documentation and analysis system (UVP, Inc., Upland, Calif.).
Data analysis.
The count of E. coli O157:H7 for each individual sample was the average of two plate counts, and this value was transformed to log10 CFU/g of feces. Positive samples detected only by enrichment were assigned a value of 0.3 log10 CFU/g to be included into the calculation. The theoretical limit of detection of our plating method was 5 CFU/g of feces but with 0.3 log10 CFU/g as the default value for positive enrichments as the intermediate value between 1 and 4 CFU/g that we could detect. The results for individual animals were used to calculate the mean and standard deviation. Fecal shedding data in the logarithm form were analyzed by using the mixed procedure model with SAS (Cary, N.C.) computer software, version 8.2. The data were entered as four different classes—room, animal, probiotic treatment, and time—at 8, 14, 2 and 9 levels, respectively. The statistical significance of the tissue sample data was calculated by using the two-sample t test method (18).
RESULTS
All 16 calves tested for natural E. coli O157:H7 were negative. Nalr E. coli O157:H7 isolates were detected in the feces of all animals at 18 h after oral inoculation at an average level of approximately 6.3 log10 CFU/g (Table 1). This number declined gradually over time, but at day 24 the animals were shedding on average 2.7 log10 CFU of E. coli O157:H7/g of feces. During the first period (P1), the level of O157:H7 was variable among each group of calves, but the average daily values for group 2 (receiving the colicinogenic mixture) were consistently smaller than those for group 1 (Table 1). However, with the exception of a difference of 1.5 log cycles observed at day 21 (P < 0.05), none of these differences were statistically significant. At the start of P2, group 2 stopped receiving the colicinogenic E. coli and group 1 began receiving the probiotic mixture at a 10-fold-greater dose than that received during P1. Group 1 had lower average numbers (≤1.1 log cycles less) of serotype O157:H7 shedding throughout P2 compared to group 2, but none of these differences were statistically significant (Table 2).
TABLE 1.
Fecal shedding of E. coli O157:H7 by calves during period 1 of the experimenta
| Group and calf no. |
E. coli O157:H7 count (log10 CFU/g) at dayb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 6 | 9 | 12 | 15 | 18 | 21 | 24 | |
| Group 1 | |||||||||
| 72 | 6.0 | 4.1 | 5.5 | ND | ND | ND | ND | ND | ND |
| 74 | 7.6 | 4.8 | 4.4 | 3.2 | 2.3 | 1.3 | 2.1 | 3.0 | 1.0 |
| 76 | 7.0 | 5.1 | 5.1 | 4.1 | 3.1 | 2.8 | 2.9 | 3.4 | 2.0 |
| 78 | 7.0 | 5.9 | 4.5 | 3.3 | 2.7 | 2.7 | 2.7 | 2.4 | 4.4 |
| 79 | 5.7 | 4.6 | 4.1 | 4.3 | 3.5 | 5.4 | 4.7 | 5.0 | 2.0 |
| 80 | 7.0 | 5.4 | 5.2 | 3.3 | 3.9 | 3.0 | 2.9 | 3.4 | 3.7 |
| 83 | 6.0 | 3.8 | 4.2 | 4.9 | 3.6 | 2.6 | 2.0 | 2.4 | 3.6 |
| 87 | 4.0 | 4.6 | 3.3 | ND | ND | ND | ND | ND | ND |
| Avg | 6.7c | 5.0c | 4.6c | 3.8 | 3.2 | 3.0 | 2.9 | 3.3 | 2.8 |
| SD | 0.7 | 0.7 | 0.5 | 0.7 | 0.6 | 1.4 | 1.0 | 0.9 | 1.3 |
| Group 2 | |||||||||
| 73 | 7.0 | 4.6 | 4.6 | 4.3 | 3.4 | 3.8 | 3.9 | 4.6 | 6.3 |
| 75 | 5.2 | 4.8 | 5.1 | 4.8 | 3.1 | 1.0 | 2.2 | 2.7 | 2.7 |
| 77 | 7.1 | 3.7 | 3.5 | 1.5 | 2.2 | + | + | + | + |
| 81 | 6.3 | 5.5 | 4.5 | 3.2 | 3.6 | 2.7 | 2.0 | 2.3 | 2.1 |
| 82 | 6.3 | 5.4 | 4.6 | 3.3 | 3.1 | 1.5 | 2.1 | − | 1.0 |
| 84 | 5.7 | 4.6 | 4.1 | 4.3 | 2.5 | 2.3 | 3.0 | 3.1 | 4.8 |
| 85 | 6.1 | 4.8 | 4.3 | 3.4 | 4.3 | 3.7 | + | 1.7 | 1.6 |
| 86 | 4.6 | 3.1 | 1.3 | 2.1 | 2.4 | 2.0 | − | − | 2.3 |
| Avg | 6.0 | 4.6 | 4.0 | 3.4 | 3.1 | 2.2 | 1.7 | 1.8 | 2.6 |
| SD | 0.9 | 0.8 | 1.2 | 1.1 | 0.7 | 1.2 | 1.4 | 1.7 | 2.0 |
The diets of group 2 calves were supplemented with 107 CFU of E. coli (the colicinogenic E. coli strains) per g.
+, detected only by enrichment and assigned an arbitrary value of 0.3 log10 CFU/g for calculation of averages; −, assigned a value of 0 log10 CFU/g for calculation of averages; ND, not detected due to death of calf.
Due to the deaths of two calves in the control group, their data were not used to calculate these averages.
TABLE 2.
Fecal shedding of E. coli O157:H7 by calves during period 2 of the experimenta
| Group and calf no. |
E. coli O157:H7 count (log10 CFU/g) at dayb:
|
||||
|---|---|---|---|---|---|
| 1 | 4 | 7 | 10 | 13 | |
| Group 1 | |||||
| 74 | − | 1.0 | + | − | − |
| 76 | 2.2 | 2.1 | 1.5 | 1.5 | 1.3 |
| 78 | 1.6 | + | + | 1.0 | + |
| 79 | 2.7 | 3.3 | 2.5 | + | − |
| 80 | 4.1 | 2.6 | 1.8 | 3.8 | 3.8 |
| 83 | 3.8 | 4.7 | 3.0 | 2.3 | 1.5 |
| Avg | 2.4 | 2.3 | 1.6 | 1.5 | 1.1 |
| SD | 1.5 | 1.6 | 1.1 | 1.4 | 1.5 |
| Group 2 | |||||
| 73 | 5.1 | 4.0 | 4.3 | 2.9 | 3.0 |
| 75 | 1.0 | + | 1.5 | 1.0 | − |
| 77 | 1.0 | + | − | 1.5 | 2.2 |
| 81 | 1.0 | 1.0 | + | + | + |
| 82 | + | 1.6 | 1.6 | 1.0 | − |
| 84 | 4.1 | 5.2 | 2.9 | − | 1.0 |
| 85 | 2.0 | 4.1 | 6.0 | 2.0 | − |
| 86 | 3.8 | 6.5 | 4.8 | 3.4 | 3.6 |
| Avg | 2.3 | 2.9 | 2.7 | 1.5 | 1.3 |
| SD | 1.8 | 2.4 | 2.2 | 1.2 | 1.5 |
The diets of group 1 calves were supplemented with 108 CFU of the eight colicinogenic E. coli strains per g.
+, detected only by enrichment and assigned an arbitrary value of 0.3 log10 CFU/g for calculation of averages; −, assigned a value of 0 log10 CFU/g for calculation of averages.
In P3, group 2 continued to receive the diet alone and group 1 resumed receiving the probiotic E. coli. With the exception of days 1 and 3, the new treatment group had consistently lower average counts of O157:H7 shedding compared to group 2 (Table 3). The differences during this period were not greater than 0.9 log cycles and were not statistically significant.
TABLE 3.
Fecal shedding of E. coli O157:H7 by calves during P3 of the experimenta
| Group and calf no. |
E. coli O157:H7 count (log10 CFU/g) at dayb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 6 | 9 | 12 | 15 | 18 | 21 | 24 | |
| Group 1 | |||||||||
| 74 | 7.3 | 4.5 | 2.3 | 3.0 | 2.2 | 2.3 | 2.6 | 2.7 | 2.5 |
| 76 | 6.0 | 5.1 | 3.4 | 3.2 | 2.0 | 3.2 | 2.5 | 2.8 | 3.3 |
| 78 | 6.1 | 5.0 | 2.6 | 1.0 | 2.2 | 1.0 | − | 1.0 | 1.7 |
| 79 | 6.2 | 4.1 | 4.0 | 2.3 | 1.0 | 2.3 | 1.0 | 2.0 | 1.3 |
| 80 | 7.0 | 3.8 | 2.8 | 2.4 | 1.6 | 2.0 | + | + | + |
| 83 | 6.3 | 4.2 | 4.5 | 2.5 | 1.8 | 1.7 | 1.3 | + | + |
| Avg | 6.5 | 4.5 | 3.3 | 2.4 | 1.8 | 2.1 | 1.3 | 1.5 | 1.6 |
| SD | 0.5 | 0.5 | 0.9 | 0.8 | 0.4 | 0.7 | 1.1 | 1.1 | 1.2 |
| Group 2 | |||||||||
| 73 | 6.5 | 4.3 | 4.7 | 5.1 | 4.5 | 4.7 | 4.7 | 4.7 | 4.4 |
| 75 | 6.8 | 4.2 | 4.4 | 2.7 | 2.3 | 2.1 | 2.0 | + | 2.0 |
| 77 | 5.8 | 4.2 | 3.7 | 2.0 | 1.5 | 1.8 | 1.0 | 1.3 | + |
| 81 | 5.4 | 3.4 | 2.3 | 2.2 | 2.3 | 3.3 | 2.2 | 1.0 | − |
| 82 | 8.5 | 4.4 | 3.6 | 2.3 | 1.6 | 1.6 | 1.6 | 1.3 | − |
| 84 | 4.8 | 3.4 | 2.5 | 2.3 | 2.3 | 2.1 | 1.0 | 5.0 | 4.0 |
| 85 | 6.5 | 4.2 | 3.7 | 4.1 | 3.8 | 4.9 | 4.4 | 4.9 | 4.5 |
| 86 | 7.3 | 4.3 | 3.2 | 2.0 | 2.2 | 2.4 | + | 1.0 | 1.6 |
| Avg | 6.4 | 4.1 | 3.5 | 2.8 | 2.6 | 2.9 | 2.1 | 2.4 | 2.1 |
| SD | 1.2 | 0.4 | 0.8 | 1.1 | 1.1 | 1.3 | 1.6 | 2 | 2 |
Diets of group 1 calves were supplemented with 108 CFU of the colicinogenic E. coli strains per g.
+, detected only by enrichment and assigned an arbitrary value of 0.3 log10 CFU/g for calculation of averages; −, assigned a value of 0 log10 CFU/g for calculation of averages.
When the daily averages of O157:H7 counts in P1 and P3 of group 2 were compared, differences that were not statistically significant were obtained (Table 4). However, if the group 1 daily average counts of serotype O157:H7 were compared to their corresponding values of P1 and P3, group 1 animals during P3 had on average 1.14 log10 cycles less than during P1 after day 6 and a maximum decline of 1.8 log10 cycles determined at day 21. The analysis of the data in this manner indicated an overall strong significant difference (P = 0.001) between P1 and P3 for group 1 that received 108 CFU of colicinogenic E. coli per g in their diet during P3 (Table 4).
TABLE 4.
Differences between daily average counts of E. coli O157:H7 and statistical significancea
| Day and overall avg | Comparison between groups
|
Comparison between P1 and P3
|
||||||
|---|---|---|---|---|---|---|---|---|
| P1
|
P3
|
Group 2
|
Group 1
|
|||||
| Diff. | P value | Diff. | P value | Diff. | P value | Diff. | P value | |
| Day | ||||||||
| 1 | 0.7 | 0.27 | −0.1 | 0.94 | 0.4 | 0.54 | 0.2 | 0.66 |
| 3 | 0.4 | 0.53 | −0.4 | 0.54 | −0.5 | 0.47 | 0.5 | 0.34 |
| 6 | 0.6 | 0.34 | 0.2 | 0.67 | −0.5 | 0.49 | 1.3 | 0.01 |
| 9 | 0.4 | 0.42 | 0.4 | 0.50 | −0.6 | 0.44 | 1.4 | 0.006 |
| 12 | 0.1 | 0.87 | 0.8 | 0.23 | −0.5 | 0.44 | 1.4 | 0.008 |
| 15 | 0.8 | 0.18 | 0.8 | 0.22 | 0.7 | 0.29 | 0.9 | 0.08 |
| 18 | 1.2 | 0.06 | 0.8 | 0.18 | 0.4 | 0.54 | 1.6 | 0.002 |
| 21 | 1.5 | 0.02 | 0.9 | 0.15 | 0.6 | 0.37 | 1.8 | 0.001 |
| 24 | 0.2 | 0.80 | 0.5 | 0.40 | −0.5 | 0.42 | 1.2 | 0.02 |
| Overall | 0.7 | 0.12 | 0.4 | 0.30 | −0.05 | 0.93 | 1.1 | 0.001 |
Shown are differences (diff.) between groups for P1 and P3 and between P1 and P3 of the same group. Differences are expressed as log10 CFU/g. Diets of group 2 calves were supplemented with 107 CFU of the colicinogenic E. coli strains per g in P1, and diets of group 1 calves were supplemented with 108 CFU of the eight colicinogenic E. coli strains per g in P3.
At the conclusion of P3, the cattle were euthanized and tissue samples were collected and analyzed for O157:H7. E. coli O157:H7 was recovered from at least one animal of each group for each tissue type, and the rumen and descending colon of group 2 calves that had not received the colicinogenic E. coli during P3 had the largest number of positives samples (7 positive samples out of 8). The differences in positives among tissues were not, however, statistically significant. Overall, 44% of the animal tissues from group 1 (treated with probiotic during P3) were positive for O157:H7, compared to 64% from group 2, and this difference was statistically significant (P < 0.04) (Table 5). For O157:H7-positive individual tissues, the rectum (P = 0.06) and spiral colon (P = 0.07) were close to being statistically significant. The group 1 calves also had a slightly lower average number of O157:H7 than group 2 animals (0.4 log10 CFU/g compared to 0.6 log10 CFU/g), but this difference was not statistically significant.
TABLE 5.
Effect of addition of colicinogenic E. coli (group 1 during P3) to feed on the average count and number of positive samples of E. coli O157:H7 in intestinal tissues collected after euthanasia of calves
| Tissue | (n = 8)
|
Group 1 (treatment) (n = 6)
|
||
|---|---|---|---|---|
| E. coli O157:H7 count (log10 CFU/g ± SD) | No. of positive animals (%) | E. coli O157:H7 count (log10 CFU/g ± SD) | No. of positive animals | |
| Rumen | 1.6 ± 1.4 | 7 | 0.9 ± 0.9 | 4 |
| Ileum | 0.5 ± 0.6 | 3 | 0.3 ± 0.6 | 4 |
| Cecum | 0.4 ± 0.6 | 4 | 0.4 ± 0.6 | 2 |
| Spiral colon | 0.7 ± 0.7 | 7 | 0.3 ± 0.5 | 3 |
| Descending colon | 0.2 ± 0.3 | 3a | 0.05 ± 0.1 | 1 |
| Rectum | 0.4 ± 0.6 | 6 | 0.4 ± 0.6 | 2 |
| Overall | 0.6 ± 0.5 | 30 (64) | 0.4 ± 0.3 | 16 (44) |
One sample was missed, and there were seven total descending colon samples for the control group.
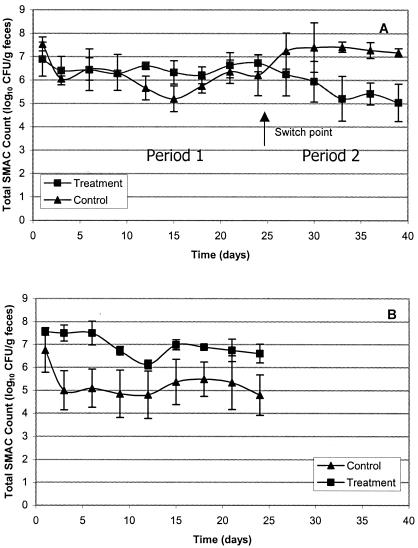
Figure 2 shows the numbers of putative GN enterobacteria capable of growing on SMAC plates during the course of the experiment. During P1, when group 2 received feed containing 107 CFU of colicinogenic E. coli per g, the total number of bacteria on SMAC plates was similar between the two groups. Group 2 shed only slightly more bacteria (0.3 log10 CFU/g) on SMAC plates than group 1, and this difference was not statistically different. During P2 and P3, when group 1 received feed containing 108 CFU of colicinogenic E. coli per g, animals shed more bacteria (1.7 log10 CFU/g) on SMAC plates than group 2, and most of these differences were statistically significant (P < 0.05).
FIG. 2.
Effect of feeding colicinogenic Escherichia coli on the fecal shedding of bacteria able to grow SMAC agar. Shown are data from (A) the first and second feeding periods and (B) the third feeding period.
SMAC colonies were selected during the experiment and tested with PCR for the presence of the colicin E7 gene. Colicin E7 strains were recovered from the feces of every animal receiving treatment throughout the study. Approximately 60% of the predominant isolates obtained from probiotic-fed animals were colicin E7-producing E. coli. At day 9 during P3, 37 days since last receiving the colicinogenic mixture, colicin E7-producing E. coli was still detected in two group 2 calves representing 35% of the predominant SMAC isolates (data not shown). This level declined gradually until day 21 and at necropsy, when E7 colicinogenic strains could not be detected from any sample. Recovered colicin E7 strains were then assessed with rep-PCR to determine their identity. E7 strains recovered at day 9 of P3 were classified into one of three isolate groupings (bovine, human, and duck). Neither sheep strain (S12 or S34) was detected. Duck strain U24 was the only isolate recovered at days 15 and 18 of P3.
DISCUSSION
The use of probiotic bacteria to inhibit E. coli O157:H7 in cattle is a promising method to control this food-borne pathogen. Previous reports have shown the potential of beneficial E. coli strains and lactic acid bacteria to reduce O157:H7 fecal shedding or prevalence in cattle (2, 22, 30-32). However, many factors that might influence the effectiveness of probiotic organisms, such as probiotic diversity, dose response, and the use of other experimental designs other than the simple control treatment protocol, have not been explored. This research project examined the effect of using a collection of strains that produced the same colicin and an animal trial that followed a crossover design.
Colicins are antimicrobial proteins produced by some E. coli strains that inhibit other E. coli strains. Colicins are produced under stress conditions and give the producing strain a competitive advantage. Surveys have shown that approximately 30% of all E. coli strains can produce a colicin (23). Approximately 25 different types of colicins have been identified, and colicins can be classified based on their modes of action. Modes of action include formation of membrane pores, DNA cleavage, RNA degradation, and inhibition of peptidoglycan synthesis.
For the present feeding study, only colicinogenic E. coli that could produce colicin E7 (DNase) was used to determine if strains that predominantly produce a single type of colicin could reduce E. coli O157:H7 in cattle. It was previously reported that among 18 previously characterized colicins, only E7 inhibited all 22 O157:H7 strains tested (24). In a follow-up publication, colicin E7 genes were detected in 8 out of a total of 14 recently isolated wild-type colicinogenic E. coli strains that had been obtained from cattle, sheep, ducks, and humans (25). Based on those observations, colicin E7-producing strains appear to be strongly inhibitory for O157:H7 and were chosen to evaluate their potential as probiotic bacteria in this study.
The rationale of the experimental design of this study was founded in the following assumptions: (i) in period 1, the comparisons between the two groups were considered as a single factorial experiment; (ii) for each group, the comparisons between periods 1 and 3 were treated as a crossover experiment; and (iii) the inclusion of period 2 had the purpose of finding the effect of providing the probiotic E. coli to animals that were already carrying E. coli O157:H7 at levels that are typically found in nature. The potential carryover effect that could have confounded the comparisons between groups at P3 and between P1 and P3 for group 2 is discussed below.
The results obtained by the selection of colicinogenic E7-producing E. coli in vitro suggested that this set of strains could be used to reduce fecal shedding of serotype O157:H7 in cattle. When both groups are compared for each specific time period, only day 21 of P1 was statistically significant. However, when the P1 and P3 for each calf group were compared, one calf group comparison was highly significant, while the other group was not. There are two possible reasons for this variation: dose response and the order of treatment.
The group 1 comparison (P1 versus P3) that had an overall significant reduction of O157:H7 included animals that were fed a larger dose of colicinogenic E. coli (108 CFU/g of feed) and first served as a control group and then as the treatment group (Table 4). The higher probiotic dose had a noticeable effect on the level of bacteria on SMAC plates during P3, compared to P1 in group 1 calves. This higher level of colicinogenic E. coli may have been needed to enhance the reduction of O157:H7. The order of treatment may have also resulted in a carryover effect for group 2 that first received the probiotic E. coli and then served as control. Although 20 days had passed from the end of P1 to the beginning of P3, E7 colicinogenic E. coli was still recovered from group 2 calves at significant levels, and its presence may have also caused a reduction effect on the numbers of serotype O157:H7 during P3. Based on these findings, it may be advisable that future animal experiments involving probiotics include subjecting all animals to the control diet in the first part of the experiment and then switching them to treatment to eliminate a possible carryover effect and to reduce the variability from animal to animal.
The significant difference between the overall presence of E. coli O157:H7 in tissue samples of control and treatment groups (64 versus 44%) as well as the consistently smaller number of positive animals for each tissue type in group 1 calves indicated that the use of the colicinogenic E. coli significantly reduced the extent of colonization by this pathogen. Comparisons among tissue sites, however, provided little indication of a specific localization of this pathogen. Previous studies that have used inoculated animals have detected almost no E. coli O157:H7 in the rumen, but they reported recovering the pathogenic strains from the rectum (13, 19). In one of those reports, it was hypothesized that E. coli O157:H7 has a particular affinity for the lymphoid follicles found in the rectoanal junction. Because our methods did not specifically test rectum samples in the region closer to the anus, the findings of this report do not necessarily contradict that hypothesis.
There have been three published papers that have reported the use of colicinogenic E. coli to reduce E. coli O157:H7 in cattle (30-32). In the first published report, six calves were inoculated with cultures of four probiotic E. coli strains, and after 2 days, the calves that were fed a probiotic and nine control calves were administered a mixture of E. coli O157:H7 strains (31). The results showed that the calves fed the colicinogenic E. coli shed significantly less (P < 0.05) E. coli O157:H7 at day 15, day 18, and afterward to the end of the trial (approximately 30 days). Three of the colicinogenic E. coli strains were included in the first anti-O157:H7 probiotic patent, and they were selected for further applications (7). In an additional report involving weaned calves, Tkalcic and coworkers confirmed the ability of a single dose of probiotic E. coli to reduce fecal shedding of serotype O157:H7 (30). They reported statistical differences with the control group in half of the measurements over the course of a 30-day feeding trial.
There are several aspects to compare between the colicinogenic E. coli calf feeding studies of Doyle's group and the present research study. The following comparisons are based on the weaned calf study by Tkalcic et al. (30), in which quantitative data were provided to allow assessments between that study and our crossover group which showed a strong overall reduction of O157:H7. The greatest differences in fecal shedding of O157:H7 between the control and treatment groups in that study was 1.75 log10 CFU/g of feces for day 16 and 1.52 log10 CFU/g of feces for day 18. The greatest O157:H7 differences between the treatment and control groups in our study were 1.6 log10 CFU/g of feces on day 21 and 1.8 log10 CFU/g of feces on day 18. These maximum differences between treatment and control periods or groups were similar. Overall, during the 24 days following O157:H7 inoculation of the calves, treatment described by Tkalcic et al. (30) resulted in an average decrease of 0.84 log10 CFU/g of feces for O157:H7, while our treatment caused an average reduction of 1.14 log10 CFU/g of feces for O157:H7 during the trial.
Another comparison between the same trials discussed above includes the number of calves that had undetectable levels of O157:H7 at day 24 of the time period. Three of six calves in the treatment group in the Tkalcic et al. (30) report were negative for O157:H7, while all six of the calves treated with probiotic in this report were still positive for O157:H7 at day 24. Procedural differences between the two studies may account for this observation. Tkalcic et al. reported an overall lower level of E. coli O157:H7 fecal shedding than our report. When the data for their control group calves and our group 1 animals during period 1 are compared, the O157:H7 fecal levels in their report were 0.5, 0.8, 1.5 and 2.2 log10 CFU/g less than those in this study at days 6, 12, 18, and 24, respectively. It would then be expected that more of their treatment calves became negative for E. coli O157:H7 because the level was already low.
Possible reasons for this difference in serotype O157:H7 levels include initial inoculum, strain variation, and diet. In this experiment, the calves were inoculated with 1011 CFU while other researchers (30, 31) used 1010 CFU of O157:H7 strains per calf. Different O157:H7 strains were used in these studies. It is possible that the strains used in the present report are better adapted to thrive in the gastrointestinal tracts of cattle.
The effect of type of diet on the fecal carriage of enterohemorrhagic E. coli has been suggested in inoculated experiments reported by several researchers, and it appears that there is a difference between forage- and corn-based diets (4, 14, 29). For their probiotic E. coli experiment, Zhao et al. (31) used a diet of alfalfa pellets and sweet feed, but its specific composition was not reported. Tkalcic et al. (30) indicated that they used a premixed roughage-concentrate dairy calf ration, but the composition was also not reported. The calves described in the present report were fed a feed mix composed predominantly of corn (73%), similar to typical beef cattle diets. The corn in the diet might have favored the permanence of E. coli O157:H7 in their intestines (15).
An additional difference between those reports that used the patented E. coli strains and the present report is the dose frequency. In those studies, the patented mix was given at the start of the experiment, but in this experiment, the colicinogenic E. coli was given every day as part of the feed (30-32). The continuous supplementation of the colicin E7-producing E. coli mix was selected in order to eliminate the potential effect of the lack of colonization capability of some of these strains. The detection of some of these strains in the group 2 calves during period 3 suggested that they could be capable of colonizing the bovine gastrointestinal tract.
In previous studies, when beneficial bacteria were selected to inhibit human pathogens in livestock, the probiotic strains were typically isolated from the same host animal species (3, 10, 12, 22, 26, 31). The rationale is that the isolates would theoretically be better able to inhabit the gastrointestinal tract of that particular species. The idea of certain isolates, especially E. coli, adapting to a specific host is supported by DNA fingerprinting research that can classify E. coli isolates to a particular animal species (6). This study found that 37 days after last receiving the colicinogenic treatment, the E7 strains originally isolated from cattle, humans, and a duck were among the predominant facultative GN bacteria. The duck strain was still isolated 46 days after the last inoculation, whereas the cattle strains were recovered only up to day 40. This finding suggests that E. coli isolated from one animal species can thrive in another.
This project found that a daily dose of 108 CFU of colicin E7-producing E. coli per g of feed can significantly reduce the fecal shedding of E. coli O157:H7 in cattle. The calf group that was first the control group and then treated with colicinogenic E. coli had a marked reduction of E. coli O157:H7 (P = 0.001). Colicin E7-producing E. coli was also able to significantly reduce the overall colonization of O157:H7 in the gastrointestinal tracts of the steers (P < 0.04). Future research should consider the use of diverse anti-O157 colicinogenic E. coli to determine if a set of different colicins enhances O157:H7 inhibition compared to the predominant colicin E7 reduction of E. coli O157:H7.
Acknowledgments
This study was funded by Renessen, LLC, Bannockburn, Ill. G.P.S. was supported by a USDA National Needs fellowship. Funding for this project was also provided by the McKnight Presidential Chair in Genomics, University of Minnesota.
We thank Les Westendorp, Marcy Brower, and Travis Ling for assistance in animal management and care.
REFERENCES
- 1.Borczyk, A. A., M. A. Karmali, H. Lior, and L. M. C. Duncan. 1987. Bovine reservoir for verotoxin-producing Escherichia coli O157:H7. Lancet i:98. [DOI] [PubMed] [Google Scholar]
- 2.Brashears, M. M., M. L. Galyean, G. H. Loneragan, J. E. Mann, and K. Killinger-Mann. 2003. Prevalence of Escherichia coli O157:H7 and performance by beef feedlot cattle given Lactobacillus direct-fed microbials. J. Food Prot. 66:748-754. [DOI] [PubMed] [Google Scholar]
- 3.Brashears, M. M., D. Jaroni, and J. Trimble. 2003. Isolation, selection, and characterization of lactic acid bacteria for a competitive exclusion product to reduce shedding of Escherichia coli O157:H7 in cattle. J. Food Prot. 66:355-363. [DOI] [PubMed] [Google Scholar]
- 4.Buchko, S. J., R. A. Holley, W. O. Olson, V. P. J. Gannon, and D. M. Veira. 2000. The effects of different grain diets on fecal shedding of Escherichia coli O157:H7 by steers. J. Food Prot. 63:1467-1474. [DOI] [PubMed] [Google Scholar]
- 5.Callaway, T. R., and F. Diez-Gonzalez. 2002. Pre-harvest intervention strategies to reduce Escherichia coli O157:H7 in cattle, p. 19-40. In S. A. Martin (ed.), Gastrointestinal microbiology in animals. Research Signpost, Kerala, India.
- 6.Dombek, P. E., L. K. Johnson, S. T. Zimmerly, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doyle, M. P., T. Zhao, B. G. Harmon, and C. A. Brown. October 1999. U.S. patent 5,965,128.
- 8.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fedorka-Cray, P. J., J. S. Bailey, N. J. Stern, N. A. Cox, S. R. Ladely, and M. Musgrove. 1999. Mucosal competitive exclusion to reduce Salmonella in swine. J. Food Prot. 62:1376-1380. [DOI] [PubMed] [Google Scholar]
- 10.Garriga, M., M. Pascual, J. M. Monfort, and M. Hugas. 1998. Selection of lactobacilli for chicken probiotic adjuncts. J. Appl. Microbiol. 84:125-132. [DOI] [PubMed] [Google Scholar]
- 11.Garthright, W. E. 1995. Appendix 2. Most probable number from serial dilutions, p. 2.01-2.05. In G. J. Jackson (ed), FDA bacteriological analytical manual. AOAC International, Washington, D.C.
- 12.Genovese, K. J., R. C. Anderson, R. B. Harvey, and D. J. Nisbet. 2000. Competitive exclusion treatment reduces the mortality and fecal shedding associated with enterotoxigenic Escherichia coli infection in nursery-raised neonatal pigs. Can. J. Vet. Res. 64:204-207. [PMC free article] [PubMed] [Google Scholar]
- 13.Grauke, L. J., I. T. Kudva, J. W. Yoon, C. W. Hunt, C. J. Williams, and C. J. Hovde. 2002. Gastrointestinal tract location of Escherichia coli O157:H7 in ruminants. Appl. Environ. Microbiol. 68:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hovde, C. J., P. R. Austin, K. A. Cloud, C. J. Williams, and C. W. Hunt. 1999. Effect of cattle diet on Escherichia coli O157:H7 acid resistance. Appl. Environ. Microbiol. 65:3233-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keen, J. E., G. A. Uhlich, and R. O. Elder. 1999. Proc. 80th Conf. Res. Workers Anim. Dis., abstr. 83.
- 16.Kogan, S., M. Doherty, and J. Gitschier. 1987. An improved method for diagnosis of genetic diseases by amplified DNA sequences. N. Engl. J. Med. 317:985-990. [DOI] [PubMed] [Google Scholar]
- 17.Lema, M., L. Williams, and D. R. Rao. 2001. Reduction of fecal shedding of enterohemorrhagic Escherichia coli O157:H7 in lambs by feeding microbial feed supplement. Small Ruminant Res. 39:31-39. [DOI] [PubMed] [Google Scholar]
- 18.Moore, D. S., and G. P. McCabe (ed.). 1999. Introduction to the practice of statistics, 3rd ed. W. H. Freeman & Co., New York, N.Y.
- 18a.National Research Council. 2000. Nutrient requirements of beef cattle, 7th ed. National Academies Press, Washington, D.C.
- 19.Naylor, S. W., J. C. Low, T. E. Besser, A. Mahajan, G. J. Gunn, M. C. Pearce, I. J. McKendrick, D. G. E. Smith, and D. L. Gally. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nisbet, D. 2002. Defined competitive exclusion cultures in the prevention of enteropathogen colonisation in poultry and swine. Antonie Leeuwenhoek 81:481-486. [DOI] [PubMed] [Google Scholar]
- 21.Nisbet, D. J., D. E. Corrier, S. C. Ricke, M. E. Hume, J. A. Byrd II, and J. R. Deloach. 1996. Maintenance of the biological efficacy in chicks of a cecal competitive-exclusion culture against Salmonella by continuous-flow fermentation. J. Food Prot. 59:1279-1283. [DOI] [PubMed] [Google Scholar]
- 22.Ohya, T., T. Marubashi, and H. Ito. 2000. Significance of fecal volatile fatty acids in shedding of Escherichia coli O157 from calves: experimental infection and preliminary use of a probiotic product. J. Vet. Med. Sci. 62:1151-1155. [DOI] [PubMed] [Google Scholar]
- 23.Riley, M. A., and D. M. Gordon. 1996. The ecology and evolution of bacteriocins. J. Ind. Microbiol. 17:151-158. [Google Scholar]
- 24.Schamberger, G., and F. Diez-Gonzalez. 2002. Selection of recently isolated colicinogenic Escherichia coli strains inhibitory against E. coli O157:H7. J. Food Prot. 65:1381-1387. [DOI] [PubMed] [Google Scholar]
- 25.Schamberger, G. P., and F. Diez-Gonzalez. 2004. Characterization of colicinogenic Escherichia coli strains inhibitory to enterohemorrhagic E. coli. J. Food Prot. 67:486-492. [DOI] [PubMed] [Google Scholar]
- 26.Schoeni, J., and M. P. Doyle. 1992. Reduction of Campylobacter jejuni colonization of chicks by cecum-colonizing bacteria producing anti-C. jejuni metabolites. Appl. Environ. Microbiol. 58:664-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, D., M. Blackford, S. Younts, R. Moxley, J. Gray, L. Hungerford, T. Milton, and T. Klopfenstein. 2001. Ecological relationships between the prevalence of cattle shedding Escherichia coli O157:H7 and the characteristics of the cattle or conditions of the feedlot pen. J. Food Prot. 64:1899-1903. [DOI] [PubMed] [Google Scholar]
- 28.Stevens, M. P., P. M. van Diemen, F. Dziva, P. W. Jones, and T. S. Wallis. 2002. Options for the control of enterohaemorrhagic Escherichia coli in ruminants. Microbiology 148:3767-3778. [DOI] [PubMed] [Google Scholar]
- 29.Tkalcic, S., C. A. Brown, B. G. Harmon, A. V. Jain, E. P. O. Mueller, A. Parks, K. L. Jacobson, S. A. Martin, T. Zhao, and M. P. Doyle. 2000. Effects of diet on rumen proliferation and fecal shedding of Escherichia coli O157:H7 in calves. J. Food Prot. 63:1630-1636. [DOI] [PubMed] [Google Scholar]
- 30.Tkalcic, S., T. Zhao, B. G. Harmon, M. P. Doyle, C. A. Brown, and P. Zhao. 2003. Fecal shedding of enterohemorrhagic Escherichia coli in weaned calves following treatment with probiotic Escherichia coli. J. Food Prot. 66:1184-1189. [DOI] [PubMed] [Google Scholar]
- 31.Zhao, T., M. P. Doyle, B. G. Harmon, C. A. Brown, P. O. E. Mueller, and A. H. Parks. 1998. Reduction of carriage of enterohemorrhagic Escherichia coli O157:H7 in cattle by inoculation with probiotic bacteria. J. Clin. Microbiol. 36:641-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao, T., S. Tkalcic, M. P. Doyle, B. G. Harmon, C. A. Brown, and P. Zhao. 2003. Pathogenicity of enterohemorrhagic Escherichia coli in neonatal calves and evaluation of fecal shedding by treatment with probiotic Escherichia coli. J. Food Prot. 66:924-930. [DOI] [PubMed] [Google Scholar]