Abstract
The production of hepatotoxic cyclic heptapeptides, microcystins, is almost exclusively reported from planktonic cyanobacteria. Here we show that a terrestrial cyanobacterium Nostoc sp. strain IO-102-I isolated from a lichen association produces six different microcystins. Microcystins were identified with liquid chromatography-UV mass spectrometry by their retention times, UV spectra, mass fragmentation, and comparison to microcystins from the aquatic Nostoc sp. strain 152. The dominant microcystin produced by Nostoc sp. strain IO-102-I was the highly toxic [ADMAdda5]microcystin-LR, which accounted for ca. 80% of the total microcystins. We assigned a structure of [DMAdda5]microcystin-LR and [d-Asp3,ADMAdda5]microcystin-LR and a partial structure of three new [ADMAdda5]-XR type of microcystin variants. Interestingly, Nostoc spp. strains IO-102-I and 152 synthesized only the rare ADMAdda and DMAdda subfamilies of microcystin variants. Phylogenetic analyses demonstrated congruence between genes involved directly in microcystin biosynthesis and the 16S rRNA and rpoC1 genes of Nostoc sp. strain IO-102-I. Nostoc sp. strain 152 and the Nostoc sp. strain IO-102-I are distantly related, revealing a sporadic distribution of toxin production in the genus Nostoc. Nostoc sp. strain IO-102-I is closely related to Nostoc punctiforme PCC 73102 and other symbiotic Nostoc strains and most likely belongs to this species. Together, this suggests that other terrestrial and aquatic strains of the genus Nostoc may have retained the genes necessary for microcystin biosynthesis.
Toxic cyanobacteria are increasingly an environmental problem as a consequence of their mass occurrences in aquatic ecosystems. Microcystins are cyclic heptapeptides produced by freshwater cyanobacteria, including several strains from the genera Anabaena, Microcystis, Planktothrix, and two strains from the genus Nostoc (39). In brackish waters strains from the genus Nodularia produce the structurally related pentapeptide nodularin (39). The production of the most common cyanobacterial toxins, microcystins, is almost exclusively limited to planktonic cyanobacteria (39). The only report of microcystin from a terrestrial cyanobacterium is from Hapalosiphon hibernicus strain BZ-3-1 (34). However, it is not clear if this strain was isolated from a soil sample or a freshwater mud sample (34).
Most microcystins are potent inhibitors of eukaryotic protein phosphatases 1 and 2A (26). Microcystins have been commonly involved in domestic and wild animal poisonings (36) and pose a threat to human health (23). Microcystins are toxic to zooplankton, and it is widely believed that they act as a grazing deterrent to zooplankton, which graze on cyanobacteria (9, 21). They are also proposed to act in cyanobacterial cell-to-cell communication (11) and as a chelator in iron scavenging (46).
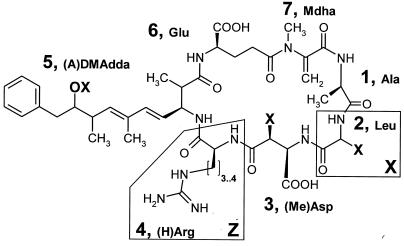
Microcystins are comprised of seven amino acids that form a ring structure. The general structure of microcystins is cyclo[-d-Ala-l-X-d-MeAsp-l-Z-Adda-d-Glu-Mdha-], of which Z and X are variable l-amino acids (Fig. 1). There are more than 65 structural variants of microcystins known (39). The most commonly reported microcystin variant (microcystin-LR) contains l-leucine (L) and l-arginine (R) in the variable X and Z positions (Fig. 1). The novel amino acid Adda (3-amino-9-methoxy-2,6,8,-trimethyl-10-phenyl-deca-4,6-dienoic acid) is found only in microcystins and nodularins (39, 47) and plays a crucial role in the inhibition of eukaryotic protein phosphatases (16). There are three structural modifications to the Adda amino acid known, geometrical isomers (6Z-Adda) (17), an acetyl-demethyl variant (ADMAdda) (47), and a demethyl variant (DMAdda) (47). The geometrical isomer of Adda is biologically inactive (17), whereas the two other modifications inhibit protein phosphatases (3, 40, 42).
FIG. 1.
General structure of [(A)DMAdda]microcystin-X(Ha)R found in Nostoc sp. strain IO-102-I. In position 5, OX is −O(CO)CH3 in ADMAdda and OH in DMAdda. In position 2, X is Leu, Leu with a modification in the side chain, an unidentified amino acid, H in Asp, and CH3 in MeAsp. In position 4, the side chain with Arg has three CH2 groups and with (H)Arg the side chain has four CH2 groups.
Microcystins and nodularins are made by nonribosomal peptide synthesis (12). Microcystin biosynthesis is directed by a synthetase encoded in a single gene cluster, which has been characterized from Microcystis aeruginosa strains K-139 (29) and PCC 7806 (44), Planktothrix agardhii strain NIVA126/8 (7), and Anabaena sp. strain 90 (37). The microcystin synthetase cluster contains genes for nonribosomal peptide synthetases, polyketide synthetase, mixed NRPS-PKS synthetases, and additional modifying enzymes (7, 37, 44). Phylogenetic analyses of the mcyA, mcyD, and mcyE genes suggested that the microcystin synthetase genes are ancient and have been inherited vertically in the cyanobacterial lineage (35).
Nostoc is a cosmopolitan cyanobacterial genus occurring in both terrestrial and aquatic ecosystems. Strains of the genus Nostoc are the most common cyanobacteria in various symbioses (13). Nostoc strains can also occur as a minor component of cyanobacterial blooms but rarely form mass occurrences (40). Nostoc collected from terrestrial habitats is used as a dietary supplement by humans in some countries, but no acute intoxications attributed to microcystins have been reported (13, 15). A Nostoc rivulare bloom in Texas caused intoxication of wild and domestic animals (8, 39). However, despite the screening of cyanobacterial cultures and strains for microcystins and other bioactive compounds during the last decade (34, 39), only two strains of the genus Nostoc, Nostoc sp. strain 152 isolated from Lake Sääskjärvi in Finland (40, 42) and Nostoc sp. strain DUN901 isolated from brackish water of Barrow Ski Club Lake in the United Kingdom (3), are known to synthesize microcystins. Of the more than 65 known microcystin variants, 10 are produced by Nostoc sp. strains 152 and DUN901, and all are acutely hepatotoxic (3, 40).
We screened Nostoc strains involved in lichen symbiosis for microcystin production. A single Nostoc strain was found to produce microcystins. We analyzed the structures of the microcystins with liquid chromatography (LC) combined with mass spectrometry (MS). We also evaluated the phylogenetic position of the microcystin-producing Nostoc strain by constructing and comparing phylogenetic trees from microcystin synthetase genes mcyA, mcyD, and mcyE and from the 16S rRNA and rpoC1 genes.
MATERIALS AND METHODS
Strain isolation and culture.
Nostoc sp. strain IO-102-I was isolated in the year 2000 from the lichen Pannaria pezizoides collected from mosses on a rock in Sysmä, in southern Finland, in 1998 by V. Haikonen (collection number of 19114) and deposited in the Botanical Museum of the University of Helsinki in Finland. The cultured strain was either involved in the lichen symbiosis or originated from the surface of the lichen. Nostoc sp. strain IO-102-I was cultured on Z8 medium lacking nitrogen (22) in continuous low light at 21°C. The culture was replated from a single filament in order to obtain a pure culture and then grown in liquid medium. Culture purity was confirmed by growth on R2A solid medium (International Diagnostics Group plc., Lancashire, United Kingdom), followed by gram-staining. Nostoc sp. strain 152 produces more than seven microcystins (42) and was included as a reference strain in toxin analyses. To obtain sufficient cyanobacterial biomass for toxin analyses, these two cyanobacterial strains were grown at a photon irradiance of 20 to 27 μmol m−2 s−1 in 2.7 liters of liquid Z8 medium aerated with filter-sterilized (Millex-FG, 0.2 μm pore size; Millipore Corp., Bedford, Mass.) compressed air.
Whole-cell extraction.
Freeze-dried cells from a 42-day-old Nostoc sp. strain IO-102-I (39 or 66 mg) and from a 21-day-old Nostoc sp. strain 152 (42 mg) culture were homogenized with a glass-bead method as follows. Cells were mixed with glass beads of sizes 425 to 600 μm and 710 to 1,180 μm and 1 ml of 85% acetonitrile (ACN). The mixture was shaken in a FastPrep (FP120; Bio 101 Savant) cell disrupter twice at speed 5.0 for 40 s and then centrifuged at 10,000 × g for 3 min. The supernatant was passed sequentially through two solid-phase extraction cartridges (StrataX Polymeric Sorbent, 30 mg; particle size, 33 μm; pore size, 85 Å; Phenomenex, Torrance, Calif.) equilibrated with 1 ml of 85% ACN and evaporated in a 100-Pa vacuum. The residue was dissolved into 200 μl of 15% ACN and passed through a 0.2-μm-pore-size filter (GHP Acrodisc; Pall Galman Laboratory, Ann Harbor, Mass.).
ELISA.
Microcystins from the 1,000-fold dilution of the whole-cell extract were screened with the competitive enzyme-linked immunosorbent assay (ELISA) by using a microcystins plate kit (Strategic Diagnostics, Inc., Newark, Del.) and detected with a microplate optical reader (iEMS Reader MF; Labsystems, Helsinki, Finland).
Protein phosphatase inhibition assay.
To test the inhibition of the alpha-isoform of type 1 protein phosphatase from rabbit skeletal muscle by the Nostoc sp. strain IO-102-I, a colorimetric inhibition assay (2) was used with minor modifications as follows. First, 10 μl of the whole-cell extract was diluted with distilled H2O to 10-, 100-, or 1,000-fold and added to the phosphatase inhibition assay. A total of 5 U of the PP1 enzyme/ml expressed in Escherichia coli (New England Biolabs, Inc., Beverly, Mass.) was dissolved in enzyme buffer. The substrate concentration was 15 mM, and incubation of the reaction before optical detection (iEMS Reader MF; Labsystems, Helsinki, Finland) was extended to 40 min.
Microcystin structures.
Whole-cell extracts were analyzed by high-pressure LC (HPLC) by injecting 10 μl of extract into the HP 1100 series modular HPLC system (Agilent Technologies, Palo Alto, Calif.) equipped with a diode array detector. A Zorbax SB-C18 column (2.1 by 50 mm; particle diameter of the stationary phase, 3.5 μm; Agilent Technologies) and a mobile phase gradient of 18% (0 min) to 20.5% (35 min) ACN-10 mM ammonium formate at a flow rate of 0.3 ml min−1 at 40°C were used. Microcystins were distinguished from other peptides based on their characteristic UV maximum absorbance of 238 nm. An ion-trap Esquire mass spectrometer (Bruker Daltonics, Bremen, Germany) directly connected to the liquid chromatograph was used to identify microcystins. Electrospray ionization was performed in positive ion mode. The capillary voltage was set to 3,270 V, the capillary exit offset was 1,300 V, the skimmer 1 potential was 58 V, and the trap drive value was 62. The nebulizer gas (N2) pressure was 40 lb/in2 (40 lb/in2 = 275,792 Pa), the drying gas (N2) flow rate was 9 liters min−1, and the drying temperature was 365°C. Spectra were recorded at a scan range of 100 to 1,500 m/z and a scanning rate of 13,000 m/z s−1. The fragmentation amplitude range was 0.6 to 1.0 in tandem MS (MS/MS) mode.
DNA amplification and sequencing.
DNA was extracted from Nostoc sp. strain IO-102-I by using the DNEasy plant kit (Qiagen, Hilden, Germany). To resolve the relationship between the microcystin synthetase genes from Nostoc sp. strain IO-102-I and other microcystin synthetase gene clusters, we amplified and sequenced the 16S rRNA gene (35) and portions of the mcyA (18), mcyD, mcyE, and rpoC1 genes as previously described (35). It was not possible to sequence the mcyA PCR product from Nostoc sp. strain IO-102-I directly due to the coamplification of nonspecific fragments. Instead, a band of the expected length was cut out from a gel, purified, and cloned by using the TOPO cloning kit (Invitrogen, Groningen, The Netherlands). Three cloned mcyA gene fragments were sequenced with two primers anchored in the pCR2.1-TOPO vector: M13F (−20) and M13R. The amplified 16S rRNA region was sequenced by using sets of internal primers (14). PCR products were cycle sequenced by using a BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) and analyzed on an ABI Prism 310 genetic analyzer (Applied Biosystems).
Phylogenetic DNA sequence analyses.
The 16S rRNA sequences, except for that from the Nostoc sp. strain IO-102-I, which was sequenced for the present study, were obtained from GenBank at the National Center for Biotechnology Information (NCBI). The 16S rRNA gene sequence alignment used to construct the tree in Fig. 2 contained 1,388 characters. The 16S rRNA and the mcyA, mcyD, mcyE, and rpoC1 gene regions for trees in Fig. 3 contained 1447, 250, 780, 769, and 682 characters, respectively. The best-fitting nucleotide substitution model was determined by testing 56 different models (33) for the 1,388-character 16S rRNA sequence alignment in PAUP 4.0b10. The model was chosen with the Modeltest 3.06 program by using the Akaike information criterion (32). The general time-reversible model was used for every data set as the best-fitting nucleotide substitution model in Bayesian phylogenetic inference applied with MrBayes 3.0b4 (19, 20). MrBayes implements a Markov Chain Monte Carlo method that runs many chains simultaneously. Four chains were performed for a million generations; every hundredth was sampled, generating a total of 10,000 trees, with each run in two separate runs with a random starting tree. Unstable initial burn-in cycles were removed (10%). A majority rule consensus tree from the stationary phase was viewed with TreeView 1.6.6 (30).
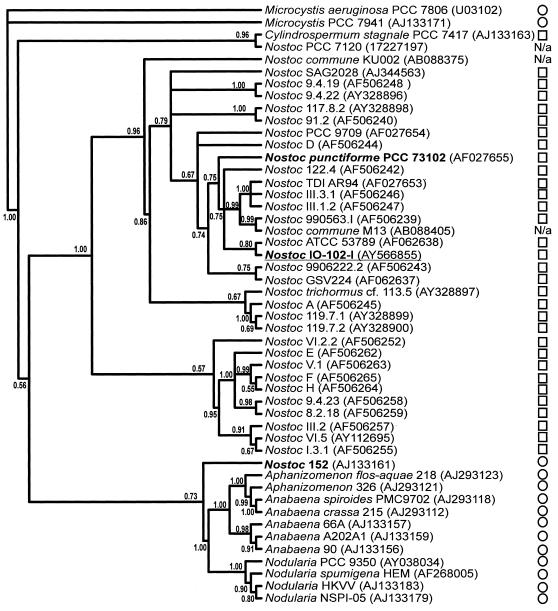
FIG. 2.
Phylogenetic position of the Nostoc sp. strain IO-102-I. A Bayesian majority rule consensus tree inferred from 16S rRNA gene sequences shows the phylogenetic position of Nostoc sp. strain IO-102-I, which falls among symbiotic Nostoc strains. Nostoc sp. strain IO-102-I is distantly related to Nostoc sp. strain 152, the closest known microcystin producer. Bayesian posterior probabilities (≤1.0) are given at the nodes. The sequence accession number is in brackets after the strain code. ○, Aquatic; □, terrestrial; N/a, datum not available.
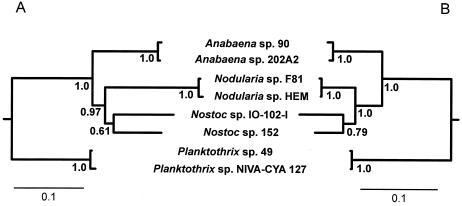
FIG. 3.
Congruence between the housekeeping genes 16S rRNA and rpoC1 and microcystin synthetase genes mcyA, mcyD, and mcyE. (A and B) Bayesian majority rule consensus tree inferred from mcyA, mcyD, and mcyE gene sequences (A) and from 16S rRNA and rpoC1 gene sequences (B). The outgroup (not shown) sequences are from Microcystis viridis strain NIES 102 and M. aeruginosa strain PCC 7806. Bayesian posterior probabilities (≤1.0) are given at the nodes.
BLAST search.
Sequence similarity searches with the mcyA, mcyB, mcyE, and mcyD genes were done with BLAST through the website of the NCBI (1).
Sequence accession numbers.
New DNA sequences from Nostoc sp. strain IO-102-I were deposited in GenBank under accession numbers AY566855 (16S rRNA), AY566856 (mcyA), AY566857 (mcyD), AY566858 (mcyE), and AY566859 (rpoC1).
RESULTS
ELISA and PP1 assays.
The immunological ELISA in which polyclonal antibodies are targeted specifically against microcystin gave a positive result for the Nostoc sp. strain IO-102-I whole-cell extract. The colorimetric protein phosphatase inhibition assay showed phosphatase inhibition by all three dilutions of the whole-cell extract. Together, this strongly suggested that Nostoc sp. strain IO-102-I produced microcystins.
Microcystin variants.
The total concentration of microcystin variants in the Nostoc sp. strain IO-102-I was 0.2 μg mg−1 (dry weight). Six microcystin peaks were detected by LC-UV/MS/MS according to their characteristic UV absorbance originating from the conjugated double bond in the Adda structure, the microcystin characteristic protonated molecular ions (M+H+), and fragment ions from the M+H+. In the LC-UV/MS analysis there were one major and five minor peaks of microcystins with molecular masses of 1,022, 980, 1,008, 1,022, 1,036, and 1,076 Da, respectively (Tables 1 and 2) . The major microcystin peak accounted for ∼80% of the total microcystins, and the minor microcystin peaks comprised 1 to 10% each of the total microcystins (2).
TABLE 1.
Assignment of main ions from microcystin peaks of Nostoc sp. strain IO-102-I in LC-MS/MS analysisa
| Fragment of [M + H]+ ion | Microcystin ionb of:
|
|||||
|---|---|---|---|---|---|---|
| [DMAdda5] MC-LR | [ADMAdda5] MC-XR | [ADMAdda5] MC-XR | [ADMAdda5] MC-LR | [D-Asp3, ADMAdda5] MC-LR | [ADMAdda5] MC-XR | |
| M + H | 981 | 1077 | 1023 | 1023 | 1009 | 1037 |
| M + H − NH3 | 964 | 1060 | 1006 | 1006 | 992 | 1020 |
| M + H − H2O | 963 | 1059 | 1005 | 1005 | 991 | 1019 |
| M + H − CO | 953 | 1049 | 995 | 995 | 981 | 1009 |
| M + H − COOH | 936 | 1032 | 978 | 978 | 964 | 992 |
| M + H − CH3COOH | 1017 | 963 | 963 | 949 | 977 | |
| Ala-Leu/X-(Me)Asp-Arg + H | 470 | 524 | 470 | 470 | 456 | 484 |
| Ala-Leu/X-(Me)Asp-Arg-(A)DMAdda + H | 769 | 865 | 811 | 811 | 797 | 825 |
| Leu/X-(Me)Asp-Arg + H | 399 | 453 | 399 | 399* | ||
| Leu/X-(Me)Asp-Arg + H − NH3 | 382 | 436 | 382 | 382 | 368 | 396 |
| (Me)Asp-Arg-(A)DMAdda-Glu-Mdha − CO + H | 769 | 811 | 811 | 797 | ||
| (Me)Asp-Arg-(A)DMAdda-Glu + H | 714 | 756 | 756 | 756 | 742 | 756 |
| (Me)Asp-Arg-NH3 + 2H | 303 | 303 | 289 | 303 | ||
| (Me)Asp-Arg + H | 286 | 286 | 286 | 286 | ||
| Arg-(A)DMAdda-Glu-Mdha-Ala-Leu/X + H | 852 | 948 | 894 | 894 | 894 | 908 |
| Arg-(A)DMAdda-Glu-Mdha-Ala-Leu/X − CO + H | 920 | 866 | 866 | 866 | 880 | |
| Arg-(A)DMAdda-Glu-Mdha-Ala + H | 781 | 781 | 781 | 781 | 781 | |
| Arg-(A)DMAdda-Glu-Mdha + H | 668 | 710 | 710 | 710 | 710 | 710 |
| Arg-(A)DMAdda-Glu-Mdha − CO + H | 640 | 682 | 682 | 682 | ||
| Arg-(A)DMAdda-Glu + H − NH3 | 568 | 610 | 610 | 610† | 610 | 610 |
| Arg-(A)DMAdda-Glu − CO + H | 557 | 599 | 599 | 599 | 599 | 599 |
| Arg-(A)DMAdda-Glu + H | 585 | 627 | 627 | 627 | 627 | 627 |
| Arg-(A)DMAdda-Glu − COOH + H | 540 | 582 | 582 | 582† | 582 | 582 |
| Arg-(A)DMAdda-Glu − CH3COOH + H | 525 | 567 | 567 | 567† | 567 | 567 |
| Arg-(A)DMAdda + H | 456 | 498 | 498 | 498 | 498 | 498 |
| Arg-(A)DMAdda + H − NH3 | 439 | 481 | 481 | 481† | 481 | 481 |
| Arg + H | 157 | 157* | ||||
| (A)DMAdda-Glu-Mdha-Ala-Leu/X-(Me)Asp + H | 825 | 921 | 867 | 867 | 853 | 881 |
| (A)DMAdda-Glu-Mdha-Ala-Leu/X + H | 696 | 792 | 738 | 738 | 738 | 752 |
| (A)DMAdda-Glu-Mdha-Ala + H | 583 | 625 | 625 | 625 | 625 | |
| (A)DMAdda + H | 342† | |||||
| (A)DMAdda + H − CH3COOH − NH3 | 265 | 265† | ||||
| Glu-Mdha-Ala-Leu/X + H | 397 | 397 | 411 | |||
| Glu-Mdha + H | 213 | 213 | 213* | |||
| Mdha-Ala-Leu/X-(Me)Asp-Arg-(A)DMAdda-NH3 + 2H | 911 | 911 | 897 | 925 | ||
| Mdha-Ala-Leu/X-(Me)Asp-Arg-(A)DMAddA + H | 852 | 948 | 894 | 894 | 880 | 908 |
| Mdha-Ala-Leu/X-(Me)Asp-Arg-(A)DMAddA − CO + H | 824 | 920 | 866 | 866 | 852 | 880 |
| Mdha-Ala-Leu-(Me)Asp-Arg-(A)DMAddA + H − CH3COOH | 792 | 834 | 834 | |||
| Mdha-Ala-Leu/X-(Me)Asp-Arg-NH3 + 2H | 570 | 624 | 570 | 570 | 556 | 584 |
| Mdha-Ala-Leu/X-(Me)Asp-Arg + H | 553 | 607 | 553 | 553 | 539 | 567 |
| Mdha-Ala-Leu/X-(Me)Asp-Arg − CO + H | 525 | 579 | 525 | 525 | 511 | 539 |
| Mdha-Ala-Leu/X-(Me)Asp + H | 397 | 397 | 397* | 383 | 411 | |
| Mdha-Ala-Leu-NH3 + 2H | 285 | 285 | 285* | 285 | ||
| Mdha-Ala-Leu + H | 268 | 268 | 268* | |||
In MS/MS fragmentation of the microcystins the protonated parent ion was [M + H]+. Most of the fragments were identified according to the method of Yuan et al. (47). The diagnostic ion of [ADMAdda5] is m/z 627 and of LR-residue is m/z 553. The most intensive peaks are in boldface.
X, unidentified amino acid in position 2; MC, microcystin; *, assignment confirmed from the fragmentation of the ion 553; †, assignment confirmed from the fragmentation of the ion 627
TABLE 2.
Microcystin variants from Nostoc sp. strains 152 and IO-102-Ia
| Microcystin variant | [M + H]+ (DA) | Rt (min) | Relative amt (%) in Nostoc strains:
|
Reference | |
|---|---|---|---|---|---|
| 152 | IO-102-I | ||||
| [DMAdda5]microcystin-LR | 981 | 2.8 | <0.5 | ∼2 | 42 |
| [ADMAdda5]microcystin-XR | 1,077 | 10.4 | 0 | <10 | |
| [ADMAdda5,MeSer7]microcystin-LR | 1,041 | 13.2 | <0.5 | ND | 42 |
| [ADMAdda5]microcystin-XR | 1,023 | 13.7 | 3 | <10 | |
| [d-Ser1,ADMAdda5]microcystin-LR | 1,039 | 13.8 | <0.5 | ND | 42 |
| [ADMAdda5]microcystin-LR | 1,023 | 16.1 | 35 | 80 | 42 |
| [ADMAdda5]microcystin-XHar | 1,037 | 17.1 | 3 | ND | |
| [ADMAdda5]microcystin-LHar | 1,037 | 19.6 | 35 | ND | 42 |
| [ADMAdda5]microcystin-XR | 1,009 | 23.7 | <0.5 | ND | |
| [ADMAdda5]microcystin-XR | 1,037 | 25.0 | 2 | ∼2 | |
| [d-Asp3,ADMAdda5]microcystin-LR | 1,009 | 25.3 | 10 | ∼2 | 42 |
| [ADMAdda5]microcystin-XHar | 1,023 | 27.7 | 1 | ND | |
| [d-Asp3,ADMAdda5]microcystin-LHar | 1,023 | 29.4 | 10 | ND | 42 |
Protonated molecular mass ion [M + H]+, retention time (Rt), and the relative amounts of the variants detected by LC-UV/MS are given. ND, not detected.
The molecular mass of the major peak, at 1,022 Da, was identical to the microcystin variants [ADMAdda5]microcystin-LR and [d-Asp3, ADMAdda5]microcystin-LHar (39). In LC-UV analysis the retention times of the microcystin variant [ADMAdda5]microcystin-LR from Nostoc sp. strain 152 and the microcystin peak at 1,022 Da from Nostoc sp. strain IO-102-I were identical (±1/10 s) in 12 sequential injections. In LC-MS/MS analyses the fragmentation patterns of the [ADMAdda5]microcystin-LR from Nostoc sp. strain 152 and the microcystin of 1,022 Da from Nostoc sp. strain IO-102-I were also in good accordance. The MS/MS spectra of m/z (ratio of molecular mass and ion charge) 1,023 ion contained most of the fragments reported previously from [ADMAdda5]microcystin-LR from Nostoc sp. strain 152 (47), including the important diagnostic ions at m/z 553 [Mdha-Ala-Leu-MeAsp-Arg + H+] and m/z 627 [Arg-ADMAdda-Glu + H+] (Table 2). The Adda-specific fragment ion m/z 135 was absent.
Another microcystin (<10%) with a molecular mass of 1,022 was also present and shared identical characteristic ions with [ADMAdda5]microcystin-LR but clearly had a smaller retention time (Table 2). The smaller retention time of this microcystin variant was due to its weaker hydrophobicity compared to the major variant 1022. The full structure of this previously unreported [ADMAdda5]microcystin variant remained unresolved.
A third microcystin (∼2%) had a mass (980 Da) identical to those of the [d-Asp3], [DMAdda5], and [Dha7] variants of microcystin-LR (39). The microcystin diagnostic ion m/z 553, [Mdha-Ala-Leu-MeAsp-Arg + H+], and the DMAdda structure specific ion m/z 585, [Arg-DMAdda-Glu + H+], were the most abundant peaks in the spectrum (Table 1). The Adda specific ion m/z 135 was missing from the spectrum. Thus, LC-MS/MS fragmentation analysis suggested that this variant was [DMAdda5]microcystin-LR.
A fourth microcystin (∼2%) had a mass (1,008 Da) identical to that of microcystin-HilR, [D-Asp3, ADMAdda5], and [D-Glu(OCH3)6] variants of microcystin-LR (39). The MS/MS spectra of the m/z 1009 ion had a fragment ion pattern similar to that reported previously (47) from the [D-Asp3, ADMAdda5] microcystin-LR, including the important diagnostic ions m/z 539 [Mdha-Ala-Leu-Asp-Arg + H+] and m/z 627 [Arg-ADMAdda-Glu + H+] (Table 1).
The remaining two minor (∼2% and <10%) microcystin variants from Nostoc sp. strain IO-102-I with molecular masses of 1,036 and 1,076 Da fragmented to the prominent ADMAdda diagnostic ion m/z 627 [Arg-ADMAdda-Glu + H+] (47) (Table 1). The diagnostic ion for [Mdha-Ala-Leu-MeAsp-Arg + H+] with an m/z of 553 was not found in these three variants (Table 1). The previously reported variant [ADMAdda5]microcystin-LHar from Nostoc sp. strain 152 had a molecular mass of 1,036 Da (39). However, these two variants had different retention times (Table 2) and different mass fragmentation patterns (47), and therefore these two microcystins do not have an identical structure (Table 2). The diagnostic ion m/z 627 (Table 1) excluded the possibility of the l-Har amino acid that gave the ion m/z 641 in this fragment (47). In this new microcystin 1036, the ions for [MeAsp-Arg-ADMAdda-Glu + H+], [MeAsp-Arg - NH2 + 2H+] and [Arg-ADMAdda-Glu-Mdha-Ala + H+] had the same m/z with the ions from [ADMAdda5]microcystin-LR, but ions from leucine-containing fragments were 14 mass units bigger (Table 1), meaning that most probably microcystin 1036 varied from [ADMAdda5]microcystin-LR in position 2 (Fig. 1). Similarly, in microcystin 1076 the ions for [MeAsp-Arg-ADMAdda-Glu + H+], [MeAsp-Arg + 2H+], and [Arg-ADMAdda-Glu-Mdha-Ala + H+] had the same m/z with the ions from [ADMAdda5]microcystin-LR, but ions from fragments containing position 2 amino acids were 54 mass units bigger (Table 1). Thus, the two partly resolved structures are ADMAdda-XR-type microcystins that contain unidentified amino acid(s), most probably in position 2 (Fig. 1). The molecular mass of 1,076 Da is new (39). The reference Nostoc sp. strain 152 produced more microcystin variants than previously reported (42).
BLAST search.
Nostoc punctiforme PCC 73102, a close relative to Nostoc sp. strain IO-102-I, contains nonribosomal peptide synthetase gene clusters (27). However, BLAST searches indicated that the microcystin synthetase genes were not present in genomes of N. punctiforme PCC 73102. The two more distant relatives, Nostoc sp./Anabaena sp. strain PCC 7120 and A. variabilis ATCC 29413, did not contain the microcystin synthetase genes either.
Phylogenetic analyses.
The 16S rRNA gene tree demonstrated that the two microcystin-producing Nostoc sp. strains 152 and IO-102-I were distantly related (Fig. 2). Among available Nostoc 16S rRNA sequences, the closest relative to Nostoc sp. strain IO-102-I was Nostoc sp. strain ATCC 53789, also isolated from lichen in Scotland (Fig. 2). Nostoc sp. strain IO-102-I formed a monophyletic group with the reference strain of N. punctiforme strain PCC 73102 and several other strains originating from symbioses. N. punctiforme strain PCC 73102 and Nostoc sp. strain IO-102-I had a 98% sequence similarity in the 1,405-nt 16S rRNA region. All of these Nostoc strains shared a recent common ancestor with other terrestrial Nostoc strains from symbiotic associations unlike microcystin-producing, planktonic Nostoc sp. strain 152, which fell outside the main Nostoc clade as a distant relative (Fig. 2). Nostoc sp. strain IO-102-I grouped with Nostoc sp. strain 152 in the tree inferred from the three microcystin synthetase genes (Fig. 3A), indicating that the closest known relative producing microcystin also shared the closest gene cluster with the terrestrial Nostoc sp. strain IO-102-I. The microcystin synthetase gene tree genes resulted in relationships that were well supported at all nodes except for the node uniting Nostoc sp. strains IO-102-I and 152 (Fig. 3A). This was consistent with strain relationships inferred from 16S rRNA gene sequences (Fig. 3B and Fig. 2). In the 1,428-nt 16S rRNA gene region there was a considerable 6% sequence difference between Nostoc sp. strains 152 and IO-102-I.
DISCUSSION
A terrestrial cyanobacterium Nostoc sp. strain IO-102-I isolated from a lichen association produced six different microcystins, of which the prominent variant was the rare and highly hepatotoxic [ADMAdda5]microcystin-LR. This variant is also known from Nostoc sp. strain 152 (40). We assigned a structure of [DMAdda5]microcystin-LR and [d-Asp3,ADMAdda5]microcystin-LR from the five minor microcystin variants. We could assign a partial structure of [ADMAdda5]-XR to the three previously unreported microcystins. ADMAdda and arginine (R) could be identified from these three partly unresolved microcystins by the ion fragmentation, but the rest of the structure could not be assigned because either the ion fragmentation did not match reported masses or the molecular mass of the variant (1,076 Da) was different. Theoretically, the variable amino acid could be other than the one we assigned if they contain amino acids with identical masses. Nostoc sp. strain IO-102-I and the reference strain Nostoc sp. strain 152 produced five identical variants; three had been assigned earlier (42), and two were found here. Nostoc sp. strain 152 strain produced all of the seven microcystins earlier reported (42) and, in addition, five unreported microcystins. One possible reason for differences in the number of variants produced by Nostoc sp. strain 152 may be that the cultivation time was twice as long in the present study (41). The third known microcystin producer from the genus Nostoc, Nostoc sp. strain DUN901, contained three microcystin variants (3) not found in Nostoc sp. strain 152 (42). The Nostoc sp. strains DUN901, IO-102-I, and 152 synthesized only the rare ADMAdda and DMAdda families of microcystins. Interestingly, ADMAdda is found only in the Nostoc strains (3, 40, 42) and the single Planktothrix agardhii strain PH-123 (24). DMAdda is found in Nostoc sp. strain 152 and a single Microcystis bloom sample (39).
The previously described microcystin variants from Nostoc sp. strain 152 (42) that Nostoc sp. strain IO-102-I also produced were hepatotoxic. The toxicity (as indicated by the 50% lethal dose) in a mouse bioassay of the dominating variant [ADMAdda5]microcystin-LR was one of highest among microcystins (39), but the total concentration of microcystins in Nostoc sp. strain IO-102-I was 10-fold smaller than in Nostoc sp. strain 152.
The finding of microcystins from terrestrial Nostoc sp. strain IO-102-I broadens the range of ecological habitats in which microcystin production has been documented. All known producers, with the exception of Hapalosiphon hibernicus BZ-3-1 originating from soil or fresh-water mud (34), are aquatic and present in freshwater and brackish water bodies (39). Thus, to our knowledge Nostoc sp. strain IO-102-I is the third Nostoc and the second terrestrial strain capable of producing microcystin. One reason for the rarity of microcystin production in the terrestrial environment may be its scattered distribution, shown here within the terrestrial Nostoc lineage. Since the clade in the 16S rRNA tree that contained Nostoc sp. strain IO-102-I also contained strains that did not produce microcystins (25), the distribution of microcystin production is not spread evenly throughout the current terrestrial Nostoc lineage. In the terrestrial Nostoc strains microcystin production was not spread throughout the phylogenetic lineage. Scattered distribution of microcystin production has also been found with Microcystis (25, 45) and Planktothrix (25) spp., but in Nostoc strains the production is likely to be even more scattered. The scattered distribution of microcystin production in cyanobacteria shows that the ability to produce secondary metabolites does not necessarily depend on how closely related the strains are. Phylogenetic congruence between the microcystin synthetase genes (mcyADE) and the housekeeping genes (16S rRNA, rpoC1) suggests that both Nostoc sp. strains IO-102-I and 152 inherited the microcystin synthetase gene cluster from a common ancestor. Since it is not known why cyanobacteria produce microcystins, it is difficult to speculate how probable the loss of the gene cluster is and if loss of the cluster is linked to habitat. The sequence differences from both the housekeeping and microcystin synthetase genes indicate that the two Nostoc strains, 152 and IO-102-I, diverged a long time ago or rapidly at some point. Nostoc sp. strain IO-102-I is in a same evolutionary lineage with symbiotic Nostoc sp. strain ATCC 53789 and N. punctiforme strain PCC 73102 with a very high sequence similarity, suggesting that it most likely belongs to that species. Another reason why microcystin production in a terrestrial environment has not been detected may be that terrestrial cyanobacteria have not been tested for microcystin production as frequently as cyanobacteria from aquatic habitats.
It is interesting that microcystins, which inhibit eukaryotic protein phosphatases 1 and 2A, were found in lichen symbiosis. It is possible that microcystin does not control fungal or algal growth directly in lichen symbiosis and therefore microcystin could be produced by cyanobacteria in symbiosis. Microcystins in vertebrates are transported across the cell membrane by bile acid carrier proteins, which are found only in hepatocytes in liver (6). Therefore, fungi might be incapable of transporting microcystins into its cells. Microcystin-LR is known to inhibit the photosynthesis of green algae (38). However, it affected algal growth at concentrations not encountered in their natural environment and did not affect the growth of fungi even at high doses (38, 39).
It is now known that the terrestrial Nostoc sp. strain IO-102-I and Hapalosiphon hibernicus strain BZ-3-1 (34) produce several toxic microcystins. Therefore, we need to consider that also terrestrial populations of cyanobacteria may represent a potential health hazard. Although no acute intoxications attributed to microcystins have been reported from areas in which Nostoc is used as human dietary supplement (13, 15), there may be a risk because microcystins act as liver tumor promoters at sublethal concentrates (28). In China, increased liver cancer in the human population has been linked to drinking water containing toxigenic cyanobacteria (4). Therefore, since there might be hazardous substances in subacute concentrations, the content of metabolites in consumed cyanobacteria should be studied. Reindeer feed on cyanobacterial lichens, e.g., on species Stereocaulon paschale, that are poorly digestible (43) but avoid cyanobacterial lichens in heavily grazed areas (10). Could the preference for green-algal lichens be due to the presence of toxic cyanobacterial secondary metabolites?
It would be worth studying the capacity of Nostoc strains to produce secondary metabolites. Sixteen metabolites with bioactivities, such as anticancer, antibiotic, antifungal, and antiviral properties, have already been found in seven Nostoc strains (5). For example, a synthetic analog derived from cryptophycin, a cytotoxic depsipeptide, isolated originally from a terrestrial Nostoc GSV224 strain, was chosen for clinical evaluation because of its antimitotic and antitumor effects (31).
To conclude, highly toxic microcystins were found in a terrestrial Nostoc strain. The toxicity of cyanobacteria therefore cannot be predicted from their habitat or from the strain identity because even closely related terrestrial cyanobacteria may vary in the production of secondary metabolites.
Acknowledgments
We thank Anu Surakka for testing protein phosphatase inhibition and Anne Rantala for 16S rRNA PCR.
This study was financially supported by grants from the Academy of Finland to K.S. (projects 53305, 201576, and 02162) and by a young scientist's grant from the Emil Aaltonen Foundation to I.O.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, J. S., and W. W. Carmichael. 1994. Use of a colorimetric protein phosphatase inhibition assay and enzyme linked immunosorbent assay for the study of microcystins and nodularins. Toxicon 32:1495-1507. [DOI] [PubMed] [Google Scholar]
- 3.Beattie, K. A., K. Kaya, T. Sano, and G. Codd. 1998. Three dehydrobutyrine-containing microcystins from Nostoc. Phytochemistry 47:1289-1292. [DOI] [PubMed] [Google Scholar]
- 4.Briand, J. F., S. Jacquet, C. Bernard, and J. F. Humbert. 2003. Health hazards for terrestrial vertebrates from toxic cyanobacteria in surface water ecosystems. Vet. Res. 34:361-377. [DOI] [PubMed] [Google Scholar]
- 5.Burja, A. M., B. Banaigs, E. Abou-Mansour, J. G. Burgess, and P. C. Wright. 2001. Marine cyanobacteria: a prolific source of natural products. Tetrahedron 57:9347-9377. [Google Scholar]
- 6.Carmichael, W. W. 1994. Toxins of cyanobacteria. Sci. Am. 270:78-86. [DOI] [PubMed] [Google Scholar]
- 7.Christiansen, G., J. Fastner, M. Erhard, T. Börner, and E. Dittmann. 2003. Microcystin biosynthesis in Planktothrix: genes, evolution and manipulation. J. Bacteriol. 185:564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson, F. F. 1959. Poisoning of wild and domestic animals by a toxic waterbloom of Nostoc rivulare Küertz. J. Am. Water Works Assoc. 51:1277-1287. [Google Scholar]
- 9.DeMott, W. R., Q. X. Zhang, and W. W. Carmichael. 1991. Effects of toxic cyanobacteria and purified toxins on the survival and feeding of a copepod and three species of Daphnia. Limnol. Oceanogr. 36:1346-1357. [Google Scholar]
- 10.den Herder, M., M. Kytöviita, and P. Niemelä. 2003. Growth of reindeer lichens and effects of reindeer grazing on ground cover vegetation in a Scots pine forest and a subarctic heathland in Finnish Lapland. Ecography 26:3-12. [Google Scholar]
- 11.Dittmann, E., M. Erhard, M. Kaebernick, C. Scheler, B. A. Neilan, H. von Döhren, and T. Börner. 2001. Altered expression of two light-dependent genes in a microcystin-lacking mutant of Microcystis aeruginosa PCC 7806. Microbiology 147:3113-3119. [DOI] [PubMed] [Google Scholar]
- 12.Dittmann, E., B. A. Neilan, M. Erhard, H. von Döhren, and T. Börner. 1997. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol. Microbiol. 26:779-787. [DOI] [PubMed] [Google Scholar]
- 13.Dodds, W. K., D. A. Gudder, and D. Mollenhauer. 1995. The ecology of Nostoc. J. Phycol. 31:2-18. [Google Scholar]
- 14.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S rRNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, K. 1998. Chinese studies on the edible blue-green alga, Nostoc flagelliforme: a review. J. Appl. Phycol. 10:37-49. [Google Scholar]
- 16.Goldberg, J., H. B. Huang, Y. G. Kwon, P. Greengard, A. C. Nairn, and J. Kuriyan. 1995. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature 376:745-753. [DOI] [PubMed] [Google Scholar]
- 17.Harada, K.-I., K. Matsuura, M. Suzuki, M. F. Watanabe, S. Oishi, A. M. Dahlem, V. R. Beasley, and W. W. Carmichael. 1990. Isolation and characterization of the minor components associated with microcystins LR and RR in the cyanobacterium (blue-green algae). Toxicon 28:55-64. [DOI] [PubMed] [Google Scholar]
- 18.Hisbergues, M., G. Christiansen, L. Rouhiainen, K. Sivonen, and T. Börner. 2003. PCR-based identification of microcystin-producing genotypes of different cyanobacterial genera. Arch. Microbiol. 180:402-410. [DOI] [PubMed] [Google Scholar]
- 19.Huelsenbeck, J. P., and F. Ronquist. 2001. MrBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 20.Huelsenbeck, J. P., F. Ronquist, R. Nielsen, and J. P. Bollback. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310-2314. [DOI] [PubMed] [Google Scholar]
- 21.Jang, M. H., K. Ha, G. J. Joo, and N. Takamura. 2003. Toxin production of cyanobacteria is increased by exposure to zooplankton. Freshwat. Biol. 48:1540-1550. [Google Scholar]
- 22.Kotai, J. 1972. Instructions for preparation of modified nutrient solution Z8 for algae, p. 1-5. Norwegian Institute for Water Research, Oslo, Norway.
- 23.Kuiper-Goodman, T., I. Falconer, and J. Fitzgerald. 1999. Human health aspects, p. 113-153. In I. Chorus and J. Bartram (ed.), Toxic cyanobacteria in water. E&FN Spon, London, Great Britain.
- 24.Laub, J., P. Henriksen, S. M. Brittain, J. Wang, W. W. Carmichael, K. L. Rinehart, and Ø. Moestrup. 2002. [ADMAdda5]-microcystins in Planktothrix agardhii strain PH-123 (cyanobacteria): importance for monitoring of microcystins in the environment. Environ. Toxicol. 17:351-357. [DOI] [PubMed] [Google Scholar]
- 25.Lyra, C., S. Suomalainen, M. Gugger, C. Vezie, P. Sundman, L. Paulin, and K. Sivonen. 2001. Molecular characterization of planktic cyanobacteria of Anabaena, Aphanizomenon, Microcystis, and Planktothrix genera. Int. J. Syst. Evol. Microbiol. 51:513-526. [DOI] [PubMed] [Google Scholar]
- 26.MacKintosh, C., K. A. Beattie, S. Klumpp, P. Cohen, and G. A. Codd. 1990. Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants. Fed. Eur. Biochem. Soc. Lett. 264:187-192. [DOI] [PubMed] [Google Scholar]
- 27.Meeks, J. C., J. Elhai, T. Thiel, M. Potts, F. Larimer, J. Lamerdin, P. Predki, and R. Atlas. 2001. An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Photosynth. Res. 70:85-106. [DOI] [PubMed] [Google Scholar]
- 28.Nishiwaki-Matsushima, R., T. Ohta, S. Nishiwaki, M. Suganuma, K. Kohyama, T. Ishikawa, W. W. Carmichael, and H. Fujiki. 1992. Liver tumor promotion by the cyanobacterial cyclic peptide toxin microcystin-LR. J. Cancer Res. Clin. Oncol. 118:420-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishizawa, T., M. Asayama, K. Fujii, K. Harada, and M. Shirai. 1999. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J. Biochem. 126:520-529. [DOI] [PubMed] [Google Scholar]
- 30.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 31.Panda, D., K. DeLuca, D. Williams, M. A. Jordan, and L. Wilson. 1998. Antiproliferative mechanism of action of cryptophycin-52: kinetic stabilization of microtubule dynamics by high-affinity binding to microtubule ends. Proc. Natl. Acad. Sci. USA 95:9313-9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 33.Posada, D., and K. A. Crandall. 2001. Selecting the best-fit model of nucleotide substitution. Syst. Biol. 50:580-601. [PubMed] [Google Scholar]
- 34.Prinsep, M. R., F. R. Caplan, R. E. Moore, G. M. L. Patterson, R. E. Honkanen, and A. L. Boynton. 1992. Microcystin-LA from a blue-green alga belonging to the Stigonematales. Phytochemistry 31:1247-1248. [Google Scholar]
- 35.Rantala, A., D. P. Fewer, M. Hisbergues, L. Rouhiainen, J. Vaitomaa, T. Börner, and K. Sivonen. 2004. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. USA 101:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ressom, R., F. S. Soong, J. Fitzgerald, L. Turczynowicz, O. El Saadi, D. Roder, T. Maynard, and I. Falconer. 1994. Health effects of toxic cyanobacteria (blue-green algae). Looking Glass Press/Commonwealth Department of Human Services and Health, Canberra, Australia.
- 37.Rouhiainen, L., T. Vakkilainen, B. Lumbye Siemer, W. Buikema, R. Haselkorn, and K. Sivonen. 2004. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Appl. Environ. Microbiol. 70:686-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh, D. P., M. B. Tyagi, A. Kumar, J. K. Thakur, and A. Kumar. 2001. Antialgal activity of a hepatotoxin-producing cyanobacterium, Microcystis aeruginosa. World J. Microb. Biot. 17:15-22. [Google Scholar]
- 39.Sivonen, K., and G. Jones. 1999. Cyanobacterial toxins, p. 41-111. In I. Chorus and J. Bartram (ed.), Toxic cyanobacteria in water. E&FN Spon, London, Great Britain.
- 40.Sivonen, K., W. W. Carmichael, M. Namikoshi, K. L. Rinehart, A. M. Dahlem, and S. I. Niemelä. 1990. Isolation and characterization of hepatotoxic microcystin homologs from the filamentous fresh-water cyanobacterium Nostoc sp. strain-152. Appl. Environ. Microbiol. 56:2650-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sivonen, K., K. Kononen, W. W. Carmichael, A. M. Dahlem, K. L. Rinehart, J. Kiviranta, and S. I. Niemelä. 1989. Occurrence of the hepatotoxic cyanobacterium Nodularia spumigena in the Baltic Sea and structure of the toxin. Appl. Environ. Microbiol. 55:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sivonen, K., M. Namikoshi, W. R. Evans, M. Fardig, W. W. Carmichael, and K. L. Rinehart. 1992. Three new microcystins, cyclic heptapeptide hepatotoxins, from Nostoc sp. strain 152. Chem. Res. Toxicol. 5:464-469. [DOI] [PubMed] [Google Scholar]
- 43.Storeheier, P. V., S. D. Mathiesen, N. J. C. Tyler, and M. A. Olsen. 2002. Nutritive value of terricolous lichens for reindeer in winter. Lichenologist 34:247-257. [Google Scholar]
- 44.Tillet, D., E. Dittmann, M. Erhard, H. von Döhren, T. Börner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC 7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7:753-764. [DOI] [PubMed] [Google Scholar]
- 45.Tillet, D., D. L. Parker, and B. A. Neilan. 2001. Detection of toxigenicity by a probe for the microcystin synthetase a gene (mcyA) of the cyanobacterial genus Microcystis: comparison of toxicities with 16S rRNA and phycocyanin operon (phycocyanin intergenic spacer) phylogenies. Appl. Environ. Microbiol. 67:2810-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Utkilen, H., and N. Gjølme. 1995. Iron-stimulated toxin production in Microcystis aeruginosa. Appl. Environ. Microbiol. 61:797-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan, M., M. Namikoshi, A. Otsuki, and K. Sivonen. 1998. Effect of amino acid side-chain on fragmentation of cyclic peptide ions: differences of electrospray ionization collision-induced decomposition mass spectra of toxic heptapeptide microcystins containing ADMAdda instead of Adda. Eur. Mass Spectrom. 4:287-298. [Google Scholar]