Abstract
The phylum Verrucomicrobia is increasingly recognized as an environmentally significant group of bacteria, particularly in soil habitats. At least six subdivisions of the Verrucomicrobia are resolved by comparative analysis of 16S rRNA genes, mostly obtained directly from environmental samples. To date, only two of these subdivisions (1 and 4) have characterized pure-culture representatives. We have isolated and characterized the first known pure-culture representative of subdivision 2. Strain Ellin428 is an aerobic heterotrophic bacterium that is able to grow with many of the saccharide components of plant biomass but does not grow with amino acids or organic acids other than pyruvate. Cells are yellow, rod-shaped, nonmotile, and gram-stain negative, and they contain peptidoglycan with direct cross-linkages of the A1γ meso-Dpm type. The isolate grows well at 25°C on a variety of standard biological media, including some used in the routine cultivation of bacteria from soil. The pH range for growth is 4.0 to 7.0. Low levels of menaquinones MK-10 and MK-11 were detected. The major cellular fatty acids are C14:0, a-C15:0, C16:1ω7c, and/or 2OH i-C15:0, and C16:0. The G+C content of the genomic DNA is 61 mol%. We propose a new genus and species, Chthoniobacter flavus gen. nov., sp. nov., with isolate Ellin428 as the type strain, and a new class for the subdivision to which it belongs, Spartobacteria classis nov. Environmental sequences indicate that the class Spartobacteria is largely represented by globally distributed, abundant, and active soil bacteria.
Using genes as phylogenetic markers has allowed microbial ecologists to survey the diversity of microorganisms in different environments without the need to cultivate them in the laboratory (23). The majority of these studies have been carried out with the 16S rRNA gene. One of the major findings from this approach has been that there are many groups of bacteria for which there are very few or no known cultivated representatives (23). This lack of cultivated strains, in turn, means that further investigations into the ecology, function, physiology, and genetics of these novel organisms are difficult, although not impossible (17). Soils in particular are inhabited by an immense diversity of bacterial groups, including order, class, and phylum level groupings that are poorly or not at all represented by cultures (4, 23).
The phylum Verrucomicrobia is one of the primary lineages of the domain Bacteria, with very few cultivated representatives (19, 23, 53). 16S rRNA genes derived from members of this phylum have been detected in soils around the world. Members of the Verrucomicrobia are estimated to comprise between 1.2 and 10.9% of the total bacteria in soil and on average represent 5.0% of all surveyed 16S rRNA genes (1, 2, 8, 10, 12, 15, 22, 30, 34, 36, 37, 39; L. Schoenborn and P. H. Janssen, unpublished data). Some libraries prepared from soil DNA lack genes indicative of members of the phylum Verrucomicrobia, but these are generally from studies with fewer than 70 analyzed sequences (e.g., references 11 and 38) so that smaller populations of verrucomicrobia, at the lower end of the abundance range, may not have been detected. In addition to being abundant, members of this phylum appear to be active members of the soil microbial community, producing 1.0 to 9.8% of the bacterial 16S rRNA in soils (3, 5, 13, 14).
The phylum Verrucomicrobia has been informally divided into five monophyletic subdivisions numbered 1 to 5 by Hugenholtz et al. (23). A sixth subdivision (subdivision 6) was proposed by Vanderkerckhove et al. (52) based on a single full-length 16S rRNA gene sequence cloned from a freshwater habitat, LD19 (58). At present, only three of these subdivisions are recognized by Bergey's Manual of Systematic Bacteriology (16), and they are accorded the rank of family: Verrucomicrobiaceae (subdivision 1), “Opitutaceae” (subdivision 4), and “Xiphinematobacteriaceae” (subdivision 2). We anticipate that these subdivisions will ultimately have to be reclassified to the rank of class to be on a phylogenetic parity with other, better-characterized phyla such as the Proteobacteria and Firmicutes. The proposed class Verrucomicrobiae (16), which we consider to be equivalent to subdivision 1, is named after the type genus represented by the aquatic bacterium Verrucomicrobium spinosum (46) and also contains four species of aquatic bacteria of the genus Prosthecobacter (18, 19). There are also characterized representatives of subdivision 4, such as Opitutus terrae and related strains, isolated from soil (6, 7, 27), and the marine bacteria “Fucophilus fucoidanolyticus” (44) and Alterococcus agarolyticus (47). The latter organism was misclassified as a member of the class Gammaproteobacteria. Only one isolate has been reported for each of the subdivisions 2 and 3, Ellin428 (28) and Ellin5102 (29), respectively, and no isolates have yet been reported for subdivisions 5 or 6.
Molecular ecological surveys suggest that members of subdivisions 2 and 3 are most widespread and abundant in soils. 16S rRNA genes originating from representatives of subdivision 2 make up most to all of the verrucomicrobial 16S rRNA genes detected in PCR-based surveys of soil bacterial communities (1, 2, 8, 10, 12, 15, 22, 30, 34, 36, 37, 39, 53; L. Schoenborn and P. H. Janssen, unpublished data). One such 16S rRNA gene, designated EA25, was estimated to originate from a species with a population size of up to 2 × 108 cells per g of soil (33) and therefore could represent 1 to 10% of all of the bacteria in that soil. The available evidence suggests that members of subdivision 2 constitute a globally distributed, abundant, and active group of soil bacteria. Given the apparent importance of this group, an understanding of their ecology and function would clearly be useful in better understanding soil biology. However, this has to date only been possible by attempting to correlate the abundance of their 16S rRNA with measurable parameters of soil and climate (3). We have recently isolated strain Ellin428 (28), the first known pure culture representative of this important group of soil bacteria, and we describe here the phenotypic properties of this isolate.
MATERIALS AND METHODS
Strain Ellin428 growth conditions.
Strain Ellin428 was isolated from a rye grass and clover pasture soil in Victoria, Australia, in an earlier study (28), and maintained in the laboratory by subculture at ∼3-monthly intervals on solid medium VL55 with 4 mM glucose (see below). Unless noted otherwise, experiments were carried out in liquid medium VL55 at pH 5.5 and 25°C with 2 mM glucose as the growth substrate. Growth experiments were performed in 10-ml aliquots of medium in 30-ml glass vials closed with plastic screw caps and incubated statically. To obtain cells for chemical analyses, 100-ml aliquots of culture were grown statically in 500-ml plastic-capped glass bottles. An inoculum of 1% (vol/vol) from a glucose-grown liquid culture was use to inoculate liquid culture experiments.
Liquid media.
Medium VL55 contained 1.95 g of 2-[N-morpholino]ethanesulfonic acid, 0.2 mmol MgSO4, 0.3 mmol CaCl2, 0.2 mmol (NH4)2HPO4, 1 ml of selenite-tungstate solution (50), and 1 ml of trace element solution SL-10 (55) per liter, and the pH was adjusted to 5.5, unless noted otherwise, with a mixture of 200 mM NaOH plus 100 mM KOH. The medium was sterilized by autoclaving, and the appropriate growth substrate was then added from concentrated stocks that were sterilized by autoclaving, in the case of polymers, or by passing them through Minisart sterile filters (Sartorius AG, Göttingen, Germany) with a pore size of 0.22 μm. Prior to use, 1 ml of vitamin solution 1 (27) and 3 ml of vitamin solution 2 (27) were added per liter of medium.
To vary the pH, medium VL55 was prepared with different additions of the NaOH-KOH mixture to obtain pH values of 4.0 to 6.5. To obtain pH values of 6.5 to 8.0, the 2-[N-morpholino]ethanesulfonic acid in the medium was replaced with 2.09 g of 3-[N-morpholino]propanesulfonic acid, and different additions of the NaOH-KOH mixture made. In all cases, the final medium pH was measured after sterilization and all additions had been made and the medium had been equilibrated at 25°C for several weeks.
Additional NaCl was added to medium VL55 as required. Sodium nitrate was added to the growth medium as required from a separately sterilized (autoclaved) 1 M stock solution.
Cultures were tested for growth under anaerobic conditions in anaerobic jars by using GasPak Plus anaerobic system envelopes (BD Diagnostic Systems, Sparks, Md.).
Solid media.
Nutrient agar, 1/100 nutrient agar, Trypticase soy agar, and 1/10 Trypticase soy agar were prepared from dehydrated nutrient broth (BD Diagnostic Systems) and Trypticase soy broth (Oxoid, Ltd., Basingstoke, United Kingdom) powders with the addition of 15 g of bacteriological agar no. 1 (Oxoid, Ltd.) per liter of medium. R2A Agar (BD Diagnostic Systems) was prepared as directed by the manufacturer. Winogradsky's salt solution agar was prepared as described by Winding et al. (56). Cold-extracted soil extract agar was prepared as described by Olsen and Bakken (41) with the nutrient solution at 1/10 of the standard concentration and with soil collected from the site from which strain Ellin428 had been isolated (28).
To make plates of medium VL55 with glucose, double-strength medium VL55 with selenite-tungstate solution and trace element solution SL-10 was prepared, autoclaved at 121°C for 20 min, and cooled to 56°C. Glucose (8 mM in double-strength medium), 2 ml of vitamin solution 1, and 6 ml of vitamin solution 2 were added per liter of double-strength medium. An equal volume of sterile 3% washed agar (54), autoclaved at 121°C for 15 min and cooled to 56°C, was then added to the medium base and mixed before dispensing the completed medium into sterile petri dishes to give single-strength medium with 4 mM glucose.
Microscopy.
Phase-contrast photomicrographs of immobilized cells (42) were made by using a Diaplan microscope (Ernst Leitz GmbH, Wetzlar, Germany) fitted with a DC200 camera (Leica Microsystems GmbH, Wetzlar, Germany), and images were obtainedd by using DC Viewer software (Leica Microsystems). Wet-mount preparations of live cells were also examined by phase-contrast microscopy. Cells stained with DAPI (4′,6′-diamidino-2-phenylindole) were prepared and viewed by epifluorescence microscopy as described by Janssen et al. (28). Staining for poly-β-hydroxybutyrate and polyphosphate inclusions was carried out as described by Rees et al. (43). Gram staining was performed by using half-strength modified Hucker's crystal violet and triple-strength Gram's iodine, with acetone as the decolorizer and dilute carbol-fuchsin as the counterstain (20). Electron microscopic examination of whole cells was carried out as described by Chin et al. (7), and cell sizes were calculated from the resulting electron micrographs. Cells were prepared for transmission electron microscopy by washing them in medium VL55 (without glucose or vitamins) and then dispersing the cell pellet in 2.5% (vol/vol) glutaraldehyde in unamended medium VL55, followed by standing for 4 h at room temperature. The cells were then rinsed three times (10 min each) in fresh unamended medium VL55 before they were fixed in 1% (wt/vol) osmium tetroxide in unamended medium VL55 for 2 h. The cells were again rinsed three times (10 min each) in fresh unamended medium VL55 before being dehydrated in increasing concentrations of ethanol consisting of 10, 30, 50, 70, 90, 100, and 100% (vol/vol) anhydrous ethanol, for 15 min at each step. After dehydration the cells were infiltrated with increasing concentrations of LR White resin (Theale, Berkshire, United Kingdom) in ethanol consisting of 25, 50, 75, and 100% (vol/vol) resin for 6 h at each step. After a second change of 100% resin, the cells were embedded in fresh resin in gelatin capsules and allowed to sink gently to the bottom to form a loose pellet. The gelatin capsule was capped to exclude air, and the resin was polymerized in an oven at 60°C for 24 h. The embedded cells in the blocks were sectioned with a diamond knife on an Ultracut R microtome (Leica Microsystems), and ultrathin sections (90 nm) were collected onto Formvar-coated 200 mesh hexagonal copper grids. The sections on grids were sequentially stained with saturated uranyl acetate for 15 min and triple lead stain for 10 min (45) and then viewed in a CM120 Biotwin transmission electron microscope (FEI Co., Eindhoven, The Netherlands) at 120 kV.
Extraction and analysis of cellular fatty acids.
Fatty acid methyl esters were analyzed at the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany) after saponification, methylation, and extraction (31) from 40 mg of lyophilized cells. The fatty acid methyl esters mixtures were separated by using Sherlock Microbial Identification System (MIS; Microbial ID, Newark, Del.), which consisted of a model 5980 gas chromatograph (Hewlett-Packard Co., Palo Alto, Calif.) fitted with a 5% phenyl-methyl silicone capillary column (0.2 mm by 25 m), a flame ionization detector, a model 7673A automatic sampler (Hewlett-Packard), and a KAYAK XA computer (Hewlett-Packard). The carrier gas was ultra-high-purity hydrogen at a column head pressure of 60 kPa; the injection volume was 2 μl; the column split ratio was 100:1; the septum purge was 5 ml/min; the column temperature was 170 to 270°C at 5°C/min; the injection port temperature was 250°C; and the detector temperature was 300°C. Peaks were automatically integrated, and identities and relative amounts were determined by using the MIS Standard Software (Microbial ID).
Analysis of isoprenoid quinones.
Isoprenoid quinones were analyzed at the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH after extraction from 500 mg of lyophilized cells in chloroform-methanol (2:1 [vol/vol]) for 2 h with a magnetic stirrer. The cell-solvent mixture was filtered to remove cell debris, and the cells were reextracted for a further 2 h. The combined extracts were reduced to dryness by evaporation at 40°C. The lipid extracts were resuspended in 200 μl of acetone. The quinones were purified by thin-layer chromatography on a Kieselgel 60F254 plastic sheet (Merck AG, Darmstadt, Germany) with hexane-diethyl ether (85:15 [vol/vol]) as the developing solvent. The plate was dried, and the quinones were visualized under UV light (254 nm). The positions of the quinones were marked and eluted from the thin-layer chromatography plate with diethyl ether. The eluate was dried, resuspended in 200 μl of isopropanol, and filtered through a DynaGard 2-μm-pore-size hollow-fiber syringe filter (Spectrum Europe BV, Breda, The Netherlands).
Five microliters of the purified extract was analyzed by a high-performance liquid chromatography (HPLC) system (Hewlett-Packard) composed of an HP-1090 HPLC pump, an HP-1090 diode array detector, an HP-Vectra computer, and HP-Chemstation software. The quinones were separated according to type and isoprene chain length at 40°C on a reversed-phase column (250 by 4.6 mm; Lichrospher 100 RP-18 endcapped; Merck AG). The eluent was acetonitrile-isopropanol (65:35 [vol/vol]) at 1 ml/min. Quinones were identified by their retention time by using bacterial quinone extracts of known composition for comparison. The type and purity of each quinone was checked on the diode array detector by running a spectrum from 200 to 400 nm at the apex of each peak.
Other chemical and biochemical characters.
Photometric examination of cell-free acetone extracts of glucose-grown cells were performed by using an Ultraspec Plus UV/Visible spectrophotometer (Pharmacia LKB Biochrom, Ltd., Cambridge, United Kingdom). The G+C content of the genomic DNA was estimated by HPLC (26). The structure of the peptidoglycan was determined by thin-layer chromatography (25) at the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH. Catalase (method 2) and oxidase activities were tested qualitatively as described by Smibert and Krieg (49).
nifH detection.
Forty microliters of reaction mixture containing 1.25× Mg-free PCR buffer (Qiagen, Pty., Ltd., Clifton Hill, Victoria, Australia), 2.5 mM MgCl2, 1.25 M betaine, 100 pmol each of the primers UMf (5′-GCIWTYTAYGGIAARGGIGG-3′) and UMr (5′-AAICCRCCRCAIACIACRTC-3′) (modified from reference 51), and a small amount of a colony from a plate, overlaid with a drop of mineral oil (Promega), was subjected to a 1-min denaturation step at 94°C before 10 nmol of each deoxynucleoside triphosphate and 2.0 U of Taq DNA polymerase (Qiagen) were added in a volume of 10 μl of water. Forty cycles of denaturation at 94°C for 30 s, annealing at 50°C for 1 min, and extension at 72°C for 30 s were followed by a final extension step at 72°C for 5 min in a Sprint Thermocycler (ThermoHybaid, Ashford, United Kingdom). This primer set has been developed to amplify a product of ca. 400 bp of the nifH gene from a wide range of bacteria (C. A. Osborne and P. H. Janssen, unpublished data). The positive controls were Sinorhizobium meliloti ICMP 3123, Desulfovibrio desulfuricans DSM 642, and Bradyrhizobium sp. strain Ellin173. Products were visualized by electrophoresis in ethidium bromide-containing agarose gels (40).
16S rRNA gene sequencing and phylogenetic analysis.
Sequencing and comparative analysis of the nearly complete 16S rRNA gene was performed essentially as described by Janssen and Hugenholtz (24). Briefly, the gene was PCR amplified directly from a cell pellet by using the broad-specificity primers 27f (28) and 1492r (32) and then sequenced by using the internal primers 519r, 357f, 1100r (32), and 1401r (21). The compiled sequence was imported into an ARB database (35), aligned, and inserted into the main ARB tree to determine an approximate phylogenetic position. A maximum-likelihood tree of the phylum Verrucomicrobia, including strain Ellin428 and 61 relatives, was constructed by using fastDNAml in ARB. Representatives of the “vadin” lineage (9, 57) were used as the outgroup for the analysis because they are the most closely related group to the Verrucomicrobia and are part of the Verrucomicrobia-Chlamydia radiation. Statistical support of interior nodes was estimated by using evolutionary distance and maximum-parsimony bootstrap resampling of the data set and by using Bayesian posterior probabilities as previously described (24). Subdivisions of the Verrucomicrobia were numbered as described previously by Hugenholtz et al. (23).
The sequence of the 16S rRNA gene of strain Ellin428 has been deposited in GenBank under the accession AY388649.
RESULTS
Cellular characteristics.
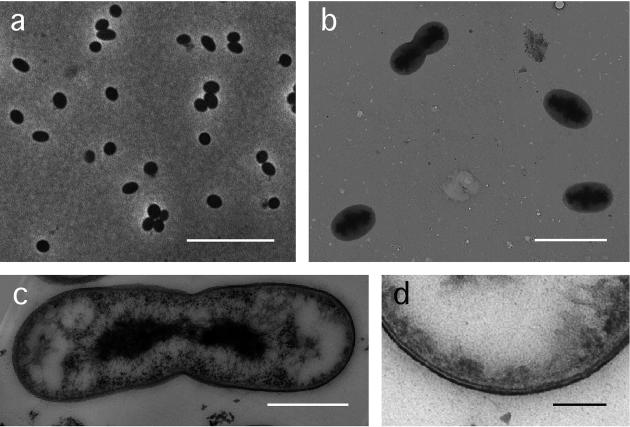
Cells of strain Ellin428 were oval in shape, and occurred singly or in pairs (Fig. 1a and b). The latter were often dividing cells, and the mode of division appeared to be by binary fission following constriction (Fig. 1b and c). The cells were 1.4 μm long (standard deviation [SD] = 0.17 [n = 30]) and 0.9 μm in diameter (SD = 0.06). Motile cells were never observed by phase-contrast microscopy of living cultures, and whole cells observed by electron microscopy did not possess flagella. No surface structures were apparent. Stains for poly-β-hydroxybutyrate and polyphosphate inclusions gave positive results.
FIG. 1.
(a) Phase-contrast photomicrograph of cells of strain Ellin428. Bar, 10 μm. (b) Electron micrograph of whole cells of strain Ellin428. Bar, 2 μm. (c) Electron micrograph of thin section of a dividing cell of strain Ellin428, showing the electron dense material that was interpreted to be DNA. Bar, 500 nm. (d) Electron micrograph of a thin section part of the cell envelope of a cell of strain Ellin428, showing the two well-stained layers separated by a poorly stained layer or region. Bar, 100 nm.
Electron microscopy of thin sections revealed that the dividing cells often contained what appeared to be condensed nuclear material that was being partitioned into the two daughter cells (Fig. 1c). This was also observed by epifluorescence microscopy of DAPI-stained cells. The longer cells and pairs of cells contained regions of intense DAPI stain in the middle of the cell, while in single cells the DAPI stain was evenly distributed throughout the cell. The cell envelope appeared to consist of a thinner inner layer separated by a nonstaining layer or region from a thicker outer layer (Fig. 1d). The Gram stain result was negative. Cells of strain Ellin428 contained peptidoglycan, with direct cross-linkages of the A1γ meso-Dpm type. The major cellular fatty acids were tetradecanoic acid (C14:0 [11% of total]), 12-methyl-tetradecanoic acid (a-C15:0 [33%]), cis-9-hexadecenoic acid and/or 2-hydroxy-13-methyl-tetradecanoic acid (C16:1ω7c and/or 2OH i-C15:0 [21%]), and hexadecanoic acid (C16:0 [15%]).
The G+C content of the genomic DNA was 60.7 mol% (SD = 0.16, n = 5).
Colonies and cell pellets of strain Ellin428 were yellow. This pigment did not diffuse into the medium and was not extracted into ethanol or chloroform. It could be extracted into acetone, and cell-free acetone extracts displayed an absorption maximum at 339 nm. The cells contained menaquinones MK-10 and MK-11 in approximately equal amounts, but only at very low levels that could not be quantified accurately.
Cultivation characteristics.
Strain Ellin428 formed soft, convex, entire yellow colonies up to 4 mm in diameter. Liquid cultures were uniformly turbid and pale yellow. Strain Ellin428 was able to grow at pH values of 4.0 (the lowest tested) to 7.0. No growth occurred at pH 7.5. NaCl added at 5 g per liter did not inhibit growth, but when it was added at 10 g per liter no growth was observed. Growth was possible at 25, 30 and 34°C but not at 37°C.
Strain Ellin428 was able to grow on plates of full-strength and 1/100-strength nutrient agar, full-strength and 1/10-strength Trypticase soy agar, R2A agar, cold-extracted soil extract agar, and Winogradsky's salt solution agar.
Metabolic characteristics.
Strain Ellin428 was able to grow aerobically in liquid culture with pyruvate, glucose, fructose, galactose, mannose, xylose, cellobiose, lactose, sucrose, galacturonate, and glucuronate (all tested individually at 2 mM) and with xylan, starch, cellulose, pectin, and alginate (all tested individually at 0.025% [wt/vol]). No growth occurred with the following when they were added as sole carbon and energy sources: formate, acetate, propionate, butyrate, lactate, malonate, succinate, fumarate, malate, tartrate, citrate, methanol, ethanol, propanol, butanol, glycerol, mannitol, glycolate, ribose, arabinose, glycine, alanine, leucine, aspartate, glutamate, lysine, valine, serine, arginine, threonine, benzoate, vanillin, catechol, betaine, or urea (all tested individually at 2 mM) or with chitin (tested at 0.025% [wt/vol]).
Growth was not possible with glucose under anaerobic conditions, even if 10 mM NaNO3 was added to liquid medium as a potential electron acceptor. Cells of strain Ellin428 were catalase and oxidase negative. A nifH gene could not be detected by PCR.
Phylogenetic position.
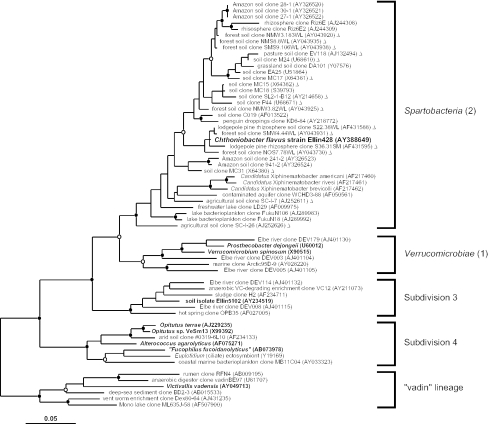
The nucleotide sequence of most (1360 bp) of the 16S rRNA gene of strain Ellin428 was determined. Based on comparative analysis of this gene sequence, strain Ellin428 is a member of subdivision 2 of the phylum Verrucomicrobia (Fig. 2). This subdivision includes three named but uncultivated nematode endosymbiont species of the candidate genus Xiphinematobacter (52). Subdivision 2 is broadly represented by cloned 16S rRNA genes from soil but also has freshwater and endosymbiont representatives. We propose that this subdivision be named the Spartobacteria to reflect its significant representation by soil bacteria (Fig. 2). The 16S rRNA gene sequence of strain Ellin428 is most closely related to partial-length environmental clone sequences derived from forest soil (98.3 to 99.5% sequence identity), is 91.5 to 92.2% identical to the 16S rRNA genes of Xiphinematobacter spp., and is 89.2% identical to Verrucomicrobium spinosum.
FIG. 2.
Maximum-likelihood dendrogram of the phylum Verrucomicrobia based on comparative analysis of 16S rRNA gene data. Subdivision numbering according to the method of Hugenholtz et al. (23) and associated class names are shown to the right of the figure. Only subdivisions with at least one cultivated representative (shown in boldface) are included in the dendrogram. Chthoniobacter flavus strain Ellin428 is shown in boldface and a larger typeface. Partial length sequences (<1,250 nucleotides), included in the maximum-likelihood inference but not in the statistical resampling, are indicated by triangles, and their positions in the Spartobacteria should be regarded as approximate. Each 16S rRNA (gene) sequence is identified by its origin, name, and GenBank accession number. Branch points supported by bootstrap resampling and posterior probabilities are indicated by solid circles (values > 90%) and open circles (values > 75%). Six representatives of the closely related “vadin” lineage (57) were used as the outgroup for the analysis. The scale bar indicates 0.05 changes per nucleotide.
DISCUSSION
General characteristics.
Strain Ellin428 is an aerobic heterotrophic bacterium that is able to grow with many of the saccharide components of plant biomass but does not appear able to grow with amino acids or organic acids other than pyruvate. It does not seem to have the genetic capacity to fix nitrogen and was unable to grow anaerobically. It appears that this bacterium is a general heterotroph that is involved in the transformation of organic carbon compounds in the soil. Whether other members of this subdivision (class Spartobacteria), detected to date as cloned 16S rRNA genes only, have the same phenotype remains to be ascertained. Although it is possible that some other members of the Spartobacteria may be endosymbionts, like Xiphinematobacter spp. (52), others are likely to be free-living soil heterotrophs such as strain Ellin428.
It is unclear why members of the Spartobacteria have not been isolated and identified in previous studies. The new isolate grew well at 25°C on a variety of standard biological media, including some used in the routine cultivation of bacteria from soil. A detailed investigation of this isolate and of other representatives, once they have been cultured, will be needed to determine why the numbers or activity of members of this group vary with changes in soil moisture content (3) and what significance, if any, this variation has for soil fertility. The demonstration that a member of this group is readily culturable should encourage more detailed study of this widespread and abundant group of soil bacteria.
Comparison of strain Ellin428 with other verrucomicrobia.
In common with the characterized members of the class Verrucomicrobiae, Verrucomicrobium spinosum (46, 48) and four species of the genus Prosthecobacter (18), strain Ellin428 has a peptidoglycan-containing cell envelope, contains menaquinones rather than ubiquinones, is an aerobe, and has yellow pigmentation. In addition, the cellular fatty acid composition is very similar to that of V. spinosum (48). It also shares with the characterized members of the Verrucomicrobiae the ability to grow only with sugars and not with amino acids or organic acids other than pyruvate (18, 46). Strain Ellin428, V. spinosum, and the four described species of the genus Prosthecobacter all have genomic DNA with G+C contents in the narrow range of 55 to 60 mol% (19, 46). However, in contrast to members of the Verrucomicrobiae, strain Ellin428 does not exhibit prosthecate structures. Phenotypically, strain Ellin428 seems very similar to members of the class Verrucomicrobiae, and these bacteria are its closest known cultivated and characterized relatives (Fig. 2). However, these closest cultivated relatives are phylogenetically distant from strain Ellin428 to a degree at which it seems justified to believe that they should belong to different classes within the phylum Verrucomicrobia. It will be interesting to explore more fundamental differences in, for example, regulatory and biosynthetic pathways between these phenotypically similar organisms. Differences could reflect the phylogenetic divergence between the groups and also the difference in ecological strategies of aquatic (Verrucomicrobiae) and soil-inhabiting (Spartobacteria) bacteria belonging to the phylum Verrucomicrobia. Strain Ellin428 is only distantly related to the cultured members of subdivision 4 (6, 7, 27, 44, 47), which have 16S rRNA genes with only 78.5 to 84.9% sequence identity to the 16S rRNA gene of strain Ellin428 (Fig. 2). Opitutus spp. and Alterococcus agarolyticus are anaerobes (7, 27, 47), whereas “Fucophilus fucoidanolyticus” is described as an aerobe that contains menaquinone-7, is motile, and grows only at sodium chloride concentrations that are too high for growth of strain Ellin428 (44). Interestingly, menaquinones rather than ubiquinones are present in the verrucomicrobia that have been analyzed to date (18, 44).
Strain Ellin428 is phylogenetically distinct from any previously reported bacterial isolates and represents a new genus and species for which we propose the name Chthoniobacter flavus.
Description of Spartobacteria classis nov.
Spartobacteria (Spar.to.bac.te′ri.a. Gr. adj spartos, sown, in relation to the Spartoi, the sown men of the Cadmus myth, who sprung from the soil; Gr. dim. neut. n. bakterion, a small rod; M.L. fem. pl. n. Spartobacteria, sown small rods).
On the basis of 16S rRNA gene sequence analyses, this group comprises one of the primary lineages in the phylum Verrucomicrobia and is equivalent to subdivision 2 (23) of the phylum. The majority of its members appear to be soil-inhabiting bacteria, while some are endosymbionts of nematodes. The Spartobacteria currently comprise two orders: the type order Chthoniobacterales, at present represented by Chthoniobacter flavus, and the order “Xiphinematobacterales,” represented by three uncultured species of the genus Xiphinematobacter (52).
Description of Chthoniobacterales ord. nov.
Chthoniobacterales (Chtho.ni.o.bac.ter.a′les. M.L. masc. n. Chthoniobacter, type genus of the order; -ales, ending to denote an order; M.L. fem. pl. n. Chthoniobacterales, the Chthoniobacter order).
The description is the same as for the genus Chthoniobacter. The order contains the type family Chthoniobacteraceae.
Description of Chthoniobacteraceae fam. nov.
Chthoniobacteraceae (Chtho.ni.o.bac.ter.a′ce.ae. M.L. masc. n. Chthoniobacter, type genus of the family; -aceae, ending to denote a family; M.L. fem. pl. n. Chthoniobacteraceae, the Chthoniobacter family).
The description is the same as for the genus Chthoniobacter. The family contains the type genus Chthoniobacter.
Description of Chthoniobacter gen. nov.
Chthoniobacter (Chtho.ni.o.bac′ter. Gr. adj. chthonios, born from the soil, also the name of one of the Spartoi of the Cadmus myth; M.L. masc. n. bacter, the equivalent of Gr. neut. n. bactrum rod or staff; M.L. masc. n. Chthoniobacter, rod from the soil). The genus description at present is the same as the description of the type species, Chthoniobacter flavus.
Description of Chthoniobacter flavus sp. nov.
Chthoniobacter flavus (fla′vus L. adj. flavus, yellow).
Cells are oval in shape, singly or in pairs, each 1.4 μm long and 0.9 μm in diameter. Cells are not motile and have no flagella. Cells contain poly-β-hydroxybutyrate and polyphosphate inclusions. Cells are Gram stain-negative and have a gram-negative cell envelope structure, with peptidoglycan with direct cross-linkages of the A1γ meso-Dpm type. The major cellular fatty acids are tetradecanoic acid, 12-methyl-tetradecanoic acid, cis-9-hexadecenoic acid and/or 2-hydroxy-13-methyl-tetradecanoic acid, and hexadecanoic acid. G+C content of the genomic DNA is 60.7 mol%. Yellow pigmentation. Cells contain menaquinones MK-10 and MK-11. Cells grow at pH values of 4 to 7 and at temperatures between 25 and 34°C but not at 37°C. Aerobic growth with sugars, sugar polymers, and pyruvate but not with other organic acids, amino acids, or alcohols. There is no growth under anaerobic conditions or with nitrate as electron acceptor. The organism is catalase and oxidase negative. There is no nifH gene. The type strain, Ellin428, was isolated from pasture soil.
Acknowledgments
We thank Simon Crawford for help with electron microscopy, Catherine A. Osborne for performing the PCR assay for the presence of nifH, and H. G. Trüper for advice on etymology.
This study was supported by a grant from the Australian Research Council.
REFERENCES
- 1.Axelrood, P. E., M. L. Chow, C. C. Radomski, J. M. McDermott, and J. Davies. 2002. Molecular characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can. J. Microbiol. 48:655-674. [DOI] [PubMed] [Google Scholar]
- 2.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckley, D. H., and T. H. Schmidt. 2001. Environmental factors influencing the distribution of rRNA from Verrucomicrobia in soil. FEMS Microbiol. Ecol. 35:105-112. [DOI] [PubMed] [Google Scholar]
- 4.Buckley, D. H., and T. M. Schmidt. 2002. Exploring the biodiversity of soil: a microbial rain forest, p. 183-208. In J. T. Staley and A.-L. Reysenbach (ed.), Biodiversity of microbial life: foundation of the earth's biosphere. Wiley-Liss, Inc., New York, N.Y.
- 5.Buckley, D. H., and T. H. Schmidt. 2003. Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ. Microbiol. 5:441-452. [DOI] [PubMed] [Google Scholar]
- 6.Chin, K.-J., D. Hahn, U. Hengstmann, W. Liesack, and P. H. Janssen. 1999. Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Appl. Environ. Microbiol. 65:5042-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin, K.-J., W. Liesack, and P. H. Janssen. 2001. Description of Opitutus terrae gen. nov., sp. nov., to accommodate new strains of the division Verrucomicrobia isolated from rice paddy soil. Int. J. Syst. Evol. Microbiol. 51:1965-1968. [DOI] [PubMed] [Google Scholar]
- 8.Chow, M. L., C. C. Radomski, J. M. McDermott, J. Davies, and P. E. Axelrood. 2002. Molecular characterization of bacterial diversity in Lodgepole pine (Pinus contorta) rhizosphere soils from British Columbia forest soils differing in disturbance and geographic source. FEMS Microbiol. Ecol. 42:347-357. [DOI] [PubMed] [Google Scholar]
- 9.Cosaro, D., M. Valassina, and D. Venditti. 2003. Increasing diversity within chlamydiae. Crit. Rev. Microbiol. 29:37-78. [DOI] [PubMed] [Google Scholar]
- 10.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis, R. J., P. Morgan, A. J. Weightman, and J. C. Fry. 2003. Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl. Environ. Microbiol. 69:3223-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felske, A., and A. D. L. Akkermans. 1998. Prominent occurrence of ribosomes from an uncultured bacterium of the Verrucomicrobiales cluster in grassland soils. Lett. Appl. Microbiol. 26:219-223. [DOI] [PubMed] [Google Scholar]
- 13.Felske, A., A. Wolterink, R. van Lis, W. M. de Vos, and A. D. L. Akkermans. 1998. Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl. Environ. Microbiol. 64:4581-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felske, A., A. Wolterink, R. van Lis, W. M. de Vos, and A. D. L. Akkermans. 2000. Response of a soil bacterial community to grassland succession as monitored by 16S rRNA levels of the predominant ribotypes. Appl. Environ. Microbiol. 66:3998-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furlong, M. A., D. R. Singleton, D. C. Coleman, and W. B. Whitman. 2002. Molecular and culture-based analyses of prokaryotic communities from an agricultural soil and the burrows and casts of the earthworm Lumbricus rubellus. Appl. Environ. Microbiol. 68:1265-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrity, G. M., J. A. Bell, and T. G. Lilburn. 2003. Taxonomic outline of the procaryotes, release 4.0. Springer, New York, N.Y.
- 17.Gray, N. D., and I. M. Head. 2001. Linking genetic identity and function in communities of uncultured bacteria. Environ. Microbiol. 3:481-492. [DOI] [PubMed] [Google Scholar]
- 18.Hedlund, B. P., J. J. Gosink, and J. T. Staley. 1996. Phylogeny of Prosthecobacter, the fusiform caulobacters: members of a recently discovered division of the Bacteria. Int. J. Syst. Bacteriol. 46:960-966. [DOI] [PubMed] [Google Scholar]
- 19.Hedlund, B. P., J. J. Gosink, and J. T. Staley. 1997. Verrucomicrobia div. nov., a new division of the Bacteria containing three new species of Prosthecobacter. Antonie Leeuwenhoek 72:29-38. [DOI] [PubMed] [Google Scholar]
- 20.Hendrikson, D. A., and M. M. Krenz. 1991. Reagents and stains, p. 1289-1314. In A. Balows, W. J. Hausler, Jr., K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 21.Heuer, H., and K. Smalla. 1997. Application of denaturing gradient gel electrophoresis and temperature gradient gel electrophoresis for studying soil microbial communities, p. 353-373. In J. D. van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker, Inc., New York, N.Y.
- 22.Holmes, A. J., J. Bowyer, M. P. Holey, M. O'Donoghue, M. Montgomery, and M. R. Gillings. 2000. Diverse, yet-to-be-cultured members of the Rubrobacter subdivision of the Actinobacteria are widespread in Australian arid soils. FEMS Microbiol. Ecol. 33:111-120. [DOI] [PubMed] [Google Scholar]
- 23.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janssen, P. H., and P. Hugenholtz. 2003. Fermentation of glycolate by a pure culture of a strictly anaerobic gram-positive bacterium belonging to the family Lachnospiraceae. Arch. Microbiol. 179:321-328. [DOI] [PubMed] [Google Scholar]
- 25.Janssen, P. H., S. Evers, F. A. Rainey, N. Weiss, W. Ludwig, C. G. Harfoot, and B. Schink. 1995. Lactosphaera gen. nov., a new genus a lactic acid bacteria, and transfer of Ruminococcus pasteurii Schink 1984 to Lactosphaera pasteurii comb. nov. Int. J. Syst. Bacteriol. 45:565-571. [DOI] [PubMed] [Google Scholar]
- 26.Janssen, P. H., W. Liesack, C. Kluge, S. Seeliger, B. Schink, and C. G. Harfoot. 1996. Sodium-dependent succinate decarboxylation by a new anaerobic bacterium belonging to the genus Peptostreptococcus. Antonie Leeuwenhoek 70:11-20. [DOI] [PubMed] [Google Scholar]
- 27.Janssen, P. H., A. Schuhmann, E. Mörschel, and F. A. Rainey. 1997. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl. Environ. Microbiol. 63:1382-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuykendall, L. D., M. A. Roy, J. J. O'Neill, and T. E. Devine. 1988. Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int. J. Syst. Bacteriol. 38:358-361. [Google Scholar]
- 32.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 33.Lee, S.-Y., J. Bollinger, D. Bezdicek, and A. Ogram. 1996. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl. Environ. Microbiol. 62:3787-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liles, M. R., B. F. Manske, S. B. Bintrim, J. Handelsman, and R. M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macrae, A., D. L. Rimmer, and A. G. O'Donnell. 2000. Novel bacterial diversity recovered from the rhizosphere of oilseed rape (Brassica napus) determined by the analysis of the 16S ribosomal DNA. Antonie Leeuwenhoek 78:13-21. [DOI] [PubMed] [Google Scholar]
- 37.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nogales, B., E. R. B. Moore, E. Llobet-Brossa, R. Rossello-Mora, R. Amann, and K. N. Timmis. 2001. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ochsenreiter, T., D. Selezi, A. Quaiser, L. Bonch-Osmolovskaya, and C. Schleper. 2003. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real-time PCR. Environ. Microbiol. 5:787-797. [DOI] [PubMed] [Google Scholar]
- 40.O'Farrell, K. A., and P. H. Janssen. 1999. Detection of verrucomicrobia in a pasture soil by PCR-mediated amplification of 16S rRNA genes. Appl. Environ. Microbiol. 65:4280-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olsen, R. A., and L. R. Bakken. 1987. Viability of soil bacteria: optimization of plate-counting techniques and comparisons between total counts and plate counts within different size groups. Microb. Ecol. 13:59-74. [DOI] [PubMed] [Google Scholar]
- 42.Pfennig, N., and S. Wagener. 1986. An improved method of preparing wet mounts for photomicrographs of microorganisms. J. Microbiol. Methods 4:303-306. [Google Scholar]
- 43.Rees, G. N., G. Vasiliadis, J. W. May, and R. C. Bayley. 1992. Differentiation of polyphosphate and poly-β-hydroxybutyrate granules in an Acinetobacter sp. isolated from activated sludge. FEMS Microbiol. Lett. 94:171-174. [DOI] [PubMed] [Google Scholar]
- 44.Sakai, T., K. Ishizuka, and I. Kato. 2003. Isolation and characterization of a fuciodan-degrading marine bacterium. Mar. Biotechnol. 5:409-416. [DOI] [PubMed] [Google Scholar]
- 45.Sato, T. 1968. A modified method for lead staining of thin sections. J. Electron Microsc. 17:158-159. [PubMed] [Google Scholar]
- 46.Schlesner, H. 1987. Verrucomicrobium spinosum gen. nov., sp. nov.: a fimbriated prosthecate bacterium. Syst. Appl. Microbiol. 10:54-56. [Google Scholar]
- 47.Shieh, W. Y., and W. D. Jean. 1998. Alterococcus agarolyticus, gen. nov., sp. nov., a halophilic thermophilic bacterium capable of agar degradation. Can. J. Microbiol. 44:637-645. [DOI] [PubMed] [Google Scholar]
- 48.Sittig, M., and H. Schlesner. 1993. Chemotaxonomic investigation of various prosthecate and/or budding bacteria. Syst. Appl. Microbiol. 16:92-103. [Google Scholar]
- 49.Smibert, R. M., and N. R. Krieg. 1994. Phenotypic characterization, p. 607-654. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 50.Tschech, A., and N. Pfennig. 1984. Growth yield increase linked to caffeate reduction in Acetobacterium woodii. Arch. Microbiol. 137:163-167. [Google Scholar]
- 51.Ueda, T., Y. Suga, N. Yahiro, and T. Matsuguchi. 1995. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J. Bacteriol. 177:1414-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanderkerckhove, T. T. M., A. Willems, M. Gills, amd A. Coomans. 2000. Occurrence of novel verrucomicrobial species, endosymbiotic and associated with parthenogenesis in Xiphinema americanum-group species (Nematoda, Longidoridae). Int. J. Syst. Evol. Microbiol. 50:2197-2205. [DOI] [PubMed] [Google Scholar]
- 53.Ward-Rainey, N., F. A. Rainey, H. Schlesner, and E. Stackebrandt. 1995. Assignment of hitherto unidentified 16S rDNA species to a main line of descent within the domain Bacteria. Microbiology 141:3247-3250. [Google Scholar]
- 54.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, N.Y.
- 55.Widdel, F., G. W. Kohring, and F. Mayer. 1983. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov. sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 134:286-294. [Google Scholar]
- 56.Winding, A., S. J. Binnerup, and J. Søorensen. 1994. Viability of indigenous soil bacteria assayed by respiratory activity and growth. Appl. Environ. Microbiol. 60:2869-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zoetendal, E. G., C. M. Plugge, A. D. L. Akkermans, and W. M. de Vos. 2003. Victivallis vadensis gen. nov., sp. nov., a sugar-fermenting anaerobe from human faeces. Int. J. Syst. Evol. Microbiol. 53:211-215. [DOI] [PubMed] [Google Scholar]
- 58.Zwart, G., R. Huismans, M. P. van Agterveld, Y. Van de Peer, P. De Rijk, H. Eenhoorn, G. Muyzer, E. J. van Hannen, H. J. Gons, and H. J. Laanbroek. 1998. Divergent members of the bacterial division Verrucomicrobiales in a temperate freshwater lake. FEMS Microbiol. Ecol. 25:159-169. [Google Scholar]