Abstract
The bacterial heat shock response is characterized by the elevated expression of a number of chaperone complexes, including the GroEL and GroES proteins. The groES and groEL genes are highly conserved among eubacteria and are typically arranged as an operon. Genome analysis of Bifidobacterium breve UCC 2003 revealed that the groES and groEL genes are located in different chromosomal regions. The heat inducibility of the groEL and groES genes of B. breve UCC 2003 was verified by slot blot analysis. Northern blot analyses showed that the cspA gene is cotranscribed with the groEL gene, while the groES gene is transcribed as a monocistronic unit. The transcription initiation sites of these two mRNAs were determined by primer extension. Sequence and transcriptional analyses of the region flanking the groEL and groES genes of various bifidobacteria revealed similar groEL-cspA and groES gene units, suggesting a novel genetic organization of these chaperones. Phylogenetic analysis of the available bifidobacterial groES and groEL genes suggested that these genes evolved differently. Discrepancies in the phylogenetic positioning of groES-based trees make this gene an unreliable molecular marker. On the other hand, the bifidobacterial groEL gene sequences can be used as an alternative to current methods for tracing Bifidobacterium species, particularly because they allow a high level of discrimination between closely related species of this genus.
The genus Bifidobacterium includes gram-positive, pleomorphic, and strictly anaerobic bacteria, which are major constituents of the intestinal microflora of humans, of other warm-blooded animals, and even of honeybees (for a review see reference 43). In recent years, bifidobacteria have been the subject of growing interest due to their possible role in the maintenance of gastrointestinal health (23, 35). For this reason various bifidobacterial strains are considered to be probiotic, and they are often added as viable bacteria to dairy products (e.g., yoghurt) or supplied in infant foods. Bifidobacterial strains have been selected on the basis of activities that could assist in maintaining an improved intestinal microflora, stimulating the immune response (2), protecting against colonization by pathogens, or reducing activities of bacterial enzymes that are associated with the development of colonic cancer (2). However, such strains must also demonstrate resilience to the adverse conditions encountered in industrial processes, such as those encountered during starter handling and food storage (freeze-drying, freezing, or spray-drying). Bifidobacteria are subjected to potentially stressful conditions not only in industrial processes but also in their natural environments, where their ability to respond rapidly to stress is essential for survival (41). The heat shock response is a universal example of a global control system designed to increase bacterial survival. The heat shock response induces the production of a large set of proteins (known as the heat shock proteins), which are generally involved in the maturation of newly synthesized proteins and in the refolding or degradation of denatured proteins (10). The heat shock response has been extensively studied in many gram-positive bacteria (Bacillus subtilis and Lactococcus lactis) and gram-negative bacteria (Escherichia coli and Agrobacterium tumefaciens). Some of the most intensively investigated heat shock proteins include the molecular chaperones GroEL and GroES, which are highly conserved in E. coli and eukaryotic cells (14). The GroEL and GroES chaperones (also known as Hsp60 and Hsp10 chaperonins) have been recognized as heat shock proteins in many bacteria, including E. coli, B. subtilis (16), A. tumefaciens (33), Streptomyces lividans (6), L. lactis (20), Lactobacillus helveticus (3), Lactobacillus johnsonii VPI 11088 (50), Streptococcus suis (4), and Pseudomonas aeruginosa (9). The genes encoding the molecular chaperones GroES and GroEL are normally organized as a monocistronic operon, while their translation products are assembled into single or double heptameric rings (10). In the presence of high-energy nucleotides, GroES forms an equimolar complex with GroEL, which binds the protein substrate (22). The release of the correctly folded protein is contingent upon ATP hydrolysis, and multiple binding and release may be necessary for a protein to reach its native conformation (51). In addition to its established role in protein folding and assembly, GroEL has recently been shown to be implicated in protection of mRNA from nuclease degradation, as well as in membrane stabilization (14, 39).
The presence of the groEL and groES genes thus makes an essential contribution to the survival of bacterial cells, and consequently these molecules are included in the category of housekeeping genes. Due in particular to their ubiquitous distribution, functional preservation, and sequence conservation, the groESL genes are considered valuable molecular markers for bacterial phylogenetic investigations (15, 24). They have been successfully used for identification of Mycobacterium, Staphylococcus, Streptococcus, Enterococcus, Ruminococcus, and Bifidobacterium species (4, 12, 17, 18, 36, 37). Several genetic approaches targeting rRNA genes (19, 25, 30, 46, 48, 49) have been used in recent years for identification of bifidobacteria. With the advent of the genomics era and polyphasic taxonomy (40), many molecular markers that are alternatives to the 16S ribosomal DNA (rDNA) sequences have now been described. Recently, alternative genes, such as those encoding elongation factor Tu (5, 44, 45) and ATPase subunits (42) and recA (21, 44), have been used to examine phylogenetic relationships and to trace bifidobacterial species.
In this study, we identified and characterized the groEL and groES loci of Bifidobacterium breve UCC 2003 and Bifidobacterium lactis LMG 18906 and explored the heat induction of these genes at transcriptional levels by Northern blot hybridization and primer extension analysis. Moreover, we evaluated the robustness of using groES and groEL sequences as molecular markers to infer the phylogeny of bifidobacterial species.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used are summarized in Table 1. All Bifidobacterium strains were grown anaerobically in MRS (Difco, Detroit, Mich.) supplemented with 0.05% l-cysteine-HCl and incubated at 37°C for 16 h.
TABLE 1.
Strains, origins, and groES and groEL sequence accession numbers
| Species | Straina | groES accession no.b | groEL accession no.b |
|---|---|---|---|
| Bifidobacterium lactis | LMG 18906 | AY586538 | AY586539 |
| Bifidobacterium animalis | ATCC 25527 | AY585250 | AY488178 |
| Bifidobacterium lactis | NCC 239 | ND | AY488182 |
| Bifidobacterium lactis | NCC 402 | ND | AY488177 |
| Bifidobacterium lactis | NCC 363 | ND | AY488176 |
| Bifidobacterium lactis | ATCC 27536 | ND | AY488181 |
| Bifidobacterium lactis | ATCC 27674 | ND | AY488179 |
| Bifidobacterium animalis | ATCC 27672 | ND | AY488183 |
| Bifidobacterium coryneforme | JCM 5819 | AY585258 | AY004275 |
| Bifidobacterium breve | UCC 2003 | AY585262 | AY585261 |
| Bifidobacterium adolescentis | JCM 1275 | AY585248 | AF210319 |
| Bifidobacterium longum | NCC 2705 | NC_004307 | NC_004307 |
| Bifidobacterium longum | JCM 7053 | ND | AY166574 |
| Bifidobacterium longum | JCM 1217 | ND | AF240578 |
| Bifidobacterium longum | JCM 7052 | ND | AY166573 |
| Bifidobacterium infantis | JCM 1222 | AY585254 | AF240577 |
| Bifidobacterium infantis | JCM 1210 | ND | AF240577 |
| Bifidobacterium suis | JCM 1269 | AY585253 | AY013248 |
| Bifidobacterium suis | JCM 7139 | ND | AY166575 |
| Bifidobacterium bifidum | JCM 1255 | AY585252 | AY004280 |
| Bifidobacterium dentium | JCM 1195 | AY585247 | AF240572 |
| Bifidobacterium catenulatum | JCM 1194 | AY585249 | AY004272 |
| Bifidobacterium pseudocatenulatum | JCM 1200 | AY585259 | AY004274 |
| Bifidobacterium angulatum | JCM 7096 | AY585256 | AF240568 |
| Bifidobacterium globosum | JCM 5820 | AY585260 | AF286736 |
| Bifidobacterium pullorum | JCM 1214 | AY585255 | AY004278 |
| Bifidobacterium magnum | JCM 1218 | AY585251 | AF240569 |
| Bifidobacterium thermophilum | JCM 1207 | AY585257 | AF240567 |
| Lactobacillus plantarum | WCFS1 | NC_004567 | NC_004567 |
| Lactobacillus gasseri | ATCC 33323 | NZ_AAA002000001 | NZ_AAA002000001 |
| Lactobacillus johnsonii | NCC 533 | NC_002662 | NC_002662 |
| Lactobacillus lactis subsp. lactis | IL1403 | NC_002662 | NC_002662 |
| Streptococcus pyogenes | M1GAS | NC_002737 | NC_002737 |
| Oenococcus oeni | MCW | NZ_AABJ00000000 | NZ_AABJ00000000 |
| Leuconostoc mesenteroides | ATCC 8293 | NZ_AABH02000054 | NZ_AABH02000054 |
| Enterococcus faecalis | V583 | NC_004668 | NC_004668 |
| Clostridium acetobutylicum | ATCC 824 | NC_003030 | NC_003030 |
| Mycobacterium bovis | AF 2122/97 | NC_002945 | NC_002945 |
| Streptomyces coelicolor | A3 | NC_003888 | NC_003888 |
| Corynebacterium glutamicum | ATCC 13032 | NC_003450 | NC_003450 |
| Listeria monocytogenes | EGD | NC_003210 | NC_003210 |
ATCC, American Type Culture Collection; LMG, Laboratorium voor Microbiologie, University of Ghent; JCM, Japanese Collection of Microorganisms.
For the strains whose genome sequences are available, the groES and groEL sequences were retrieved from the complete bacterial genome. ND, not determined.
DNA isolation.
Genomic DNA was extracted by using the protocol described in a previous study (47).
DNA amplification and cloning of the groES gene.
PCR was used to amplify the groES gene in all Bifidobacterium strains investigated. DNA fragments that were approximately 200 bp long corresponding to the groES gene were amplified by using the oligonucleotides gro-1 (5′-CTCACACCGTTGGAAG-3′) and gro-2 (5′-GN(CA)GGAGACGATGAGGTA-3′). The resultant amplicons represent the most conserved central part of the groES gene. Each PCR mixture (50 μl) contained a reaction cocktail consisting of 20 mM Tri-HCl, 50 mM KCl, each deoxynucleoside triphosphate at a concentration of 200 μM, 50 pmol of each primer, 1.5 mM MgCl2, 1 U of Taq DNA polymerase (Gibco BRL, Paisley, United Kingdom), and 25 ng of DNA template. Each PCR cycling profile consisted of an initial denaturation step of 3 min at 95°C, followed by amplification for 30 cycles as follows: denaturation for 30 s at 95°C, annealing for 30 s at 51°C, and extension for 1 min at 72°C. The resulting amplicons were separated on a 1.5% agarose gel, and this was followed by ethidium bromide staining. PCR fragments were purified with a PCR purification spin kit (Genomed, Löhne, Germany) and were subsequently sequenced.
DNA sequencing and phylogenetic study.
Nucleotide sequencing of both strands obtained from PCR amplicons was performed by MWG-Biotech AG (Ebersberg, Germany). The primers used were gro-1 and gro-2 labeled with IRD800 (MWG Biotech). The groES genes of all Bifidobacterium strains determined in this study, as well as those available in the GenBank database, were used for comparison. Sequence data assembly and analysis were performed by using the DNASTAR software (version 5.05; DNASTAR, Madison, Wis.). Sequence alignments were done by using the MultiAlign program and Clustal W (38). Phylogenetic calculations, including distance calculations and generation of phylogenetic trees, were performed by using the PHYLIP package (8). Trees were calculated by the neighbor-joining method as implemented in the neighbor module of PHYLIP. DNA distances were calculated with dnadist by using the maximum-likelihood option. Protein distances were calculated with protdist by using the PAM matrix of amino acid substitution (8). The robustness of the results was assessed by resembling with substitution, commonly referred to as bootstrapping. Branch length estimates (from dnadist or protdist) were superimposed on the consensus tree by using the fitch module within PHYLIP. Also, dendrograms from gene sequences were constructed by using the Clustal X program and were visualized with the TreeView program.
Bioinformatic analysis.
Secondary structure prediction was performed with MFOLD, version 3.1 (55). Isoelectric points were predicted with the European Molecular Biology Open Software Suite (EBI).
Southern hybridization.
Ten micrograms of bacterial DNA was digested to completion by using restriction endonuclease EcoRI, EcoRV, or XbaI as recommended by the supplier (Roche, Mannheim, Germany). These restriction enzymes were chosen because no restriction sites were observed within the amplified groES and groEL gene fragments. Southern blotting of agarose gels was performed on Hybond N+ membranes (Amersham, Little Chalfont, United Kingdom) by using the method outlined by Sambrook and Russell (29). The filters were hybridized with groES and groEL probes which were labeled with α-32P by use of the Random Primed DNA labeling system (Roche) and a DNA template extracted from B. breve strain UCC 2003. Subsequent prehybridization, hybridization, and autoradiography were carried out as described by Sambrook and Russell (29).
RNA isolation and Northern blot analysis.
B. breve UCC 2003 cells were grown to an optical density at 600 nm of 0.6, at which point the culture was held at 37°C or shifted to 43, 47, or 50°C. At the initial time, at 25, 50, 100, and 150 min, and at 15 h a 30-ml sample was collected from each culture and briefly centrifuged to harvest cells. Total RNA was isolated by using macaloid acid and was treated with DNase (Roche, East Sussex, United Kingdom). The initial Northern blot analysis of the groEL-groES activity of bifidobacteria was carried out by using 15-μg aliquots of RNA. The RNA was separated in a 1.5% agarose-formaldehyde denaturing gel, transferred to a Zeta-Probe blotting membrane (Bio-Rad, Hertfordshire, United Kingdom) as described by Sambrook and Russell (29), and fixed by UV cross-linking with a Stratalinker 1800 (Stratagene, La Jolla, Calif.). By using PCR amplicons obtained with primers targeting the groEL, groES, and cspA genes, all the genes were radiolabeled (29). Prehybridization and hybridization were carried out at 65°C in 0.5 M NaHPO4 (pH 7.2)-1.0 mM EDTA-7.0% sodium dodecyl sulfate (SDS). Following 18 h of hybridization, the membranes were rinsed twice for 30 min at 65°C in 0.1 M NaHPO4 (pH 7.2)-1.0 mM EDTA-1% SDS and twice for 30 min at 65°C in 0.1 mM NaHPO4 (pH 7.2)-1.0 mM EDTA-0.1% SDS and then exposed to X-OMAT autoradiography film (Eastman Kodak, Rochester, N.Y.). Autoradiographs were analyzed with ImaGene 5.1 (BioDiscovery).
Primer extension analysis.
The 5′ ends of the RNA transcripts were determined as follows. Separate primer extension reactions were conducted with 15-μg aliquots of RNA isolated as described above and mixed with 1 pmol of labeled primer IRD800 (MWG Biotech) and 2 μl of buffer H [2 M NaCl, 50 mM piperazine-N,N′-bis(ethanesulfonic acid) (PIPES), pH 6.4]. The mixture was denatured by incubation at 90°C for 5 min and then hybridized for 60 min at 42°C. After addition of 5 μl of 1 M Tris-HCl (pH 8.2), 10 μl of 0.1 M dithiothreitol, 5 μl of 0.12 M MgCl2, 20 μl of a mixture containing each deoxynucleoside triphosphate at a concentration of 2.5 mM, 0.4 μl (5 U) of reverse transcriptase (Sigma, St. Louis, Mo.), and 49.6 μl of double-distilled water, the enzymatic reaction mixture was incubated at 42°C for 2 h. The reaction was stopped by adding 250 μl of an ethanol-acetone mixture (1:1), and the reaction mixture was incubated at −70°C for 15 min was and then centrifuged at 10,000 rpm for 15 min in a model S417C centrifuge (Eppendorf, Hamburg, Germany). The pellets were dissolved in 4 μl of distilled water and mixed with 2.4 μl of loading buffer from a sequencing kit (Thermosequenase; fluorescence labeled; Amersham, Buckinghamshire, United Kingdom). The primer extension product was subjected to electrophoresis on an 8% polyacrylamide-urea gel along with sequencing reaction mixtures from reactions that were conducted by using the same primers employed for the primer extension and detected by using a LiCor sequencer (MWG Biotech). The following synthetic oligonucleotides were used: cspA-prom (5′-GATCACCCTCGTACAGCATC-3′) and groES-prom (5′-GATGCGGTCTGAGTCTCG-3′), located at positions 91 to 110 and 933 to 955 in the corresponding nucleotide sequences.
Slot blot hybridization of the groEL and groES mRNAs.
Twenty-five micrograms of total RNA was alkali denatured, transferred to Zeta-Probe blotting membranes (Bio-Rad Laboratories) with a Bio-Dot SF microfiltration apparatus (Bio-Rad) as specified by the manufacturer, and subjected to one UV auto-cross-linking cycle with the UV Stratalinker 1800 (Stratagene). Prehybridization and hybridization were carried out at 65°C in 0.5 M NaHPO4 (pH 7.2)-1.0 mM EDTA-7.0% SDS with the same [α-32P]dATP-labeled, groEL- and groES PCR-generated probes.
Nucleotide sequence accession numbers.
The GenBank accession numbers for partial Bifidobacterium groES gene sequences generated in this study are as follows: B. bifidum JCM 1255, AY585252; B. infantis JCM 1222, AY585254; B. catenulatum JCM 1194, AY585249; B. pseudocatenulatum JCM 1200, AY585259; B. adolescentis JCM 1275, AY585248; B. animalis ATCC 25527, AY585250; B. lactis LMG 18906, AY586538; B. suis JCM 1269, AY585253; B. coryneforme JCM 5919, AY585258; B. dentium JCM 1195, AY585247; B. angulatum JCM 7096, AY585256; B. thermophilum JCM 1207, AY585255; B. magnum JCM 1218, AY585251; B. globosum JCM 5820, AY585260; and B. pullorum JCM 1214, AY585255. The nucleotide sequence data for the groEL and groES loci of B. breve UCC 2003 have been deposited in the GenBank database under accession numbers AY585261 and AY585262, respectively, and the nucleotide sequence data for the groEL and groES loci of B. lactis LMG 18906 have been deposited in the GenBank database under accession numbers AY586539 and AY586538, respectively.
RESULTS
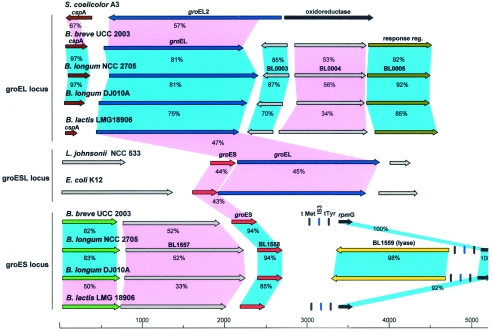
Within the framework of the B. breve UCC 2003 genome sequencing project, the nucleotide sequences of the groEL and groES regions were determined. The sequences encoded by two open reading frames (ORFs) located in distant DNA regions displayed high levels of similarity with the deduced amino acid sequences of GroEL (81% identical amino acids) and GroES (94% identical amino acids) from B. longum NCC 2705, which led to precise assignment of the B. breve UCC 2003 groEL and groES genes. The groEL gene starts with the canonical start codon AUG and encodes a predicted protein consisting of 541 amino acids, whereas the groES gene starts at the alternative start codon GUG and encodes a deduced protein consisting of 97 amino acids. Comparison of the B. breve UCC 2003 groEL and groES products with proteins deposited in the publicly available databases revealed high levels of sequence similarity with GroEL and GroES chaperonins from other high-G+C-content gram-positive bacteria (e.g., Mycobacterium and Streptomyces). The complete nucleotide sequences of the groES and groEL loci in B. breve and B. longum were determined, and the proposed organization of the two regions is shown in Fig. 1.
FIG. 1.
Comparison of the groEL and groES loci in B. breve UCC 2003 with the corresponding loci in different bacteria. Each arrow indicates an ORF. The length of the arrow is proportional to the length of the predicted ORF. Corresponding genes are indicated by the same color. Red indicates the groES gene; blue indicates the groEL gene; brown indicates the cspA gene; dark green indicates a gene encoding a putative response regulator; brilliant green indicates a gene encoding a putative transport regulator; yellow indicates a gene encoding a putative lyase; black indicates a gene encoding the putative ribosomal protein L33; grey indicates a gene encoding a hypothetical protein. The putative function of the protein is indicated above each arrow. Genes exhibiting ≥70% amino acid similarity are linked by blue shading, and genes exhibiting ≤69% amino acid similarity are linked by violet shading. The levels of amino acid identity, expressed as percentages, are indicated.
We also analyzed the groEL and groES gene composition of a phylogenetically distant taxon, B. lactis LMG 18906. Thus, screening of a clone library of B. lactis LMG 18906 revealed the presence of two clones, whose groEL and groES gene products exhibited significant amino acid homology with the groEL and groES gene products of B. breve UCC 2003. By using a PCR amplification strategy it was possible to extend the region surrounding the groEL and groES genes of B. lactis LMG 18906.
The deduced amino acid sequences encoded by the B. breve UCC 2003 groEL and groES loci were aligned with those of B. lactis LMG 18906, B. longum NCC 2705, B. longum DJO10A, Streptomyces coelicolor A3, L. johnsonii NCC 533, and E. coli K-12 (Fig. 1). In B. breve UCC 2003 the groEL gene is located directly downstream of the cspA gene (encoding a predicted major cold shock protein) and upstream of a hypothetical ORF. Comparative analysis of the cspA-encoded product with proteins in the databases revealed a high degree of similarity with several cold shock proteins from various high-G+C-content bacteria and also a significant level of similarity with CspA of E. coli. Furthermore, CspA of B. breve shared extensive features with other CspA proteins, including an acidic isoelectric point, the presence of the RNP-1 motif (KGFGFIQP), and the absence of cysteine residues. The predicted B. breve UCC 2003 CspA protein contains the consensus cold shock domain that has been described as being highly conserved among CspA homologs; this protein is thought to be involved in the binding to DNA or RNA (26).
The protein comparison showed that the proteins most similar to B. breve CspA were those from B. longum strains NCC 2705 and DJO10A. In contrast, CspA from B. lactis LMG 18906 exhibited high levels of similarity with CspA proteins of many low-G+C-content bacteria (L. lactis, Lactobacillus plantarum, and Clostridium actetobutylicum). The groES gene is preceded by a gene encoding a hypothetical protein and is followed by an rpmG gene that encodes the putative ribosomal protein L33. Interestingly, two tRNA genes specific for Met (CAT anticodon) and Tyr (GTA anticodon) were located in the intergenic region between the groES and rpmG genes. Furthermore, an insertion (IS)-like element belonging to the IS3 family was identified between the two tRNA genes, and it was identical to an IS-like element (ISBlo3a) identified in the genome of B. longum NCC 2705 (31). The DNA region spanning the tRNA genes and the IS-like sequences showed a level of similarity of more than 80% in the bifidobacterial strains used.
The analysis of the genome sequences of B. longum NCC 2705 and DJO10A revealed similar physical locations for the groEL and groES genes, which were similar to those observed in B. breve UCC 2003. We found that the groES and groEL genes were located in different chromosome regions and not on a contiguous DNA segment.
The overall genetic structure of the cspA-groEL region was highly conserved among bifidobacteria. In fact, PCR amplification with a primer pair targeting conserved DNA sequences within the cspA and groEL genes yielded the expected amplicons for all bifidobacterial species used. Subsequently, sequencing of these amplicons confirmed the conserved cspA-groEL organization (data not shown). Surprisingly, this genetic organization of the groEL locus does not resemble the organization of any other groEL operon described so far. The only exception to this finding is S. coelicolor A3, which has a similar cspA-groEL gene arrangement, but the similarity at the amino acid level with the homologous proteins of B. breve UCC 2003 was low. Interestingly, S. coelicolor A3 contains two copies of the groEL genes, and only groEL2 is located next to the cspA gene, whereas groEL1 is adjacent to a groES gene (Fig. 1 and data not shown).
Heat induction of the B. breve UCC 2003 groEL gene.
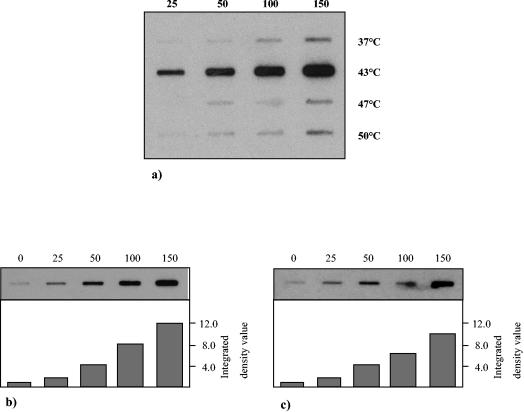
To evaluate the heat shock response in B. breve UCC 2003 and to determine the most effective temperature for subsequent groESL induction experiments, a slot blot hybridization procedure was used to test RNA which was isolated from B. breve UCC 2003 cultures grown for different lengths of time at temperatures ranging from 37 to 50°C. Based on the intensity of the hybridization signal, the highest expression of the groEL gene in this temperature range occurred at 43°C (Fig. 2a). To verify this finding and to calculate the extent of heat induction, RNA was isolated from heat-treated cultures of B. breve and used as a target in an RNA slot blot analysis with radiolabeled probes for the groEL and groES genes. The levels of groES and groEL mRNAs were induced approximately 8- and 12-fold, respectively, when bacterial cells were subjected to heat stress for 150 min (Fig. 2b and c).
FIG. 2.
Heat shock induction of the B. breve UCC 2003 groEL and groES loci. Total RNA was isolated from B. breve UCC 2003 following exposure to various temperatures for specific times and was analyzed by slot blot hybridization. (a) All slots, each of which contained 25 μg of RNA from cells incubated for up to 150 min at a temperature from 37 to 50°C, were probed with 32P-labeled PCR products corresponding to the groEL gene. (b) All slots, each of which contained 25 μg of RNA from cells incubated for up to 150 min at 43°C, were probed with 32P-labeled PCR products corresponding to the groEL gene. (c) All slots, each of which contained 25 μg of RNA from cells incubated for up to 150 min at 43°C, were probed with 32P-labeled PCR products corresponding to the groES gene. The numbers above the slot blots indicate the incubation times (in minutes), while the temperatures are indicated on the right in panel a.
Transcription analysis of groEL and groES loci.
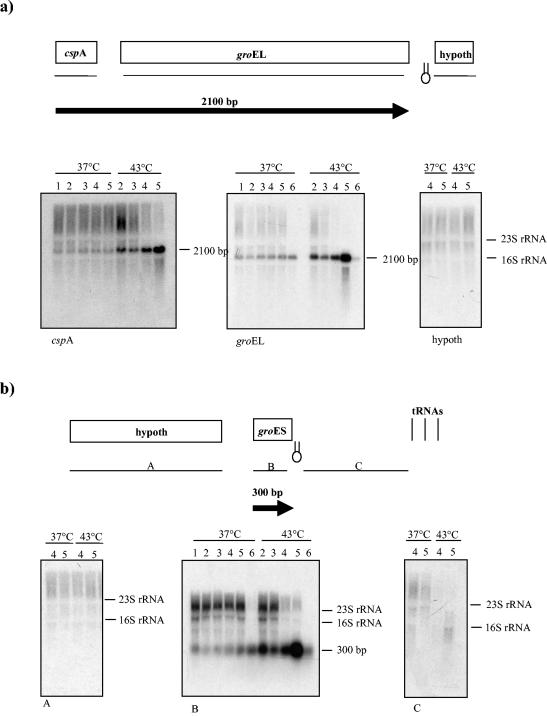
Northern hybridization experiments were performed in order to determine whether the groEL and groES genes were cotranscribed with their flanking genes. Total RNA was extracted from culture of B. breve UCC 2003 grown at 37°C or under heat stress conditions (43°C). The transcription of the groEL gene was investigated by Northern blotting by using an internal groEL probe. A 2.1-kb transcript was detected in RNA extracted from 37 and 43°C samples. The shift to heat shock conditions (43°C) clearly increased the strength of expression of the 2.1-kb transcript (Fig. 3a). When a probe spanning the cspA gene was used in Northern blot hybridization experiments, a 2.1-kb signal was still detected. The cspA gene had kinetics of activation similar to that of the groEL gene. The transcription levels of both genes increased upon a temperature shift and reached the maximum value at 150 min. This result indicated that the cspA and groEL genes form a monocistronic unit. When Northern hybridization was performed with a probe corresponding to the ORF following the groEL gene, no transcripts were detected, suggesting that this gene is not part of the groEL locus. An inverted repeat was observed in the region immediately downstream of the groEL gene, which may serve as the terminator sequence (Fig. 3a). Northern hybridization with a specific groES probe yielded a 0.3-kb signal whose size did not change after heat shock (Fig. 3b). However, the levels of groES-specific mRNA increased upon heat shock. Northern analysis of the DNA sequences surrounding the groES gene with RNA extracted from unstressed and heat-shocked cells did not reveal any transcripts, indicating that these genes do not belong to the groES locus. Analysis of the nucleotide sequence of the groES locus revealed that the groES gene was delimited at its 3′ end by a strong inverted repeat that may function as a terminator sequence (Fig. 3b).
FIG. 3.
Northern blot analysis of the B. breve UCC 2003 groEL (a) and groES (b) loci. The mRNAs isolated from cultures maintained under normal or heat shock conditions were probed with PCR fragments corresponding to the groEL, groES, and cspA genes and genes located in intergenic or upstream regions. Schematic representations of the transcription maps of the groEL and groES loci are included. All predicted ORFs are indicated and are annotated with their database matches. The locations of the probes used are indicated by the lines below the gene maps. The transcripts are indicated by arrows, and the arrows point to the 3′ end of the mRNA. The estimated size of each transcript is indicated. Hairpins indicate possible rho-independent terminators. The transcripts are positioned with respect to the genome map shown above. The DNA probes used for hybridization are indicated as thin lines below the genome map. Each blot contained mRNA extracted from B. breve UCC 2003 maintained under normal or heat shock conditions. Lane 1, RNA isolated from a culture at the beginning of the experiments; lane 2, RNA isolated from a culture at 25 min upon a temperature shift; lane 3, RNA isolated from a culture at 50 min upon a temperature shift; lane 4, RNA isolated from a culture at 100 min upon a temperature shift; lane 5, RNA isolated from a culture at 150 min upon a temperature shift; lane 6, RNA isolated from a culture after 15 h upon a temperature shift. The estimated length of the transcript corresponding to the hybridization signal is indicated. hypoth, hypothetical open reading frame.
Identification of transcription start sites of the groEL and groES loci.
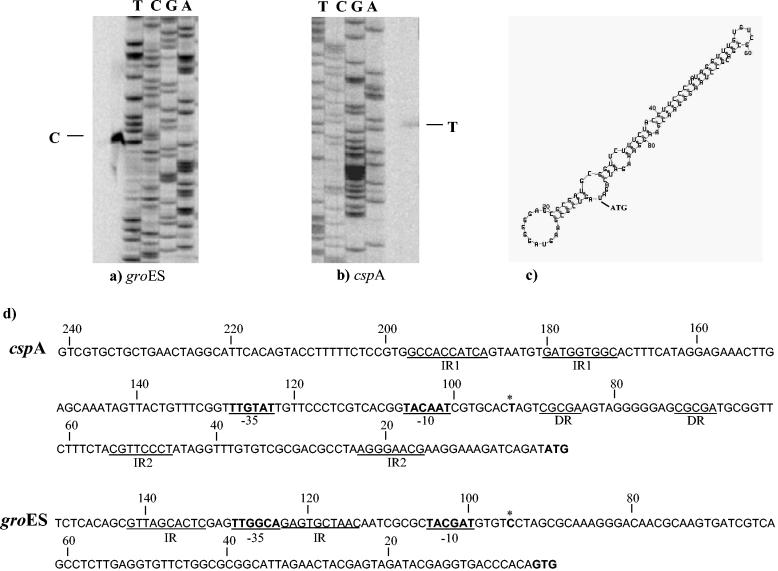
To determine the general characteristics and organization of the cspA and groES promoters, transcription initiation sites were mapped by primer extension. Primer extension experiments were performed by using total RNA isolated from heat-shocked and unshocked cells. The transcriptional start sites were identified upstream of the assumed start codons of the cspA and groES genes (Fig. 4a and b). Analysis of the putative promoter regions revealed a potential promoter-like sequence having a putative −10 hexamer and −35 box (Fig. 4d). The same 5′ terminus was found for the transcripts synthesized at 37°C (data not shown). Thus, the cspA and groES promoters are functional under stressed and unstressed conditions. Notably, in the groES promoter sequence two inverted repeats (GTTAGCACTC) were detected in the region surrounding the −35 box. These inverted repeats varied by only one mismatch from a regulatory structure termed CIRCE (33), which has been demonstrated to be involved in expression of the groES gene in many bacteria. In the cspA promoter region a 10-bp inverted repeat (GCCACCATCA) was detected upstream of the −35 box, and an 8-bp inverted repeat (CGTTCCCT) and a 5-bp direct repeat were found in the untranslated leader sequences. The promoter region of cspA and groES revealed the presence of long 92- and 93-bp untranslated leader sequences, respectively. Computer analysis of the secondary structure of the untranslated leader sequence of the putative cspA promoter predicted the formation of an extensive hairpin-like structure (Fig. 4c). This feature is shared with several cold shock genes, and it has been shown to play a crucial role in mRNA stability and in the translation efficiency of the cspA mRNA (53, 54).
FIG. 4.
Determination of B. breve UCC 2003 cspA and groES gene transcription start sites by primer extension analysis. (a and b) Primer extension results obtained by using oligonucleotides targeting the 5′ ends of the groES and cspA genes. (c) Computer prediction of the secondary structure of the untranslated leader sequence of mRNA. (d) Comparison of the putative promoter sequences for the cspA and groES genes. Boldface type and underlining indicate the −10 and −35 putative hexamers; boldface type with an asterisk indicates the transcription start point; boldface type without asterisks indicates the start codon. DR, direct repeats; IR1 and IR2, inverted repeats.
Investigation of the copy number of the groEL and groES genes in the genomes of different bifidobacterial species.
Many species of bacteria (including high-G+C-content gram-positive bacteria) contain several genes homologous to groEL (33). To determine whether the members of the genus Bifidobacterium also contain multiple copies of the groEL and groES genes, the amplified groEL and groES DNAs were used as probes in Northern blot experiments and hybridized to genomic DNAs of 12 bifidobacterial species digested with the enzymes EcoRI, EcoRV, and XbaI (data not shown). Each of the bifidobacterial strains examined yielded a single band, and the bands were different sizes (ranging from 1,600 to 5,100 bp for the groEL probe and from 1,800 to 5,000 bp for the groES probe), suggesting that only single copies of the groEL and groES genes are present in all of these genomes. These findings were also confirmed by sequence analysis of the entire genomes of B. breve UCC 2003 (S. Leahy, J. A. M. Munoz, G. F Fitzgerald, D. G. Higgins, and D. van Sinderen, unpublished data), B. longum NCC 2705 (31), and B. longum DJO10A (GenBank accession numbers NZ_AABM02000001 to NZ_AABM02000120 [DOE Joint Genome Institute]), each of which harbors a unique copy of the groEL and groES genes.
Sequencing and phylogenetic analysis based on groEL and groES sequences.
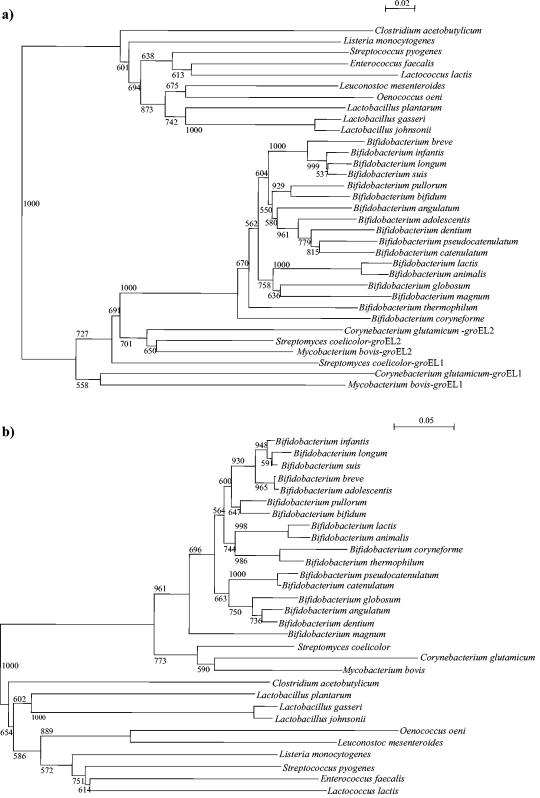
Several groES sequences retrieved from public database were aligned and compared. Two conserved regions were identified, and two PCR primers (gro-1 and gro-2) amplifying a 200-bp region were designed. This primer pair allowed amplification of the central part of the groES gene from 16 Bifidobacterium strains. The sequence alignments of the groES genes were used to examine the phylogenetic relationships of the bifidobacterial species included in our analysis, as well as other strains belonging to different genera representing the high- and low-G+C-content gram-positive bacteria (Fig. 5b). A phylogenetic tree was also generated by using partial groEL sequences retrieved from publicly available databases. The groEL-based tree was designed by using the same set of strains that were employed for the groES gene-based tree (Fig. 5). In order to evaluate the reliability of the branching of the trees, a bootstrap analysis was performed. Both trees showed that the gram-positive bacteria form two groups based on the different G+C contents: the low-G+C-content bacteria (the genera Lactobacillus, Lactococcus, Streptococcus, Listeria, Clostridium, Oenococcus, Leuconostoc, and Enterococcus) and the high-G+C-content bacteria (Bifidobacterium, Mycobacterium, Corynebacterium, and Streptomyces). The genera Mycobacterium, Streptomyces, and Corynebacterium have been described as organisms that contain more than one copy of the groEL gene (33). Phylogenetic analysis of these bacteria based on the partial groEL gene sequence showed that all groEL2 genes clustered together, while the groEL1 sequences resulted in a separate cluster. The bifidobacterial groEL genes branched with the groEL2 gene from high-G+C-content gram-positive bacteria, suggesting a common origin.
FIG. 5.
Phylogenetic trees obtained by using the groEL (a) and groES (b) genes. The bar scales indicate phylogenetic distances. Bootstrap values are indicated for a total of 1,000 replicates. The trees were calculated by the neighbor-joining method as implemented in the neighbor module of PHYLIP.
Comparison of the two phylogenetic trees shows that there are many branching discrepancies (Fig. 6). The phylogenetic positions of many bifidobacterial species were bound to be very different in the two trees. In order to improve the accuracy of our phylogenetic estimates, we traced trees using different methods. The tree topologies obtained had similar hierarchical arrangements (data not shown). A phylogenetic tree was also constructed on the basis of the 16S rDNA sequences available in databases by using the same set of strains that were employed to construct the groES- and groEL-based trees. The phylogenetic positions of bifidobacterial species based on groEL sequences were generally in agreement with those determined by using the 16S rDNA sequences but were not in agreement with the groES-based phylogeny (data not shown). These findings were also confirmed by the relationship between the pairwise distances of the 16S rRNAs and the synonymous distances for the groEL and groES genes. In fact, the correlation between the genetic distances of the 16S rDNA sequences and the distances of the groES genes was very low (r = 0.26), whereas there was a significant correlation (r = 0.94) between the genetic distances of the16S rDNA sequences and those of the groEL sequences.
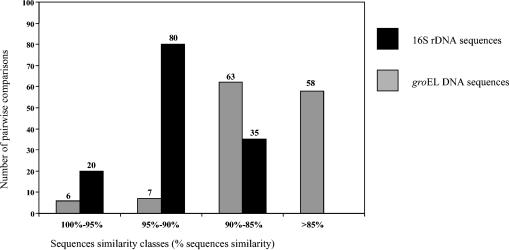
FIG. 6.
Histograms showing the distribution of the chaperonin 60 gene (grey bars) and 16S rRNA (solid bars) pairwise DNA sequence identities for bifidobacteria. A total of 135 pairwise comparisons were used for each gene.
Comparison between 16S rRNA distances and groEL gene distances.
The pairwise distances (for 135 comparisons) between groEL gene sequences were calculated by the maximum-likelihood procedure (8) and were compared to the distances between the corresponding 16S rDNA sequences. The results are presented as a histogram in Fig. 6. The chaperonin groEL gene sequences of bifidobacteria were found to be significantly more distant from each other than the 16S rDNA sequences were; the average pairwise groEL gene distance was 0.16, compared to an average 16S rRNA distance of 0.084. The chaperonin groEL gene sequences also showed a greater diversity; there were 7 distances of 95 to 90% in the chaperonin groEL gene distance matrix, compared to 80 such distances in the 16S rRNA distance matrix, again indicating that the chaperonin groEL gene sequences are more discriminating.
When we aligned the partial groEL gene sequences of bifidobacterial species included in Table 1, we noticed that most of the base substitutions in the groEL sequences were synonymous; i.e., they did not result in amino acid changes. In addition, attention was focused on closely related taxa, like B. lactis and B. animalis strains or B. longum, B. suis, and B. infantis strains. Twelve nucleotide substitutions were observed when the groEL sequences of B. lactis and B. animalis strains were compared, but only two of these substitutions contributed to an amino acid substitution. In parallel, 11 synonymous nucleotide substitutions were noticed for the groEL gene sequences of B. longum, B. suis, and B. infantis strains. Interestingly, many of these base differences were thymine or adenine in B. longum and cytosine in B. infantis and B. suis. In addition, most of the base substitutions were adenine or thymine in B. lactis and cytosine or guanine in B. animalis.
DISCUSSION
The GroEL and GroES chaperones have been extensively studied in low-G+C-content gram-positive bacteria, whereas very little is known about these proteins in high-G+C-content gram-positive bacteria, like bifidobacteria. In the present study we genetically characterized the individual transcription units containing the groEL and groES genes of B. breve UCC 2003. In contrast to other high-G+C-content gram-positive bacteria, which may contain multiple copies of chaperonin-encoding genes (33), bifidobacterial genomes were shown to contain just a single groEL homolog.
The genetic organization of the groEL and groES loci in B. breve, as well as in B. longum and B. lactis, is without precedent in the bacterial world, which could be of great interest from an evolutionary point of view. The only exception to this finding is S. coelicolor A3, which has a similar cspA-groEL gene arrangement; however, unlike the situation in bifidobacteria, the S. coelicolor A3 cspA gene is located downstream of the groEL gene, which might indicate that there are different gene regulation mechanisms for the cspA-groEL locus in these species. The bifidobacterial groEL and groES genes are not organized as a monocistronic operon, while a short ORF is located upstream of the groEL gene, which encodes a protein which exhibits significant amino acid homology with the cspA gene product of E. coli (52).
Interestingly, we showed that in bifidobacterial genomes the groEL and groES genes are located in two different chromosome regions, and an ISBlo3a element was identified adjacent to the groES chaperonin-encoding gene. The significance of this genomic arrangement is unknown, but a similar organization has been reported for IS1223 and groES in L. johnsonii (50).
Phylogenetic analysis of bifidobacterial species based on the groES and groEL genes indicates that these genes evolved differently. The data presented here show that the phylogenetic position of bifidobacteria based on GroEL-encoding sequences is generally in agreement with 16S rRNA-based phylogeny but not with groES-based phylogeny. The discrepancies in the branching orders suggest that the groES gene in bifidobacteria might have been acquired through horizontal transfer in one lineage at an earlier stage of divergence. Another possible scenario which might explain this gene inventory situation may be that an early incomplete operon duplication generated a second copy of the groEL gene in the Actinobacteria, which subsequently came under the control of the promoter of a cspA-like gene. While the environmental Actinobacteria maintained the original groEL gene in an operon with groES, the groEL copy was lost by deletion early in speciation of bifidobacteria since all of these organisms seem to lack this gene.
Since the groES and groEL sequences have been used for phylogenetic purposes (4, 12, 14, 17, 18, 36, 37), the existence of the groES gene originating from horizontal gene transfer or by duplication followed by a deletion event may alter the phylogeny when this gene is used for such an analysis. Hence, this finding, together with the fact that the groES gene evolved separately and distantly from the classical molecular marker, makes the Hsp10-encoding gene an unreliable molecular evolutionary clock for inferring the phylogeny of bifidobacteria. Conversely, the GroEL-encoding gene fulfils all of the prerequisites for being a suitable phylogenetic marker, such as a wide distribution, very high genetic stability, and no exchangeability among lineages by horizontal gene transfer (24). In this study, we confirmed the robustness of the groEL genes as a molecular marker, as proposed by Jian et al. (18). Our results showed that the groEL sequences provide superior discrimination between closely related strains (e.g., B. animalis and B. lactis or B. longum, B. infantis, and B. suis) compared to the 16S rRNA sequences and at the same time produce results which closely parallel those of a 16S rRNA phylogenetic analysis. Interestingly, most of the nucleotide differences between these closely related taxa might be a consequence of the spontaneous deamination of cytosine. A similar finding was described for mutation of the tuf and recA gene sequences of B. lactis and B. animalis (44) and for mutation of the 16S rDNA of Lactobacillus delbrueckii in relation to its speciation (11). These sequence signatures can be used directly to design specific PCR primers or as a target for specific restriction enzymes that provide species-specific restriction fragment length polymorphism patterns. Use of the groEL sequences, as well as the tuf (5, 44, 45), recA (44), and atpD genes (42), as phylogenetic markers for bifidobacteria has the advantage that the amino acid sequences can be used to infer bacterial phylogenies, which avoids the problems of rRNAs and likely overestimation of the relatedness of taxa with similar nucleotide differences, a lack of independence of substitution patterns at different sites, and bias resulting from different G+C contents (7, 27).
The bacterial heat stress response is a very complicated mechanism, which involves a large arsenal of proteins (41). For many bacteria it has been demonstrated that exposure to high temperature and the subsequent protein denaturation are followed by an increase in the amount of Hsp60 and Hsp10 (3, 6, 10, 20, 50). Identification of the genetic basis of heat resistance for industrially applicable bifidobacterial strains that are more resistant to high temperatures during food manufacture (e.g., spray-drying) is highly desirable.
This study showed that in bifidobacteria the groEL and groES genes are strongly induced during growth at 43°C, which is in agreement with the observation that the activity of Hsp10 and Hsp60 in related high-G+C-content bacteria is enhanced upon heat shock (32, 34). Interestingly, transcription of the groEL-groES genes from B. breve occurs at a normal growth temperature and is increased significantly upon a shift to a higher temperature. The level of groEL transcription seems to be considerably higher than the level of groES transcription. A similar observation was described for S. lividans, in which one of the two copies of the groEL gene (groEL2) showed a higher level of heat induction (6). In the latter case it was suggested that groEL2 chaperone activity might not require the presence of a cochaperonin. It may be that a similar situation occurs in B. breve.
We demonstrated that the CspA- and GroEL-encoding genes are cotranscribed and belong to the same transcription unit. Primer extension experiments precisely mapped the start of the transcript in the cspA-groEL operon. A 2.1-kb transcript derived from the cspA promoter covers all of the cspA-groEL operon of B. breve UCC 2003. Cotranscription of the cspA and groEL genes might suggest a common function for these genes following environmental stresses. Like E. coli CspA and B. subtilis CspB (13, 52), it is possible that CspA of B. breve acts as an RNA chaperone or has a role in ensuring protein synthesis from the groEL gene. In fact, groEL transcripts of B. breve contain a large number of high-energy RNA secondary structures, which can reduce its translation level. Hence, it may be that CspA acts as an RNA chaperone by preventing the formation of extensive secondary structures along groEL mRNAs. Moreover, cold shock proteins have been described as functioning as molecular chaperones that act upon the structure of preexisting polypeptides by assisting the refolding of denatured proteins in a concerted action with GroEL and GroES chaperonins (15). Therefore, CspA of B. breve might act as a molecular chaperonin by interacting with the GroEL chaperonin in assisting protein folding. A likely ancient function of the CspA protein was binding to nucleic acids. It is possible that the genes came under the control of different promoters during the course of evolution of gram-positive and gram-negative bacteria, leading to the acquisition of mutations which resulted in CspA becoming a true cold shock regulatory protein in the evolutionarily younger proteobacteria, whereas it became a heat shock regulatory protein in bifidobacteria.
The heat shock proteins are highly conserved, whereas control of their expression is highly variable among organisms, even among various bacteria (32). Expression of the groEL and groES genes in many bacteria has been reported to be governed by the widespread HrcA-CIRCE control system (1, 54). HrcA, a repressor protein, negatively regulates transcription of the groES-groEL genes by binding to a DNA element called CIRCE (for controlling inverted repeat of chaperone expression) upon heat shock. Sequence analysis of the promoter region of the groEL locus did not reveal any consensus CIRCE operator sequence, whereas the groES promoter region contains a nearly perfect consensus CIRCE sequence. Control of chaperone expression by the HrcA-CIRCE system has been postulated to be more ancient than the σ32-dependent transcription of heat shock genes (28); consequently, the presence of CIRCE sequences in the groES promoter region of bifidobacteria is another sign supporting the ancient presence of a traditional groESL operon in the ancestors of all bifidobacteria, whose descendants (the extant bifidobacteria) maintained the CIRCE sequence but lost the groEL gene.
Interestingly, in the genome sequence of B. longum NCC 2705 a heat shock sigma factor has been identified (31), which might be implicated in regulation of the expression of the cspA-groEL and groES loci.
A better understanding of the mechanisms of heat stress resistance or other adaptive responses and the associated cross-protection is expected to lead to full exploitation of fitter bifidobacteria for industrial processes (32). In this context, future genome and transcriptome analyses should increase the genetic information available and shed new light on the perception of, and the response to, stress by bifidobacteria.
Acknowledgments
This work was financially supported by Enterprise Ireland (grant BR/1998/202), by the Higher Education Authority Programme for Research in Third Level Institutions, and by the Science Foundation Ireland Alimentary Pharmabiotic Centre located at University College Cork.
We thank S. Leahy, J. A. M. Munoz, and H. G. Higgins for sharing unpublished data with us.
REFERENCES
- 1.Baldini, R. L., M. Avedissian, and S. L. Gomes. 1998. The CIRCE element and its putative repressor control cell cycle expression of the Caulobacter crescentus groESL operon. J Bacteriol. 180:1632-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bengmark, S. 1998. Ecological control of the gastrointestinal tract. The role of probiotic flora. Gut 42:2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broadbent, J. R., C. J. Orberg, and L. Wei. 1998. Characterization of the Lactobacillus helveticus groELS operon. Res. Microbiol. 149:247-253. [DOI] [PubMed] [Google Scholar]
- 4.Brousseau, R., J. E. Hill, G. Prefontaine, S. H. Goh, J. Harel, and S. M. Hemmingsen. 2001. Streptococcus suis serotype characterization by analysis of chaperonin 60 gene sequences. Appl. Environ. Microbiol. 67:4828-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavagnat, F., M. Haueter, J. Jimeno, and M. G. Casey. 2002. Comparison of partial tuf gene sequences for the identification of lactobacilli. FEMS Microbiol. Lett. 217:177-183. [DOI] [PubMed] [Google Scholar]
- 6.de Leon, P., S. Marco, C. Isiegas, A. Marina, J. L. Carrascosa, and R. P. Mellado. 1997. Streptomyces lividans groES, groEL1 and groEL2 genes. Microbiology 143:3563-3571. [DOI] [PubMed] [Google Scholar]
- 7.Eisen, J. A. 1995. The recA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of recAs and 16S rRNAs from the same species. J. Mol. Evol. 41:1105-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 9.Fujita, M., A. Amemura, and H. Aramaki. 1998. Transcription of the groESL operon in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 163:237-242. [DOI] [PubMed] [Google Scholar]
- 10.Georgopoulus, C., and W. J. Welch. 1993. Role of the major heat shock proteins as molecular chaperones. Annu. Rev. Cell Biol. 9:601-634. [DOI] [PubMed] [Google Scholar]
- 11.Germond, J. E., L. Lapierre, M. Delley, B. Mollet, G. E. Felis, and F. Dellaglio. 2003. Evolution of the bacterial species Lactobacillus delbrueckii: a partial genomic study with reflections on prokaryotic species concept. Mol. Biol. Evol. 20:93-104. [DOI] [PubMed] [Google Scholar]
- 12.Goh, S. H., S. Potter, J. O. Wood, S. M. Hemmingsen, R. P. Reynolds, and A. W. Chow. 1996. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J. Clin. Microbiol. 34:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graumann, P., and M. A. Marahiel. 1998. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 23:286-290. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, R. S. 1995. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and origin of eukaryotic cells. Mol. Microbiol. 15:1-11. [DOI] [PubMed] [Google Scholar]
- 15.Guy, C. L. 1990. Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41:187-223. [Google Scholar]
- 16.Hecker, M., W. Schumann, and U. Volker. 1996. Heat-shock and general response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 17.Hill, J. E., R. P. Seipp, M. Betts, L. Hawkins, A. G. Van Kessel, W. L. Crosby, and S. M. Hemmingsen. 2002. Extensive profiling of a complex microbial community by high-throughput sequencing. Appl Environ Microbiol. 68:3055-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jian, W., L. Zhu, and X. Dong. 2001. New approach to phylogenetic analysis of the genus Bifidobacterium based on partial HSP60 gene sequences. Int. J. Syst. Evol. Microbiol. 51:1633-1638. [DOI] [PubMed] [Google Scholar]
- 19.Kaufmann, P., A. Pfefferkorn, M. Teuber, and L. Meile. 1997. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl. Environ. Microbiol. 63:1268-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilstrup, M., S. Jakobsen, K. Hammer, and F. K. Vogensen. 1997. Induction of heat shock proteins Dnak, GroEL, and GroES by salt stress in Lactococcus lactis. Appl. Environ. Microbiol. 63:1826-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kullen, M., L. J. Brady, and D. J. O'Sullivan. 1997. Evaluation of using a short region of the recA gene for rapid and sensitive speciation of dominant bifidobacteria in the human large intestine. FEMS Microbiol. Lett. 154:377-383. [DOI] [PubMed] [Google Scholar]
- 22.Langer, T., G. Pfeifer, J. Martin, W. Baumeister, and F. U. Hartl. 1992. Chaperonin-mediated protein folding: GroES binds to one end of the GroEL cylinder, which accommodates the protein substrate within its central cavity. EMBO J. 11:4757-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lievin V, I. Pfeiffer, S. Hudault, F. Rochat, D. Brassart, J. R. Neeser, and A. L. Servin. 2000. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut 47:646-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludwig, W., and K. H. Schleifer. 1999. Phylogeny of bacteria beyond the 16S rRNA standard. ASM News 65:752-757. [Google Scholar]
- 25.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto, K., and A. P. Wolffe. 1998. Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol. 8:318-323. [DOI] [PubMed] [Google Scholar]
- 27.Palys, T., L. K. Nakamura, and M. Chan. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145-1156. [DOI] [PubMed] [Google Scholar]
- 28.Rosen, R., and E. Z. Ron. 2002. Proteome analysis in the study of the bacterial heat-shock response. Mass Spectrom. Rev. 21:244-265. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Satokari, R. M., Vaughan, E., Akkermans, A. D. L., Saaerela, M., and W. M. De Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt, G., and R. Zink. 2000. Basic features of stress response in three species of bifidobacteria: B. longum, B. adolescentis, and B. breve. Int. J. Food Microbiol. 55:41-45. [DOI] [PubMed] [Google Scholar]
- 33.Segal, G., and E. Z. Ron. 1996. Regulation and organization of the groE and dnaK operons in eubacteria. FEMS Microbiol. Lett. 138:1-10. [DOI] [PubMed] [Google Scholar]
- 34.Servant, P., and P. Mazodier. 2001. Negative regulation of the heat shock response in Streptomyces. Arch. Microbiol. 176:237-242. [DOI] [PubMed] [Google Scholar]
- 35.Tannock, G. W. 1994. The acquisition of the normal microflora of the gastrointestinal tract, p. 1-16. In S. A. Gibson (ed.), Human health: the contribution of microorganisms. Springer, London, United Kingdom.
- 36.Teng, L. J., P. R. Hsueh, J. C. Tsai, P. W. Chen, J. C. Hsu, H. C. Lai, C. N. Lee, and S. W. Ho. 2002. groESL sequence determination, phylogenetic analysis, and species differentiation for viridans group streptococci. J. Clin. Microbiol. 40:3172-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teng, L. J., P. R. Hsueh, Y. H. Wang, H. M. Lin, K. T. Luh, and S. W. Ho. 2001. Determination of Enterococcus faecalis groESL full-length sequence and application for species identification. J. Clin. Microbiol. 39:3326-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Török, Z., P. Goloubinoff, I. Horváth, N. M. Tsvetkova, A. Glatz, G. Balogh, V. Varvasovszki, D. A. Los, E. Vierling, J. H. Crowe, and L. Vígh. 2001. Synechocystis HSP17 is an amphitropic protein that stabilizes heat-stressed membranes and binds denatured proteins for subsequent chaperone-mediated refolding Proc. Natl. Acad. Sci. USA 98:3098-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandamme, P., B. Pot, M. Gillis, P. de Vos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van de Gouchte, M., P. Serror, C. Chervaux, T. Smokvina, S. D. Ehrlich, and E. Manguin. 2002. Stress responses in lactic acid bacteria. Antonie Leeuwenhoek 82:187-216. [PubMed] [Google Scholar]
- 42.Ventura, M., C. Canchaya, D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Bifidobacterium lactis DSM 10140: identification of the atp (atpBEFHAGDC) operon and analysis of its genetic structure, characteristics, and phylogeny. Appl. Environ. Microbiol. 70:3110-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ventura, M., D. van Sinderen, G. F. Fitzgerald, and R. Zink. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Leeuwenhoek, in press. [DOI] [PubMed]
- 44.Ventura, M., and R. Zink. 2003. Comparative sequence analysis of the tuf and recA genes and restriction fragment length polymorphism of the internal transcribed spacer region sequences supply additional tools for discriminating Bifidobacterium lactis from Bifidobacterium animalis. Appl. Environ. Microbiol. 69:7517-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ventura, M., C. Canchaya, V. Meylan, T. R. Klaenhammer, and R. Zink. 2003. Analysis, characterization, and loci of the tuf genes in Lactobacillus and Bifidobacterium and their direct application for species identification. Appl. Environ. Microbiol. 69:6908-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ventura, M., V. Meylan, and R. Zink. 2003. Identification and tracing of Bifidobacterium species by enterobacterial repetitive intergenic consensus sequences. Appl. Environ. Microbiol. 69:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ventura, M., and R. Zink. 2002. Rapid identification, differentiation, and proposed new taxonomic classification of Bifidobacterium lactis. Appl. Environ. Microbiol. 68:6429-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ventura, M., R. Reniero, and R. Zink. 2001. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl. Environ. Microbiol. 67:0-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ventura, M., M. Elli, R. Reniero, and R. Zink. 2001. Molecular microbial analysis of Bifidobacterium isolates from different environments by the species-specific amplified ribosomal DNA restriction analysis (ARDRA). FEMS Microbiol. Ecol. 36:113-121. [DOI] [PubMed] [Google Scholar]
- 50.Walker, D. C., H. S. Girgis, and T. R. Klaenhammer. 1999. The groESL chaperone operon of Lactobacillus johnsonii. Appl. Environ. Microbiol. 65:3033-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weissman, J. S., Y. Kashi, A. Fenton, and A. L. Horwich. 1994. GroEL-mediated protein folding proceeds by multiple rounds of binding and release of nonnative forms. Cell 78:693-702. [DOI] [PubMed] [Google Scholar]
- 52.Yamanaka, K., L. Fang, and M. Inouye. 1998. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol. Microbiol. 27:247-256. [DOI] [PubMed] [Google Scholar]
- 53.Yamanaka, K., M. Mitta, and M. Inouye. 1999. Mutation analysis of the 5′ untranslated region of the cold shock cspA mRNA of Escherichia coli. J. Bacteriol. 181:6284-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuber, U., and W. Schumann. 1994. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J. Bacteriol. 176:1353-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]