Abstract
Many species of gram-negative bacteria communicate by synthesizing, secreting, and responding to N-acylhomoserine lactones (AHLs), a mechanism termed quorum sensing. Several investigations have characterized numerous AHL-degrading enzymes (AiiA lactonases) encoded by environmental isolates of Bacillus spp. The Burkholderia thailandensis quorum system is comprised of at least three AHL synthases (AHSs) and five transcriptional regulators belonging to the LuxIR class of proteins. Expression of the Bacillus anthracis (Ames strain) AiiA lactonase in B. thailandensis completely abolished the accumulation of N-decanoylhomoserine lactone (C10-HSL) and N-octanoylhomoserine lactone (C8-HSL), reduced N-hexanoylhomoserine lactone (C6-HSL) levels, altered both swarming and twitching motility, caused a significant increase in generation time, and affected carbon metabolism. In contrast, heterologous expression of the Bacillus cereus strain A24 AiiA lactonase in B. thailandensis reduced the concentrations of C6-HSL, C8-HSL, and C10-HSL to nondetectable levels; altered both swarming and twitching motility; and caused fluctuations in carbon utilization. Individual disruption of the B. thailandensis AHSs, specifically disruption of the btaI1 and btaI3 genes, which encode the proteins that direct the synthesis of C8-HSL and C6-HSL, respectively, caused the hyper-beta-hemolysis of sheep erythrocytes on blood agar plates. In contrast, AHL cleavage in B. thailandensis by the Bacillus AiiA lactonases failed to enhance beta-hemolytic activity. The results of this study demonstrate that heterologous expression of Bacillus sp. AiiA lactonases in B. thailandensis reduced AHL accumulation, affected both swarming and twitching motility, increased generation time, altered substrate utilization, and prevented the beta-hemolysis of sheep erythrocytes.
Many gram-negative bacteria utilize N-acylhomoserine lactones (AHLs) in a cell density-dependent manner, referred to as quorum sensing (QS), to control gene expression (14). When an adequate AHL concentration is reached, the signaling molecules are bound by LuxR proteins, which in turn either induces or represses gene expression. QS networks have been identified in dozens of bacterial species and have been shown to be involved in various physiological process (8, 12, 19, 36).
Burkholderia thailandensis is a gram-negative bacterium that is genetically and biochemically similar to Burkholderia pseudomallei (the causative agent of melioidosis) and Burkholderia mallei (the etiologic agent of glanders) (3, 16). Although the bacterium is genetically related to B. pseudomallei, B. thailandensis is an avirulent Burkholderia species that does not cause disease in humans. Comparative analysis of the B. pseudomallei and B. thailandensis 16S rRNAs revealed differences in 15 nucleotides, which justified the classification of a new Burkholderia species (3). The most striking difference between these organisms is their ability to cause acute disease in a Syrian hamster model. An intraperitoneal challenge of <101 CFU of B. pseudomallei causes mortality in hamsters within 3 to 5 days, in contrast to a dose of >106 CFU for B. thailandensis (3).
Several investigations have recently identified functional QS networks in Burkholderia species (2, 5, 17, 26, 27). In Burkholderia cepacia, QS negatively regulates ornibactin biosynthesis and positively induces protease and N-octanoylhomoserine lactone (C8-HSL) production (26, 27). Recently, an additional pair of luxIR genes (cciIR) were identified in B. cepacia and shown to be involved in pathogenicity (2). Burkholderia vietnamiensis produces multiple AHL molecules, with the predominant AHL being N-decanoylhomoserine lactone (C10-HSL) and with C8-HSL and N-hexanoylhomoserine lactone (C6-HSL) being produced to a lesser extent (5, 17). Our laboratory has recently identified a complex QS network encoded by B. thailandensis that is comprised of at least three AHL synthases and five putative transcriptional regulators homologous to LuxIR proteins, which were shown to affect swarming and twitching motility, lipase production, beta-hemolysis, and carbon metabolism (38).
Considering the diverse microbial species that use QS for the regulation of numerous physiological processes, applications targeted at disrupting this mode of intraspecies communication would be of significant interest to the medical, environmental, and agricultural communities. In the marine environment, Delisea pulchra (a microalga) produces brominated furanones that are hypothesized to compete with bacterial AHLs for the binding sites within putative LuxR proteins (29). In some culture environments, AHL signaling molecules can be utilized as sole carbon and energy sources by species of Pseudomonas, Variovorax, and Ralstonia (22, 24, 28). Xu et al. recently demonstrated that porcine kidney acylase I deacylates both N-butanoylhomoserine lactone (C4-HSL) and C8-HSL into l-homoserine (41). This acylase I-mediated deacylation of C4-HSL and C8-HSL also reduced biofilm formation in an aquarium water model (41). Enzymatic cleavage, specifically lactone ring hydrolysis, of signaling molecules by numerous Bacillus species has also been reported (9-11). These enzymes, termed AiiA lactonases, hydrolyze the lactone bond within the AHL moiety, thus changing the relative conformational structure of the signaling molecule, which prevents binding to the LuxR transcriptional regulator. Heterologous expression of the Bacillus cereus strain A24 AiiA lactonase in Pseudomonas aeruginosa PA01 reduced AHL accumulation, altered swarming motility, and decreased the expression and secretion of numerous virulence factors, including elastase, rhamnolipid, hydrogen cyanide, and pyocyanin (30). In Erwinia carotovora, a plant pathogen, aiiA expression reduced the synthesis of pectolytic enzymes and reduced pathogenicity in several plant models of infection (11).
This study analyzes the effect of C6-HSL, C8-HSL, and C10-HSL cleavage on twitching and swarming motility, beta-hemolysis, and carbon utilization by the expression of Bacillus sp. AiiA lactonases in B. thailandensis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All bacterial strains and plasmids used in this study are described in Table 1. B. thailandensis, Agrobacterium tumefaciens A136, and Escherichia coli were cultured in Luria-Bertani (LB) broth or on LB agar plates at 30 or 37°C, as required.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli | ||
| SM10 | Mobilizing strain; RP4 tra genes; Kmr | 35 |
| TOP10 | Used for cloning and blue-white screening | Invitrogen |
| S17-1 | thi pro hsdR recA; chromosomal RP4; Tra+; Smr Spr | 35 |
| A. tumefaciens bioreporter strain A136 | Lacks the Ti plasmid | 13 |
| B. thailandensis | ||
| DW503 | Derived from E264; Δ(amrR-oprA)(Kms Gms Sms); rpsL (Smr) | 4 |
| RU01 | DW503 containing pBHR1; Kmr | This study |
| RU02 | DW503 containing pBA01; Kmr | This study |
| RU03 | DW503 containing pME6000; Tcr | This study |
| RU04 | DW503 containing pME6863; Tcr | This study |
| BTRJ1 | DW503 derivative; 11::pRU11; Gmr | 38 |
| BTRJ3 | DW503 derivative; 13::pRU13; Gmr | 38 |
| BTRJ6 | DW503 derivative; R3::pRUR3; Gmr | 38 |
| Plasmids | ||
| pPCR2.1-TOPO | TA cloning vector; Kmr Apr | Invitrogen |
| pBHR1 | Mobilizable broad-host-range vector; Kmr Cmr | MoBiTee |
| pME6000 | Broad-host-range cloning vector; Tcr | 30 |
| pME6863 | pME6000 carrying the aiiA gene from B. cereus strain A24; Tcr | 30 |
| pBA01 | pBHR1 containing the B. anthracis (Ames strain) aiiA gene | This study |
Cloning of B. anthracis aiiA.
Genomic DNA for PCR amplification of the Bacillus anthracis (Ames strain) aiiA lactonase gene was purified by using the Puregene yeast and gram-positive bacterium kit according to the instructions of the manufacturer (Gentra Systems, Minneapolis, Minn.). The B. anthracis aiiA gene was PCR amplified by using the primer set (forward, 5′-ATG GGA TCC ATG ACA GTA AAG AAG CTT TAT-3′; reverse, 5′-GTC GAA TTC CTC AAC AAG ATA CTC CTA ATG-3′) reported by Dong et al. (9) and amplified as follows: one cycle of 94°C for 5 min; 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min; and a final 7-min extension at 72°C. All PCRs were performed with the FailSafe PCR system with buffer E (Epicentre Technologies, Madison, Wis.), and amplicons were subcloned into pCR2.1-TOPO (Invitrogen, Carlsbad, Calif.). Ligations were transformed into One Shot chemically competent E. coli (Invitrogen) and screened on LB plates containing 25 μg of kanamycin (Sigma, St. Louis, Mo.) per ml and 50 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) (Sigma) per ml. For heterologous expression in B. thailandensis, the B. anthracis aiiA gene was PCR amplified as described above with the forward primer 5′-CCG GAA TTC CGC ATG ACA GTA AAG AAG CTT TAT TTC G-3′ and reverse primer 5′-CAT GCC ATG GCA TGC TCA ACA AGA TAC TCC TAA TGA TGT-3′. (Boldface represents the EcoRI and NcoI sites incorporated into the oligonucleotides for directional cloning.) The resulting amplicon was digested with EcoRI and NcoI and cloned in frame into similarly digested pBHR1 by using the Fast-Link DNA ligation kit (Epicentre Technologies). Plasmid DNA was purified using the Wizard Plus Miniprep DNA kit (Promega, Madison, Wis.), electroporated into E. coli S17-1, and mobilized into B. thailandensis, and transconjugants were selected on LB plates containing 25 μg of kanamycin per ml and 15 μg of polymyxin per ml (Sigma) as described previously (37). The expression vectors pME6000 and pME6863 containing the B. cereus strain A24 aiiA lactonase gene were generously forwarded to our laboratory from Clemson University and mobilized into B. thailandensis as described above.
Analysis of AHL-degrading activity by B. thailandensis expressing the AiiA lactonases.
Extraction of AHLs from culture supernatants and preparative thin-layer chromatography (TLC) were performed as described by Shaw et al. (34). Extracts were reconstituted in 1 ml of acetonitrile (Sigma) and separated (60:40 [vol/vol] methanol solution) by spotting 10 μl of a 1:10 dilution of each extract onto a reversed-phase C18 plate (Analtech Inc., Newark, Del.). TLC plates were dried and overlaid with 30 ml of top agar (0.75% LB agar) containing 5 ml of A. tumefaciens A136 (from an overnight culture) and X-Gal. The plates were incubated at 30°C for 48 h or until adequate color development was achieved.
Motility and exoproduct secretion.
Protease and lipase production were examined by using methods described by DeShazer et al. (7). To assay for beta-hemolysis, colonies were transferred onto 5% sheep blood agar plates and incubated for 48 to 72 h at 37°C. Twitching motility and swarming motility were examined by using methods described by Reimmann et al. (30).
Growth kinetics and substrate utilization of B. thailandensis expressing the B. anthracis and B. cereus AiiA lactonases.
From a glycerol stock of RU01, -02, -03, or -04, 50 μl was inoculated into 3 ml of LB broth containing the appropriate antibiotics and incubated for 24 h at 37°C. For growth curve analysis, the optical density at 600 nm from the 3-ml overnight cultures was measured, and 25 μl was used to inoculate 250 ml of LB broth. The optical density at 600 nm was monitored hourly over a 15-h time course. At each time point, serial 10-fold dilutions were prepared and appropriate dilutions were plated onto LB plates containing selective antibiotics.
To examine whether carbon utilization was affected by AHL cleavage, BiOLOG (Hayward, Calif.) phenotypic arrays were employed. Each derivative strain (with the exception of RU02, which was grown for 48 h) was cultured on R2A agar plates (BiOLOG) with the appropriate antibiotics for 24 h at 37°C. Cellular densities were adjusted in IF-0 fluid (BiOLOG), and plates were inoculated with 100 μl of the cell suspension. Plates were incubated at 37°C for 24 to 48 h or until sufficient color development was reached. For growth in M9 minimal medium (31), 50 μl of wild-type B. thailandensis and RU04 from an overnight culture in LB broth was pelleted (14,000 rpm for 2 min with a Spectrafuge 16M [National Labnet Co., Edison, N.J.]), washed three times in phosphate-buffered saline, resuspended in 50 μl of phosphate-buffered saline, and inoculated into M9 minimal medium with and without d-arabinose as the sole carbon source.
RESULTS
Cloning of the B. anthracis aiiA gene and expression in B. thailandensis.
The B. anthracis aiiA lactonase gene was PCR amplified by using primers described by Dong et al. (9) and directionally cloned into the broad-host-range expression vector pBHR1 to produce pBA01. With pBA01 as the template, the B. anthracis aiiA lactonase gene was reamplified and sequenced to ensure fidelity during initial PCR amplification for subcloning. Nucleotide and amino acid sequence alignments between the B. anthracis aiiA and other closely related lactonase genes were performed (data not shown), and homologies were identical to the findings of Lee et al. (25). The B. cereus strain A24 aiiA lactonase gene was cloned and genetically characterized as described by Reimmann et al. (30).
Lactonase expression in B. thailandensis reduces AHL accumulation.
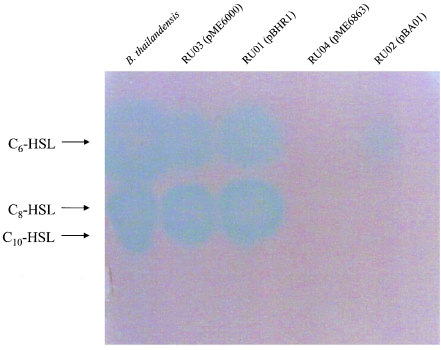
To examine the effect of aiiA lactonase expression on AHL accumulation in B. thailandensis, a semiquantitative approach was employed, incorporating soft-agar TLC overlays containing the bioreporter strain A. tumefaciens A136. Heterologous expression of the B. cereus AiiA lactonase in B. thailandensis reduced C6-HSL, C8-HSL, and C10-HSL to undetectable levels, while the B. anthracis AiiA lactonase reduced the level of C6-HSL but completely prevented the accumulation of C8-HSL and C10-HSL (Fig. 1, lanes 4 and 5). The concentrations of C6-HSL, C8-HSL, and C10-HSL in overnight culture supernatants have not been determined for wild-type B. thailandensis. However, the limits of detection for C6-HSL, C8-HSL, and C10-HSL by A. tumefaciens A136 are approximately 300, 2.4, and 100 pmol, respectively (34), indicating that the expression of these Bacillus sp. AiiA lactonases in B. thailandensis reduces the concentration of these signaling molecules below the threshold of detection for A. tumefaciens A136 (Fig. 1, lanes 4 and 5). Maintenance of each broad-host-range expression vector (pME6000 and pBHR1) in wild-type B. thailandensis had no effect on AHL production (Fig. 1, lanes 2 and 3).
FIG. 1.
Expression of the B. anthracis and B. cereus AiiA lactonases reduces AHL accumulation in B. thailandensis culture supernatants. TLC plates were overlaid with the bioreporter strain A. tumefaciens A136 and incubated at 30°C for 48 h. Each lane contains 10 μl (1:10 dilution) of the corresponding AHL extract. Strain genotypes are listed in Table 1.
Swarming and twitching motilities in B. thailandensis are affected by aiiA expression.
Several investigations have shown that QS regulates motility in gram-negative bacteria (1, 15, 23, 30). Expression of pBA01 (RU02) in B. thailandensis reduced swarming motility, while heterologous expression of pME6863 (RU04) resulted in a hyperswarming phenotype (Fig. 2, row 1, panels B and C). RU04(pME6863) translocation on swarm plates began at 12 h, and after 24 to 30 h the entire plate was colonized (Fig. 2, row 1, panel C). RU04(pME6863) grew in a rhizoid fashion with semiumbonate colony elevations. In contrast, RU02(pBA01) exhibited a reduction in swarming motility and displayed a colony morphology similar to that of wild-type B. thailandensis. Disruption of the btaI1 gene (luxI homologue) resulted in a similar phenotype observed by expression of the B. cereus AiiA lactonase in B. thailandensis (Fig. 2, row 1, panel D). Maintenance of the broad-host-range expression vector pBHR1 or pME6000 in B. thailandensis had no affect on swarming motility (data not shown).
FIG. 2.
AHL cleavage in B. thailandensis affects swarming and twitching motility in addition to the beta-hemolysis of sheep erythrocytes. Row 1, swarming motility; row 2, twitching motility; row 3, beta-hemolytic activity. DW503 represents wild-type B. thailandensis. Strain genotypes are listed above each image and are described in Table 1. The B. thailandensis luxI genes are listed as btaI, while the luxR alleles are listed as btaR.
Expression of pBA01 (RU02) and pME6863 (RU04) in B. thailandensis also resulted in altered colony morphologies on twitching motility plates. The uniform concentric ring produced by wild-type B. thailandensis was not observed with RU02(pBA01) and RU04(pME6863) (Fig. 2, row 2, panels B and C). The colony morphology of RU02(pBA01) was semiround with a protruding lobate region extending at the peripheral colony margin (Fig. 2, row 2, panel B). In contrast, RU04(pME6863) grew in an irregular and spreading pattern with hilly elevations, displayed an extensive wrinkling phenotype, and produced a diffusible pale orange pigment (Fig. 2, row 2, panel C). Similarly, mutagenesis of the btaR3 allele (luxR homologue) caused a similar twitching phenotype observed with RU04(pME6863). Twitching motility was not altered by the maintenance of pBHR1 or pME6000 (broad-host-range expression vectors) in B. thailandensis (data not shown).
Lactonase expression in B. thailandensis prevents hyperhemolysis of sheep erythrocytes.
Haussler et al. recently purified and characterized a hemolytic rhamnolipid synthesized by B. pseudomallei (20). Interestingly, B. thailandensis produces a rhamnolipid that is structurally and biochemically analogous to the B. pseudomallei cytotoxic and hemolytic exolipid (38). To determine whether expression of the Bacillus sp. AiiA lactonases in B. thailandensis affected beta-hemolytic activity, each derivative strain was tested on sheep blood agar plates. Expression of pBA01 (RU02) and pME6863 (RU04) in B. thailandensis had no effect on beta-hemolytic activity, whereas individual mutagenesis of the btaI1 and btaI3 genes (luxI mutant strains), which encode the proteins that direct the synthesis of C6-HSL and C8-HSL, resulted in hyperhemolytic phenotypes on sheep blood agar plates (Fig. 2, row 3, panels D to E). Even following prolonged incubation periods (5 to 7 days), no visible beta-hemolysis was observed for RU02(pBAO1) or RU04(pME6863) (data not shown).
Expression of the B. anthracis and B. cereus aiiA in B. thailandensis affects growth and carbon utilization.
During analysis of the effect of AHL cleavage on swarming and twitching motility in B. thailandensis, a notable AHL-dependent growth phenotype was observed for RU02(pBA01) (Table 2). To further confirm this growth reduction, the generation times of wild-type B. thailandensis and RU02(pBA01) were calculated throughout exponential growth in LB broth. Cleavage of C10-HSL and C8-HSL by expression of pBA01 (RU02) in B. thailandensis caused a pronounced increase in generation time (243 min) in comparison to that of wild-type B. thailandensis (48 min) (Table 2). In addition, the duration of logarithmic growth of RU02(pBA01) in comparison to wild-type B. thailandensis was reduced, as were the relative cell yields (data not shown). Further, the transition from lag-phase to logarithmic growth for RU02(pBA01) compared to wild-type B. thailandensis was increased (data not shown). Maintenance of the B. cereus AiiA lactonase (RU04) or the broad-host-range expression vectors pBHRI (RU01) and pME6000 (RU03) imposed no inhibitory effect on B. thailandensis growth (data not shown).
TABLE 2.
Phenotypes affected by heterologous expression of aiiA lactonases in B. thailandensis
| Substrate | Phenotype for straina:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| DW503 | RU01 | RU02 | RU03 | RU04 | BTRJ1 | BTRJ3 | BTRJ6 | |
| C6-HSL | D | D | D | D | ND | D | ND | D |
| C8-HSL | D | D | ND | D | ND | ND | D | D |
| C10-HSL | D | D | ND | D | ND | D | D | D |
| Swarming motility | NA | NA | M | NA | M | M | NA | NA |
| Twitching motility | NA | NA | M | NA | M | NA | NA | A |
| β-Hemolysis | NA | NA | NA | NA | NA | A | A | A |
| Generation time | NA | NA | A | NA | NA | NA | NA | NA |
| α-Hydroxybutyric acid | G | NG | NG | NG | NG | G | G | G |
| l-Threonine | G | NG | NG | NG | NG | G | G | G |
| l-Alanylglycin | G | NG | NG | NG | NG | G | G | G |
| d-Xylose | G | G | G | NG | NG | G | G | G |
| P-Hydroxy phenyl acetic acid | G | G | NG | G | NG | G | G | G |
| d-Trehalose | G | NG | G | NG | NG | G | G | G |
| d-Glucosaminic acid | G | G | G | G | NG | G | G | G |
| α-Ketobutyric acid | G | G | G | G | NG | G | G | G |
| l-Aspartic acid | G | G | NG | G | NG | G | G | G |
| l-Fucose | G | G | NG | G | G | G | G | G |
| Acetic acid | G | G | NG | G | G | G | G | G |
| α-Ketoglutaric acid | G | G | NG | G | G | G | G | G |
| Glucose-1-phosphate | G | G | NG | G | G | G | G | G |
| Citric acid | G | G | NG | G | G | G | G | G |
| l-Serine | G | G | NG | G | G | G | G | G |
| Gelatin | G | NG | G | NG | G | G | G | G |
| l-Isolcucine | G | NG | NG | NG | G | G | G | G |
| dl-Camitine | G | NG | NG | NG | G | NG | G | G |
| d-Arabinose | NG | NG | NG | NG | G | NG | G | NG |
| d-Arabitol | G | G | NG | G | G | G | G | G |
| Xylitol | G | G | NG | G | G | G | G | G |
| β-Hydroxybutyric acid | G | G | NG | G | G | G | G | G |
| Malonic acid | G | G | NG | G | G | G | G | G |
| l-Phenylalanine | G | G | NG | G | G | G | G | G |
| M9 minimal medium with d-arabinose | NG | NG | NG | NG | G | NT | NG | NT |
D, detected; ND, not detected; M, colony morphology altered; NA, not affected; A, affected; G, growth; NG, no growth; NT, not tested. Strain genotypes are listed in Table 1.
To further assess this growth characteristic, BiOLOG phenotypic microarrays were utilized to determine if carbon acquisition or metabolism was affected by AiiA lactonase expression in B. thailandensis. On BiOLOG PM1 plates, heterologous expression of the B. cereus (RU04) and B. anthracis (RU02) AiiA lactonases in B. thailandensis prevented growth on l-aspartic acid and P-hydroxy phenyl acetic acid (Table 2). Numerous substrates unique to RU02(pBA01), which heterologously expresses the B. anthracis AiiA lactonase and does not accumulate C8-HSL and C10-HSL (Fig. 1, lane 5), were identified on PM1 plates, including l-fucose, acetic acid, glucose-1-phosphate, citric acid, l-serine, and α-ketoglutaric acid (Table 2). As with PM1 plates, fluctuations in carbon metabolism were observed for RU02(pBA01) on the PM2 carbon source plates, including d-arabitol, xylitol, β-hydroxybutyric acid, malonic acid, and l-phenylalanine (Table 2). Of the 192 substrates analyzed, only d-arabinose appears to be negatively regulated [RU04(pME6863)]; that is, QS in B. thailandensis confers the ability to utilize a carbon source (Table 2). To further confirm this phenotype, wild-type B. thailandensis and RU04(pME6863) were cultured in M9 minimal broth containing d-arabinose as the sole carbon source. Consistent with the results obtained on PM2 plates, only RU04(pME6863) was able to metabolize d-arabinose as a sole carbon source (Table 2). A summary of the relevant phenotypes analyzed in this study is given in Table 2.
DISCUSSION
This study analyzed the affect of heterologous expression of Bacillus sp. AiiA lactonases in B. thailandensis and found that these AHL-cleaving enzymes significantly reduced the accumulation of C6-HSL, C8-HSL, and C10-HSL; affected bacterial motility; increased cellular generation time; and caused fluctuations in carbon metabolism and transport. The inhibition of AHL accumulation in B. thailandensis by maintenance of pBA01 (RU02) was similar to the cleavage pattern observed by expression of the B. cereus AiiA lactonase (RU04). The B. anthracis AiiA lactonase efficiently cleaved both C8-HSL and C10-HSL in B. thailandensis; however, trace amounts of C6-HSL were still detected by using TLC bioreporter overlays (Fig. 1, lane 5). In contrast, expression of the B. cereus AiiA lactonase in B. thailandensis completely abolished the accumulation of C10-HSL, C8-HSL, and C6-HSL (Fig. 1, lane 4). These findings are consistent with the results reported by Lee et al., which demonstrated that numerous Bacillus thuringiensis strains produce biologically active AiiA lactonases that vary in enzymatic activity (25). Despite the detection of C6-HSL in RU02(pBA01) cell supernatants, these findings demonstrate that expression of AiiA lactonases in B. thailandensis reduces AHL accumulation (Fig. 1, lanes 4 and 5). The exact mechanism for AHL cleavage mediated by the B. cereus strain A24 and B. anthracis AiiA lactonases has not been deciphered and was not within the scope of this investigation. However, considering the extensive homology between these AHL-cleaving enzymes and other AiiA lactonases, it is likely that the reduction in AHL accumulation reported in this study is the result of enzymatic lactone ring hydrolysis within the AHL molecule (9-11).
Like B. pseudomallei, B. thailandensis is capable of bacterial motility, and disruption (individual gene mutagenesis) of the B. thailandensis QS network resulted in defective motility phenotypes (38). As observed with the BTRJ1 (luxI) and BTRJ6 (luxR) QS mutants, expression of pBA01 (RU02) and pME6863 (RU04) in B. thailandensis also caused altered swarming and twitching motility phenotypes (Fig. 2, rows 1 and 2). The relative diameter of the swarm zone between RU02 (which does not accumulate C8-HSL and C10-HSL) and wild-type B. thailandensis was visibly different, possibly resulting from the growth phenotype observed by pBA01 expression (Fig. 2, row 1, panels A and B). However, this reduced swarm zone was still present after prolonged incubation periods (data not shown). In contrast, RU04(pME6863) demonstrated a hyperswarming phenotype as indicated by colonization of the entire swarm plate within 16 h. Interestingly, BTRJ1 (btaI1), which is a luxI mutant and deficient in C8-HSL biosynthesis, produced a phenotype similar to that of RU04(pME6863) on swarm plates (Fig. 2, row 1, panel D). The exact mechanisms for these defects in swarming and twitching motility remain to be determined. Preliminary analysis of flagellum and pillus biosynthesis (by electron microscopy) implies that the motility defects are independent of these cellular structures required for swarming and twitching motility (data not shown). However, these findings suggest that the accumulation of C8-HSL by B. thailandensis either directly or indirectly affects swarming motility.
As with swarming, alterations in twitching motility were observed by heterologous expression of pBA01 (RU02) and pME6863 (RU04) in B. thailandensis. Most notable was the phenotype observed for RU04(pME6863). Colony symmetry, morphology, and pigmentation of RU04 were similar to the phenotype observed for BTRJ6 (which contains an insertionally disrupted btaR3 gene, a luxR homologue) on twitching plates. Interestingly, BTRJ6 (btaR3) is a transcriptional regulator mutant, and previous analysis of AHL biosynthesis in B. thailandensis indicated that C6-HSL, C8-HSL, and C10-HSL accumulation was not affected by disruption of any of the five encoded LuxR homologues (38). Curiously, disruption of the B. thailandensis btaI1, btaI2, and btaI3 luxI homologues failed to cause a phenotype similar to that of RU04(pME6863) on twitching plates (data not shown). These findings suggest that the altered colony morphologies observed for B. thailandensis on twitching motility plates are linked to the accumulation of multiple signaling molecules in conjunction with the BtaR3 (LuxR homologue) transcriptional regulator. However, further analysis of this phenotype at the genetic level will be necessary before any definitive conclusion can be reached.
Mutations in the btaI1 and btaI3 luxI homologues, which encode the proteins that direct the synthesis of C8-HSL and C6-HSL, result in elevated beta-hemolytic activity on sheep blood agar plates (Fig. 2, row 3, panels D and E). Likewise, disruption of numerous luxR homologues encoded by B. thailandensis (btaR1, btaR2, btaR3, and btaR5) also causes hyper-beta-hemolysis (38). Interestingly, and despite efficient AHL cleavage, expression of pBA01 (RU02) and pME6863 (RU04) failed to induce hyper-beta-hemolysis of sheep erythrocytes as observed by individual mutagenesis of the B. thailandensis btaI1 and btaI3 genes (Fig. 2, row 3, panels D to E). The mechanism(s), in relation to QS and this hyper-beta-hemolysis, remains to be determined; however, these findings suggest that enhanced beta-hemolytic activity observed with mutagenesis of the B. thailandensis btaR3 and btaR5 luxR homologues is independent of AHL accumulation. This hypothesis is further supported by the observation that disruption of the btaR3 and btaR5 genes has no effect on C6-HSL, C8-HSL, andC10-HSL biosynthesis in B. thailandensis (38). The failure of RU02(pBA01) and RU04(pME6863) to lyse sheep erythrocytes may be the result of residual concentrations of AHL molecules, as seen for RU02, that are not detected by using the bioreporter strain A. tumefaciens A136 (Fig. 1, lane 5). It is also possible that B. thailandensis carries an additional luxI gene(s), which produces signaling molecules not recognized by A. tumefaciens A136, not recovered in our in silico analysis.
QS has been shown to affect the growth of various bacterial species belonging to the Rhizobium genus; however, the cellular mechanisms contributing to these reduction in growth remain to be determined (6, 18, 21, 32, 40). Interestingly, AHL cleavage in B. thailandensis, specifically C8-HSL and C10-HSL (Fig. 1, lane 5), by heterologous expression of pBA01 (RU02) caused an increase in generation time for RU02 (243 min) compared to wild-type B. thailandensis (48 min) and also extended the transition from lag-phase to logarithmic growth by 3 h (Table 2). Individual mutagenesis of the B. thailandensis btaI1, btaI2, and btaI3 genes, which encode the proteins that synthesize C6-HSL, C8-HSL, and C10-HSL, respectively, failed to cause this increase in generation time, as did expression of the broad-host-range vector pBHR1 (data not shown). To further confirm these findings, and to eliminate the possibility of plasmid segregation despite antibiotic selection, serial dilutions were plated onto selective and nonselective (without kanamycin) LB plates during growth analysis (data not shown), in addition to performing colony PCR (10 isolates from each medium type) with the B. anthracis aiiA gene-specific primers. As anticipated, the proportions of recoverable bacteria on each medium type were identical, indicating stable maintenance of pBA01 (RU02). Several factors may contribute to this observed growth phenotype following AHL cleavage, with the most obvious being the metabolic burden of constitutive AiiA lactonase expression in RU02(pBA01). However, this increase in generation time is unique to RU02, and RU04(pME6863), which also constitutively expresses a homologous AiiA lactonase, exhibits an generation time analogous to that of wild-type B. thailandensis (data not shown). Additional parameters that may cause growth inhibition include defects in DNA translation, negative regulation (overproduction) of sigma factors, and, a focus of this study, substrate utilization. To determine if QS was linked to carbon acquisition and metabolism, BiOLOG phenotypic arrays were employed. Table 2 illustrates that AHL cleavage in B. thailandensis causes fluctuations in either the transport or catabolism of numerous carbon sources. Although to a lesser extent, individual mutagenesis of each of the B. thailandensis QS alleles also affected substrate utilization (38). Interestingly, expression of the B. cereus AiiA lactonase (RU04) in B. thailandensis demonstrated that AHL accumulation negatively regulates (confers the ability to utilize a carbon source) the protein(s) involved in the metabolism of d-arabinose (Table 2). Similar findings at the transcriptional level in P. aeruginosa PA01 have linked QS to substrate metabolism and transport (33, 39). In fact, Schuster et al. demonstrated that disruption of the P. aeruginosa PA01 lasIR and rhlIR QS networks both positively and negatively affected the transcription of various genes involved in cellular metabolism (33). The mechanism of this growth inhibition remains to be determined; however, these findings demonstrate that the B. thailandensis QS system is involved in the metabolism of various carbon sources. It is conceivable that B. thailandensis utilizes QS in the environment to reduce energy needed for substrate transport and catabolism when sufficient carbon sources (i.e., glucose versus arabinose) are present.
It seems that QS in B. thailandensis represents a complex gene regulatory network. This study demonstrates that AiiA lactonase expression in B. thailandensis reduces the accumulation of the signaling molecules C6-HSL, C8-HSL, and C10-HSL and bacterial motility; increases cellular generation time; and causes fluctuations in carbon metabolism. Stable heterologous expression of an AiiA lactonase(s) in Burkholderia species will possibly eliminate the need to generate multideletion mutants in a single organism that synthesizes numerous AHLs and may facilitate vaccine development for gram-negative bacterial pathogens that utilize QS to regulate virulence gene expression.
Acknowledgments
I extend a special thank you to Melanie Ulrich and Katheryn Kenyon for reviewing the manuscript and to Donald J. Chabot for providing purified B. anthracis genomic DNA for aiiA lactonase gene cloning. I additionally thank David DeShazer for scientific and technical discussions regarding this investigation.
The research described herein was sponsored by the Medical Biological Defense Research Program, U.S. Army Medical Research and Material Command.
Opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the U.S. Army.
REFERENCES
- 1.Atkinson, S., J. P. Throup, G. S. Stewart, and P. Williams. 1999. A hierarchical quorum-sensing system in Yersinia pseudotuberculosis is involved in the regulation of motility and clumping. Mol. Microbiol. 33:1267-1277. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, A., P. A. Sokol, J. Parkhill, and E. Mahenthiralingam. 2004. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brett, P. J., D. DeShazer, and D. E. Woods. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48:317-320. [DOI] [PubMed] [Google Scholar]
- 4.Burtnick, M., A. Bolton, P. Brett, D. Watanabe, and D. Woods. 2001. Identification of the acid phosphatase (acpA) gene homologues in pathogenic and non-pathogenic Burkholderia spp. facilitates TnphoA mutagenesis. Microbiology 147:111-120. [DOI] [PubMed] [Google Scholar]
- 5.Conway, B. A., and E. P. Greenberg. 2002. Quorum-sensing signals and quorum-sensing genes in Burkholderia vietnamiensis. J. Bacteriol. 184:1187-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels, R., D. E. De Vos, J. Desair, G. Raedschelders, E. Luyten, V. Rosemeyer, C. Verreth, E. Schoeters, J. Vanderleyden, and J. Michiels. 2002. The cin quorum sensing locus of Rhizobium etli CNPAF512 affects growth and symbiotic nitrogen fixation. J. Biol. Chem. 277:462-468. [DOI] [PubMed] [Google Scholar]
- 7.DeShazer, D., P. J. Brett, M. N. Burtnick, and D. E. Woods. 1999. Molecular characterization of genetic loci required for secretion of exoproducts in Burkholderia pseudomallei. J. Bacteriol. 181:4661-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donabedian, H. 2003. Quorum sensing and its relevance to infectious diseases. J. Infect. 46:207-214. [DOI] [PubMed] [Google Scholar]
- 9.Dong, Y. H., A. R. Gusti, Q. Zhang, J. L. Xu, and L. H. Zhang. 2002. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl. Environ. Microbiol. 68:1754-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, Y. H., L. H. Wang, J. L. Xu, H. B. Zhang, X. F. Zhang, and L. H. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 11.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 13.Fuqua, C., and S. C. Winans. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glessner, A., R. S. Smith, B. H. Iglewski, and J. B. Robinson. 1999. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J. Bacteriol. 181:1623-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godoy, D., G. Randle, A. J. Simpson, D. M. Aanensen, T. L. Pitt, R. Kinoshita, and B. G. Spratt. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 41:2068-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotschlich, A., B. Huber, O. Geisenberger, A. Togl, A. Steidle, K. Riedel, P. Hill, B. Tummler, P. Vandamme, B. Middleton, M. Camara, P. Williams, A. Hardman, and L. Eberl. 2001. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst. Appl. Microbiol. 24:1-14. [DOI] [PubMed] [Google Scholar]
- 18.Gray, K. M., J. P. Pearson, J. A. Downie, B. E. Boboye, and E. P. Greenberg. 1996. Cell-to-cell signaling in the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum: autoinduction of a stationary phase and rhizosphere-expressed genes. J. Bacteriol. 178:372-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardman, A. M., G. S. Stewart, and P. Williams. 1998. Quorum sensing and the cell-cell communication dependent regulation of gene expression in pathogenic and non-pathogenic bacteria. Antonie Leeuwenhoek. 74:199-210. [DOI] [PubMed] [Google Scholar]
- 20.Haussler, S., M. Nimtz, T. Domke, V. Wray, and I. Steinmetz. 1998. Purification and characterization of a cytotoxic exolipid of Burkholderia pseudomallei. Infect. Immun. 66:1588-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, X., W. Chang, D. L. Pierce, L. O. Seib, J. Wagner, and C. Fuqua. 2003. Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J. Bacteriol. 185:809-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, J. J., J. I. Han, L. H. Zhang, and J. R. Leadbetter. 2003. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 69:5941-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohler, T., L. K. Curty, F. Barja, C. van Delden, and J. C. Pechere. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leadbetter, J. R., and E. P. Greenberg. 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182:6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, S. J., S. Y. Park, J. J. Lee, D. Y. Yum, B. T. Koo, and J. K. Lee. 2002. Genes encoding the N-acyl homoserine lactone-degrading enzyme are widespread in many subspecies of Bacillus thuringiensis. Appl. Environ. Microbiol. 68:3919-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewenza, S., and P. A. Sokol. 2001. Regulation of ornibactin biosynthesis and N-acyl-l-homoserine lactone production by CepR in Burkholderia cepacia. J. Bacteriol. 183:2212-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, Y. H., J. L. Xu, J. Hu, L. H. Wang, S. L. Ong, J. R. Leadbetter, and L. H. Zhang. 2003. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 47:849-860. [DOI] [PubMed] [Google Scholar]
- 29.Rasmussen, T. B., M. Manefield, J. B. Andersen, L. Eberl, U. Anthoni, C. Christophersen, P. Steinberg, S. Kjelleberg, and M. Givskov. 2000. How Delisea pulchra furanones affect quorum sensing and swarming motility in Serratia liquefaciens MG1. Microbiology 146:3237-3244. [DOI] [PubMed] [Google Scholar]
- 30.Reimmann, C., N. Ginet, L. Michel, C. Keel, P. Michaux, V. Krishnapillai, M. Zala, K. Heurlier, K. Triandafillu, H. Harms, G. Defago, and D. Haas. 2002. Genetically programmed autoinducer destruction reduces virulence gene expression and swarming motility in Pseudomonas aeruginosa PAO1. Microbiology 148:923-932. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Schripsema, J., K. E. de Rudder, T. B. van Vliet, P. P. Lankhorst, E. de Vroom, J. W. Kijne, and A. A. van Brussel. 1996. Bacteriocin small of Rhizobium leguminosarum belongs to the class of N-acyl-l-homoserine lactone molecules, known as autoinducers and as quorum-sensing cotranscription factors. J. Bacteriol. 178:366-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 36.Swift, S., J. A. Downie, N. A. Whitehead, A. M. Barnard, G. P. Salmond, and P. Williams. 2001. Quorum sensing as a population-density-dependent determinant of bacterial physiology. Adv. Microb. Physiol. 45:199-270. [DOI] [PubMed] [Google Scholar]
- 37.Ulrich, R. L., and D. DeShazer. 2004. Type III secretion: a virulence factor delivery system essential for the pathogenicity of Burkholderia mallei. Infect. Immun. 72:1150-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulrich, R. L., H. B. Hines, N. Parthasarathy, and J. A. Jeddeloh. Mutational analysis and biochemical characterization of the Burkholderia thailandensis DW503 quorum-sensing network. J. Bacteriol. 186:4350-4360. [DOI] [PMC free article] [PubMed]
- 39.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkinson, A., V. Danino, F. Wisniewski-Dye, J. K. Lithgow, and J. A. Downie. 2002. N-Acyl-homoserine lactone inhibition of rhizobial growth is mediated by two quorum-sensing genes that regulate plasmid transfer. J. Bacteriol. 184:4510-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, F., T. Byun, H. J. Deussen, K. R. Duke, and H. J. Dussen. 2003. Degradation of N-acylhomoserine lactones, the bacterial quorum-sensing molecules, by acylase. J. Biotechnol. 101:89-96. [DOI] [PubMed] [Google Scholar]