Abstract
The microbial community of the human colon contains many bacteria that produce lactic acid, but lactate is normally detected only at low concentrations (<5 mM) in feces from healthy individuals. It is not clear, however, which bacteria are mainly responsible for lactate utilization in the human colon. Here, bacteria able to utilize lactate and produce butyrate were identified among isolates obtained from 10−8 dilutions of fecal samples from five different subjects. Out of nine such strains identified, four were found to be related to Eubacterium hallii and two to Anaerostipes caccae, while the remaining three represent a new species within clostridial cluster XIVa based on their 16S rRNA sequences. Significant ability to utilize lactate was not detected in the butyrate-producing species Roseburia intestinalis, Eubacterium rectale, or Faecalibacterium prausnitzii. Whereas E. hallii and A. caccae strains used both d- and l-lactate, the remaining strains used only the d form. Addition of glucose to batch cultures prevented lactate utilization until the glucose became exhausted. However, when two E. hallii strains and one A. caccae strain were grown in separate cocultures with a starch-utilizing Bifidobacterium adolescentis isolate, with starch as the carbohydrate energy source, the l-lactate produced by B. adolescentis became undetectable and butyrate was formed. Such cross-feeding may help to explain the reported butyrogenic effect of certain dietary substrates, including resistant starch. The abundance of E. hallii in particular in the colonic ecosystem suggests that these bacteria play important roles in preventing lactate accumulation.
Lactic acid is produced by many of the microorganisms that colonize the digestive tract of animals and humans. In pure culture, l-lactate and/or d-lactate is the major product of lactic acid bacteria, including lactobacilli, bifidobacteria, enterococci, and streptococci and can also be produced by strict anaerobes such as Eubacterium spp. that are abundant in the human gastrointestinal (GI) tract (4, 36). l-Lactic acid may also enter the gut from host tissues (29). Despite this, lactate is seldom detected as a major fermentation product of mixed anaerobic communities in human feces or in gut contents under normal conditions. This is assumed to reflect lactate utilization by other bacterial species, but in the human gut, the identity of these bacteria is largely unknown. Lactate has been reported to accumulate in feces from individuals who have undergone gut resections (short bowel syndrome) (32) or who are suffering from ulcerative colitis, at concentrations up to 100 mM (29, 54), although in individuals with no apparent disease, fecal lactate is usually less than 5 mM. The mechanism of disease development in ulcerative colitis is uncertain, but colonic microbial imbalance may be a contributing factor, and colonic infusion of donor human intestinal microbes from healthy donors has been shown to reverse the condition in selected patients (6). The consequences of d-lactate accumulation in short bowel syndrome can be serious, leading to neurotoxicity and cardiac arythmia (11, 53).
In the rumen and the pig GI tract, it is well established that species such as Megasphaera, Selenomonas, and Veillonella are capable of utilizing lactate and converting it largely to acetate and propionate, and in the case of Megasphaera, also butyrate (12, 23, 30). Abnormal nutritional conditions resulting from an oversupply of soluble carbohydrate can, however, lead to runaway production of lactate in the rumen, leading to the serious condition lactic acidosis (48). Although Megasphaera and Veillonella-related organisms have been reported from the human GI tract (5, 19), molecular studies indicate that their populations may be quite variable between individuals (24, 27, 49). Meanwhile, recent reports suggest that significant amounts of lactate may be converted into butyrate in the human colonic system (7). Here we report for the first time the isolation of bacteria from human feces that can convert lactate largely into butyrate. Some of these isolates belong to a new species, and some belong to Eubacterium hallii, which accounts for 2 to 3% of the human fecal community (22), and Anaerostipes caccae (46). Cross-feeding of l-lactate is demonstrated in cocultures between a starch-degrading species, Bifidobacterium adolescentis, and the newly identified lactate utilizers. It is suggested that this type of cross-feeding might contribute to the reported butyrogenic effect of certain dietary substrates, such as resistant starch in the human colon (13, 33, 40, 50, 56, 58).
MATERIALS AND METHODS
Bacterial isolations and growth media.
The human fecal bacterial strains used in this study are described in Table 1. Details of the isolation of most of these strains have been described previously (4, 34, 46). The lactate-utilizing strains listed from reference 34 came from a single adult female donor and were isolated with anaerobic isolation media that contain dl-lactate as sole added energy source or that were designed for the selective isolation of Selenomonas strains from the rumen (20, 34). Out of 57 colonies picked from 10−8 dilutions of fecal samples, none produced significant (>5 mM) propionate, but 17 colonies produced butyrate, and 5 of these were able to use lactate for growth. ART1 and ART92 were isolated as follows from fecal samples from two different healthy adult volunteers who had not received antibiotics in the previous 6 months. Whole stools were collected, and 1 g was mixed in 9 ml of anaerobic diluent (4, 26). Decimal serial anaerobic dilutions were prepared, and 0.5 ml was inoculated into roll tubes by the Hungate technique, under 100% CO2 (10). A total of 264 colonies were initially picked (24 per sample) from the 10−8 dilutions of samples from 11 different donors. Colonies were regrown in YCFA medium (18) containing lactate only, lactate plus glucose, or no addition unless stated otherwise. Fermentation products were analyzed after 24 h growth. Sixty-one of these isolates that produced either butyrate or propionate (>5 mM) or utilized lactate (>5 mM) were repurified, and their fermentation behavior was retested. Strain ART1 was from roll tubes of Ss medium (2) modified to contain 35 mM dl-lactate as the added carbon source. ART92 was from roll tubes of rumen fluid-based M2 medium (described in reference 26, as modified in reference 37) with dl-lactate (35 mM) as the added carbon source. The majority of isolates recovered from isolation media containing dl-lactate were not lactate utilizers, presumably because they were able to use other compounds, including amino acids and peptides present in the medium, as carbon and nitrogen sources. Ethical approval for this work was granted by the Grampian Research Ethics Committee (project no. 00/00133).
TABLE 1.
Bacterial isolates included in this study
| Species and strain | Subject | Closest species (% identity)a | Closest 16S rRNA sequence (% identity)a | Accession no. | Reference(s) |
|---|---|---|---|---|---|
| Lactate utilizers | |||||
| A. caccae L1-92T | 1 | A. caccae (100) | AJ270487 | 4, 46 | |
| P2 | 2 | A. caccae (99) | 46 | ||
| L2-7 | 1 | E. hallii (96) | HuCA15 (98) | AJ270490 | 4 |
| SL6/1/1 | 3 | E. hallii (97) | HuCA15 (99) | AY305317 | 34 |
| SM6/1 | 3 | E. hallii (98) | p2001-s959-5 (99) | AY305318 | 34 |
| ART1 | 4 | E. hallii (98)b | This study | ||
| SS2/1 | 3 | C. indolis (94) | Adhufec25 (99) | AY305319 | 34 |
| SSC/2 | 3 | C. indolis (94) | Adhufec25 (99) | AY305320 | 34 |
| ART92 | 5 | C. indolis (94) | Adhufec25 (99) | This study | |
| Other isolates | |||||
| R. intestinalis L1-82T | 1 | R. intestinalis (100) | AJ312385 | 4, 15, 17 | |
| A2-183 | 6 | R. intestinalis (97) | Adhufec225 (99) | AJ270482 | 4 |
| A2-194 | 6 | R. intestinalis (94) | HuCB37 (99) | AJ270473 | 4 |
| A1-86 | 6 | E. rectale (99) | AJ270475 | 4 | |
| L2-50 | 1 | E. ruminantium (93) | A19 (96) | AJ270491 | 4 |
| A2-165 | 6 | F. prausnitzii (100) | AJ270469 | 4, 16, 17 | |
| L2-32 | 1 | B. adolescentis (99) | Bifidobacterium sp. strain PL1 (99) | AY305304 | 4 |
Percent identity in 16S rRNA sequence (1,400 bases unless otherwise stated).
Indicates partial 16S rRNA sequences (833 bases).
B. adolescentis L2-32, used in the coculture experiments, was isolated previously by Barcenilla et al. (3).
16S rRNA sequencing of new isolates and phylogenetic relationships.
Cell pellets from 1 ml of culture were resuspended in 50 μl of sterile distilled H2O and served as templates for PCRs (0.5 μl per 50 μl of PCR mixture). 16S rRNA sequences were amplified with a universal primer set, corresponding to positions 8 to 27 (27f, forward primer, AGAGTTTGATCMTGGCTCAG) and 1491 to 1511 (rP2, reverse primer, ACGGCTACCTTGTTACGACTT) of the Escherichia coli numbering system (8, 57) with an MgCl2 concentration of 1.5 mM. PCR amplifications were performed as described previously (34). The amplified PCR products were purified with QIA quick columns (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions and directly sequenced with a capillary sequencer (Beckman) with primers 27f, rP2, 519f (CAGCMGCCGCGGTAATWC), 519r (GWATTACCGCGGCKGCTG) (corresponding to positions 518 to 535 of the Escherichia coli numbering system), 926f (AAACTCAAAKGAATTGACGG), and 926r (CCGTCAATTCMTTTRAGTTT) (corresponding to positions 906 to 925). Similarity of the 16S rRNA sequences from the isolates to those from other organisms was compared with all sequence data in GenBank, using the BLAST algorithm (1).
Acidic fermentation product and H2 analyses.
Short chain fatty acid (SCFA) production and utilization, including dl-lactate measurements, were determined by capillary gas chromatography (GC) (41), and H2 was analyzed by packed-column GC (42). All values reported refer to means of triplicate cultures.
l-Lactate and glucose analyses.
l-Lactate concentrations were determined enzymatically, and glucose concentrations were determined by the glucose oxidase method (51). Analyses were conducted in a robotic clinical analyzer (Kone analyzer; Konelab Corporation, Espoo, Finland). d-Lactate concentrations were assessed as the difference between total dl-lactate and the l-lactate measurements. All values reported refer to means of triplicate cultures.
Coculture of lactate utilizers with B. adolescentis.
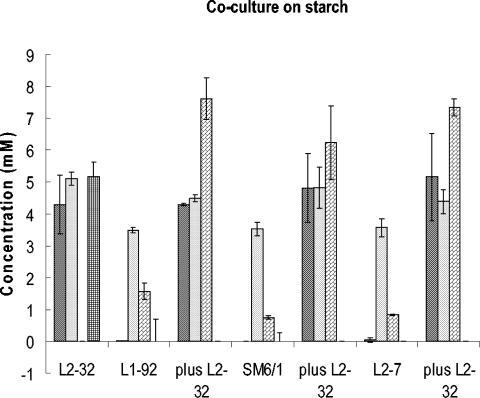
Three lactate-utilizing strains, A. caccae L1-92 and two strains of E. hallii (SM6/1 and L2-7), were incubated alone and in coculture with B. adolescentis L2-32 on YCFA medium modified to contain reduced Casitone (0.1%) and 0.2% soluble starch as an added energy source. The inoculated tubes were incubated for 24 h at 37°C. B. adolescentis L2-32 was enumerated on Mann-Ragosa-Sharpe (MRS) medium containing 2.0% agar with a final concentration of 67.5 mM propionate, and the three butyrate-producing strains were enumerated with appropriate dilutions in triplicate roll tubes (26) of M2 medium containing 35 mM dl-lactate. Three independent repeats of this experiment (one of which is shown in Fig. 3) gave the same outcome.
FIG. 3.
Coculture between lactate-utilizing bacteria and B. adolescentis L2-32. The concentrations of SCFA, formate ( ), acetate (
), acetate ( ), butyrate (
), butyrate ( ), and l-lactate (
), and l-lactate ( ) are shown after 24 h of growth in YCFA medium with 0.2% starch as an energy source (values for acetate, which was initially present at 33 mM in the medium in all cases, are shown on a 10-fold reduced scale). Butyrate production by A. caccae L1-92 and by E. hallii L2-7 and SM6/1 was stimulated by coculture with B. adolescentis L2-32, while l-lactate disappeared from the cocultures. Means and standard deviations are shown based on results from triplicate cultures.
) are shown after 24 h of growth in YCFA medium with 0.2% starch as an energy source (values for acetate, which was initially present at 33 mM in the medium in all cases, are shown on a 10-fold reduced scale). Butyrate production by A. caccae L1-92 and by E. hallii L2-7 and SM6/1 was stimulated by coculture with B. adolescentis L2-32, while l-lactate disappeared from the cocultures. Means and standard deviations are shown based on results from triplicate cultures.
RESULTS
Identification of lactate-utilizing, butyrate-producing bacteria from human feces.
Four butyrate-producing, lactate-utilizing bacteria (defined as using >5 mM lactate during 24 h of incubation) were isolated initially from 10−8 dilutions of fecal samples from one adult female donor, as described in Materials and Methods. These isolates were found to belong to clostridial cluster XIVa, based on 16S rRNA sequencing. Lactate utilization had not previously been reported among human colonic bacteria belonging to clostridial cluster XIVa; since this represents one of the most abundant bacterial groups in human feces (21, 27, 49), it was considered that these bacteria could play a significant role in lactate metabolism in the human colon. A further set of isolations was therefore performed as described in Materials and Methods. This resulted in a further two confirmed isolates, obtained from 10−8 dilutions of fecal samples from different individuals, that utilized lactate and produced butyrate. Again these isolates were shown to belong to clostridial cluster XIVa based on their 16S rRNA sequences, and they were closely related to the four initial isolates (Table 1, subjects 3, 4, and 5).
Representatives of other butyrate-producing genera previously shown to be abundant in human feces (4, 28) were also tested for their ability to utilize lactate. Eubacterium rectale A1-86, Roseburia intestinalis L1-82 (15), Roseburia sp. strains A2-183 and A2-194 and the Eubacterium ruminatium-like strain L2-50 (all clostridial cluster XIVa) failed to show significant lactate utilization, and very limited utilization (<3 mM) was found for Faecalibacterium prausnitzii A2-165 (16), which belongs to clostridial cluster IV (results not shown). The ability to utilize lactate, however, was found in E. hallii L2-7 and in the two known strains of A. caccae (Tables 2 and 3).
TABLE 2.
Lactate utilization by human fecal isolates
| Strain/closest species | Change in fermentation product concn (mM) on YCFA medium plus 35 mM dl-lactatea
|
Change in product concn (mM) on YCFA mediumab
|
Carbon recovery (%)c | |||||
|---|---|---|---|---|---|---|---|---|
| Butyrate | l-Lactate | d-Lactate | Formate | Acetate | Butyrate | Acetate | ||
| E. hallii SL6/1/1 | 21.06 ± 1.06 | −15.29 ± 0.10 | −14.29 ± 0.51 | 0.0 ± 0.0 | −18.51 ± 0.96 | 1.42 ± 1.42 | −4.96 ± 3.26 | 90.5 |
| E. hallii SM6/1 | 25.70 ± 0.43 | −12.70 ± 0.50 | −18.70 ± 1.06 | 0.03 ± 0.03 | −21.20 ± 2.08 | 1.42 ± 0.05 | −2.61 ± 2.36 | 98.8 |
| E. hallii L2-7 | 22.58 ± 0.76 | −15.50 ± 0.0 | −16.50 ± 0.0 | 1.09 ± 1.55 | −14.77 ± 0.93 | 0.60 ± 0.03 | −1.58 ± 1.73 | 98.3 |
| A. caccae L1-92 | 23.35 ± 1.16 | −13.66 ± 0.85 | −14.91 ± 0.87 | −0.05 ± 0.01 | −21.98 ± 2.45 | 1.99 ± 0.09 | −2.35 ± 2.03 | 94.4 |
| A. caccae P2 | 25.01 ± 1.0 | −15.50 ± 1.0 | −16.50 ± 1.0 | 1.03 ± 1.0 | −21.19 ± 1.0 | 1.92 ± 1.00 | 3.88 ± 1.00 | 95.4 |
| SS2/1 | 12.98 ± 0.19 | 0.57 ± 0.40 | −15.49 ± 1.01 | 0.18 ± 0.02 | −12.51 ± 1.27 | 2.24 ± 0.26 | −4.25 ± 4.68 | 94.7 |
| SSC/2 | 13.49 ± 0.0 | −0.1 ± 0.78 | −9.51 ± 6.1 | 0.36 ± 0.0 | −12.12 ± 0.0 | 2.37 ± 0.09 | −0.16 ± 1.32 | 119.3 |
Change in concentration compared with uninoculated controls during 24 h of incubation at 37°C. Values are means of triplicate cultures from one experiment and are shown ± standard deviation.
No significant change in formate or lactate occurred in basal YCFA medium.
Calculated as described in Table 3 footnotes.
TABLE 3.
Carbon and electron balances for A. caccae L1-92 and E. hallii L2-7
| Parameter | No. of carbons or electronsa or mmol of substrate liter−1 consumedb
|
No. of carbons or electrons or mmol of product liter−1 formedb
|
% Recovery | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | Acetate | Lactate (d + l) | Sum | Formate | Acetate | Butyrate | H2 | CO2c | Sum | ||
| No. of carbons/mol | 6 | 2 | 3 | 1 | 2 | 4 | 0 | 1 | |||
| No. of electrons/mol | 24 | 8 | 12 | 2 | 8 | 20 | 2 | 0 | |||
| L1-92 (A. caccae) | |||||||||||
| Grown on medium + dl-lactated | |||||||||||
| Δ Concn | 29.8 | (17.2 + 23.7) | 0.1 | 35.6 | 13.0 | 40.9 | |||||
| Δ Carbon | 59.6 | 122.7 | 182.3 | 0.1 | 142.4 | 40.9 | 183.3 | 100.5 | |||
| Δ Electrons | 238.4 | 490.8 | 729.2 | 0.2 | 712.4 | 26.0 | 738.4 | 101.3 | |||
| Grown on medium + glucose | |||||||||||
| Δ Concn | 10.4 | 2.3 | 2.2 | 11.5 | 8.6 | 18.6 | |||||
| Δ Carbon | 62.4 | 6.9 | 69.3 | 2.2 | 46.0 | 18.6 | 66.8 | 96.4 | |||
| Δ Electrons | 249.6 | 18.4 | 268 | 8.8 | 230.0 | 17.2 | 256 | 95.5 | |||
| L2-7 (E. hallii) | |||||||||||
| Grown on medium + dl-lactate | |||||||||||
| Δ Concn | 26.1 | (16.0 + 22.4) | 0.4 | 29.6 | 15.5 | 38.4 | |||||
| Δ Carbon | 52.2 | 115.2 | 167.4 | 0.4 | 118.4 | 38.4 | 156.8 | 93.7 | |||
| Δ Electrons | 208.8 | 460.8 | 669.6 | 0.8 | 592.0 | 31.0 | 623 | 93.2 | |||
| Grown on medium + glucose | |||||||||||
| Δ Concn | 10.2 | 2.0 | 2.7 | 8.0 | 12.0 | 18.4 | |||||
| Δ Carbon | 61.2 | 61.2 | 2.0 | 5.4 | 32.0 | 18.4 | 57.8 | 94.4 | |||
| Δ Electrons | 244.8 | 244.8 | 4.0 | 21.6 | 160.0 | 24.0 | 209.6 | 85.6 | |||
References to electrons refer to electrons released upon formal oxidation to CO2.
Experimental values are means of data from triplicate cultures after 24 h of growth. For L2-7, data are from the experiment shown in Fig. 1; standard deviations (also for the L1-92 experiment) were as indicated in the legend to Fig. 1.
The value for carbon dioxide is calculated on the assumption that every 1 mol of lactate taken up (since formate production is minimal) releases 1 mol of CO2, while 1 mol of glucose utilized releases 2 mol of CO2 or formate: hence CO2 is calculated as (2 × glucose) − formate.
YCFA medium was supplemented as shown above with either 33 mM dl-lactate or 10 mM glucose.
The phylogenetic relationships of these nine lactate-utilizing, butyrate-producing strains based on their 16S rRNA sequences are summarized in Table 1. Four strains (from three individuals) are related to E. hallii, two (from two individuals) are A. caccae, and three (from two individuals) are distantly related to Clostridium indolis.
Regulation and stereospecificity of lactate utilization.
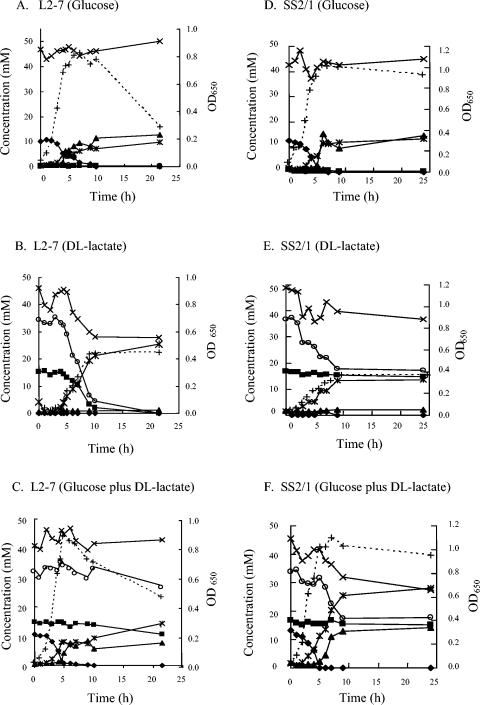
Three E. hallii-related strains and the two A. caccae strains were able to use both the d and l isomers of lactate during 24 h of growth on dl-lactate medium (Table 2). The fourth E. hallii-related strain, ART1 (not shown), also used both the d and l isomers, while the remaining three isolates, two of which are shown in Table 2, showed a strong preference for d-lactate. Addition of glucose to the medium decreased lactate utilization by the strains listed in Table 2. In order to investigate this effect further, time courses were followed for selected strains grown on glucose, lactate, or glucose and lactate (Fig. 1). E. hallii L2-7, when grown with dl-lactate, used all of the added lactate together with some acetate, producing more than 20 mM butyrate (Fig. 1B). Less butyrate, but a significant amount of formate, was produced during growth on glucose or on glucose plus lactate, and lactate utilization was almost abolished by the presence of glucose (Fig. 1C). Levels of hydrogen production in 24 h were 12 μmol ml−1 for growth on glucose, 15.5 μmol ml−1 for growth on lactate, and 10.9 μmol ml−1 for growth on glucose plus lactate. Similar results were obtained for the E. hallii-related strain SM6/1 and for A. caccae L1-92, except that these strains showed a greater ability to use lactate once glucose had been exhausted, following inoculation into glucose-plus-lactate medium (results not shown). The C. indolis-related strain SS2/1 was able to use d-lactate, but not l-lactate, during growth on dl-lactate (Fig. 1E) or following glucose exhaustion in lactate-plus-glucose medium (Fig. 1F). Again formate was not a significant product when lactate was the sole energy source; 4.7 μmol of hydrogen ml−1 was formed in 24 h on dl-lactate.
FIG. 1.
Time course of SCFA formation and growth in batch culture of E. hallii L2-7 (A, B, C) of the C. indolis-related strain SS2/1 (D, E, F) on media containing dl-lactate, glucose, or dl-lactate plus glucose. Millimolar concentrations of substrates utilized and products formed are shown for glucose (⧫), l-lactate (▪), formate (▴), acetate (×), butyrate ( ), and dl-lactate (○). Growth (+) is measured as optical density at 650 nm (OD650). All values are the means of results from triplicate cultures. Standard deviations (not shown) were typically around 5% of the mean value.
), and dl-lactate (○). Growth (+) is measured as optical density at 650 nm (OD650). All values are the means of results from triplicate cultures. Standard deviations (not shown) were typically around 5% of the mean value.
Carbon and electron balances.
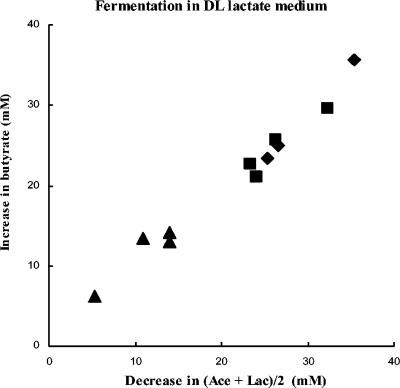
Carbon and electron balances were calculated for growth on lactate or on glucose following measurement of hydrogen production, as illustrated for one strain each of E. hallii and A. caccae in Table 3. Carbon and electron balances were close to 100%, suggesting that the major fermentation products are accounted for. For the strains listed in Table 2, there was an approximate 1:1 relationship between the moles of butyrate formed and half the moles of acetate plus lactate removed from the medium during growth (Fig. 2). Calculated carbon recoveries for growth on dl-lactate ranged from 90.5 to 119.3%, with butyrate accounting for >95% of the non-CO2 carbon recovered (Table 2).
FIG. 2.
Relationship between butyrate formation and net removal of acetate and lactate from the medium during 24 h of growth on dl-lactate medium. Data are largely from Tables 2 and 3 for E. hallii strains L2-7, SM6/1 and SL6/1/1 (▪), A. caccae strains L1-92 and P2 (⧫), and C. indolis-related strains SSC/2 and SS2/1 (▴). Values from two independent experiments are included for L2-7, L1-92, SSC/2, and SS2/1.
Potential for cross-feeding.
In most human diets, resistant starch is considered to be the most important energy source for microbial growth in the large intestine (50). The major amylolytic species in the human colon are generally considered to be Bacteroides and Bifidobacterium spp. (35, 45). Bifidobacteria produce acetate and lactate from carbohydrate substrates, typically in the molar ratio of 3:2, together with formate. Since the lactate utilizers used for the coculture studies either did not utilize starch or utilized it weakly as a growth substrate in pure culture (data not shown), it was of interest to coculture them with a starch-degrading Bifidobacterium strain in order to establish whether they could remove the lactate formed. The amylolytic B. adolescentis strain L2-32 was used for these experiments. As shown in Fig. 3, coculture with any one of three lactate utilizers tested, with starch as the growth substrate, resulted in complete conversion of the l-lactate formed by B. adolescentis L2-32 into butyrate. This corresponded with an increase in viable cell numbers of the lactate utilizers in the presence of B. adolescentis L2-32, as determined by selective plating (as described in Materials and Methods). Viable counts after 24 h of growth for A. caccae L1-92, E. hallii SM6/1, and E. hallii L2-7 were, respectively, 2.4 × 108, 1.0 × 107 and 8.0 × 106 CFU ml−1 in the absence of B. adolescentis and 1.7 × 109, 6.8 × 108 and 5.4 × 109 CFU ml−1 in the presence of B. adolescentis L2-32. Growth of B. adolescentis L2-32 was unaffected by coculture (mean of 4.3 × 108 CFU ml−1). In addition to lactate and acetate, products of starch hydrolysis that escape uptake by B. adolescentis L2-32 may also have contributed to the growth of the lactate utilizers.
DISCUSSION
Conversion of lactate to butyrate is thought to be significant in the human colon (7, 31), but there have been few attempts to define the bacteria that might be responsible. We report here the recovery and identification of three groups of lactate-utilizing bacteria from human feces that produce butyrate as their main fermentation end product. Since the isolates reported here come from 10−8 dilutions of fecal samples from five different individuals and since related strains were recovered from more than one individual, these bacteria represent numerically significant components of the human fecal microbiota. Two of these groups of lactate utilizers are represented by the known species E. hallii and A. caccae that belong to clostridial cluster XIVa. E. hallii-related sequences have been reported to account for up to 3.6% of 16S rRNA sequence diversity in human feces, as measured by fluorescent in situ hybridization (22); lactate utilization has not been reported before in this species, possibly because acetate would have been absent from the test medium (38). The remaining three lactate-utilizing isolates considered here also belong to clostridial cluster XIVa and represent a new species distantly related to C. indolis. Representatives of other abundant groups of butyrate-forming human colonic bacteria (17), R. intestinalis, E. rectale, and F. prausnitzii, did not utilize lactate to any significant extent. We can conclude that lactate utilization is a specific attribute of only a few species within clostridial cluster XIVa and is not a property common to all butyrate-producing bacteria.
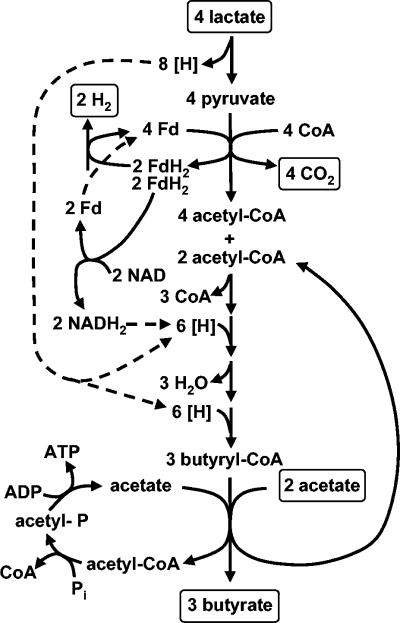
When grown on medium containing dl-lactate, in the absence of glucose, E. hallii and A. caccae strains carried out a net conversion of acetate and lactate to butyrate. In molar terms, the butyrate produced was approximately equal to half of the acetate plus lactate lost from the medium (Fig. 2). A metabolic scheme that approximates the observed stoichiometries for growth on lactate (Tables 2 and 3) is shown in Fig. 4. Carbon sources other than lactate and acetate derived from the medium may contribute to biosynthesis and growth in these bacteria, but apparently have little impact on the overall carbon balance during growth on dl-lactate, for which values were close to 100% (Tables 2 and 3). The conversion shown in Fig. 4 implies that H arising from the pyruvate-to-acetyl coenzyme A step (43) can be used in butyrate synthesis via ferredoxin/NADH exchange, as reported by Saint-Amans et al. (44) in Clostridium butyricum. When growing on glucose, many of the strains produced both formate and butyrate, whereas formate was never a major product of lactate utilization (Table 2). The reasons for this metabolic shift are not known but could reflect, for example, the activation or induction of a pyruvate formate lyase during growth on glucose.
FIG. 4.
Scheme for butyrate formation from lactate in E. hallii and A. caccae.
E. hallii and A. caccae strains were the only ones found to be able to utilize both the d and l isomers of lactic acid. The strains distantly related to C. indolis used only d-lactate, suggesting that they lack an l-lactate dehydrogenase capable of producing pyruvate from l-lactate and also lack a racemase capable of interconverting l- and d-lactate.
In time course experiments with media containing 10 mM glucose, lactate utilization occurred in all strains tested only as the glucose became depleted. Thus, in general, the availability of glucose delayed the utilization of lactate, as is observed in propionate producers (12). The repression of lactate utilization by high glucose concentrations in batch cultures does not necessarily imply significant repression in vivo, however, as monosaccharide concentrations are normally very low in the large intestine (36). A coculture experiment was therefore designed in order to create a nutritional situation that is more likely to occur in vivo. This showed that lactate-utilizing strains of E. hallii and A. caccae were able to efficiently convert l-lactate produced from starch by B. adolescentis strain L2-32 into butyrate. Therefore, it is apparent that starch breakdown products released by the B. adolescentis L2-32 did not achieve concentrations sufficient to abolish lactate assimilation by the lactate utilizers. Cross-feeding of lactate has also been demonstrated in vitro between Bifidobacterium longum and Eubacterium limosum, resulting mainly in acetate formation, but with some butyrate also being formed (31).
This type of cross-feeding could be significant in vivo. There is substantial evidence to indicate that resistant starch promotes butyrate formation by the human colonic microbial community (9, 14, 50, 58). The two major groups that have been considered to be active starch degraders, bifidobacteria and bacteroides (36, 45, 55), however, do not produce butyrate. While it is clear that some recently isolated butyrate producers are also amylolytic (3), their ecological role is as yet unclear and a recent study failed to detect increases in Eubacterium-related, butyrate-producing populations in response to starch in a rat model (47). It seems likely, however, that the increase in butyrate formation in these studies involves cross-feeding of fermentation products formed by other non-butyrate-producing species. This type of explanation has also been proposed to explain the butyrogenic effects of other substrates such as gluconate in the pig gut (52). The conversion of lactate to butyrate has been demonstrated in incubations with mixed human fecal bacteria (7), and we propose that the bacteria such as those identified here may play a significant role in converting lactate into butyrate in the colonic ecosystem.
The bacterium assumed to be mainly responsible for lactate utilization in the rumen and in the large intestine of pigs and rodents is Megasphaera elsdenii (12, 23, 25, 52). This species can produce propionate, butyrate, acetate, and valerate from lactate, depending on the strain and on the pH, with propionate being formed as a major product via the acrylate pathway (12). In humans, some cultural studies report M. elsdenii to be present in feces (e.g., reference 24), whereas others have failed to report it or have found it in only a minority of subjects (e.g., references 19 and 39). Other bacteria belonging to clostridial cluster IX that are reported to convert lactate into propionate, such as Veillonella and Selenomonas, have also been isolated from human feces (19, 24, 39). There is little evidence, however, to indicate whether propionate, butyrate, or acetate is the main product of lactate utilization in the human large intestine and also which bacterial groups contribute most to preventing lactate accumulation.
The present investigation has now identified several abundant, but previously unconsidered, groups of lactate-utilizing bacteria belonging to clostridial cluster XIVa from human feces for which butyrate is a major product. Since we screened only five donors in one geographical location, it is not ruled out that more extensive surveys would reveal additional phylogenetic groups of lactate-utilizing, butyrate-producing bacteria from the human gut. Such bacteria potentially could make a highly significant contribution to metabolic balance within the large intestine that will be examined in future work.
Acknowledgments
This work is supported by the Scottish Executive Environment and Rural Affairs Department.
We are grateful to Rustam Aminov for helpful discussions. Thanks go to M. Blaut for A. caccae strain P2, Kenneth Young for technical help, Pauline Young for DNA sequence analysis, and Maureen Annand for glucose and l-lactate analyses.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Atlas, R. M. 1997. Selenomonas selective medium (SS medium), p. 1245. In L. C. Parks (ed.), Handbook of microbiological media, 2nd ed. CRC Press, Cleveland, Ohio.
- 3.Barcenilla, A. 1999. Diversity of the butyrate-producing microflora of the human gut. Ph.D. thesis. The Robert Gordon University, Aberdeen, United Kingdom.
- 4.Barcenilla, A., S. E. Pryde, J. C. Martin, S. H. Duncan, C. S. Stewart, C. Henderson, and H. J. Flint. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66:1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benno, Y., K. Endo, H. Miyoshi, T. Okuda, H. Koishi, and T. Mitsuoka. 1989. Effect of rice fiber on human fecal microflora. Microbiol. Immunol. 33:435-440. [DOI] [PubMed] [Google Scholar]
- 6.Borody, T. J., E. F. Warren, S. Leis, R. Surace, and O. Ashman. 2003. Treatment of ulcerative colitis using fecal bacteriotherapy. J. Clin. Gastroenterol. 37:42-47. [DOI] [PubMed] [Google Scholar]
- 7.Bourriaud, C., S. Akoka, S. Goupry, R. Robins, C. Cherbut, and C. Michel. 2002. Butyrate production from lactate by human colonic microflora. Reprod. Nutr. Dev. 42(Suppl. 1):S55. [Google Scholar]
- 8.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brouns, F., B. Kettlitz, and E. Arrigoni. 2002. Resistant starch and the “butyrate revolution.” Trends Food Sci. Technol. 13:251-261. [Google Scholar]
- 10.Bryant, M. P. 1972. Commentary on the Hungate technique for cultivation of anaerobic bacteria. Am. J. Clin. Nutr. 25:1324-1328. [DOI] [PubMed] [Google Scholar]
- 11.Chan, L., J. Slater, J. Hasbargen, D. N. Herndon, R. I. Veech, and S. Wolf. 1994. Neurocardiac toxicity of racemic D, L-lactate fluids. Int. Physiol. Behav. Sci. 29:383-394. [DOI] [PubMed] [Google Scholar]
- 12.Counotte, G. H. M., R. A. Prins, R. H. A. M. Janssen, and M. J. A. deBie. 1981. Role of Megasphaera elsdenii in the fermentation of dl-[2-13C]lactate in the rumen of dairy cattle. Appl. Environ. Microbiol. 42:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cummings, J. H. 1995. Short chain fatty acids, p. 101-130. In G. R. Gibson and G. T. MacFarlane (ed.), Human colonic bacteria: role in nutrition, physiology and pathology. CRC Press, Boca Raton, Fla.
- 14.Cummings, J. H., E. R. Beatty, S. M. Kingman, S. A. Bingham, and H. N. Englyst. 1996. Digestion and physiological properties of resistant starch in the human large bowel. Br. J. Nutr. 75:733-737. [DOI] [PubMed] [Google Scholar]
- 15.Duncan, S. H., G. L. Hold, A. Barcenilla, C. S. Stewart, and H. J. Flint. 2002. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. E vol. Microbiol. 52:1-6. [DOI] [PubMed] [Google Scholar]
- 16.Duncan, S. H., G. L. Hold, H. J. M. Harmsen, C. S. Stewart, and H. J. Flint. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify the species into a new genus, Faecalibacterium gen. nov. Int. J. Syst. E vol. Microbiol. 52:2141-2146. [DOI] [PubMed] [Google Scholar]
- 17.Duncan, S. H., A. Barcenilla, C. S. Stewart, S. E. Pryde, and H. J. Flint. 2002. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl. Environ. Microbiol. 68:5186-5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duncan, S. H., K. P. Scott, A. G. Ramsay, H. J. M. Harmsen, G. W. Welling, C. S. Stewart, and H. J. Flint. 2003. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Appl. Environ. Microbiol. 69:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finegold, S. M., V. L. Sutter, and G. E. Mathison. 1983. Normal indigenous flora, p. 3-31. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, New York, N.Y.
- 20.Flint, H. J., and J. Bisset. 1990. Genetic diversity in Selenomonas ruminantium isolated from the rumen. FEMS Microbiol. Ecol. 73:351-360. [Google Scholar]
- 21.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harmsen, H. J. M., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashizume, K., T. Takamitsu, K. Yamada, H. Koyama, and K. Ushida. 2003. Megasphaera elsdenii JCM1772T normalises hyperlactate production in the large intestine of fructooligosaccharide-fed rats by stimulating butyrate production. J. Nutr. 133:3187-3190. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi, H., M. Sakamoto, and Y. Benno. 2002. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46:535-548. [DOI] [PubMed] [Google Scholar]
- 25.Hino, T., and S. Kuroda. 1993. Presence of lactate dehydrogenase and lactate racemase in Megasphaera elsdenii grown on glucose or lactate. Appl. Environ. Microbiol. 59:255-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hobson, P. N. 1969. Rumen bacteria. Methods Microbiol. 3B:133-149. [Google Scholar]
- 27.Hold, G. L., S. E. Pryde, V. J. Russell, E. Furrie, and H. J. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33-39. [DOI] [PubMed] [Google Scholar]
- 28.Hold, G. L., A. Schwiertz, R. I. Aminov, M. Blaut, and H. J. Flint. 2003. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 69:4320-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hove, H., I. Nordgaard Andersen, and P. B. Mortensen. 1994. Fecal DL-lactate concentration in 100 gastrointestinal patients. Scand. J. Gastroenterol. 29:255-259. [DOI] [PubMed] [Google Scholar]
- 30.Hungate, R. E. 1979. Evolution of a microbial ecologist. Annu. Rev. Microbiol. 33:1-20. [DOI] [PubMed] [Google Scholar]
- 31.Kanauchi, O., Y. Fujiyama, K. Mitsuyama, Y. Araki, T. Ishii, T. Nakamura, Y. Hitomi, K. Agata, T. Saiki, A. Andoh, A. Toyonaga, and T. Bamba. 1999. Increased growth of Bifidobacterium and Eubacterium by germinated barley foodstuff, accompanied by enhanced butyrate production in healthy volunteers. Int. J. Mol. Med. 3:175-179. [DOI] [PubMed] [Google Scholar]
- 32.Kaneko, T., Y. Banda, H. Kurihara, K. Satomi, K. Nonoyama, and N. Matsuura. 1997. Fecal microflora in a patient with short-bowel syndrome and identification of dominant lactobacilli. J. Clin. Microbiol. 35:3181-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Blay, G., C. Michel, H. M. Blottiere, and C. Cherbut. 1999. Enhancement of butyrate production in the rat caecocolonic tract by long-term ingestion of resistant potato starch. Br. J. Nutr. 82:419-426. [DOI] [PubMed] [Google Scholar]
- 34.Louis, P., S. H. Duncan, S. I. McCrae, J. Millar, M. S. Jackson, and H. J. Flint. 2004. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacteriol. 186:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macfarlane, G. T., and H. N. Englyst. 1986. Starch utilization by the human large intestinal microflora. J. Appl. Bacteriol. 60:195-201. [DOI] [PubMed] [Google Scholar]
- 36.Macfarlane, G. T., and G. R. Gibson. 1996. Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine, p. 269-318. In R. I. Mackie, B. R. White, and R. E. Isaacson (ed.), Gastrointestinal microbiology, vol. 1. Chapman and Hall, London, United Kingdom. [Google Scholar]
- 37.Miyazaki, K., J. C. Martin, R. Marinsek-Logar, and H. J. Flint. 1997. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis B14). Anaerobe 3:373-381. [DOI] [PubMed] [Google Scholar]
- 38.Moore, W. E. C., and L. V. Holdeman Moore. 1986. Genus Eubacterium. Prevot 1938, 294AL, p. 1364-1365. In P. H. A. Sneath (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams and Wilkins, Baltimore, Md. [Google Scholar]
- 39.Moore, W. E. C., and L. H. Moore. 1995. Intestinal floras of populations that have a high risk of colon cancer. Appl. Environ. Microbiol. 61:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pryde, S. E., S. H. Duncan, G. L. Hold, C. S. Stewart, and H. J. Flint. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217:133-139. [DOI] [PubMed] [Google Scholar]
- 41.Richardson, A. J., A. G. Calder, C. S. Stewart, and A. Smith. 1989. Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography. Lett. Appl. Microbiol. 9:5-8. [Google Scholar]
- 42.Rumney, C., S. H. Duncan, C. Henderson, and C. S. Stewart. 1995. Isolation and characteristics of a wheatbran degrading butyrivibrio from human faeces. Lett. Appl. Microbiol. 20:232-236. [DOI] [PubMed] [Google Scholar]
- 43.Russell, J. B., and R. J. Wallace. 1997. Energy yielding and energy-consuming reactions, p. 246-282. In P. N. Hobson and C. S. Stewart (ed.), The rumen microbial ecosystem, 2nd ed. Blackie, London, United Kingdom.
- 44.Saint-Amans, S., L. Girbal, J. Andrade, K. Aherns, and P. Soucaille. 2001. Regulation of carbon and electron flow in Clostridium butyricum VPI 3266 grown on glucose-glycerol mixtures. J. Bacteriol. 183:1748-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salyers, A. A., S. E. H. West, J. R. Vercellotti, and T. D. Wilkins. 1977. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl. Environ. Microbiol. 34:529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwiertz, A., G. L. Hold, S. H. Duncan, P. A. Lawson, M. D. Collins, H. J. Flint, and M. Blaut. 2002. Anaerostipes caccae gen. nov., sp. nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Syst. Appl. Microbiol. 25:46-51. [DOI] [PubMed] [Google Scholar]
- 47.Schwiertz, A., U. Lehmann, G. Jacobasch, and M. Blaut. 2002. Influence of resistant starch on the SCFA production and cell counts of butyrate-producing Eubacterium spp. in the human intestine. J. Appl. Bacteriol. 93:157-162. [DOI] [PubMed] [Google Scholar]
- 48.Slyter, L. L. 1976. Influence of acidosis on rumen function. J. Anim. Sci. 43:910-929. [DOI] [PubMed] [Google Scholar]
- 49.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Topping, D. L., and P. M. Clifton. 2001. Short chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81:1031-1064. [DOI] [PubMed] [Google Scholar]
- 51.Trinder, P. 1969. Determination of glucose in blood using glucose oxidase with an alternative oxygen acceptor. Ann. Clin. Biochem. 6:24-27. [Google Scholar]
- 52.Tsukuhara, T., H. Koyama, M. Okada, and K. Ushida. 2002. Stimulation of butyrate production by gluconic acid in batch culture of pig cecal digesta and identification of butyrate-producing bacteria. J. Nutr. 132:2229-2234. [DOI] [PubMed] [Google Scholar]
- 53.Vella, A., and G. Farrugia. 1998. D-lactic acidosis: pathological consequences of saphrophytism. Mayo Clin. Proc. 73:451-456. [DOI] [PubMed] [Google Scholar]
- 54.Vernia, P., R. Caprilli, G. Latella, F. Barbetti, F. M. Magliocca, and M. Cittadini. 1988. Fecal lactate and ulcerative colitis. Gastroenterology 95:1564-1568. [DOI] [PubMed] [Google Scholar]
- 55.Wang, X., P. L. Conway, I. L. Brown, and A. J. Evans. 1999. In vitro utilization of amylopectin and high-amylose maize (amylomaize) starch granules by human colonic bacteria. Appl. Environ. Microbiol. 65:4848-4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weaver, G. A., J. A. Krause, T. L. Miller, and M. J. Wolin. 1992. Cornstarch fermentation by the colonic microbial community yields more butyrate than does cabbage fiber fermentation—cornstarch fermentation rates correlate negatively with methanogenesis. Am. J. Clin. Nutr. 55:70-77. [DOI] [PubMed] [Google Scholar]
- 57.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolin, M. J., T. L. Miller, S. Yerry, Y. Zhang, S. Bank, and G. A. Weaver. 1991. Changes of fermentation pathways of fecal microbial communities associated with a drug treatment that increases dietary starch in the human colon. Appl. Environ. Microbiol. 65:2807-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]