Abstract
Changes in soil microbial community structure and diversity may reflect environmental impact. We examined 16S rRNA gene fingerprints of bacterial communities in six agroecosystems by PCR amplification and denaturing gradient gel electrophoresis (PCR-DGGE) separation. These soils were treated with manure for over a century or different fertilizers for over 70 years. Bacterial community structure and diversity were affected by soil management practices, as evidenced by changes in the PCR-DGGE banding patterns. Bacterial community structure in the manure-treated soil was more closely related to the structure in the untreated soil than that in soils treated with inorganic fertilizers. Lime treatment had little effect on bacterial community structure. Soils treated with P and N-P had bacterial community structures more closely related to each other than to those of soils given other treatments. Among the soils tested, a significantly higher number of bacterial ribotypes and a more even distribution of the bacterial community existed in the manure-treated soil. Of the 99 clones obtained from the soil treated with manure for over a century, two (both Pseudomonas spp.) exhibited 100% similarity to sequences in the GenBank database. Two of the clones were possible chimeras. Based on similarity matching, the remaining 97 clones formed six major clusters. Fifty-six out of 97 were assigned taxonomic units which grouped into five major taxa: α-, β-, and γ-Proteobacteria (36 clones), Acidobacteria (16 clones), Bacteroidetes (2 clones), Nitrospirae (1 clone), and Firmicutes (1 clone). Forty-one clones remained unclassified. Results from this study suggested that bacterial community structure was closely related to agroecosystem management practices conducted for over 70 years.
Understanding the complex microbial community in the soil environment has proven to be a challenging task because of the vast diversity and the enormity of the population inhabiting the environment. Recent development of culture-independent molecular techniques has brought us hope for in-depth understanding of the soil black box (16, 30). Unfortunately, changes in microbial community structure and diversity due to seasonal and temporal variations in nutrient or physical conditions are slow and gradual, making it difficult to interpret the data and obtain conclusive results (2, 30). Extensive studies demonstrated perturbation of microbial community equilibrium populations by changes in environmental conditions and soil management practices (25, 30). The challenge is how to quantify the changes and link them to the corresponding ecosystems. In this study, we have taken advantage of a unique opportunity to examine soil bacterial communities in different agroecosystems following a long-term winter wheat experiment with regular manure applications for over a century and chemical fertilizer applications for over 70 years.
It has long been recognized that the activity of soil microorganisms plays an intrinsic role in residue decomposition, nutrient cycling, and crop production. Understanding microbial community structure shifts following implementation of various land use and management systems may lead to development of best management practices for agroecosystems. Previous investigation of these agroecosystems showed that microbial biomass carbon (C) and activities of dehydrogenase and enzymes involved in phosphorus transformations were significantly higher in soil treated with manure for a century than in inorganic-fertilizer-treated soils (24). The objective of this study was to evaluate bacterial community structure and diversity in these soils by PCR-denaturing gradient gel electrophoresis (PCR-DGGE) analysis and cloning and sequencing of PCR-amplified 16S rRNA gene fragments using soil microbial community DNA as the template.
MATERIALS AND METHODS
Soil samples.
Composite (18 cores) surface soil samples (0 to 10 cm depth) were taken in January 2000 from plots of a long-term continuous winter wheat (Triticum aestivum L.) experiment in central Oklahoma. Detailed descriptions of the experimental site and soils have been reported (24). Briefly, the experiment was initiated on a Kirkland (fine, mixed, thermic Udertic Paleustolls) silt loam with a mean particle size distribution of 37.5% sand, 40% silt, and 22.5% clay. The manure treatment plot and an untreated control were initiated in 1899, and the chemical fertilizer treatment plots were initiated in 1929. Six treatment plots (treatment with manure; P; N and P [NP], N, P, and K [NPK]; and N, P, and K plus lime [NPKL]; and an untreated control) were investigated. Cattle manure from a feedlot had been applied every 4 years since 1899 at 269 kg of N ha−1 (approximately 13,450 kg of manure ha−1) and incorporated into the soil immediately following application. The most recent cattle manure application was made in September 1999. Chemical fertilizer plots received annual applications of 67 kg of N, 14.6 kg of P, and 28 kg of K ha−1 in the forms of NH4NO3, Ca(H2PO4)2, and KCl, respectively, before winter wheat was planted in October. The field-moist soil samples were passed through a 2-mm sieve and stored at 4°C. Before DNA extraction, soils were incubated for 2 weeks at 23°C and 60% water holding capacity.
Extraction of total community DNA and nucleic acid amplification.
Total soil DNA was extracted with an UltraClean soil DNA kit (Mo Bio Laboratories, Inc., Solana Beach, Calif.). It was reported that PCR primers BF1092 (5′-AAGTCCCGTAACGAGCGCAA-3′) (34) and U1392GC with 40 bases of a GC clamp (5′-CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCCACGGGCGGTGTGTAC-3′) (9, 18, 21) maximized recovery of soil bacterial communities. These two primers correspond to Escherichia coli positions 1092 to 1111 and 1392 to 1406, respectively. Theoretically, the resulting PCR products should be 354 bp, of which 314 bp are from the 16S rRNA genes of members of the domain Bacteria, including the highly variable V9 region (11). Each 100-μl PCR mixture contained 50 ng of soil DNA extracts, 0.6 μM (each) primer, 200 μM PCR nucleotide mixture (Fisher Scientific Inc., Pittsburgh, Pa.), 1.75 mM MgCl2, 1.5 μg of bovine serum albumin, and 3.2 U of Taq DNA polymerase in PCR buffer A (Promega Chemicals, Madison, Wis.). PCR was performed by a modified procedure described by Ferris et al. (9). Briefly, it included an initial denaturation of 2 min at 94°C, followed by nine cycles consisting of 1 min of denaturation at 94°C, 1 min of annealing at 56°C with a touchdown of 1°C per cycle, and 2 min of extension at 72°C. Then, 22 additional cycles, each consisting of 40 s at 94°C, 40 s at 47°C, and 2 min at 72°C, were performed. The final extension at 72°C was 8 min. PCR products were checked for the expected size and quantified on 2.5% agarose gels.
DGGE analysis of rRNA genes.
The PCR amplicons were separated with a DCode universal mutation detection system (Bio-Rad, Inc., Hercules, Calif.). A 6.5% acrylamide gel with a 30 to 55% parallel denaturing gradient was prepared with a Hoefer SG100 gradient maker (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). Denaturant (100%) contained 7 M urea and 40% deionized formamide. The gel was polymerized for at least 4 h. Approximately 3 μg of PCR products of the expected size was loaded in each well. The gel was run for 5 h at 200 mV in 1× Tris-acid-EDTA buffer at 60°C. The image of the gel was obtained with a Kodak (New Haven, Conn.) 1D scientific imaging system and a Kodak DC 290 zoom digital camera. DGGE banding patterns were analyzed with GeneProfiler (Scanalytics Inc., Fairfax, Va.).
Isolation and sequencing of dominant DGGE bands.
Some dominant DGGE bands were excised and eluted by incubation in 20 μl of sterilized distilled water overnight at 4°C. The following day, 10 μl of liquid from each tube was used as the template to reamplify the band of interest with the primers previously described and a slightly different PCR mixture and conditions. Briefly, each 50-μl PCR mixture contained 10 μl of eluted DNA from the DGGE band, 0.2 μM (each) primer, 200 μM PCR nucleotide mixture (Fisher Scientific Inc.), 1.5 mM MgCl2, 1 μl of dimethyl sulfoxide, and 1.5 U Taq of DNA polymerase in PCR buffer A (Promega Chemicals). PCR included an initial denaturation of 3 min at 94°C, followed by 31 cycles consisting of 1 min of denaturation at 94°C, 30 s of annealing at 60°C, and 30 s of extension at 72°C, followed by a final extension of 8 min at 72°C. PCR products were checked for purity and expected size and quantified as described above.
The reamplified products were purified with the UltraClean PCR cleanup DNA purification kit (Mo Bio Laboratories, Inc.) to remove primers and short oligonucleotides and were cloned into plasmids with a TOPO TA cloning kit (Invitrogen Life Technologies). Plasmid DNAs were isolated from randomly selected clones and screened for inserts of the expected sizes. Following confirmation of their DGGE positions under the conditions previously stated, the plasmid DNAs were sequenced with primers (M13 reverse/forward) on an ABI PRISM 3700 DNA analyzer (Applied Biosystems, Foster City, Calif.).
Analysis of the DGGE community profiles.
Banding patterns of three replicated experiments were analyzed by a GeneProfiler database manager with TreeCon software (LI-COR, Inc.). The lower limit for band detection was set by the band peak height threshold of 0.1% of the total detected optical density in the lane. The total number of different bands was determined for the samples being compared, and then each sample was scored based on the presence or absence of each band in its profile compared with the profile of each of the other samples. This provided a comparison of detected microbial community structures among soils, but not the relative dominance of each detected ribotype in that community. The Shannon diversity indices were calculated to reflect species richness and evenness of the bacterial community (29). Species evenness of the community was further estimated with Pielou's diversity (J) (26). Similarity among treatments was determined by pairwise matching to determine matching bands by comparing two treatments at a time. The band sharing coefficient (S; also termed the Dice similarity coefficient) (7) was used as a measure of similarity between two given treatments; it was determined from the equation S = 2Ns/NT, where Ns is the number of shared bands in samples A and B and NT is the total number of detected bands in samples A and B. Relatedness of bacterial communities was determined by determining similarity coefficients for bands common to two samples. Two bands were common if they migrated the same distance on a gel. Based on the number, intensity values, and positions of detected bands, cluster analysis and statistical analysis were performed. Comparisons of relative band intensity within lanes were made. Significant difference among treatment means was determined by the least significant difference test at a P value of <0.05. Percentage data were normalized by arc sine transformation before analysis (10). Bacterial species richness in a community was indicated by the total number of bands detected, and species distribution evenness was evaluated by determining the intensity, mobility, and distribution of bands in the rRNA gene fingerprint.
rRNA gene cloning and sequencing, and phylogenetic analysis.
The PCR products obtained for DGGE analysis from the manure-treated soil were also directly cloned into PCR 2.1-TOPO plasmids by the procedure recommended by the manufacturer. Ninety-four clones were randomly selected and sequenced.
Sequence similarity searches were conducted using the BLAST network service (1) of the GenBank database to identify the nearest relatives of the partially sequenced 16S rRNA genes and the excised dominant bands. Sequences were screened for chimeras by the CHECK-CHIMERA program of the Ribosome Database Project (http://rdp.cme.msu.edu/cgis/chimera.cgi?su=SSU) and by BLAST alignment. Sequences yielding a bell-shaped histogram, matching <93% with sequences in the database, and showing different closest relatives from two sides of a breakpoint were considered chimeras and were removed from the data set. Phylogenetic analyses were conducted with Biology WorkBench, version 3.2 (San Diego Supercomputer Center; http://workbench.sdsc.edu). The phylogenetic tree of the aligned sequences was constructed with a maximum-likelihood tree algorithm.
Nucleotide sequence accession numbers.
A total of 105 nucleotide sequences obtained in this study have been sent to the National Center for Biotechnology Information database. The accession numbers for the 11 excised band sequences are AY391554 to AY391564, and the 94 randomly selected clone sequences are assigned AY391617 to AY391710.
RESULTS
DGGE analysis of bacterial community.
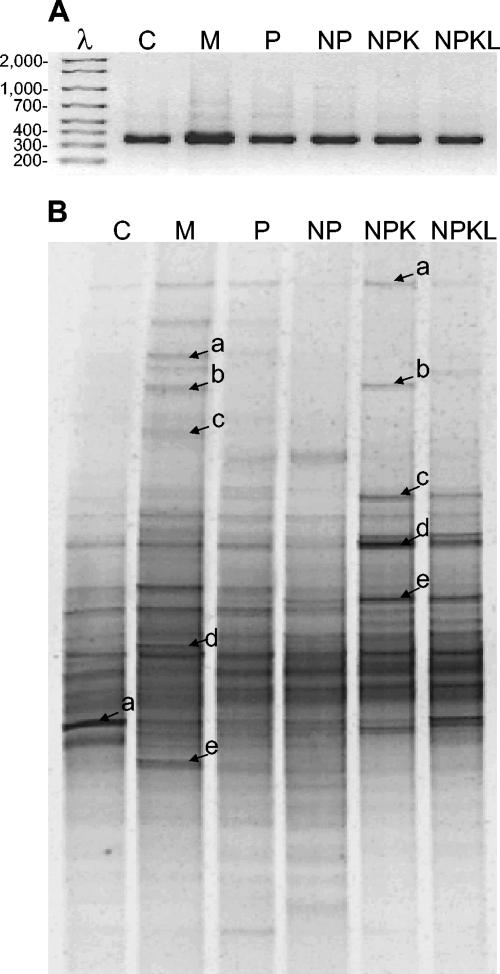
Bacterial species richness and evenness are demonstrated by DGGE banding patterns of 16S rRNA genes amplified by PCR with the bacterium-specific primer and a universal primer containing a GC clamp (Fig. 1). The DGGE patterns from the six soils were distinctly different and were reproducible according to DGGE banding patterns generated in three repeated experiments. Numerous discrete DGGE bands, resulting from differences between the 16S rRNA gene sequences of different bacterial species, were apparent (Fig. 1). Each band represents at least one unique ribotype. A significantly higher number of ribotypes was detected in the manure-treated soil, while the least number of ribotypes was detected in the control soil (Table 1). The different banding patterns among the soils tested suggest that each soil contained different bacterial community structure and diversity.
FIG. 1.
(A) A 2.5% agarose gel electrophoresis of PCR-amplified 16S rRNA gene fragments. (B) DGGE analysis of PCR-amplified 16S rRNA genes. Each lane was loaded with 3 μg of DNA. Bands marked were excised and sequenced. C, control; M, manure treatment.
TABLE 1.
Total bands detected on a DGGE gel with 6.5% acrylamide and a 30 to 55% parallel denaturing gradient
| Treatment | No. of bands detected in:
|
Avga | ||
|---|---|---|---|---|
| Rep I | Rep II | Rep III | ||
| Control | 14 | 16 | 16 | 15.3 ± 0.7c |
| Manure | 29 | 23 | 29 | 27.0 ± 2.0a |
| P | 21 | 19 | 22 | 20.7 ± 0.9b |
| NP | 20 | 21 | 21 | 20.7 ± 0.3b |
| NPK | 23 | 18 | 20 | 20.3 ± 1.5b |
| NPKL | 21 | 18 | 18 | 19.0 ± 1.0b |
Means of three replicates (Rep) ± standard errors. Different letters indicate significantly different means (P < 0.05) according to the least significant difference test.
When the band sharing coefficients between treatments were compared, bacterial community structures in P- and NP-treated soils showed the highest similarity, with an average band sharing coefficient of 0.68, followed by NPK- and NPKL-treated soils, with an average band sharing coefficient of 0.64 (Table 2). The bacterial community structure in the manure-treated soil was most similar to that of the control soil (0.56) and least similar to that of the NPKL-treated soils (0.41) (Table 2).
TABLE 2.
Average band sharing coefficient of DGGE bands detected in three repeated experiments between the paired treatments
| Treat- ment | Mean avg band sharing coefficient ± SE for:
|
||||
|---|---|---|---|---|---|
| Manure | P | NP | NPK | NPKL | |
| Control | 0.56 ± 0.10 | 0.50 ± 0.04 | 0.50 ± 0.07 | 0.47 ± 0.06 | 0.46 ± 0.09 |
| Manure | 0.51 ± 0.03 | 0.43 ± 0.05 | 0.45 ± 0.05 | 0.41 ± 0.11 | |
| P | 0.68 ± 0.06 | 0.47 ± 0.05 | 0.44 ± 0.06 | ||
| NP | 0.49 ± 0.05 | 0.44 ± 0.07 | |||
| NPK | 0.64 ± 0.03 | ||||
The Shannon diversity index for the bacterial community in the manure-treated soil (1.19 ± 0.07) was significantly higher than that for the bacterial community in the untreated control (0.97 ± 0.12). Compared to that for the manure-treated soil, lower Shannon diversity indices were obtained for soils treated with P (1.10 ± 0.10). NP (1.11 ± 0.05), NPK (1.16 ± 0.12), and NPKL (1.11 ± 0.04), though the differences were not statistically significant. However, the indices for evenness of the bacterial community were not significantly different among control soil (0.81 ± 0.078) and soil given manure (0.84 ± 0.039), P (0.83 ± 0.058), NP (0.85 ± 0.032), NPK (0.89 ± 0.033), and NKPL (0.87 ± 0.025) treatments.
The distribution evenness of a community is also indicated by dominance of individual ribotypes in a community, as reflected by the relative band intensities on a DGGE gel. The higher the intensity, the more dominant is the bacterial ribotype corresponding to that band. According to the intensity of the three most intense bands in each community, expressed as percentages of total DNA intensity in the lane, the most even distribution of bacterial ribotypes occurred in the manure-treated soil, with the top three ribotypes comprising about 40% of the total bacterial community detected (data not shown). The top three dominant ribotypes in the control soil comprised, by average, 56% of the total bacterial community detected, a value significantly higher than that for the manure-treated soil. In fact, one ribotype distinctly dominated the bacterial community of the control soil (Fig. 1), comprising, by average, 28% of the total community (data not shown). The top three dominant ribotypes in P-, NP-, NPK-, and NPKL-treated soils comprised, by average, 47, 45, 42, and 43% of the total bacterial community detected, respectively.
Seven of 11 excised dominant DGGE bands (marked in Fig. 1) had sequences most similar to unknown uncultured bacteria in the GenBank database (Table 3). Three of these 11 excised band sequences belong to Proteobacteria, and one belongs to Acidobacteria. The sequence of the most dominant bacterial community in the control soil exhibited 99% similarity to sequences of Sinorhizobium spp. and Agrobacterium spp. In general, these 11 bacterial ribotypes belong to a diverse group of bacteria with distant phylogenetic relationships, but with little soil specificity. Among them, NPKd and NPKe were closely related to one another, as were Ma and Mb.
TABLE 3.
Tentative identification of dominant DGGE bands by sequencing the excised bands and BLAST analysisa
| Band name | Sequence size (bp) | Accession no. | Closest relative (accession no.) | Alignment, % similarity | Score | Taxonomic affiliation |
|---|---|---|---|---|---|---|
| Ca | 314 | AY391554 | Sinorhizobium sp. and Agrobacterium sp. (e.g., AJ300187 and L39882) | 313/314, 99 | 615 | α-Proteobacteria |
| Ma | 300 | AY391555 | Uncultured soil bacterium (AF507686) | 290/292, 99 | 563 | Unknown |
| Mb | 301 | AY391556 | Uncultured soil bacterium (AF507686) | 289/292, 98 | 555 | Unknown |
| Mc | 317 | AY391557 | Uncultured soil bacterium (AF507740) | 312/317, 98 | 589 | Unknown |
| Md | 315 | AY391558 | Uncultured acidobacterium (AY177759) | 310/315, 98 | 585 | Acidobacteria |
| Me | 302 | AY391559 | Uncultured soil bacterium (e.g., AF423305 and AF423295) | 294/294, 100 | 583 | Unknown |
| NPKa | 313 | AY391560 | Uncultured bacterium (AB075101) | 306/313, 97 | 565 | Unknown |
| NPKb | 301 | AY391561 | Uncultured bacterium (AJ318200) | 282/292, 96 | 500 | Unknown |
| NPKc | 314 | AY391562 | Uncultured soil bacterium (AY326617) | 304/314, 96 | 543 | Unknown |
| NPKd | 309 | AY391563 | Massilia timonae (e.g., U54470 and AY157761) | 306/309, 99 | 589 | β-Proteobacteria |
| NPKe | 309 | AY391564 | β-Proteobacterium (AY278883) | 306/309, 99 | 589 | β-Proteobacteria |
Sequences were aligned with the closest relatives (highest score) in the GenBank database with BLAST. Bands correspond to those excised from the DGGE gel (Fig. 1). The percentage of similarity was calculated without taking gaps into account. The part of the total sequence used for alignment is indicated by the alignment data. Nucleotide sequences can be accessed by using the accession numbers at http://www.ncbi.nlm.nih.gov.
Bacterial diversity in soil treated for a century with manure.
Phylogenetic analysis of 99 sequences, including 94 randomly selected clones and 5 excised bands, from a DGGE gel from the manure-treated soil is shown in Fig. 2. Based on similarity matching (≥98%) of these sequences with those in the GenBank database, 17 clones were tentatively identified as members of Proteobacteria, Bacteroidetes-Sphingobacteria, Nitrospirae, and Firmicutes (Table 4). Two of the clones (M2 and M19) are possible chimeras. These two clones showed not only typical bell-shaped histograms from the CHECK-CHIMERA program but also low percentages of matching with sequences in the database. Several other clones also showed typical bell-shaped histograms but matched over 95% with sequences in the database, including several sequences of culturable bacteria. For example, the sequence of M50 (AY391666) matched 96% with the Nitrosomonas sp. strain Nm86 16S rRNA gene (AY123798). These clones are less likely to be chimeras. The remaining 80 clones were most similar to unclassified or uncultured bacterial 16S rRNA genes or showed less than 98% matching with sequences in the GenBank database.
FIG. 2.
Phylogenetic analyses of 99 sequences obtained from the soil treated for a century with manure, including 94 randomly selected clones and 5 excised bands (boldface and italics) from a DGGE gel (Fig. 1). Sequences were designated by the prefixes M (indicating manure treatment), followed by replicate numbers (randomly selected clones) or letters (excised bands). Three ribotypes were repeated three or four times (boxed) in the randomly selected clones. Clones exhibiting ≥98% sequence similarity were used as references for the corresponding taxonomic-unit assignments.
TABLE 4.
Summary of the phylogenetic assignments of 99 rDNA clones from soil treated for a century with manure
| Major taxon | Phylogenetic assignment based on:
|
|
|---|---|---|
| BLAST search (≥98% matching) | BioWorkBench cluster analysis | |
| Proteobacteria | ||
| α-Proteobacteria | 9 | 18 |
| β-Proteobacteria | 2 | 10 |
| γ-Proteobacteria | 2 | 8 |
| Bacteroidetes-Sphingobacteria | 2 | 2 |
| Acidobacteria | 0 | 16 |
| Firmicutes | 1 | 1 |
| Nitrospirae | 1 | 1 |
| Unclassified groups (possible chimeras, 2) | 80 | 41 |
Cluster analysis of clone sequences revealed six major clusters with numerous diverse groups (Fig. 2). One of the major clusters was not closely related to any sequences in the database, indicating a potential novel bacterial group. Among the 99 clones analyzed, 56 (58%) were assigned taxon affiliations and 41 were unclassified (Fig. 2). The Acidobacteria were the second-most-dominant group (16.5%). The sequences in this group matched ≥97% with sequences of uncultured acidobacteria in the database. When the 16S rRNA gene sequence of Acidobacterium capsulatum was included in the analysis, it was clustered closely with Md (data not shown). The third major group belonged to Bacteroidetes. Two clones (M1 and M81) in this group exhibited 99% similarity to the culturable bacteria Pedobacter sp. strain LMG 10342 (AF329966) and Pedobacter sp. strain LMG9525 (AF329962). In addition, two closely related clones (M13 and M56) also exhibited similarity to sequences of Bacteroidetes, but at lower percentages (92 and 89%, respectively). Among the 94 randomly selected clones, three ribotypes belonging to γ-Proteobacteria and Acidobacteria were selected three or four times (Fig. 2).
Five excised dominant bands from the DGGE gel in the manure-treated soil (Fig. 1) were distributed in three groups, the Acidobacteria (Md) group, an unclassified group that is closely related to Bacteroidetes (Mc), and an unclassified group that is a major cluster in the phylogenetic tree (Ma, Mb, and Me) (Fig. 2).
DISCUSSION
DGGE analysis demonstrated different banding patterns of bacterial community structures as well as species richness and evenness in the agroecosystems evaluated. Some of the discrete DGGE bands were further purified and sequenced to reveal specific differences in the bacterial community. Among the numerous recently emerged molecular methods for microbial community analysis, DGGE is one of few that holds this potential and has also been shown to be a sensitive method for microbial community analyses in compost-amended soils (13) and bacterial biofilms (17). Compared to amplified ribosomal DNA restriction analysis (ARDRA), the patterns of DGGE are more useful for a direct comparison of structural diversity among different microbial communities from different natural sites or environments. In ARDRA, each potential ribotype may be represented by multiple bands, and the patterns are affected by restriction enzymes used. In addition, ARDRA is not as sensitive as DGGE in detecting differences in microbial community structure (23).
DGGE banding patterns from soils of different agroecosystems obtained in this study indicated that bacterial community structure was affected by soil management practices. Shifts in bacterial community structure following adaptation of soil management practices have been reported by other studies (5, 6, 25, 30). The observed impact was greater for long-term soil management practices than for short-term land use or plant community composition (6). In this study, the similarity in community structures in NPK- and NPKL-treated soils, as demonstrated by DGGE banding patterns and by band sharing coefficients, suggests that recognizable bacterial community structures exist in relation to field management following over 70 years of management practices. This finding is consistent with the conclusions reported by Buckley and Schmidt (6).
While the significance of the microbial community shifts following soil management practices remains to be recognized, it is evident that organic amendment, in general, enriches the soil bacterial community and promotes diversity and a more even distribution of bacterial species within the community. Among treatments evaluated in this study, the number of bands (ribotypes) detected and the Shannon diversity index were highest for the manure-treated soil, in which the sum of the three most dominant bacterial ribotypes comprised the lowest percentage of the total community, indicating the most even distribution. These results are in agreement with those obtained by evaluating the culturable bacterial populations (23). They also support the studies of Øvreås and Torsvik (22), who reported that the bacterial community in an organic soil was more diverse and more evenly distributed than that in a sandy soil. Enrichment of the bacterial community following manure treatment has also been shown by enhanced microbial biomass C contents (12, 24), soil enzyme activities (24), N flush (27), and gram-negative bacterial populations (25).
The bacterial community in the soil treated with manure for a century was further evaluated by sequencing 94 randomly selected clones and 11 excised dominant bands from a DGGE gel. Five major taxa, α-, β-, and γ-Proteobacteria, Acidobacteria, Bacteroidetes, Nitrospirae, and Firmicutes, were identified. These results deviate from those reported by Smit et al. (30), who found that Acidobacteria, Proteobacteria, Nitrospira, Cyanobacteria, and green sulfur bacteria were the main bacterial divisions in a manure-treated winter wheat field in The Netherlands. Three of the five divisions, Proteobacteria, Acidobacteria, and Nitrospira, were dominant in this study.
The most dominant bacterial group was Proteobacteria, which is consistent with reports by several other studies related to agricultural agroecosystems (4, 30, 33). In this study, Proteobacteria comprised 37% of the 99 sequences obtained, a percentage that closely resembled that reported by Smit et al. (30), who showed that Proteobacteria comprised 35% of the bacterial community in an agroecosystem in The Netherlands. This may reflect the similarities of the two agroecosystems; both were winter wheat fields treated with manure. In contrast, only 16% of 122 bacterial clones from a clover-grass pasture in southern Wisconsin (4) and only 18% of 51 clones from the clay fraction of an animal manure-treated soil (28) were Proteobacteria.
The Acidobacteria were the second-most-dominant group based on cluster analysis, a result which was further evidenced by sequence analysis of excised DGGE bands and by the fact that two of the three repeatedly selected clones belonged to Acidobacteria. The dominance of Acidobacteria in a bacterial community is not surprising. Following a survey of 43 environmental samples, Barns et al. (3) concluded that members of the division Acidobacteria are as genetically and metabolically diverse, environmentally widespread, and ecologically important as the division Proteobacteria.
Dominance of Firmicutes-Bacillus in agroecosystems has been reported in California soils (13, 33). Among the five major taxa revealed in this study, Bacteroidetes-Sphingobacteria are well-known rumen bacteria that are commonly found in the gastrointestinal tracts of warm-blooded animals. The presence of this major group of bacteria in soil from a clover-grass pasture also was reported (4). With increasing concerns about adaptation of fecal bacteria in the environment following manure application (14), these findings merit additional research effort.
Results from this study suggested that bacterial diversity in agricultural soils was substantially different from that in uncultivated soils in a geothermal heating environment. Phylogenetic analysis of 39 clones from a soil in Yellowstone National Park suggested that the majority belonged to Acidobacteria (51%) and Planctomyces (18%) (20). Acidobacteria also dominated Arizona woodland soils (8).
Ratios between the ribotype numbers of Proteobacteria and Acidobacteria in an ecosystem might reflect the nutrient status in the soils tested (30). The reported ratios ranged from 0.16 in an oligotrophic soil (8) to 0.87 in a high-input agricultural soil (16). In a soil in The Netherlands, this ratio was 0.46 (30), and in the manure-treated soil evaluated in this study, it was 0.44, demonstrating another consistency of the data obtained from these two studies.
Although rRNA gene sequence-based analysis of microbial communities has been widely used to reveal bacterial diversity in the soil environment (4, 19, 30, 35), a limited number of sequences can be obtained and analyzed. The randomly selected clones represent dominant ribotypes in the bacterial community tested. Few of the DGGE-excised band sequences were similar to those of the randomly selected clone sequences, indicating that a vast diversity of bacterial ribotypes existed in the community. This discrepancy may also suggest that limited information on bacterial community structure and diversity can be derived from analysis of a limited number of randomly selected clones. Following analysis of 99 rRNA gene sequences in the manure-treated soil, only two matched 100% and a small percentage (about 17.5%) of the clones matched ≥98% with sequences in the GenBank database, indicating limitations of our knowledge of environmental microbial ecology as a whole. Similarly, 7 out of 11 excised bands from a DGGE gel remain unknown, further suggesting that a considerable portion of the bacterial community is largely unknown. It has been estimated that 80 to 99% of the microorganisms in soil are unidentified due to limitations in culture techniques (4). Less than 1% of the cells observed by direct counting can be recovered using standard cultivation methods (2, 15, 31, 32).
In summary, the soil bacterial community was impacted considerably by management practices of the agroecosystems studied. The distinct bacterial community structures in different agroecosystems reflected soil management practices conducted for over 70 years. Manure application increased soil bacterial diversity and resulted in a more even distribution than that in soil given inorganic fertilizer treatments and in an untreated control soil.
Acknowledgments
This work was supported in part by the USDA Special Grants Program and by the Oklahoma Agricultural Experimental Station (OAES) under projects h-OKLO2509, h-OKL02394, and h-OKLO2460.
This paper was approved for publishing by the director of OAES.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borneman, J., P. W. Skroch, K. M. O'Sullivan, J. A. Palus, N. G. Rumjanek, J. L. Jansen, J. Nienhuis, and E. W. Triplett. 1996. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl. Environ. Microbiol. 62:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckley, D. H., and T. M. Schmidt. 2001. Exploring the biodiversity of soil—a microbial rain forest, p. 183-208. In J. T. Staley and A. L. Reysenbach (ed.), Biodiversity of microbial life: foundation of Earth's biosphere. Wiley-Liss, Inc., New York, N.Y.
- 6.Buckley, D. H., and T. M. Schmidt. 2003. Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ. Microbiol. 5:441-452. [DOI] [PubMed] [Google Scholar]
- 7.Dice, L. R. 1945. Measures of the amount of ecologic association between species. Ecology 26:297-302. [Google Scholar]
- 8.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez, K. A., and A. A. Gomez. 1984. Statistical procedures for agricultural research, 2nd ed. John Wiley & Sons, New York, N.Y.
- 11.Gray, M. W., D. Sankoff, and R. J. Cedergren. 1984. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 12:5837-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopkins, D. W., and R. S. Shiel. 1996. Size and activity of soil microbial communities in long-term experimental grassland plots treated with manure and inorganic fertilizers. Biol. Fertil. Soils 22:66-70. [Google Scholar]
- 13.Ibekwe, A. M., S. K. Papiernik, J. Y. Gan, S. R. Yates, C. H. Yang, and D. E. Crowley. 2001. Impact of fumigants on soil microbial communities. Appl. Environ. Microbiol. 67:3245-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamieson, R. C., R. J. Gordon, K. E. Sharples, G. W. Stratton, and A. Madani. 2002. Movement and persistence of fecal bacteria in agricultural soils and subsurface drainage water: a review. Can. Biosyst. Eng. 44:1.1-1.9. [Google Scholar]
- 15.Liles, M. R., B. F. Manske, S. B. Bintrim, J. Handelsman, and R. M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers, R. M., S. G. Fischer, L. S. Lerman, and T. Maniatis. 1985. Nearly all single base substitutions in DNA fragments joined to a GC-clamp can be detected by denaturing gradient gel electrophoresis. Nucleic Acids Res. 13:3131-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nogales, B., E. R. B. Moore, E. Llobet-Brossa, R. Rossello-Mora, R. Amann, and K. N. Timmis. 2001. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norris, T. B., J. M. Wraith, R. W. Castenholz, and T. R. McDermott. 2002. Soil microbial community structure across a thermal gradient following a geothermal heating event. Appl. Environ. Microbiol. 68:6300-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen, G. J., D. J. Lane, S. J. Giovannoni, and N. R. Pace. 1986. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40:337-365. [DOI] [PubMed] [Google Scholar]
- 22.Øvreås, L., and V. Torsvik. 1998. Microbial diversity and community structure in two different agricultural soil communities. Microb. Ecol. 36:303-315. [DOI] [PubMed] [Google Scholar]
- 23.Parham, J. A., S. P. Deng, H. N. Da, H. Y. Sun, and W. R. Raun. 2003. Long-term cattle manure application in soil. II. Effect on soil microbial populations and community structure. Biol. Fertil. Soils 38:209-215. [Google Scholar]
- 24.Parham, J. A., S. P. Deng, W. R. Raun, and G. V. Johnson. 2002. Long-term cattle manure application in soil. I. Effect on soil phosphorus levels, microbial biomass C, and dehydrogenase and phosphatase activities. Biol. Fertil. Soils 35:328-337. [Google Scholar]
- 25.Peacock, A. D., M. D. Mullen, D. B. Ringelberg, D. D. Tyler, D. B. Hedrick, P. M. Gale, and D. C. White. 2001. Soil microbial community responses to dairy manure or ammonium nitrate applications. Soil Biol. Biochem. 33:1011-1019. [Google Scholar]
- 26.Pielou, E. C. 1966. The measurement of diversity in different types of biological collections. J. Theoret. Biol. 13:131-144. [Google Scholar]
- 27.Ritz, K., R. E. Wheatley, and B. S. Griffiths. 1997. Effects of animal manure application and crop plants upon size and activity of soil microbial biomass under organically grown spring barley. Biol. Fertil. Soils 24:372-377. [Google Scholar]
- 28.Sessitsch, A., A. Weilharter, M. H. Gerzabek, H. Kirchmann, and E. Kandeler. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon, C. E. 1948. A mathematical theory of communication. Bell Syst. Technol. J. 27:379-423. [Google Scholar]
- 30.Smit, E., P. Leeflang, S. Gommans, J. van den Broek, S. van Mil, and K. Wernars. 2001. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 67:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torsvik, V., J. Goksøyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torsvik, V., R. Sorheim, and J. Goksøyr. 1996. Total bacterial diversity in soil and sediment communities—a review. J. Ind. Microbiol. 17:170-178. [Google Scholar]
- 33.Valinsky, L., G. D. Vedova, A. J. Scupham, S. Alvey, A. Figueroa, B. Yin, R. J. Hartin, M. Chrobak, D. E. Crowley, T. Jiang, and J. Borneman. 2002. Analysis of bacterial community composition by oligonucleotide fingerprinting of rRNA genes. Appl. Environ. Microbiol. 68:3243-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, J. Z., M. E. Davey, J. B. Figueras, E. Rivkina, D. Gilichinsky, and J. M. Tiedje. 1997. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 143:3913-3919. [DOI] [PubMed] [Google Scholar]