Abstract
The occurrence and distribution of magnetotactic bacteria (MB) were studied as a function of the physical and chemical conditions in meromictic Salt Pond, Falmouth, Mass., throughout summer 2002. Three dominant MB morphotypes were observed to occur within the chemocline. Small microaerophilic magnetite-producing cocci were present at the top of the chemocline, while a greigite-producing packet-forming bacterium occurred at the base of the chemocline. The distributions of these groups displayed sharp changes in abundance over small length scales within the water column as well as strong seasonal fluctuations in population abundance. We identified a novel, greigite-producing rod in the sulfidic hypolimnion that was present in relatively constant abundance over the course of the season. This rod is the first MB that appears to belong to the γ-Proteobacteria, which may suggest an iron- rather than sulfur-based respiratory metabolism. Its distribution and phylogenetic identity suggest that an alternative model for the ecological and physiological role of magnetotaxis is needed for greigite-producing MB.
Magnetotactic bacteria (MB) are motile gram-negative bacteria whose directional swimming behavior is affected by the Earth's geomagnetic and external magnetic fields. They contain highly ordered intracellular chains of magnetic iron minerals, either magnetite (Fe3O4), greigite (Fe3S4) (33), or in one case, both (7). Magnetite MB (MMB) are found in both freshwater and marine environments, while greigite MB (GMB) appear to be unique to marine systems. Both MMB and GMB are present in salt marsh sediments, stratified salt ponds, estuarine basins in the Eastern coastal United States (5-7, 32, 33, 51), coastal marshes along the California coast (E. F. DeLong, personal communication), and other similar habitats worldwide, as well as deep-sea sediments (43). This widespread distribution of marine MB suggests that they are globally more abundant than freshwater MB.
All known MB share the following characteristics: motility by means of flagella, negative tactic responses to atmospheric concentrations of oxygen, optimal growth under microaerophilic or anaerobic conditions, and a respiratory metabolism (51). MB are typically found at or below the oxycline in sediments and water columns and can reach significant population densities: the freshwater MMB “Magnetotacticum bavaricum” (proposed name) can reach 7 × 105 cells per cm3 at the oxycline in freshwater sediments (48). Many, if not most, groups of MB that have been observed in the environment have not been grown in pure culture in the laboratory. Fewer than 10 strains are available in pure culture, and most of those are magnetite-producing α-Proteobacteria of the genus Magnetospirillum from freshwater environments. A few strains of marine MMB are available in culture (3, 45, 51). These strains are all microaerophiles when growing on O2, and some can grow anaerobically on N2O (3). All are capable of oxidizing but not reducing inorganic sulfur compounds, with the exception of the freshwater sulfate-reducing rod Desulfovibrio magneticus (45). GMB have not yet been isolated in axenic cultures.
Magnetotaxis occurs among several distinct phylogenetic lineages. Molecular analysis has shown that freshwater MMB are generally affiliated with the α-Proteobacteria (48-50). Exceptions are Desulfovibrio magneticus of the δ-Proteobacteria (22) and M. bavaricum in the Nitrospira phylum (52). The phylogeny of GMB is unknown, with the exception of one uncultivated packet-forming GMB called the many-celled magnetotactic prokaryote (MMP), which has been identified as one of the δ-Proteobacteria by fluorescent in situ hybridization (FISH) (13).
The ecology of GMB remains almost completely uncharacterized. Although several studies have examined mechanisms of biomineralization in GMB (18, 44), little information is available concerning their distribution, population structure, or geochemical significance. The composition of greigite (Fe3S4) and the abundance of magnetosomes in GMB suggest that they may play a significant role in marine iron and sulfur cycling in coastal areas. GMB can accumulate picograms of solid Fe and S per cell, transforming dissolved complexes into reactive minerals. As cells die and lyse, these minerals may accumulate in the sediment and provide reactive surfaces for further microbial (47) and chemical transformations, eventually leading to pyrite formation. Pyrite formation is the end point of the reductive portion of the sulfur cycle, suggesting that there is a biogeochemically important role for GMB. The energy-yielding reactions used by GMB are also unknown. The phylogenetic affiliation of MMP with the δ-Proteobacteria (13) suggests a sulfate- or iron-reducing metabolism. Greigite is metastable under the reducing conditions favored by MMP and should transform to pyrite (9), indicating that GMB must actively control the intracellular redox environment.
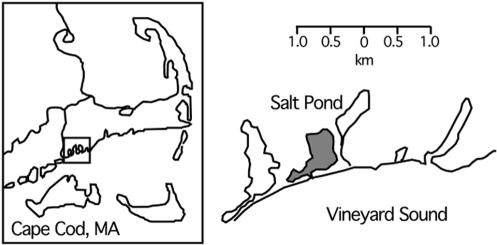
We have conducted studies on the ecology and biogeochemical role of GMB in Salt Pond, a small seasonally stratified (monomictic) coastal salt pond in Falmouth, Mass. (Fig. 1). Small saline lakes provide compact physiochemical gradients that are ideal for the study of microbial distributions as a function of redox chemistry. Studies of permanently and seasonally stratified lakes have shown characteristic changes in microbial communities across the interface between euxinic bottom waters and oxic surface waters (11, 40, 41, 54, 55). The oxycline (metalimnion) of these stratified lakes typically shows elevated cell counts, peaks in dissolved and particulate iron, and large populations of anoxic phototrophs where light is available (e.g. see reference 55). The basic water column chemistry of Salt Pond is similar to that of larger enclosed marine systems, such as the Pettaquamscutt Estuary, R.I. (38), the Black Sea (36), and Framvaren Fjord, Norway (61). It has been used for previous studies on chemical cycling (26, 28, 53, 59) and is both accessible and easy to sample. The rapid changes in water column chemistry with the onset, persistence, and breakdown of water column stratification offered the opportunity to examine MB populations in relation to iron, sulfide, and oxygen levels.
FIG. 1.
Sample site at Salt Pond, Falmouth, Mass. The location of Salt Pond is shown relative to other coastal features.
Previous measurements of nanophase magnetic minerals in Salt Pond showed a peak at the oxycline (4), suggesting the presence of MB in significant numbers. Our numerous unpublished observations indicated the presence of a significant MB community comprised of at least four different morphotypes at and slightly below the metalimnion. Magnetically responsive protists were also observed at the oxycline (8). MB appeared to be associated with peaks in dissolved and particulate iron that were also present at the oxycline. Peaks of particulate Fe(III) are often observed immediately above marine chemoclines due to the upward flux of Fe(II) into oxygenated waters (36, 61).
Here we report the results of studies aimed at (i) describing the distribution of different MB morphotypes and phylotypes with respect to physical and chemical environmental conditions and (ii) determining how marine MB impact chemical cycling in this stratified water column. This report describes changes in the marine MB population structure and water chemistry in Salt Pond during summer 2002.
MATERIALS AND METHODS
Site description.
Salt Pond is a shallow, brackish, seasonally stratified kettle hole pond in Falmouth, Mass. (Fig. 1). It is approximately 4.6 to 5.8 m deep, with a surface area of 0.29 km2 (53), and is surrounded by 0.02 km2 of salt marsh (10). The pond has a permanent tidal exchange with Vineyard Sound, and freshwater input comes from runoff, rainwater, and groundwater. The average daily tidal amplitude is approximately 0.6 m (http://co-ops.nos.noaa.gov/data_res.html). The pond becomes stratified beginning in early summer and develops sulfidic bottom waters. As of mid-October, thermal stratification begins to break down. The epilimnion of Salt Pond is highly productive: Wakeham et al. (59) measured chlorophyll at concentrations of 70 μg liter−1 in mid-August.
Physical and chemical sampling.
Salt Pond was sampled from a small aluminum rowboat. Salinity, temperature, and oxygen data were measured by use of a YSI 85 profiler (Yellow Springs Inc., Yellow Springs, Ohio) in the field to determine the location of the oxycline. The YSI electrode was fixed to the modified intake of a Geopump peristaltic pump (Geotech Environmental Equipment, Inc., Denver, Colo.), allowing simultaneous water sampling and profiling. Water samples were taken every 0.3 m above and below the oxycline and every 0.15 m within the oxycline. Water was collected in autoclaved 1-liter Nalgene bottles. The pH was measured on board by use of an Orion 250A field pH meter (Orion Research Inc., Beverly, Mass.). Water for iron and sulfide analyses was withdrawn directly from the pump tubing with 1-ml syringes.
Chemical analysis.
Iron was measured by the Ferrozine method as described previously (58). Sterile 1.5-ml microcentrifuge tubes were prepared with 100 μl of 10 mM Ferrozine in 100 mM ammonium acetate. Five hundred microliters of unfiltered water was placed in each tube. The tubes were stored in the dark until analysis immediately upon return to the lab, 1 to 2 h after collection. Initial absorbance (A1) values at 562 nm were measured with a UV-Vis spectrophotometer, followed by a 10-min reduction step with 150 μl of 1.4 M hydroxylamine HCl in 2 M HCl. Fifty microliters of 10 M ammonium acetate buffer at pH 9.5 was added, and the final absorbance (A2) was measured. Standard curves were constructed from a 200-ppm ferrous iron standard in sulfuric acid (LabChem Inc., Pittsburgh, Pa.) diluted in a 2% NaCl solution. A1 corresponded to [Fe(II)], and A2 corresponded to [Fe(II) + Fe(III)]. The concentration of Fe(III) was determined by calculating the difference.
Sulfide was analyzed according to the Cline method (12). In the field, 0.5-ml samples of water were placed into microcentrifuge tubes containing 1 ml of 2.6% zinc acetate solution. Upon our return to the lab approximately 1 to 2 h later, 200 μl of N,N-dimethyl-p-phenylenediamine sulfate (DPDS; 0.3% in 5.5 N HCl) and 200 μl of ferric chloride (0.0115 M in 0.6 N HCl) were added, and the samples were incubated for 30 min. The absorbance was measured at 665 nm. Standard curves were constructed by using dilutions of a 100 mM sodium sulfide solution fixed in 2.6% zinc acetate.
Sample handling.
Within 2 h of collection, bulk water samples were withdrawn from the bottles for fixation and DNA extraction. MB were then concentrated by placing the south pole of a bar magnet against the side of the bottle. After 1 h, MB were harvested next to the magnet with a Pasteur pipette. Both bulk water and MB-enriched samples for DNA extraction were concentrated by centrifugation at 10,000 × g for 15 min. The supernatants were decanted, and the resulting pellets were stored at −80°C. Samples for cell counting were fixed in 4% paraformaldehyde overnight and then filtered onto 25-mm-diameter black Millipore Isopore polycarbonate filters (pore size, 0.2 μm). The filters were rinsed with a sterile 2% NaCl solution, air dried, and stored in Parafilm-sealed petri dishes at −20°C until further analysis.
Microscopy.
A 100-μl drop from the enrichment was placed on a glass coverslip, and the south pole of a bar magnet was placed near the drop. MB accumulated at the edge of the drop closest to the bar magnet and were examined at a magnification of ×40 under a Zeiss Axiovert 100 inverted microscope by differential interference contrast light microscopy for evaluation of the MB morphotypes. In some cases, MB were further enriched by using a 10-μl glass capillary tube to remove liquid from the edge of a drop, where high densities of MB had accumulated (capillary samples). These samples were frozen at −20°C immediately.
Microbiological and molecular analyses.
Based on initial chemical and microbiological data, three representative time points (June 26, August 2, and October 9) were chosen for further analysis. To obtain total cell counts, we stained the filters with a 1-μg/ml DAPI (4′,6′-diamidino-2-phenylindole) solution for 3 min, rinsed them in a 2% NaCl solution, and examined them with an inverted Zeiss Axiovert 100 microscope equipped with a mercury arc lamp and appropriate filters. Seven fields were counted per filter based on previously published statistical recommendations (23). The mean and standard error of total cell numbers were calculated for each filter.
DNAs from magnetically enriched and bulk water samples were extracted by use of a standard cetyltrimethylammonium bromide-isopropanol precipitation protocol (1). For denaturing gradient gel electrophoresis (DGGE), initial PCR amplification of the 16S rRNA gene was carried out with the 341F-GC clamp and 907R primer pair by use of a touchdown protocol (46). After quantification in an agarose gel by use of a low-molecular-weight standard (Invitrogen), amplification products were run in a 20 to 70% denaturing gradient gel for 18 h at 100 V (Bio-Rad Dcode system). Magnetically enriched and bulk water samples from the same depth were run in adjacent lanes. Gels were stained in a 1:10,000 solution of Sybr Green I (Molecular Probes) in 1× Tris-acetate-EDTA for 1 h and then visualized on a transilluminator system. Individual bands of interest were excised.
DNAs were eluted by overnight incubation of the acrylamide bands in sterile MilliQ water at 4°C. PCRs were performed with the band DNAs and the 341F (non-GC) and 907R primer pair. The protocol used (46) was as follows: (i) initial denaturation for 5 min at 94°C, (ii) 25 cycles of 94°C (1 min), 55°C (1 min), and 72°C (3 min), and (iii) extension for 7 min at 72°C. The products were cloned into the pCR2.1-TOPO vector (Invitrogen). Minipreps were performed by use of a Wizard SV kit (Promega), and plasmids were quantified in an agarose gel by the use of mass ladders (Invitrogen). Alkaline lysis minipreps were also done in a 96-well format on a RevPrep Orbit instrument at the Josephine Bay Paul Center, Marine Biological Laboratory (MBL), Woods Hole, Mass. Sequencing reactions (1/8 reactions from an ABI v3.1 BigDye kit [Applied Biosystems]) were run on an ABI 3730xl capillary sequencer, also at MBL.
Clone libraries were also constructed directly from four capillary samples with high MB levels. One microliter of each sample was used directly in a PCR. The samples were amplified by PCR with the bacterial primers 341F (non-GC) and 1492R. The PCR protocol used was as follows: (i) initial denaturation at 94°C for 2 min, (ii) 25 cycles of 94°C (1 min), 47°C (1 min), and 72°C (3 min), and (iii) extension for 7 min at 72°C. The products were cloned and sequenced as described above.
PCRs with the capillary samples were also performed by a multiplex approach with a reverse primer specific for green sulfur bacteria (GSB) and with eubacterial primers (314F and 1492R). These reactions were used to enrich samples for non-Chlorobium sequences (see below). The reaction mixtures contained primer 8F or 341F at 0.5 μM, primer 1492R at 0.25 μM, and primer 1144R (GSB specific) (2) at 0.25 μM. The optimal annealing temperature for the production of two bands was determined by using the temperature gradient feature of the Bio-Rad iCycler. The PCR protocol used was 30 cycles of 94°C (1 min), 49°C (1 min), and 72°C (3 min), followed by extension for 7 min at 72°C. The amplification reactions produced two bands of different lengths, one from the 314F and 1144R primer pair (specific to GSB) and the other from the 314F-1492R set (all Bacteria). Bands corresponding to the 8F-1492R or 341F-1492R sequences were excised from 1% agarose gels in Tris-acetate-EDTA, and DNAs were isolated with Qiagen gel purification kits. The products were cloned and sequenced as described above. Several samples of interest were sequenced from both directions by the use of universal M13F and M13R primers.
Chromatograms were edited and contigs were assembled with the Staden software package (http://www.mrc-lmb.cam.ac.uk/pubseq/). Sequences were imported into a preexisting 16S rRNA database provided with ARB (31) and were aligned with the ARB editor. Bootstrap parsimony and maximum likelihood trees were constructed with the Phylip package (http://evolution.genetics.washington.edu/phylip.html).
FISH was performed on 10-well slides coated with a gelatin solution (0.075% gelatin, 0.01% chromium potassium sulfate dodecahydrate). Five microliters of paraformaldehyde-fixed sample stored in 1:1 phosphate-buffered saline-ethanol was spotted into each well, air dried, and dehydrated in an ethanol series (50, 80, and 98%; 3 min each). A dual stain was performed with the probe GAM42A (Cy3), targeting 23S RNA (34), and the nonsense probe NON338 (fluorescein) (60) as a control for the nonspecific uptake of the labeled probe. Fixed cells of Vibrio parahaemolyticus were used as a positive control for probe GAM42A, and cells of the α-proteobacterium MV-1 were used as a negative control. Hybridization with 35% formamide and a subsequent wash were performed according to previously described protocols (42). Images were captured from an inverted Zeiss Axiovert 100 microscope with a digital camera and were processed in Adobe Photoshop.
Electron microscopy.
Transmission electron microscopy (TEM) was performed at the MBL Central Microscope Facility with a Zeiss 10CA microscope operating at 80 kV. Selected area electron diffraction measurements were performed at the MIT Center for Materials Research with a JEOL model 200CX microscope operating at 200 kV. The microscope camera length was calibrated with a sample of thallous chloride.
Nucleotide sequence accession numbers.
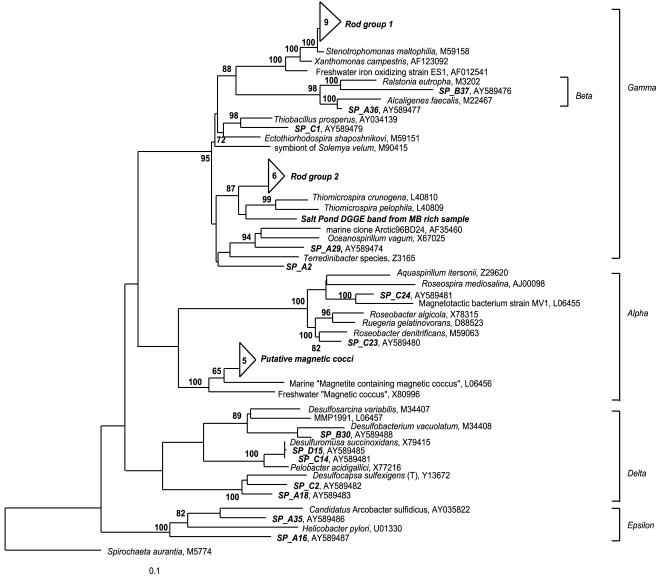
The sequences obtained in this study were deposited under the following accession numbers: AY587877 to AY587881 (putative magnetic cocci cluster), AY587194 to AY587198 (Thiomicrospira-related cluster), AY587199 to AY587207 (Stenotrophomonas-related cluster), and AY589474 to AY589488 (individual clones, shown in Fig. 6).
FIG. 6.
Phylogenetic relationship of several novel 16S rRNA sequences in the Proteobacteria obtained from Salt Pond samples that were highly enriched in different types of magnetotactic bacteria. The tree was obtained with a maximum parsimony analysis of sequences of >1,000 bp, with 200 bootstrap replicates, by using Phylip software. Short sequences were added to the tree by using the interactive parsimony tool of ARB, maintaining the previous topology. Spirochaeta aurantia was used as the outgroup. Nodes with >60% bootstrap support are marked. The tree topology was also supported by maximum likelihood and distance analyses. The two groups of clones in the gamma proteobacteria (rod group 1 and rod group 2), as well as the sequences labeled SP_A and SP_B, were obtained from capillary samples that were highly enriched in the large greigite-producing rod. An additional cluster of sequences (putative magnetic cocci) was obtained from capillary samples that were highly enriched in magnetococci. Sequences labeled SP_C and SP_D were obtained from capillary samples that were highly enriched in MMP.
RESULTS
Water chemistry.
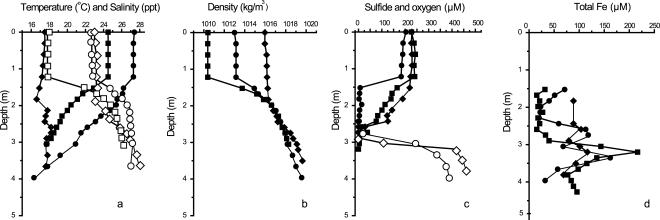
Figure 2 shows the compiled data for water column chemistry in June, August, and October. Density profiles through the water column showed that stratification was most pronounced in June and August. The chemocline of Salt Pond occurred between 1.5 and 3.4 m on all sampling dates. The chemocline reached 0.6 m wide, and particulate iron was highest at the base of the oxycline, where MB were most abundant. The total precipitation was 7.8 cm in June, with freshwater input likely resulting in a low density and high oxygen level in the epilimnion. July was hot and dry, with solar radiation at 5% above the average and total precipitation of only 1.7 cm, the lowest in 36 years (http://www.whoi.edu/climate/2002). Solar heating, low freshwater input, and biological oxygen consumption likely resulted in the warm, dense, low-oxygen epilimnion observed in early August.
FIG. 2.
Chemistry of stratified Salt Pond water column. Shown are chemical profiles from early (squares), mid (circles), and late (diamonds) season sampling dates. Early season, June 26; midseason, August 2; late season, October 9. (a) Temperature (closed symbols) and salinity (open symbols); (b) water column density; (c) oxygen (closed symbols) and sulfide (open symbols) concentrations; (d) dissolved and particulate iron concentration. June 26 sulfide measurements are not shown due to a possible sampling error.
The chemocline was widest on August 2, when oxygen and sulfide were below detection limits between 1.5 and 2.4 m. A warm, less saline epilimnion overlay a cool, saline hypolimnion on all sampling dates. The pH declined from highs of 8.5 to 9 in the oxic epilimnion to approximately 7 in the sulfidic hypolimnion. Peaks in pH and oxygen measurements at a depth of approximately 0.9 m corresponded to blooms of oxygenic phototrophs in the epilimnion. Total iron (particulate plus dissolved) concentrations peaked at the oxycline on all sampling dates. Hypolimnion sulfide concentrations remained steady, at 400 to 500 μM, between August and October. The rise in sulfide was typically steep below the oxycline, with increases from 0 to 500 μM in little more than 1 m.
Microscopic analysis of MB.
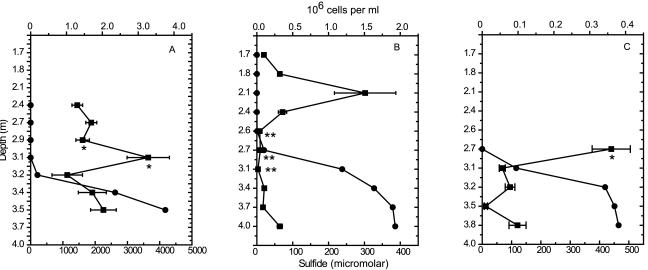
The total numbers of DAPI-stained cells closely tracked the oxycline. Cell numbers peaked at the oxycline on all sampling dates (Fig. 3), corresponding with visual observations of increased turbidity and a change in water color to purple-brown. Cell numbers immediately above and below the oxycline were similar. Maximum cell numbers were observed in June, and October counts were about an order of magnitude lower.
FIG. 3.
Total cell numbers for each sampling date obtained by DAPI staining and epifluorescence microscopy. (A) June 26; (B) August 2; (C) October 9. Counts are shown as squares, and sulfide profiles are shown as circles. Error bars, ±1 standard error of the means. Cell numbers peaked at the oxycline. The location of the largest MB population for each sampling date is marked with * (cocci) or ** (MMP). MMP were the most numerous at 2.7 m on August 2 (B). Note the differences in scales between the graphs.
The abundance, distribution, and types of MB present shifted with the degree of stratification and total cell number. MB were observed at their highest density immediately above, within, and immediately below the oxycline. Microscopic observations suggested that they were most abundant when stratification was greatest (August). Three readily distinguished morphologies (morphotypes) of MB were dominant: magnetite-producing cocci, greigite-producing MMP, and a large, slow-moving rod. A small rod (0.5 by 1 μm) was observed at similar depths as the MMB cocci, and a chain-forming organism (with each cell being about 2 μm) was observed in the lower part of the oxycline at similar depths to the GMB MMP. Light micrographs and TEM images of the three dominant morphotypes are shown in Fig. 4 and Fig. 5. Each MB morphotype showed tactic and motile responses that were unique to the morphotype.
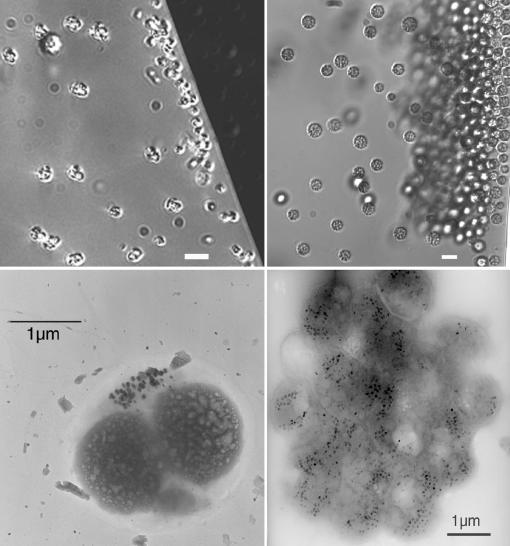
FIG. 4.
Depth stratification of MB in Salt Pond. Top and bottom images correspond to the same cell type, as shown by differential contrast light microscopy (top) and TEM (bottom). Left, magnetite-producing cocci collected from the top of the oxycline; right, greigite-producing MMP collected from the bottom of the oxycline. Scale bars are shown for each image. Bars, 5 μm for differential interference contrast images.
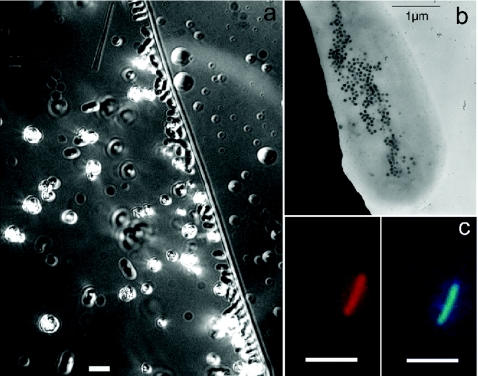
FIG. 5.
Greigite-producing rod found in sulfidic bottom waters of Salt Pond. (a) Differential interference contrast image. Bar, 5 μm. The diagonal line is the edge of the drop. (b) TEM image showing intracellular greigite crystals (black dots). (c) FISH with GAM42A probe (Cy3, left) and DAPI stain (right) of the same cell. Bars, 5 μm.
Cocci and MMP.
The cocci (MMB) occurred at the top of the oxycline. They exhibited a rapid bouncing motion back and forth at the drop edge. MMP (GMB) were found at the bottom of or slightly below the oxycline in areas of low sulfide, and they swam by spinning counterclockwise when moving away from the drop edge and clockwise when moving towards it. MMP and cocci were found at significantly different mean sulfide levels (P < 0.05 by a t test, unequal variances). The cocci were most abundant at sulfide levels of 0 to 10 μM. MMP were most abundant at sulfide concentrations of <40 μM but occurred in samples with sulfide levels up to 1 mM. MMP and cocci fluctuated in abundance throughout the season. Cocci were most abundant early and late in the season, while MMP were most abundant during periods of intense stratification in midsummer. The MMP abundance reached an estimated 103 ml−1 at the oxycline in August, which was the highest number of MMP observed in the water column during the entire summer.
Novel rod-shaped GMB.
We observed morphologically distinct, large, slow-moving magnetotactic rods in sulfidic regions of Salt Pond that were approximately 5 μm long and 3 μm wide (Fig. 5). These organisms swam very slowly to the drop edge, where they accumulated without exhibiting the back-and-forth swimming behavior displayed by both the cocci and MMP. Sulfide concentrations in samples with abundant rods averaged 327 μM, compared with 116 μM for MMP and 19.2 μM for cocci, and the rods were never found in water with sulfide concentrations of <100 μM. Unlike the cocci and MMP, the rods did not show sharp changes in abundance with depth or season, although they were most abundant in mid- to late summer. We further characterized the slow-moving rod by TEM (Fig. 5), sequencing of the 16S ribosomal DNA, and FISH (see below). TEM analysis revealed that the rod contained numerous mineral crystals similar to the magnetosomes observed in other organisms (21). The crystals appear as irregularly shaped black dots in Fig. 5b. A single crystal electron diffraction pattern obtained from the crystals corresponded to that of greigite (data not shown).
Sequencing results.
DGGE profiling (data not shown) showed a marked decrease in bacterial diversity for the midseason sampling compared to the early and late seasons. Based on microscopy, MMP were by far the most abundant MB type during the period of low diversity, while cocci were the most abundant morphotype present in the early and late seasons. Sequencing results confirmed that the dominant amplicon in both bulk and magnetically enriched samples was related to various Chlorobiales organisms. We then used the multiplex PCR approach described above to eliminate this dominant amplicon from the cloning and sequencing steps, thereby increasing the probability of obtaining MB sequences from magnetically enriched samples.
A diverse set of sequences were obtained from capillary samples that were highly enriched in MMP (Table 1; Fig. 6). A group of five novel sequences was closely related to a marine magnetotactic coccus of the α-Proteobacteria, and one was closely related to the marine magnetotactic vibrio MV-1 (13). Several δ-proteobacterial sequences were also identified in the MMP-rich sample, but no sequences identical to previously sequenced MMP clones (13) were found. GenBank accession numbers for individual clones are shown in Fig. 6.
TABLE 1.
Distribution of clones obtained from sequencing of capillary samples that were highly enriched in different types of MBa
| Phylogenetic grouping | % MB in sample set
|
||
|---|---|---|---|
| Enriched in large GMB rods (multiplex) | Enriched in MMP (multiplex) | Enriched in MMP (nonmultiplex) | |
| Alpha proteobacteria | 0 | 14 | 13 |
| Gamma proteobacteria | 55 | 9 | 0 |
| Beta proteobacteria | 6.5 | 0 | 0 |
| Delta proteobacteria | 6.5 | 9 | 10 |
| Epsilon proteobacteria | 6.5 | 0 | 0 |
| CFBb | 10 | 9 | 3.2 |
| Chlorobiales | 13 | 23 | 74 |
| Chloroflexi | 0 | 4.5 | 0 |
| Cyanobacteria | 0 | 4.5 | 0 |
| CV | 3.2 | 14 | 0 |
| OP11 | 0 | 9 | 0 |
| Planctomycetales | 0 | 4.5 | 0 |
Multiplex PCR was used to screen out Chlorobiales sequences.
CFB, Cytophaga-Flexibacter-Bacteroides.
We obtained several 16S ribosomal DNA sequences clustering in the γ-Proteobacteria from samples that were highly enriched in the slow-moving rod (Fig. 6) and amplified by multiplex PCR. These separated into two major groups: the first contains clones with 90% similarity to Thiomicrospira sp., and the second contains clones with 99% similarity to Stenotrophomonas maltophilia. Combined, these two groups accounted for 42% of the clones obtained from the samples that were highly enriched in large magnetotactic rods. Additionally, the sequence of a DGGE band obtained from a magnetically enriched sample clustered with Thiomicrospira spp. (Fig. 6). The remaining clones were widely distributed among the Bacteria (Table 1).
FISH.
To confirm that the newly identified large GMB rod is associated with the gamma subclass of the Proteobacteria, we performed FISH on samples that were highly enriched in this morphotype, using the γ-proteobacterium-specific GAM42a probe (34). The large GMB rod FISH results are shown in Fig. 5c.
DISCUSSION
We observed a marked stratification in the distribution of three different types of MB at and below the chemocline in meromictic Salt Pond, which we hypothesize was due to their as yet unknown respiratory requirements. A schematic cartoon which summarizes these distributions within Salt Pond is shown in Fig. 7. Small, fast-moving MMB cocci were found at the top of the oxycline, GMB MMP were found at the base of and slightly below the oxycline, and a novel GMB rod was found in the sulfidic hypolimnion (Fig. 4 and 5). This distribution remained constant throughout the summer, although the numbers of cocci and MMP fluctuated widely. Peaks in abundance of the cocci coincided with peaks in total cell numbers, while peaks in MMP abundance coincided with low total cell numbers below the oxycline (Fig. 3).
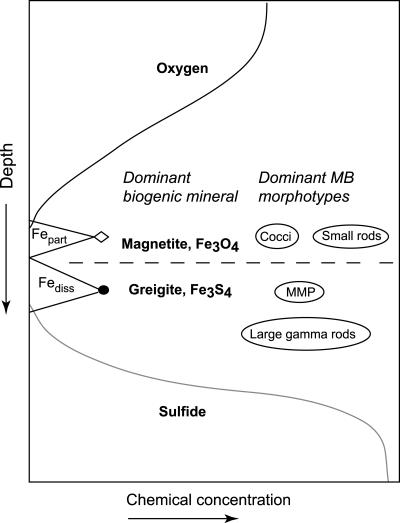
FIG. 7.
Schematic of MB distribution observed in Salt Pond, showing approximate locations of magnetite- and greigite-producing MB relative to a typical stratified water column chemical profile. Fepart, total particulate iron; Fediss, total dissolved iron.
Major factors determining the distribution of bacteria in a stratified salt pond include the location and width of the chemocline as well as the proximity and concentrations of electron donors and acceptors. Motile microbial populations are often finely layered around the chemocline with respect to these properties (e.g., see reference 55). In particular, many known colorless sulfur bacteria can closely track shifting oxygen-sulfide gradients (39) without the need for magnetotaxis. Populations of magnetotactic cocci were strongly layered in a similar fashion, with orders of magnitude changes in abundance over a depth of 30 cm.
It is likely that the small magnetotactic cocci we observed were microaerophilic sulfide oxidizers. Their position on top of the chemocline, coinciding with maximum total cell counts, suggests that they prefer the oxygen-sulfide interface. They were most abundant in the early and late season samplings, when oxygen was higher in the epilimnion and the chemocline was narrowest (Fig. 2). Additionally, the few cultivated marine MMB are all autotrophic sulfide-oxidizing microaerophiles (51). We recovered several sequences from Salt Pond that are closely related to these strains (Fig. 6).
MMP also appears to be a gradient organism, but of a different type. MMP always occurred at the base of the chemocline and never at depths with detectable oxygen. It was most abundant in midsummer, when the chemocline was widest (Fig. 2), but also occurred in small numbers in the early and late seasons. In August, the MMP concentration reached an estimated 103 per ml within a 50-cm window directly below the chemocline and dropped off abruptly outside it (Fig. 3B). Within that window, an order of magnitude shift in MMP abundance occurred within 15 cm.
It is not certain which environmental stimuli the MMP uses to position itself within the chemocline. The MMP may be chemotactic to an iron or sulfur compound, since it precipitates an iron-sulfur mineral and appears to be most closely related to the sulfate reducer Desulfosarcina variabilis (13). Fe(III) concentrations peaked at the depth with the most MMP abundance, and the sulfide concentration was <1 μM. We did not measure intermediate sulfur compounds, but studies on other euxinic water bodies showed that they typically peak at the oxygen-sulfide interface (summarized in reference 61). It is possible that MMP responded to gradients in one of these compounds. Interestingly, while δ-proteobacterial clones were identified in MMP-rich samples (Fig. 6), the sequences we recovered are distinct from the one previously reported for this organism (13). Recently, however, we have identified sequences similar to those that were previously reported (13) from an MMP-rich sample obtained from a salt marsh (S. L. Simmons and K. J. Edwards, unpublished data). This recent result suggests that the absence of MMP sequences from the Salt Pond samples may be due to the preferential amplification of other sequences or to some phylogenetic differences between salt marsh and pond MMPs. The clones identified in the present study also support the suggestion of an active chemocline sulfur cycle typical of meromictic salt lakes (reviewed in reference 39). We identified clones from organisms that are capable of sulfate and iron reduction (Desulfobacteriaceae), sulfur disproportionation (Desulfocapsa sulfexigens [15]), and the reduction of either elemental sulfur or iron (Desulfuromusa succinoxidans [27, 29]).
DGGE analysis suggested that the chemocline population was dominated by various species of GSB, mainly nonmotile Chlorobiales, while various Cytophaga/Bacteroides species were the dominant hypolimnion population detected by DGGE. Both DGGE and PCR with general primers were an ineffective means of identifying MB; Chlorobiales were the dominant amplicon even in the samples that were extracted by capillary tubes from the microscope stage, where we could verify that the template was dominated by MB. Often in molecular studies, when a specific group of microorganisms is targeted such as we did for MB, specific rather than general primers targeting the group of interest will be used (e.g., δ-proteobacterial primers for sulfate-reducing bacteria). However, studies of MB to date indicate that they are not a phylogenetically or physiologically coherent group, precluding the use of primers that target specific phylotypes. The multiplex PCR approach we used here allowed us to eliminate the masking amplicons (Table 1) and to obtain MB sequences.
The rod-shaped GMB identified in this study, in part through the multiplex method, is the first magnetotactic bacterium that appears to be a member of the γ-Proteobacteria (Fig. 6). It is clearly not a gradient organism, as it was found in low densities in the sulfidic hypolimnion on multiple sampling dates. Previous studies have shown TEM images of organisms similar to this rod (21), but its phylogeny was not known. The placement of this new GMB into the γ subclass of the Proteobacteria may suggest that it is involved primarily in iron cycling. Respiration that uses metals, particularly iron, is widespread among the γ-Proteobacteria. Emerson and Moyer (14) identified two strains of freshwater microaerobic Fe-oxidizing bacteria and several environmental clones with 95% similarity to S. maltophilia. Stenotrophomonas is a diverse genus with isolates from soil, water, and medical sources (20). The previously described iron-oxidizing strain ES-1 (14) is closely related to one cluster of clones from the rod-shaped GMB identified in this study (Fig. 6). Additionally, several γ-Proteobacteria isolated from anoxic soils and marine sediments are capable of dissimilatory metal reduction (16, 30, 35, 37).
The second cluster of putative GMB clones has about 90% 16S rRNA similarity to Thiomicrospira crunogena and Thiomicrospira pelophila. A DGGE band from a sample enriched in GMB also clustered with the Thiomicrospira species, providing additional support for the presence of the rod GMB in the γ-Proteobacteria. Both Thiomicrospira species are obligate sulfur chemolithotrophs that use oxygen as an electron acceptor (24). Notably, T. pelophila was isolated from sulfide-rich mud and has a high tolerance for sulfide (25). The novel GMB was also found in water with high sulfide concentrations. However, the spiral cells of T. pelophila are quite a bit smaller than the greigite rods.
The in situ hybridization of samples that were highly enriched in greigite rods with the GAM42 probe targeting γ-Proteobacteria (34) showed positive hybridization to cells of the right size and shape (Fig. 5). Further work with probes designed from these two clusters is necessary to determine the phylogenetic position of the rod-shaped GMB.
It remains unclear why cells in highly sulfidic waters should be magnetotactic. Frankel et al. (17) proposed that magnetotactic cocci and other microaerophilic MMB found at the chemocline in Salt Pond use magnetotaxis to more rapidly locate optimal levels of O2. If the rod-shaped GMB use magnetotaxis to optimize their position with respect to some physical or chemical gradient, we have not yet identified it. Unlike the magnetic cocci and the MMP, the rod GMB were never observed near the chemocline and were not concentrated in layers of high cell density. They did not appear to be most abundant at a specific iron concentration. Measurements taken the following summer (unpublished data) showed that the hypolimnion light level is about 0.01 microeinstein, suggesting that the GMB do not respond to light gradients. It is possible that they respond to gradients of some as yet unidentified electron acceptor or donor.
Magnetotaxis may also be the relatively rare consequence of a more common form of prokaryotic iron metabolism. Metal inclusions of various kinds have been found in nonmagnetotactic γ-Proteobacteria. Glasauer et al. (19) identified membrane-bound crystalline Fe inclusions in Shewanella putrefaciens grown with ferrihydrite as an electron acceptor. Vainshtein et al. (57) identified magnet-sensitive particles in Ectothiorhodospira shaposhnikovii which were rich in Fe but did not contain sulfur. In a separate study, Vainshtein et al. (56) found magnet-sensitive inclusions of various types in prokaryotic cells of diverse phylogenies when they were grown in medium containing 1 mM iron, suggesting some kind of detoxification function. The inclusions were not ordered crystals and incorporated various other unspecified organic compounds. Further studies are necessary to determine whether GMB utilize ferrous or ferric iron in dissimilatory metabolism as well as how such a metabolism might be linked to intracellular greigite precipitation.
The clear stratification in populations of marine MB with respect to the chemocline suggest that each type of MB is adapted to different chemical and physical gradients within the water column. It is likely that iron and sulfur compounds play a key role in determining the population dynamics of GMB. We sampled Salt Pond in a subsequent summer in an attempt to better determine these gradients. While the phylogeny and geochemical environment of MB provide clues about their ecological role, additional studies are needed to better define the predominant electron donors and acceptors used by MB as well as the relationship between respiration and intracellular mineral formation.
Acknowledgments
We thank L. Hirt, S. Selgrade, D. Rogers, and C. Santelli for their assistance. The manuscript was greatly improved with input from E. Webb and three anonymous reviewers.
This work was partially funded by a grant from the Woods Hole Oceanographic Institution Reinhart Coastal Research Center to S.L.S. and K.J.E. S.L.S. was supported by a National Defense Science and Engineering graduate fellowship. D.A.B. was supported by National Science Foundation grant EAR-0311950 and National Aeronautics and Space Administration (NASA) Johnson Space Center grant NAG 9-1115.
Footnotes
This paper is WHOI contribution 11168.
REFERENCES
- 1.Achenbach, L. A., J. Carey, and M. T. Madigan. 2001. Photosynthetic and phylogenetic primers for detection of anoxygenic phototrophs in natural environments. Appl. Environ. Microbiol. 67:2922-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., et al. (ed.). 2003. Current protocols in molecular biology. [Online.] Wiley Interscience, New York, N.Y. http://www.mrw2.interscience.wiley.com/cponline.
- 3.Bazylinski, D. A., R. B. Frankel, and H. W. Jannasch. 1988. Anaerobic magnetite production by a marine magnetotactic bacterium. Nature 334:518-519. [Google Scholar]
- 4.Bazylinski, D. A., and B. M. Moskowitz. 1997. Microbial biomineralization of magnetic iron minerals: microbiology, magnetism, and environmental significance, p. 181-224. In J. Banfield and K. Nealson (ed.), Geomicrobiology: interactions between microbes and minerals, vol. 35. Mineralogical Society of America, Washington, D.C.
- 5.Bazylinski, D. A., and R. B. Frankel. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217-230. [DOI] [PubMed] [Google Scholar]
- 6.Bazylinski, D. A., and R. B. Frankel. 1992. Production of iron sulfide minerals by magnetotactic bacteria in sulfidic environments, p. 147-159. In H. C. Skinner and R. W. Fitzpatrick (ed.), Biomineralization processes, iron, manganese, vol. 21. Catena, Reiskirchen, Germany. [Google Scholar]
- 7.Bazylinski, D. A., R. B. Frankel, B. R. Heywood, S. Mann, J. W. King, P. L. Donaghay, and A. K. Hanson. 1995. Controlled biomineralization of magnetite (Fe3O4) and gregite (Fe3S4) in a magnetotactic bacterium. Appl. Environ. Microbiol. 61:3232-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bazylinski, D. A., D. R. Schlezinger, B. H. Howes, R. B. Frankel, and S. S. Epstein. 2000. Occurrence and distribution of diverse populations of magnetic protists in a chemically stratified coastal salt pond. Chem. Geol. 169:319-328. [Google Scholar]
- 9.Berner, R. 1967. Thermodynamic stability of sedimentary iron sulfides. Am. J. Sci. 265:773-785. [Google Scholar]
- 10.Cape Cod Commission. 2001. Cape Cod atlas of tidally restricted salt marshes. Cape Cod Commission, Barnstable, Mass.
- 11.Casamayor, E. O., H. Schafer, L. Baneras, C. Pedros-Alió, and G. Muyzer. 2000. Identification of and spatiotemporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 66:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 13.DeLong, E. F., R. B. Frankel, and D. A. Bazylinski. 1993. Multiple evolutionary origins of magnetotaxis in bacteria. Science 259:803-806. [DOI] [PubMed] [Google Scholar]
- 14.Emerson, D., and C. Moyer. 1997. Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl. Environ. Microbiol. 63:4784-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finster, K., W. Liesack, and B. Thamdrup. 1998. Elemental sulfur and thiosulfate disproportionation by Desulfocapsa sulfoexigens sp. nov., a new anaerobic bacterium isolated from marine surface sediment. Appl. Environ. Microbiol. 64:119-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis, C. A., A. Y. Obraztsova, and B. M. Tebo. 2000. Dissimilatory metal reduction by the facultative anaerobe Pantoea agglomerans SP1. Appl. Environ. Microbiol. 66:543-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frankel, R. B., D. A. Bazylinski, M. S. Johnson, and B. L. Taylor. 1997. Magneto-aerotaxis in marine coccoid bacteria. Biophys. J. 73:994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frankel, R. B., D. A. Bazylinski, and D. Schüler. 1998. Biomineralization of magnetic iron minerals in magnetotactic bacteria. J. Supramol. Sci. 5:388-390. [Google Scholar]
- 19.Glasauer, S., S. Langley, and T. J. Beveridge. 2002. Intracellular iron minerals in a dissimilatory iron-reducing bacterium. Science 295:117-119. [DOI] [PubMed] [Google Scholar]
- 20.Hauben, L., L. Vauterin, E. Moore, B. Hoste, and J. Swings. 1999. Genomic diversity of the genus Stenotrophomonas. Int. J. Syst. Bacteriol. 49:1749-1760. [DOI] [PubMed] [Google Scholar]
- 21.Heywood, B. R., D. A. Bazylinski, A. J. Garrett-Reed, S. Mann, and R. B. Frankel. 1990. Controlled biosynthesis of greigite (Fe3S4) in magnetotactic bacteria. Naturwissenschaften 77:536-538. [Google Scholar]
- 22.Kawaguchi, R., J. Burgess, T. Sakaguchi, H. Takeyama, R. Thornhill, and T. Matsunaga. 1995. Phylogenetic analysis of a novel sulfate-reducing bacterium, RS-1, demonstrates its membership of the delta-Proteobacteria. FEMS Microbiol. Lett. 126:277-282. [DOI] [PubMed] [Google Scholar]
- 23.Kirchman, D., J. Sigda, R. Kapuscinski, and R. Mitchell. 1982. Statistical analysis of the direct count method for enumerating bacteria. Appl. Environ. Microbiol. 44:376-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuenen, J. G., L. A. Robertson, and O. H. Tuovinen. 21. May 1999, revision date. The genera Thiobacillus, Thiomicrospira, and Thiosphaera. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. [Online.] Springer-Verlag, New York, N.Y. http://link.springer-ny.com/link/service/books/10125/.
- 25.Kuenen, J. G., and H. Veldkamp. 1972. Thiomicrospira pelophila, gen. nov., sp. nov., a new obligately chemolithotrophic colourless sulfur bacterium. Antonie Leeuwenhoek 38:241-256. [DOI] [PubMed] [Google Scholar]
- 26.Lee, C., and N. Jorgensen. 1995. Seasonal cycling of putrescine and amino acids in relation to biological production in a stratified coastal salt pond. Biogeochemistry 29:131-157. [Google Scholar]
- 27.Liesack, W., and K. Finster. 1994. Phylogenetic analysis of 5 strains of gram-negative, obligately anaerobic, sulfur-reducing bacteria and description of Desulfuromusa gen. nov., including Desulfuromusa-Kysingii sp. nov., Desulfuromusa-Bakii sp. nov., and Desulfuromusa-Succinoxidans sp. nov. Int. J. Syst. Bacteriol. 44:753-758. [Google Scholar]
- 28.Lohrenz, S. E., C. D. Taylor, and B. H. Howes. 1987. Primary production of protein. II. Algal protein metabolism and its relation to particulate organic matter composition in the surface mixed layer. Mar. Ecol. Prog. Ser. 40:175-183. [Google Scholar]
- 29.Lonergan, D. J., H. L. Jenter, J. D. Coates, E. J. Phillips, T. M. Schmidt, and D. R. Lovley. 1996. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J. Bacteriol. 178:2402-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovley, D. R. 1 December 2000, revision date. Dissimilatory Fe(III)- and Mn(IV)-reducing prokaryotes. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, release 3.4 [Online.] Springer-Verlag, New York, N.Y. http://link.springer-ny.com/link/service/books/10125/.
- 31.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mann, S., N. H. C. Sparks, and R. G. Board. 1990. Magnetotactic bacteria: microbiology, biomineralization, paleomagnetism and biotechnology. Adv. Microb. Physiol. 31:125-181. [DOI] [PubMed] [Google Scholar]
- 33.Mann, S., N. H. C. Sparks, R. B. Frankel, D. A. Bazylinski, and H. W. Jannasch. 1990. Biomineralization of ferrimagnetic greigite (Fe3S4) and iron pyrite (FeS2) in a magnetotactic bacterium. Nature 343:258-261. [Google Scholar]
- 34.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Scheifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 35.McLean, J., and T. J. Beveridge. 2001. Chromate reduction by a pseudomonad isolated from a site contaminated with chromated copper arsenate. Appl. Environ. Microbiol. 67:1076-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray, J., L. Codispoti, and G. Friedrich. 1995. Oxidation-reduction environments: the suboxic zone in the Black Sea, p. 157-176. In C. Huang et al. (ed.), Aquatic chemistry, vol. 244. American Chemical Society, Washington, D.C. [Google Scholar]
- 37.Nevin, K. P., K. T. Finneran, and D. R. Lovley. 2003. Microorganisms associated with uranium bioremediation in a high-salinity subsurface sediment. Appl. Environ. Microbiol. 69:3672-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Sullivan, D. W., A. K. Hanson, Jr., and D. R. Kester. 1997. The distribution and redox chemistry of iron in the Pettaquamscutt Estuary. Estuar. Coast. Shelf Sci. 45:769-788. [Google Scholar]
- 39.Overmann, J., and H. van Gemerden. 2000. Microbial interactions involving sulfur bacteria: implications for the ecology and evolution of bacterial communities. FEMS Microbiol. Rev. 24:591-599. [DOI] [PubMed] [Google Scholar]
- 40.Peduzzi, S., M. Tonolla, and D. Hahn. 2003. Isolation and characterization of aggregate-forming sulfate-reducing and purple sulfur bacteria from the chemocline of meromictic Lake Cadagno, Switzerland. FEMS Microbiol. Ecol. 45:29-37. [DOI] [PubMed] [Google Scholar]
- 41.Peduzzi, S., M. Tonolla, and D. Hahn. 2003. Vertical distribution of sulfate-reducing bacteria in the chemocline of Lake Cadagno, Switzerland, over an annual cycle. Aquat. Microb. Ecol. 30:295-302. [Google Scholar]
- 42.Pernthaler, J., F. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 43.Petermann, H., and U. Bleil. 1993. Detection of live magnetotactic bacteria in deep-sea sediments. Earth Planet. Sci. Lett. 117:223-228. [Google Scholar]
- 44.Pósfai, M., P. R. Buseck, D. A. Bazylinski, and R. B. Frankel. 1998. Reaction sequence of iron sulfide minerals in bacteria and their use as biomarkers. Science 280:880-883. [DOI] [PubMed] [Google Scholar]
- 45.Sakaguchi, T., A. Arakaki, and T. Matsunaga. 2002. Desulfovibrio magneticus sp. nov., a novel sulfate-reducing bacterium that produces intracellular single-domain-sized magnetite particles. Int. J. Syst. Evol. Microbiol. 52:215-221. [DOI] [PubMed] [Google Scholar]
- 46.Schafer, H., and G. Muyzer. 2001. Denaturing gradient gel electrophoresis in marine microbial ecology. Methods Microbiol. 30:425-468. [Google Scholar]
- 47.Schippers, A., and B. Jorgensen. 2002. Biogeochemistry of pyrite and iron sulfide oxidation in marine sediments. Geochim. Cosmochim. Acta 66:85-92. [Google Scholar]
- 48.Spring, S., R. Amann, W. Ludwig, K. Schleifer, H. van Gemerden, and N. Petersen. 1993. Dominating role of an unusual magnetotactic bacterium in the microaerobic zone of a freshwater sediment. Appl. Environ. Microbiol. 59:2397-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spring, S., R. Amann, W. Ludwig, K.-H. Schleifer, and N. Petersen. 1992. Phylogenetic diversity and identification of nonculturable magnetotactic bacteria. Syst. Appl. Microbiol. 15:116-122. [Google Scholar]
- 50.Spring, S., R. Amann, W. Ludwig, K.-H. Schleifer, D. Schüler, K. Poralla, and N. Petersen. 1994. Phylogenetic analysis of uncultured magnetotactic bacteria from the alpha-subclass of Proteobacteria. Syst. Appl. Microbiol. 17:501-508. [Google Scholar]
- 51.Spring, S., and D. Bazylinski. 1. December 2000, revision date. Magnetotactic bacteria. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, release 3.4. [Online.] Springer Verlag, New York, N.Y. http://link.springer-ny.com/link/service/books/10125/.
- 52.Spring, S., and K. H. Schleifer. 1995. Diversity of magnetotactic bacteria. Syst. Appl. Microbiol. 18:147-153. [Google Scholar]
- 53.Taylor, C., J. Molongoski, and S. Lohrenz. 1983. Instrumentation for the measurement of phytoplankton production. Limnol. Oceanogr. 28:781-787. [Google Scholar]
- 54.Tonolla, M., A. Demarta, S. Peduzzi, D. Hahn, and R. Peduzzi. 2000. In situ analysis of sulfate-reducing bacteria related to Desulfocapsa thiozymogenes in the chemocline of meromictic Lake Cadagno (Switzerland). Appl. Environ. Microbiol. 66:820-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tonolla, M., S. Peduzzi, D. Hahn, and R. Peduzzi. 2003. Spatio-temporal distribution of phototrophic sulfur bacteria in the chemocline of meromictic Lake Cadagno (Switzerland). FEMS Microbiol. Ecol. 43:89-98. [DOI] [PubMed] [Google Scholar]
- 56.Vainshtein, M., N. Suzina, E. Kudryashova, and E. Ariskina. 2002. New magnet-sensitive structures in bacterial and archaeal cells. Biol. Cell 94:29-35. [DOI] [PubMed] [Google Scholar]
- 57.Vainshtein, M., N. Suzina, and V. Sorokin. 1997. A new type of magnet-sensitive inclusions in cells of photosynthetic purple bacteria. Syst. Appl. Microbiol. 20:182-186. [Google Scholar]
- 58.Viollier, E., P. W. Inglett, K. Hunter, A. N. Roychoudhury, and P. Van Cappellen. 2000. The ferrozine method revisited: Fe(II)/Fe(III) determination in natural waters. Appl. Geochem. 15:785-790. [Google Scholar]
- 59.Wakeham, S. G., B. L. Howes, J. W. H. Dacey, R. P. Schwarzenbach, and J. Zeyer. 1987. Biogeochemistry of dimethylsulfide in a seasonally stratified coastal salt pond. Geochim. Cosmochim. Acta 51:1675-1684. [Google Scholar]
- 60.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 61.Zopfi, J., T. G. Ferdelman, B. B. Jorgensen, A. Teske, and B. Thamdrup. 2001. Influence of water column dynamics on sulfide oxidation and other major biogeochemical processes in the chemocline of Mariager Fjord (Denmark). Mar. Chem. 74:29-51. [Google Scholar]