Abstract
Ultramicrobacteria (cell volume < 0.1 μm3) are the numerically dominant organisms in the plankton of marine and freshwater habitats. Flagellates and other protists are assumed to be the most important predators of these ultramicrobacteria as well as of larger planktonic bacteria. However, due to controversial observations conducted previously, it is not clear as to whether fractions of the ultramicrobacteria are resistant to flagellate predation. Furthermore, it is not known if closely related bacteria vary significantly in their sensitivity to flagellate predation. We investigated the sensitivity of ultramicrobacteria affiliated with the cosmopolitan Polynucleobacter cluster to grazing by Spumella-like nanoflagellates. Laboratory grazing experiments with four closely related (≥99.6% 16S rRNA gene sequence similarity) bacteria and three closely related (100% 18S rRNA gene sequence similarity) flagellates were performed. In comparison to larger bacteria, predation on the ultramicrobacterial Polynucleobacter strains was weak, and the growth of the predating flagellates was slow. Specific clearance rates ranged between 0.14 × 105 and 2.8 × 105 units of predator size h−1. Feeding rates strongly depended on the flagellate and bacterial strain (P < 0.001). Grazing mortality rates of the three flagellate strains investigated varied for the same prey strain by up to almost fourfold. We conclude that (i) ultramicrobacteria affiliated with the Polynucleobacter cluster are not protected from grazing, (ii) strain-specific variations in grazing sensitivity even between closely related bacteria are high, and (iii) strain-specific differences in predator-prey interaction could be an important factor in the evolution and maintenance of microbial microdiversity.
It has been postulated that size-selective grazing of protists tends to select the small bacteria that are found to dominate in most natural waters (6, 23). Even though the ingestion of virus particles (15) and high-molecular-weight polysaccharides (30) has been demonstrated, it is undoubted that smaller bacteria experience lower grazing pressure than larger bacteria (1, 13, 14, 23, 29). For the smallest bacteria, i.e., ultramicrobacteria (<0.1 μm3), whether complete grazing protection is due to their small size or insufficient capturing devices of their predators is still in discussion. For larger bacterivores, such as cladocerans and some ciliate species, it is well accepted that the smallest bacteria escape grazing. In contrast, nanoflagellates are generally assumed to also graze on small bacteria (14, 25). Even flagellate species that usually prey on larger bacteria are able to ingest the smallest particles, i.e., virus-like particles (15), high-molecular-weight polysaccharides (30), and artificial particles less than 0.4 μm in diameter (unpublished data). On the other hand, grazing protection against Ochromonas has been demonstrated for an ultramicrobacterial strain (19).
It is thus still controversial whether ultramicrobacteria are subject to nanoflagellate predation and to what extent findings on grazing resistance can be generalized among functional groups (3, 19, 34). The debate is still open because, until recently, only a few ultramicrobacterial strains have been isolated and grazing experiments with such isolates have not been performed. Furthermore, specific investigations and field studies were often restricted to broad taxonomic or so-called functional groups (e.g., Spumella-like flagellates) (33) due to the paucity of characteristic morphological features (28). It is unclear to what extent grazing interactions can be generalized among closely related strains, and it has been suggested that investigations of feeding interactions on the level of rough functional groups may not be adequate (3, 7). Comparative studies of flagellate feeding processes indicated that there are significant species-specific differences on a broader taxonomic scale (2, 4, 5, 9, 11). Grazing resistance in general and strain-specific variation have not been investigated thus far for very closely related bacteria and flagellates.
We investigated the grazing sensitivity of several closely related strains (0.05 to 0.07 μm3; sequence similarity, ≥99.6%) of ultramicrobacteria affiliated with the Polynucleobacter D subcluster (17). Furthermore, strain-specific differences in the predator-prey interactions were investigated with these closely related bacteria and closely related strains of Spumella-like flagellates (chrysomonads; 23 to 60 μm3; sequence similarity, 100%). Based on these sequence similarities, it is highly likely that the strains belong to one bacterial and one flagellate species. For a direct comparison to the feeding interactions of larger organisms, i.e., typical laboratory organisms, the flagellate strain Ochromonas sp. strain DS and the bacterial strains Brevundimonas diminuta and Listonella pelagia were included in the study as reference organisms. Specifically, we tested (i) whether ultramicrobacteria affiliated with the Polynucleobacter cluster are resistant to grazing by flagellate predators and (ii) whether the grazing vulnerability of these ultramicrobacteria differs significantly between closely related strains.
MATERIALS AND METHODS
Organisms.
Four bacterial strains affiliated with the monophyletic D subcluster of the Polynucleobacter group (betaproteobacteria; sequence similarity ≥99.6%) (17) and, as reference organisms, the alphaproteobacterium Brevundimonas diminuta LMG 2089T and the gammaproteobacterium Listonella pelagia CB5 (GenBank synonym Vibrio pelagius) (16, 18) were used as prey organisms (Table 1). Bacteria were grown on inorganic basal medium supplemented with 1 g of nutrient broth, Soyotone peptone, and yeast extract per liter (19).
TABLE 1.
Size and morphology of bacterial strains used in the short-term (experiment 1) and long-term (experiment 2) grazing experiments
| Strain | Morphology | Mean cell vol (μm3) ± SEM
|
|
|---|---|---|---|
| Expt 1 | Expt 2 | ||
| MWH-TaW3 | C shaped | 0.063 ± 0.010 | 0.072 ± 0.010 |
| MWH-NR1 | C shaped | 0.055 ± 0.011 | 0.073 ± 0.011 |
| MWH-VicM1 | C shaped | 0.048 ± 0.010 | 0.058 ± 0.010 |
| MWH-MoIso2 | C shaped | 0.057 ± 0.013 | |
| B. diminuta | Rods | 0.309 ± 0.175 | 0.246 ± 0.091 |
| L. pelagia | Rods | 1.150 ± 0.469 | |
Four axenically cultured bacterivorous chrysomonads were used as predators in pairwise predator-prey experiments. The axenic strains were cultivated on a diet of heat-killed bacteria and not, as usual, on organic media. The flagellates were therefore precultured phagotrophically and not osmotrophically in order to culture them under conditions as close to their natural conditions as possible. We are aware that axenic conditions are different from natural conditions, but the setup used allowed us to exclude the side effects of other organisms that were present. As we focused on the general grazing sensitivity of the ultramicrobacteria investigated and on strain-specific differences between closely related organisms, it was primarily important to produce experimental conditions with only one predator and prey present.
The closely related chrysomonad strains (monophyletic group, sequence similarity 100%; J. Boenigk et al., submitted for publication) used in this study were: strain JBM10 (cell volume, 45.7 ± 19.5 μm3), isolated from an artificial pond in Mondsee, Austria; strain JBC07 (60.1 ± 30.0 μm3), isolated from Lake Taihu, China; and strain JBNZ41 (23.3 ± 10.1 μm3), isolated from a natural pond in New Zealand, all three distantly related to Poterioochromonas malhamensis (94.9% 18S rRNA gene sequence similarity [Boenigk et al., submitted]), and, as a reference organism, Ochromonas sp. strain DS (147.2 ± 64.5 μm3), isolated from Lake Constance (18), were used in the experiments. All of the flagellate strains were cultured in an inorganic basal medium (19) and fed with heat-killed Listonella pelagia CB5; 48 h prior to the experiments, subcultures of the flagellates were adapted to the experimental conditions, i.e., 16°C and food concentrations of 3 × 106 to 6 × 106 bacteria ml−1.
All organisms were grown to the early stationary phase before the experiments began. Size measurements of the bacteria were made from 4′,6′-diamidino-2-phenylindole (DAPI)-stained cells at the beginning of the experiments with the Lucia software package (19). Even though absolute size estimates for fluorescently stained bacterial strains may be biased, relative size estimates for bacterial strains are robust (18) and are largely independent of the growth phase of the bacteria (18). At least 100 cells were measured. The C-shaped ultramicrobacteria resemble curved rod-shaped bacteria, and therefore the curved cell length and the cell diameter were determined, and bacterial cell volume was calculated based on the formula for rod-shaped bacteria (1). The length and width of the flagellates were measured live at ×1,000 magnification from a video screen with an inverted videomicroscope. Cell volume was calculated by assuming an ellipsoid cell shape. Statistical analyses were carried out with the software package SigmaStat 2.03.
Short-term grazing experiments.
Experiments were conducted in 100-ml Erlenmeyer flasks at 16°C in the dark. All possible predator-prey combinations with the four ultramicrobacterial strains and Brevundimonas diminuta LMG 2089T as the prey and the four flagellate strains as the predators were tested. In addition, treatments containing bacteria only were used as a control. All experiments were performed in triplicate. The inocula of 4 × 106 bacteria ml−1 and 2,500 flagellates ml−1 were adjusted at the onset of the experiment. Subsamples of 1 ml were taken at 0, 6, 12, 16, 20, and 24 h and subsequently fixed with formaldehyde (2% final concentration). Even though flagellates may egest food vacuole content upon fixation (31), the estimated error is much less than 1% of the absolute number of bacteria present and is therefore considered negligible. For bacterial enumeration, 200 μl was stained with Syto 13 (Molecular Probes) for 45 min. Bacteria were counted in a diluted sample with the FACSCalibur cell cytometer (Becton Dickinson). For flagellate enumeration, 800 μl was stained with DAPI (final concentration, 15 μg ml−1) for 30 min, filtered onto black Nuclepore filters, and counted under an epifluorescence microscope with UV excitation.
Bacterial net growth rates were calculated from the slope of log-transformed bacterial abundances versus time with the subsamples from 6 h onwards to exclude the initial lag phase from the analysis. The grazing mortality rate (G) was calculated by subtracting the bacterial growth rate in the experiment from the bacterial growth rate in the control treatment. Clearance rates (C) were calculated by the formula C (nanoliters per flagellate per day) = G × 106/nFlag, where where nFlag is the mean flagellate abundance during the experiment. Ingestion rates were calculated from clearance rates and bacterial abundances; the community clearance rate was calculated from the grazing mortality rate and bacterial abundance.
Long-term growth experiments.
Experiments were conducted in 100-ml Erlenmeyer flasks at 16°C in the light and in triplicate. These experiments were run with the flagellates JBC07 and JBNZ41 as predators and the ultramicrobacteria MWH-NR1, MWH-TaW3, and MWH-VicM1 as well as the larger bacteria Listonella pelagia CB5 and Brevundimonas diminuta LMG 2089T (not tested for JBNZ41) as prey. The inocula of 6.5 × 106 to 9 × 106 bacteria ml−1 and 5,000 flagellates ml−1 were adjusted at the onset of the experiment. Subsamples of 5 ml were taken at 0, 24, 48, 72, and 144 h and subsequently fixed with formaldehyde (2% final concentration). For the enumeration of bacteria and flagellates, 1.7 ml was stained with DAPI (final concentration, 15 μg ml−1) for 30 min, filtered onto black Nuclepore filters, and counted under an epifluorescence microscope with UV excitation. The growth rates of the flagellates were calculated from the slope of log-transformed flagellate abundances versus time with the linear part of the slope only, i.e., days 1 to 3.
RESULTS
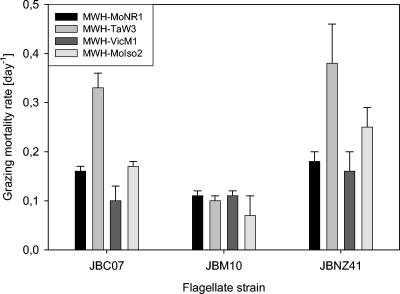
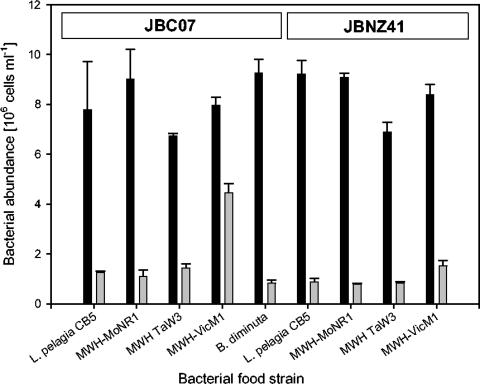
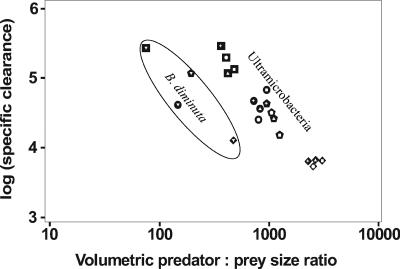
All flagellate strains investigated were able to graze on all the ultramicrobacterial strains even though the grazing mortality rates were below 0.4 day−1 (Fig. 1). In the long-term grazing experiment, the ultramicrobacteria were grazed down to an abundance of 0.8 × 106 to 4.5 × 106 bacteria ml−1, which also indicated a low grazing sensitivity of the ultramicrobacteria investigated (Fig. 2). Predator and prey size were important factors in explaining observed specific interactions, i.e., grazing mortality, clearance, and growth rates (Fig. 3).
FIG. 1.
Grazing mortality rates of the Spumella-like flagellates grazing on four strains of ultramicrobacteria affiliated with the Polynucleobacter cluster.
FIG. 2.
Abundance of bacterial strains, at the start (0 h; black bars) as well as the end (158 h; grey bars) of the long-term grazing experiment with flagellate strains JBC07 and JBNZ41. Towards the end of the experiment, flagellate abundances were already decreasing. This indicates that the flagellates were not able to graze the bacteria below 0.8 × 106 to 2.2 × 106 bacteria ml−1 and that grazing at these bacterial concentrations did not even support the basic metabolic costs of maintaining the flagellate population.
FIG. 3.
Specific clearance versus predator-prey size ratio based on volume. Flagellate strains are represented by the large black symbols: JBC07 (pentagon), JBM10 (circle), JBNZ41 (square), and Ochromonas sp. strain DS (diamond). Bacterial strains are represented by small white symbols: MWH-NR1 (triangle), MWH-TaW3 (+), MWH-MoIso2 (circle), MWH-VicM1 (square), and B. diminuta (upside-down triangle).
Grazing mortality and feeding rates.
Grazing mortality and clearance rates were calculated from the short-term experiment. The grazing mortality rate (Table 2) was found to be significantly different depending on the flagellate strain and the bacterial strain (two-way analysis of variance, P < 0.001 in both cases). In addition, there was a significant interaction between the flagellate and bacterial strains (P < 0.001). In contrast to our assumption that grazing mortality is generally higher for larger bacteria, we observed the highest grazing mortality and clearance rates for feeding on the large bacterium B. diminuta only for the flagellate strains JBC07 and Ochromonas sp. strain DS (Tables 2 and 3). The flagellate JBM10 did not show strong differences in the clearance rate except for a significant difference between the bacterial strains MWH-VicM1 and MWH-MoIso2, and the corresponding bacterial grazing mortality rates for JBM10 were not significantly different for the different bacterial strains (Tukey test, P > 0.7 in all cases). For the flagellate JBNZ41, significantly higher bacterial grazing mortality rates were observed for feeding on the ultramicrobacterial strain MWH-TaW3 compared to all other bacterial strains (Tukey test, P ≤ 0.003 in all cases; Table 3) and correspondingly also the highest clearance feeding on MWH-TaW3. In general, the flagellate JBNZ41 cleared the ultramicrobacterial strains MWH-MoIso2 and MWH-TaW3 with significantly higher rates than all the other flagellates (Tukey test, P ≤ 0.002 in all cases). JBNZ41 also had relatively high clearance rates on the two other ultramicrobacteria, MWH-MoNR1 and MWH-VicM1, even though this was not significant (Tables 2 and 3).
TABLE 2.
Feeding rates of Spumella-like flagellates feeding on ultramicrobacteriaa
| Strain | Parameter | JBC07 | JBNZ41 | JBM10 | Ochromonas sp. strain DS |
|---|---|---|---|---|---|
| MWH-MoNR1 | Grazing mortality (d−1) | 0.16 ± 0.01 | 0.18 ± 0.02 | 0.11 ± 0.01 | 0.08 ± 0.01 |
| Clearance (nl ind−1 h−1) | 1.6 ± 0.4 | 2.7 ± 0.4 | 1.7 ± 0.2 | 1.7 ± 0.2 | |
| Ingestion (bact ind−1 h−1) | 5.9 ± 1.5 | 10.4 ± 1.5 | 6.7 ± 0.7 | 7.1 ± 0.7 | |
| Community grazing (cells ml−1 h−1) | 2.4 ± 0.2 × 104 | 2.8 ± 0.3 × 104 | 1.7 ± 0.2 × 104 | 1.3 ± 0.2 × 104 | |
| Growth (d−1) | 0.28 ± 0.03 | 0.41 ± 0.10 | ND | ND | |
| MWH-TaW3 | Grazing mortality (d−1) | 0.33 ± 0.03 | 0.38 ± 0.08 | 0.10 ± 0.01 | 0.08 ± 0.02 |
| Clearance (nl ind−1 h−1) | 2.6 ± 0.3 | 6.6 ± 2.0 | 2.2 ± 0.4 | 1.7 ± 0.4 | |
| Ingestion (bact ind−1 h−1) | 9.2 ± 1.0 | 24.3 ± 7.8 | 8.5 ± 1.6 | 6.6 ± 1.6 | |
| Community grazing (cells ml−1 h−1) | 4.8 ± 0.4 × 104 | 5.7 ± 1.4 × 104 | 1.5 ± 0.2 × 104 | 1.2 ± 0.3 × 104 | |
| Growth (d−1) | 0.12 ± 0.03 | 0.47 ± 0.12 | ND | ND | |
| MWH-VicM1 | Grazing mortality (d−1) | 0.10 ± 0.03 | 0.16 ± 0.04 | 0.11 ± 0.01 | 0.07 ± 0.03 |
| Clearance (nl ind−1 h−1) | 0.9 ± 0.4 | 3.1 ± 0.9 | 3.2 ± 0.7 | 1.7 ± 1.1 | |
| Ingestion (bact ind−1 h−1) | 4.1 ± 2.0 | 13.5 ± 4.6 | 13.9 ± 3.3 | 7.4 ± 4.9 | |
| Community grazing (cells ml−1 h−1) | 1.9 ± 0.5 × 104 | 2.8 ± 0.8 × 104 | 2.0 ± 0.2 × 104 | 1.3 ± 0.5 × 104 | |
| Growth (d−1) | 0.06 ± 0.07 | 0.31 ± 0.11 | ND | ND | |
| MWH-MoIso2 | Grazing mortality (d−1) | 0.17 ± 0.01 | 0.25 ± 0.04 | 0.07 ± 0.04 | 0.06 ± 0.04 |
| Clearance (nl ind−1 h−1) | 1.9 ± 0.2 | 4.5 ± 0.4 | 1.2 ± 0.5 | 1.4 ± 0.8 | |
| Ingestion (bact ind−1 h−1) | 9.9 ± 0.9 | 22.5 ± 2.0 | 6.7 ± 3.0 | 7.7 ± 4.6 | |
| Community grazing (cells ml−1 h−1) | 3.6 ± 0.1 × 104 | 5.0 ± 0.8 × 104 | 1.6 ± 0.8 × 104 | 1.3 ± 0.8 × 104 | |
| B. diminuta | Grazing mortality (d−1) | 0.67 ± 0.09 | 0.26 ± 0.04 | 0.11 ± 0.03 | 0.16 ± 0.04 |
| Clearance (nl ind−1 h−1) | 7.0 ± 0.7 | 6.2 ± 1.6 | 1.9 ± 0.2 | 3.3 ± 1.8 | |
| Ingestion (bact ind−1 h−1) | 25.6 ± 2.4 | 23.9 ± 6.1 | 7.9 ± 1.0 | 14.3 ± 5.3 | |
| Community grazing (cells ml−1 h−1) | 7.7 ± 3.1 × 104 | 4.1 ± 6.0 × 104 | 1.8 ± 0.5 × 104 | 2.8 ± 0.7 | |
| Growth (d−1) | 0.81 ± 0.11 | ND | ND | ND | |
| L. pelagia | Growth (d−1) | 0.51 ± 0.04 | 0.47 ± 0.17 | ND | ND |
Grazing mortality, clearance, ingestion, and community grazing rates were calculated from the short-term experiment, and the growth rate was calculated from the long-term experiment. Please note that the calculation of the ingestion rate strongly depends on the actual food concentration; clearance and grazing are therefore more robust measures at the realized food concentrations. Ingestion rates were, however, exemplarily calculated for the realized food concentration after 6 h in the short-term experiment, 2.9 × 106 to 5.4 × 106 bacteria ml−1. ind, individual; bact, bacteria; ND, not determined.
TABLE 3.
Tukey test for pairwise comparisons of the parameters of grazing mortality (upper right) and clearance (lower left) between bacterial strains within flagellates after applying analysis of variancea
| Flagellate | Strain |
P
|
||||
|---|---|---|---|---|---|---|
| MWH-MoNR1 | MWH-TaW3 | MWH-VicM1 | MWH-MoIso2 | B. diminuta | ||
| JBC07 | MWH-MoNR1 | <0.001* | 0.333 | 1.000 | <0.001* | |
| MWH-TaW3 | 0.623 | <0.001* | <0.001* | <0.001* | ||
| MWH-VicM1 | 0.827 | 0.118 | 0.231 | <0.001* | ||
| MWH-MoIso2 | 0.993 | 0.858 | 0.581 | <0.001* | ||
| B. diminuta | <0.001* | <0.001* | <0.001* | <0.001* | ||
| JBNZ41 | MWH-MoNR1 | <0.001* | 0.970 | 0.223 | 0.093 | |
| MWH-TaW3 | <0.001* | <0.001* | <0.001* | <0.003* | ||
| MWH-VicM1 | 0.983 | <0.001* | 0.062 | 0.021* | ||
| MWH-MoIso2 | 0.083 | 0.023* | 0.242 | 0.992 | ||
| B. diminuta | <0.001* | 0.970 | <0.001* | 0.101 | ||
| JBM10 | MWH-MoNR1 | 0.998 | 1.000 | 0.802 | 1.000 | |
| MWH-TaW3 | 0.917 | 0.989 | 0.928 | 0.992 | ||
| MWH-VicM1 | 0.172 | 0.603 | 0.701 | 1.000 | ||
| MWH-MoIso2 | 0.969 | 0.587 | 0.043* | 0.729 | ||
| B. diminuta | 0.993 | 0.993 | 0.352 | 0.831 | ||
| Ochromonas sp. strain DS | MWH-MoNR1 | 1.000 | 1.000 | 0.985 | 0.051 | |
| MWH-TaW3 | 1.000 | 1.000 | 0.983 | 0.053 | ||
| MWH-VicM1 | 1.000 | 1.000 | 0.989 | 0.046* | ||
| MWH-MoIso2 | 0.991 | 0.991 | 0.995 | 0.014* | ||
| B. diminuta | 0.117 | 0.119 | 0.104 | 0.043* | ||
Significant differences are indicated by an asterisk and boldface type.
Significance of predator and prey size and specific clearance.
Within a flagellate strain, the clearance rates (Table 2) on ultramicrobacteria were positively correlated with prey size only for strain JBC07 (correlation, F = 504,6; P = 0.002; adjusted R2 = 0.857). A similar trend was observed for strain JBNZ41 even though this was not significant (correlation, F = 4.762; P = 0.161; adjusted R2 = 0.556). For strain JBM10 and Ochromonas sp. strain DS, no significant correlation was found (JBM10: F = 0.842, P = 0.456, adjusted R2 = 0.000; DS: F = 0.037, P = 0.865, adjusted R2 = 0.000).
Specific clearance was, however, generally correlated negatively with the predator- prey size ratio (linear regression on log-log transformed data: regression coefficient, −0.897; F = 91.573; P < 0.001; adjusted R2 = 0.606). This correlation was even stronger when the analysis was restricted to the ultramicrobacteria (linear regression on log-log transformed data: regression coefficient, −1.491; F = 238.196; P < 0.001; adjusted R2 = 0.835; Fig. 3).
Table 2 indicates that all of the interactions cannot be explained by the predator-prey size ratio but some interactions are specific. For instance, strain JBM10 showed the highest clearance rate when feeding on MWH-VicM1, strain JBNZ41 for MWH-TaW3 and B. diminuta, and strains JBC07 and Ochromonas sp. strain DS for B. diminuta. In general, clearance rates of a flagellate strain feeding on ultramicrobacteria differed more than twofold for each of the flagellate strains JBC07, JBNZ41, and JBM10. Only Ochromonas sp. strain DS showed similar clearance rates for all of the ultramicrobacterial strains.
Growth rates.
Growth rates were calculated from the long-term experiment. Growth rates of both flagellates tested, JBC07 and JBNZ41, were positive for all of the bacterial strains tested (Table 2) but were significantly different depending on the flagellate strain as well as on the bacterial strain (two-way analysis of variance, P < 0.001 for both factors). The growth experiment also indicates that the flagellates were able to predate successfully on the ultramicrobacteria and that bacterial food concentrations of 6 × 106 to 9 × 106 bacteria ml−1 allow the positive growth of the flagellates. However, growth rates were low for flagellates feeding on ultramicrobacteria, i.e., 0.06 to 0.47 day−1 corresponding to a doubling time of 1.5 to 11 days. Rough calculations indicate that ingested bacterial biovolume within a doubling period correspond to one to three times the flagellates cell volume. Reliable feeding rates can only be calculated from the short-term experiment and reliable growth rates only from the long-term experiment. Therefore, the above estimates are probably underestimations as a bacterial food concentration, and consequently the expected ingestion rate was slightly higher in the long-term experiment.
In addition to the general support of flagellate growth, strain-specific interactions were also significant (two-way analysis of variance, P = 0.011). The growth rates of the flagellate JBC07 were significantly higher when feeding on the large control bacteria B. diminuta and L. pelagia in comparison to all ultramicrobacterial strains (Tukey test, P < 0.01 in all cases). Only regarding the ultramicrobacterial strains was the growth of the flagellate JBC07 significantly higher for the largest strain, MWH-NR1, compared to the smallest, MWH-VicM1 (Tukey test, P = 0.009). In contrast to the flagellate JBC07, there was no significant difference in the growth of the flagellate JBNZ41 feeding on the different bacteria (one-way analysis of variance, P = 0.227; Table 2).
DISCUSSION
Spumella-like heterotrophic nanoflagellates are able to predate on ultramicrobacteria affiliated with Polynucleobacter. Grazing and growth of closely related small Spumella-like flagellates feeding on different ultramicrobacterial strains affiliated with the Polynucleobacter D subcluster was tested. One goal of this study was to prove whether Spumella-like heterotrophic nanoflagellates can successfully predate on these ultramicrobacteria, and to estimate the potential mortality of these ultramicrobacteria due to predation by those flagellates. All of the flagellates tested successfully predated on all of the tested bacterial strains. In contrast to our assumption, even the relatively large Ochromonas sp. strain DS predated successfully on all of the investigated ultramicrobacterial strains. Thus, the bacterial strains were still in the edible size range of the flagellates. This corresponds to findings that the smallest bacteria and virus-like particles are successfully ingested by nanoflagellates (15, 30).
Small prey size, however, is thought to provide at least relative grazing protection. Several studies support this assumption, i.e., selection for larger strains (6, 25) or actively growing/dividing cells (29). This theory is further supported by the fact that ultramicrobacteria often dominate pelagic bacterial communities in terms of abundance (24, 27). Even though these bacteria may not exhibit high growth rates (17) they are not suppressed, but rather dominate pelagic bacterial communities. Regarding Ochromonas sp. strain DS, grazing protection has already been demonstrated for an ultramicrobacterial actinobacterial strain (19). In contrast, the ultramicrobacteria affiliated with Polynucleobacter were not completely grazing protected.
Feeding on the investigated ultramicrobacteria resulted in low growth rates of the flagellates and low specific clearance rates. The clearance rates of Spumella-like nanoflagellates have been reported to vary between 0.1 and more than 100 nl h−1 and mostly are in the range of a few nanoliters per hour, corresponding to a specific clearance of 1 × 105 to 5 × 105 units of predator volume h−1 (for an overview see 2, 8, 11). Observed clearance rates in our experiments were low, 0.9 to 6.6 nl individual−1 h−1 corresponding to a specific clearance of 0.15 × 105 to 2.8 × 105 units of predator volume h−1. Low grazing mortality of the ultramicrobacteria is further supported by the observation that flagellates failed to decrease the investigated ultramicrobacterial strains below 0.8 × 106 to 4.4 × 106 bacteria ml−1 (long-term grazing experiment). Thus, at such abundances grazing mortality and bacterial growth seem to be balanced. It is therefore likely that the investigated bacterial strains are grazed at typical field abundances but grazing pressure is similar or even lower than bacterial growth possibly allowing for a stable population even under medium to high grazing pressure.
In conclusion, all of the tested strains of ultramicrobacteria were successfully predated by the Spumella-like flagellates, but bacterial grazing mortality is significantly lower than for larger bacteria. This was consistently found in both long and short-term grazing experiments.
Ultramicrobacteria affiliated with Polynucleobacter are near the lower prey size limit for the investigated flagellates and the predator-prey size ratio is crucial for the grazing sensitivity of these bacteria.
Predator and prey size is generally an important variable for feeding ecology (11, 13, 14, 25, 32). It is, however, less clear to what extent prey size is a significant factor for the selection of similar-sized organisms as factors other than size may override the effects of prey size. Our data support a strong positive correlation between the prey size and clearance rate, generally supporting the model presented by Gonzalez (14). This model already predicts high size selection efficiency for flagellates feeding on ultramicrobacteria. Our data contradict, however, a basic assumption of size selective feeding, i.e., that clearance increases with predator and prey size (10, 25). In contrast, clearance rates generally decreased with predator size (Fig. 3) and the control flagellate (Ochromonas) showed similar low clearance rates on all ultramicrobacteria. This indicates that the observed size selection of the Spumella-like flagellates feeding on the investigated ultramicrobacteria depends on factors other than the propelled water volume or contact probability as proposed by the theoretical models (10, 25). Two other aspects related to predator and prey size have to be considered, i.e., the absolute size of the predator regarding the effects of scaling of metabolic rates, as well as the relative size of predator and prey regarding the realized food size range.
Scaling of metabolic rates.
Even though absolute feeding rates tend to increase with body size, specific feeding rates tend to decrease when organisms covering a wide size span are compared. In general, the scaling exponent has been estimated to be approximately −0.25 or somewhat less for a wide range of planktonic organisms including flagellates (21, 22). In contrast, we observed that the absolute feeding rates decreased with body size corresponding to a strong negative correlation between predator size and specific clearance rates (scaling exponent of −1.5). In our experiments, the prey organisms were ultramicrobacteria and we therefore speculate that the strong correlation in our study is due to a decreasing ability to deal with such small prey items with the increasing size of the predator.
Realized food size range.
Optimal predator-prey size ratio with respect to the equivalent spherical diameter has been compiled by Hansen et al. (20). They reported the optimal size ratio to range from 1:1 for interception feeding dinoflagellates to 50:1 for filter feeding cladocerans. For flagellates, an optimal predator-prey equivalent spherical diameter size ratio of 3:1 was calculated. Regarding the lower prey size range, ratios of around 7:1 have been estimated for Spumella and Ochromonas (Pfandl et al., submitted) as well as for the dinoflagellate Gyrodinium spirale (20). These studies suggest that interception feeding flagellates can in fact ingest such small prey, but only from around a predator-prey equivalent spherical diameter size ratio of 7:1 to 8:1, corresponding to a volumetric predator-prey size ratio of around 500:1, to where prey capture, and thereafter feeding begins to become inefficient.
In our study, the equivalent spherical diameter predator-prey size ratio of the Spumella-like flagellates feeding on the ultramicrobacterial strains ranged between 7:1 and 11:1, supporting the assumption that predator prey size ratios in our experiment are far from being optimal. These size ratios as well as the strong correlation between predator prey size ratio and specific clearance strongly support the hypothesis that flagellates feeding on ultramicrobacteria are preying near their lower prey size limit. Our data also indicate that prey volume or prey mass are somehow related to the decreasing feeding efficiency, not prey length. As previously suggested (34; Pfandl et al., submitted) mechanical contact strength, i.e., momentum, of the predator prey contact might not be sufficiently strong for the small ultramicrobacteria in each contact event to induce a capture reaction by the flagellate.
In conclusion, the predator-prey size ratio significantly contributes to the observed variability in the predation vulnerability of ultramicrobacteria. Even for the closely related and similar sized organisms investigated in our study, we found a surprisingly strong effect of predator and prey size even though the size ratio did not fully explain all of the strain-specific differences.
Grazing patterns are strain specific even between closely related organisms.
Both of the ultramicrobacterial strains investigated (16S rRNA gene sequence similarity > 99.6%) and the Spumella-like flagellates (18S rRNA gene sequence similarity 100%) used in this study are closely related to each other, and possibly belong to the same bacterial and flagellate species, respectively. Regarding the investigated ultramicrobacteria, their cell size seems to be strain specific, even though it is flexible within relatively small size ranges and closely related strains, and therefore they potentially differ with respect to grazing vulnerability. Even the small size differences between the bacteria and flagellates, respectively, resulted in pronounced differences in grazing vulnerability of the bacteria (Fig. 2), indicating a niche differentiation between the closely related strains.
Even though Ochromonas is much larger compared to the small Spumella-like flagellates we found Ochromonas to be able to predate on the investigated ultramicrobacteria. In contrast to the successful grazing of all nanoflagellates (including Ochromonas sp. strain DS) on ultramicrobacteria affiliated to the Polynucleobacter cluster we previously found Ochromonas sp. strain DS to be unable to predate and grow on a strain of Actinobacteria which was very similar in size and morphology (i.e., C shaped) (19). Factors other than size and morphology could additionally therefore be of importance for the observed strain-specific interactions. Accordingly, if size is the only character determining bacterial grazing mortality, the grazing mortality rates for the large bacterium B. diminuta should be expected to be higher than what was found in our study. Even though we cannot quantify the strain specific interactions related to factors other than size, the different findings imply that niche differentiation according to prey size, but also to prey characters other than size is realized in the investigated model organisms, i.e., Spumella-like flagellates feeding on ultramicrobacteria affiliated with the Polynucleobacter cluster. Field studies usually do not allow the separation of such similar organisms with respect to morphology and size. Future studies with high taxonomic resolution may therefore provide further evidence for high intraspecific variability even within very closely related members of the same species. If this pattern turns out to be general, it would have crucial implications for analyzing feeding interactions in microbial food webs.
Potential role of strain-specific differences in predator-prey interaction in the evolution and maintenance of microbial microdiversity.
Microbial microdiversity is known as molecular diversity within a microbial species or among closely related yet undescribed organisms (26). Coexistence of microorganisms differing only in one or a few sequence positions of their small-subunit rRNA genes was documented repeatedly (12, 26) and seems to be a common phenomenon in microbial populations. Moore et al. linked the observed 16S rRNA microdiversity of a Prochlorococcus population to the physiological diversity of their light adaptation (26). We demonstrated significant differences in sensitivity of the same prey strain to predation by a closely related predator strain. The flagellate JBNZ41, for instance, led in comparison to JBM10 to an almost fourfold higher grazing mortality rate of the bacterial strain MWH-TaW3 (Fig. 1). This indicates that specific predator-prey interactions may be another driving force in the evolution and maintenance of microbial microdiversity, as well as in the niche separation of closely related microorganisms.
Conclusions.
C-shaped ultramicrobacteria affiliated with the Polynucleobacter cluster proved to be vulnerable to grazing by Spumella-like flagellates even though grazing-related mortality is much lower than for larger bacteria. Regarding the usually low maximal growth rates of these ultramicrobacteria (17), they cannot be considered in general to be grazing protected. On the other hand, predator-specific differences in grazing vulnerability of the investigated bacterial strains were observed. These strain-specific differences may allow for certain protection from grazing by some flagellate strains. Spumella-like flagellates feeding upon the investigated ultramicrobacteria are supposed to feed near the lower prey size limit. Our study highlights the importance of microdiversity in predator-prey interactions and supports the general idea of niche differentiation even between closely related and similar-sized organisms with respect to their feeding ecology.
Acknowledgments
We thank the Austrian Science Fund (FWF) for financial support (FWF projects P15655 and P15940).
REFERENCES
- 1.Andersson, A., U. Larsson, and Å. Hagström. 1986. Size-selective grazing by a microflagellate on pelagic bacteria. Mar. Ecol. Prog. Ser. 33:51-57. [Google Scholar]
- 2.Boenigk, J., and H. Arndt. 2000. Comparative studies on the feeding behavior of two heterotrophic nanoflagellates: the filter-feeding choanoflagellate Monosiga ovata and the raptorial-feeding kinetoplastid Rhynchomonas nasuta. Aquat. Microb. Ecol. 22:243-249. [Google Scholar]
- 3.Boenigk, J., and H. Arndt. 2002. Bacterivory by heterotrophic flagellates: community structure and feeding strategies. Anthony van Leeuwenhoek 81:465-480. [DOI] [PubMed] [Google Scholar]
- 4.Boenigk, J., C. Matz, K. Jürgens, and H. Arndt. 2002. Food concentration dependent regulation of food selectivity of interception feeding bacterivorous nanogflagellates. Aquat. Microb. Ecol. 27:195-202. [Google Scholar]
- 5.Caron, D. A. 1987. Grazing of attached bacteria by heterotrophic microflagellates. Microb. Ecol. 13:203-218. [DOI] [PubMed] [Google Scholar]
- 6.Chrzanowski, T. H., and K. Šimek. 1990. Prey-size selection by freshwater flagellated protozoa. Limnol. Oceanogr. 35:1429-1436. [Google Scholar]
- 7.Cleven, E. J., and T. Weisse. 2001. Seasonal succession and taxon specific bacterial grazing rates of heterotrophic nanoflagellates in Lake Constance. Aquat. Microb. Ecol. 23:147-161. [Google Scholar]
- 8.Eccleston-Parry, J. D., and B. S. C. Leadbeater. 1994. A comparison of the growth kinetics of 6 marine heterotrophic nanoflagellates fed with one bacterial species. Mar. Ecol. Prog. Ser. 105:167-177. [Google Scholar]
- 9.Eccleston-Parry, J. D., and B. S. C. Leadbeater. 1994. The effect of long-term low bacterial density on the growth-kinetics of 3 marine heterotrophic nanoflagellates. J. Exp. Mar. Biol. Ecol. 177:219-233. [Google Scholar]
- 10.Fenchel, T. 1984. Suspended marine bacteria as a food source, p. 301-315. In M. J. Fasham (ed.), Flows of energy and materials in marine ecosystems. Plenum Press, New York, N.Y.
- 11.Fenchel, T. 1986. The ecology of heterotrophic microflagellates. Adv. Microb. Ecol. 9:57-97. [Google Scholar]
- 12.García-Martínez, J., and F. Rodríguez-Valera. 2000. Microdiversity of uncultured marine prokaryotes: the SAR11 cluster and the marine Archaea of Group I. Mol. Ecol. 9:935-948. [DOI] [PubMed] [Google Scholar]
- 13.González, J. M., E. B. Sherr, and B. F. Sherr. 1990. Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl. Environ. Microbiol. 56:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González, J. M. 1996. Efficient size-selective bacterivory by phagotrophic nanoflagellates in aquatic ecosystems. Mar. Biol. 126:785-789. [Google Scholar]
- 15.González, J. M., and C. A. Suttle. 1993. Grazing by marine nanoflagellates on virus-sized particles-ingestion and digestion. Mar. Ecol. Prog. Ser. 94:1-10. [Google Scholar]
- 16.Hahn, M. W. 1997. Experimentelle Untersuchungen zur Interaktion von bakterivoren Nanoflagellaten mit pelagischen Bakterien. Ph.D. thesis. Technische Universität Braunschweig, Braunschweig, Germany.
- 17.Hahn, M. W. 2003. Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Appl. Environ. Microbiol. 69:5248-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn, M. W., and M. G. Höfle. 1998. Grazing pressure by a bacterivorous flagellate reverses the relative abundance of Comamonas acidovorans PX54 and Vibrio strain CB5 in chemostat coculture. Appl. Environ. Microbiol. 64:1910-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn, M. W., H. Lünsdorf, Q. Wu, M. Schauer, M. G. Höfle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as Actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen, B., P. K. Bjørnsen, and P. J. Hansen. 1994. The size ratio between planktonic predators and their prey. Limnol. Oceanogr. 39:395-403. [Google Scholar]
- 21.Hansen, P. J., P. K. Bjørnsen, and B. H. Hansen. 1997. Zooplankton grazing and growth: Scaling within the 2-2,000 mm body size range. Limnol. Oceanogr. 42:687-704. [Google Scholar]
- 22.Hemmingsen, A. M. 1960. Energy metabolism as related to body size and respiratory surfaces, and its evolution. Rep. Steno Mem. Hosp. Nord. Insulin Lab. 8:1-110. [Google Scholar]
- 23.Holen, D. A., and M. E. Boraas. 1991. The feeding behaviour of Spumella sp. as a function of particle size: Implications for bacterial size in pelagic systems. Hydrobiologia 220:73-88. [Google Scholar]
- 24.Loferer-Kröβbacher, M., J. Klima, and R. Psenner. 1998. Determination of bacterial cell dry mass by transmission electron microscopy and densitometric image analysis. Appl. Environ. Microbiol. 65:688-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monger, B. C., and M. R. Landry. 1991. Prey-size dependency of grazing by free-living marine flagellates. Oecologia 74:239-248. [Google Scholar]
- 26.Moore, L. R., Rocap, G., and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 27.Pernthaler, J., A. Alfreider, T. Posch, S. Andreatta, and R. Psenner. 1997. In situ classification and image cytometry of pelagic bacteria from a high mountain lake (Gossenköllersee, Austria). Appl. Environ. Microbiol. 63:4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preisig, H. R., N. Vørs, and G. Hällfors. 1991. Diversity of heterotrophic heterokont flagellates, p. 361-399. In D. J. Patterson and J. Larsen (ed.), The biology of free-living heterotrophic flagellates. Clarendon Press, Oxford, England.
- 29.Sherr, B. F., E. B. Sherr, and J. McDaniel. 1992. Effect of protistan grazing on the frequency of dividing cells in bacterioplankton assemblages. Appl. Environ. Microbiol. 58:2381-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherr, E. B. 1988. Direct use of high molecular weight polysaccharide by heterotrophic flagellates. Nature 335:348-351. [Google Scholar]
- 31.Sieracki, M. E., L. W. Haas, D. A. Caron, and E. J. Lessard. 1987. Effect of fixation on particle retention by microflagellates: underestimation of grazing rates. Mar. Ecol. Prog. Ser. 38:251-258. [Google Scholar]
- 32.Šimek, K., J. Vrba, and P. Hartman. 1994. Size-selective feeding by Cyclidium sp. on bacterioplankton and various sizes of cultured bacteria. FEMS Microbiol. Ecol. 14:157-167. [Google Scholar]
- 33.Weitere, M., and H. Arndt. 2003. Structure of the heterotrophic flagellate community in the water column of the river Rhine (Germany). Eur. J. Protistol. 39:287-300. [Google Scholar]
- 34.Wu, Q. L., J. Boenigk, and M. W. Hahn. 2004. Successful predation of filamentous bacteria by a nanoflagellate challenges current models of flagellate bacterivory. Appl. Environ. Microbiol. 70:332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]