Abstract
Dehalococcoides sp. strain BAV1 couples growth with the reductive dechlorination of vinyl chloride (VC) to ethene. Degenerate primers targeting conserved regions in reductive dehalogenase (RDase) genes were designed and used to PCR amplify putative RDase genes from strain BAV1. Seven unique RDase gene fragments were identified. Transcription analysis of VC-grown BAV1 cultures suggested that bvcA was involved in VC reductive dechlorination, and the complete sequence of bvcA was obtained. bvcA was absent in Dehalococcoides isolates that failed to respire VC, yet was detected in four of eight VC-respiring mixed cultures.
Vinyl chloride (VC) is a toxic and carcinogenic priority pollutant that threatens drinking water quality in most industrialized countries (16). Naturally occurring transformation reactions acting on chlorinated solvents such as tetrachloroethene (PCE) and trichloroethene (TCE), which are abundant groundwater pollutants (25, 29, 34, 36), serve as a major source of environmental VC. Additional environmental VC originates from landfills (2, 5), polyvinyl chloride production facilities (16), and abiotic formation in soils (15). Bioremediation approaches that rely on the activity of bacterial populations that use chlorinated compounds as growth-supporting electron acceptors (i.e., chlororespiration) have been implemented successfully in the field (8, 17, 22). Recent efforts identified the key populations responsible for reductive dechlorination and detoxification of VC to ethene as members of the Dehalococcoides (6, 7, 11), a deeply branching group on the bacterial tree most closely affiliated with the Chloroflexi (1, 23). 16S rRNA gene-based PCR approaches were designed to detect (9, 12, 18) and quantify (6, 10, 11) members of this group. Such approaches are critical for assessing VC-contaminated sites, monitoring bioremediation efforts, and establishing cause-effect relationships (17, 22). Unfortunately, Dehalococcoides strains with different dechlorination activities have similar or identical 16S rRNA gene sequences (10). Dehalococcoides sp. strain CBDB1 dechlorinates chlorobenzenes and some polychlorinated dibenzodioxin congeners but fails to grow with PCE and TCE as electron acceptors (1, 4). Dehalococcoides ethenogenes 195 (23, 24) and Dehalococcoides sp. strain FL2 grow with polychlorinated ethenes as electron acceptors but cannot grow with VC, and Dehalococcoides sp. strain BAV1 is the only isolate known to respire all DCE isomers and VC (10). Despite their metabolic differences, some of these populations share a 16S rRNA gene sequence with more than 99.9% similarity (11). Identifying genes directly involved in the dechlorination process of interest could overcome these limitations and complement 16S rRNA gene-based approaches.
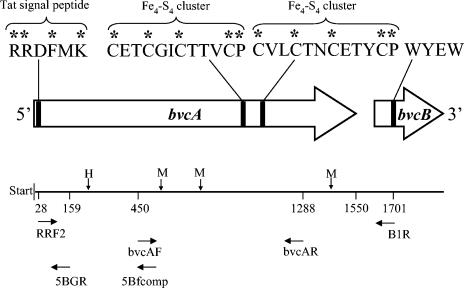
A few RDase sequences involved in partial reductive dechlorination of PCE and chlorinated aromatic compounds have been obtained (20, 21, 26, 27, 30, 31, 33), but the genes involved in VC reduction are unknown. Associated with each RDase gene is a B gene, which encodes a hydrophobic protein with transmembrane helices believed to anchor the RDase to the membrane (20, 21, 26, 31, 33). We used available sequence information to design a procedure to PCR amplify putative RDase genes of Dehalococcoides spp. and to identify RDase genes implicated in VC reductive dechlorination. Multiple alignments of full-length protein and DNA sequences of TceA (GenBank accession numbers AAN85590, AAN85588, and AAF73916A) and putative RDase genes identified in the genome of D. ethenogenes strain 195 (35) were constructed by using ClustalW and ClustalX (32). Conserved amino acid sequences were identified and used to design degenerate PCR primers (Fig. 1). The forward primer RRF2 (5′-SHMGBMGWGATTTYATGAARR-3′) was designed to target the RRXFXK motif, which is part of the Tat protein export pathway (3). This motif near the amino terminus is highly conserved but not unique to RDases. For this reason, Regeard et al. (28) designed degenerate primer pairs targeting conserved RDase internal regions, excluding the RRXFXK motif, that yielded relatively short amplicons of a maximum size of about 900 bp. Since our goal was to specifically amplify putative Dehalococcoides RDase genes and obtain their complete sequences, we took advantage of the conserved twin arginine motif, and identified a conserved motif, WYEW, internal to the Dehalococcoides B genes for the design of the degenerate reverse primer B1R (5′-CHADHAGCCAYTCRTACCA-3′). The expected size of amplicons generated with these primers ranged from 1,500 to 1,700 bp. Abbreviations for degenerate nucleotides positions are as follows: R = A or G; K = G or T; M = A or C; S = C or G; W = A or T; Y = C or T; B = C, G, or T; D = A, G, or T; V = A, C, or G; H = A, C, or T.
FIG. 1.
Arrangement of the bvcA gene and the corresponding B gene, bvcB. Also shown are conserved dehalogenase features including the Tat signal peptide RRDFMK, and two Fe4-S4 clusters near the C-terminal end. Amino acids conserved in RDases are labeled with an asterisk. Primer binding sites used for amplification of putative RDase genes (primers RRF2 and B1R), detection of bvcA (primers bvcAr and bvcAF), and chromosome walking for capturing the entire coding region of bvcA (primers 5bfcomp and 5BG) are indicated. H and M, HhaI and MspI restriction sites, respectively.
DNA from VC-dechlorinating pure and mixed cultures was extracted as described previously (11) and used as a template for amplification with degenerate primers RRF2 and B1R. PCRs were performed in total volumes of 30 μl as described previously (10, 11), except that the annealing temperature was 48°C and the primer and MgCl2 concentrations were 0.5 μM and 3.0 mM, respectively. Amplicons generated with primers RRF2 and B1R were purified with a QIAquick PCR purification kit (Qiagen, Valencia, Calif.) and cloned by using commercial kits (TOPO or TA cloning kit; Invitrogen, Carlsbad, Calif.). Recombinant E. coli clones were screened by verifying the correct insert size (approximately 1,700 bp) by direct PCR with primers targeting the pCR2.1 cloning vector flanking the inserted fragment (37). A total of seven clones were recovered in the clone library generated with DNA from the VC-dechlorinating Bachman mixed culture (11). Restriction fragment length polymorphism analysis using the enzymes MspI and HhaI (Promega Biosciences, San Luis Obispo, Calif.) identified five clone types with distinct reductive dehalogenase-homologous inserts, designated rdhA1BAV1 to rdhA5BAV1. Plasmid DNA from the five recombinant clones containing the different inserts was extracted with a Qiaprep spin miniprep kit (Qiagen) and partially sequenced with vector-specific primers by using an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.). In a second clone library established with strain BAV1 pure-culture DNA, 54 clones were recovered, and two additional putative RDase sequences were identified, i.e., rdhA6BAV1 and rdhA7BAV1. No clones harboring rdhA3BAV1, rdhA4BAV1, or rdhA5BAV1 were identified in the BAV1 clone library, but subsequent PCR analysis (conditions as described in reference 11 except that 2 mM MgCl2 was used) with primer pairs specifically targeting each of the rdhA1BAV1 to rdhA7BAV1 sequences (Table 1) demonstrated the presence of all putative RDase fragments in isolate BAV1 and in the Bachman mixed culture from which BAV1 was isolated (10). BLASTX analysis revealed that all fragments were most similar to TceA, the TCE RDase in D. ethenogenes strain 195. DNA sequences were translated with the TRANSLATE program (http://us.expasy.org/tools/dna.html) into amino acid sequences to examine for known RDase motifs. Alignments using ClustalX (32) revealed that the seven amino acid sequences had two iron sulfur clusters but were different from each other and from all previously reported RDases.
TABLE 1.
Specific primers used in this study
| Primera | Sequence (5′→3′) | Gene targeted | Position relative to bvcA startb |
|---|---|---|---|
| bavrdA1F | GTACCGATGATGATTCACG | rdhA1BAV1 | 504 |
| bavrdA1R | AGCCATACATGTCCCGCAA | rdhA1BAV1 | 1377 |
| bavrdA2F | TGCAAGCAGGTTCCCAT | rdhA2BAV1 | 467 |
| bavrdA2R | GGCTTGATGTTAAACCC | rdhA2BAV1 | 1305 |
| bavrdA3F | GATTATGCTTTGTTTGGG | rdhA3BAV1 | 406 |
| bavrdA3R | TTAGAACAACCACCAGGC | rdhA3BAV1 | 1367 |
| bavrdA4F | ATGCCATGTATTCGGTC | rdhA4BAV1 | 412 |
| bavrdA4R | TCAACCCTCCAGCCTTTA | rdhA4BAV1 | 1305 |
| bavrdA5F | GTTAATGTTGCCAAGGCT | rdhA5BAV1 | 527 |
| bavrdA5R | CATGGTCTTTTCCATATTGGC | rdhA5BAV1 | 1491 |
| bvcAF | TGCCTCAAGTACAGGTGGT | rdhA6BAV1/bvcA | 450 |
| bvcAR | ATTGTGGAGGACCTACCT | rdhA6BAV1/bvcA | 1288 |
| bavrdA7F | AAACTGCTCAGGGTTG | rdhA7BAV1 | 463 |
| bavrdA7R | TTGCCCGGAACACTGTA | rdhA7BAV1 | 1339 |
| 5Bfcomp | ACCACCTGTACTTGAGGCA | rdhA6BAV1/bvcA | 468 |
| 5BGR | ACCCGACAAAGAACTGGTTTCG | rdhA6BAV1/bvcA | 138 |
PCR amplification used an annealing temperature of 51°C except for bvcAR and bvcAF (52°C), 5Bfcomp (55°C), and 5BGR (57°C).
Nucleotide position based on the complete bvcA gene sequence.
Expression analysis and identification of a VC RDase.
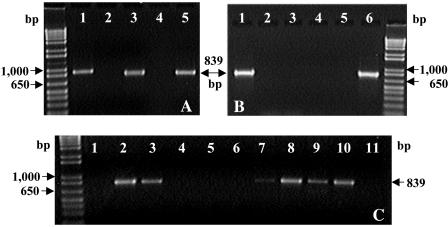
To identify the gene involved in VC reductive dechlorination, total RNA from VC-grown BAV1 cultures (10) was extracted with an RNeasy extraction kit (Qiagen) according to the manufacturer's recommendations with the following modifications. A cell pellet was suspended in 100 μl of lysozyme digestion buffer (30 mM Tris-HCl, 1 mM EDTA [pH 8.0], 15 mg of lysozyme per ml), 20 μl of proteinase K (25 mg/ml), and 10 μl of achromopeptidase (7,500 U/ml). The suspension was mixed and incubated at room temperature for 10 min, before 50 μl of 0.1% Triton X-100 was added, and the mixture was shaken vigorously for 10 s. Lysis buffer (350 μl) (RNeasy) was added, and the lysate was transferred to a MicroRNA bead tube (Mo Bio Laboratories, Carlsbad, Calif.) and shaken horizontally on a Vortex mixer at maximum speed for 10 min. DNA was removed by two consecutive treatments with RNase-free DNase (Qiagen). All solutions used for RNA extraction and reverse transcription-PCR (RT-PCR) were prepared with diethyl pyrocarbonate-treated water. RT-PCR was performed with the two-step RT-PCR Sensiscript kit (Qiagen) to produce cDNA. The reaction mix contained 1 mM random hexamer primers (Promega) and 5 to 50 ng of extracted RNA in a total volume of 20 μl and was incubated for 3 h at 37°C. PCR amplification with primers RRF2 and B1R using cDNA obtained from VC-grown BAV1 cells as a template yielded a PCR fragment of approximately 1,700 bp, indicating that the putative RDase gene and the associated B gene are cotranscribed. A cDNA clone library was established, and restriction fragment length polymorphism and sequence analyses of six clones confirmed that all the cloned fragments were identical to rdhA6BAV1. Transcription of the putative VC RDase genes was explored in more detail by using specific primer pairs targeting each putative RDase gene (Table 1). PCRs using cDNA generated from VC-grown BAV1 cultures as the template and the primer pair bvcAF-bvcAR (Table 1) yielded amplicons of the correct size of 839 bp (Fig. 2A), and sequence analysis confirmed their identity to rdhA6BAV1. To test if the other six RDase genes were expressed at lower levels, 1 μl of the amplified PCR product generated from cDNA with primer pair RRF2-B1R was used for a subsequent nested PCR with the specific primer pairs listed in Table 1, and all amplicons were sequenced. These results suggest that genes contributing to fragments rdhA1BAV1, rdhA3BAV1 to rdhA5BAV1, and rdhA7BAV1 were also expressed in VC-grown BAV1 cells, but at lower levels than rdhA6BAV1. rdhA2BAV1 was not transcribed at detectable levels. Although the involvement of these putative RDases in VC-reductive dechlorination cannot be excluded, our findings suggest that the gene that contains the fragment rdhA6BAV1 encodes a VC-reductive dechlorinase in strain BAV1.
FIG. 2.
(A) PCR amplification of the putative VC RDase gene with the specific primers bvcAR and bvcAF and templates generated from VC-grown BAV1 cultures. Shown are the DNA size marker 1Kb Plus (Invitrogen) (left lane), BAV1 genomic DNA (lane 1), BAV1 total RNA (lane 2), BAV1 cDNA (lane 3), H2O (lane 4), and plasmid DNA containing the rdhA6BAV1 gene fragment (lane 5). (B) Specificity of primers (bvcAR/bvcAF) targeting the putative VC RDase gene bvcA. Shown are the DNA size marker 1Kb Plus (Invitrogen) (right lane), plasmid DNA containing rdhA6BAV1 (lane 1), H2O (lane 2), genomic DNA from strain FL2 (lane 3), D. ethenogenes (lane 4), strain CBDB1 (lane 5), and strain BAV1 (lane 6). (C) Detection of bvcA in VC-dechlorinating mixed cultures using primers bvcAR and bvcAF. Shown are the DNA size marker 1Kb Plus (Invitrogen) (left lane), H2O (lane 1), plasmid DNA containing rdhA6BAV1 (lane 2), and genomic DNA from the Bachman enrichment culture (lane 3), the Au Sable culture (lane 4), the Père Marquette culture (lane 5), the Red Cedar culture (lane 6) (19), the Hydrite culture (lane 7), the Minerva culture (lane 8), Bio-Dechlor Inoculum (lane 9), KB-1 (lane 10), and the Victoria culture (lane 11).
Chromosome walking and assembling the bvcA coding sequence.
Since the putative RDase gene fragment rdhA6BAV1 lacked approximately 30 bp on the 5′ end of the gene, the gene fragment was extended by using the TOPO Walker kit from Invitrogen. BAV1 genomic DNA was digested with the restriction enzymes PstI and SacI. Extension and PCR amplification used primers 5Bfcomp and 5BGR and the PCR conditions described above (Table 1 and Fig. 1). The missing upstream portion of the putative RDase gene was obtained and aligned with the previously obtained rdhA6BAV1 sequence, and the coding region was determined by using Frameplot (http://www.nih.go.jp/∼jun/cgi-bin/frameplot.pl) (14). The complete gene was designated bvcA, and the chromosomal organization of the bvcA region is shown in Fig. 1. The deduced coding sequence of bvcA is 1,550 nucleotides long, which translates into a 516-amino-acid protein. The BvcA protein sequence contained the twin arginine motif (RRXFXK) in the form RRDFMK. A second incomplete open reading frame for the putative B gene bvcB was found 51 nucleotides downstream of the bvcA stop codon, TAA. A comparison of the translated amino acid sequence BvcA with putative RDases found on other Dehalococcoides sp. genomes (13, 35) showed that sequence identity did not exceed 39% at the protein level. These findings indicate that bvcA is a unique gene not present in other described Dehalococcoides isolates. BvcA shared the highest similarity of 39% with TceA, the RDase responsible for TCE dechlorination in D. ethenogenes strain 195 (20). The biochemistry of Dehalococcoides RDases is of great interest, but detailed investigations are hampered by difficulties in obtaining sufficient biomass. This procedure designed to obtain the complete sequence of bvcA is applicable to the capture of other Dehalococcoides RDase genes and will assist future efforts aimed at heterologous expression to allow in-depth investigations of these interesting enzyme systems.
Implications for bioremediation monitoring.
Current efforts to assess and monitor chloroethene-contaminated sites rely on nucleic acid-based approaches targeting Dehalococcoides 16S rRNA genes (9, 12, 17, 22). A major limitation of the 16S rRNA gene approach lies in the similarity of 16S rRNA genes from Dehalococcoides populations exhibiting different dechlorination activities (10). In order to explore if bvcA is a suitable gene target to distinguish the VC-respiring strain BAV1, bvcA-targeted primers bvcAF and bvcAR (Table 1) were tested with genomic DNA from other Dehalococcoides isolates and VC-dechlorinating, Dehalococcoides-containing mixed cultures. A correct-size amplicon (839 bp) was generated with isolate BAV1 genomic DNA but not with genomic DNA from D. ethenogenes strain 195, strain FL2, or strain CBDB1, none of which have been reported to grow with VC (Fig. 2B). Additionally, bvcA was detected in four of eight Dehalococcoides-containing, ethene-producing mixed cultures (Fig. 2C). bvcA was present in cultures KB-1 and Bio-Dechlor Inoculum, two commercially available ethene-producing enrichment cultures that have been successfully used in bioaugmentation approaches (17, 22). Also, bvcA was identified in two ethene-producing enrichment cultures derived from chloroethene-contaminated aquifer materials (i.e., the Minerva site in Ohio and the Hydrite Chemical site in Wisconsin). bvcA, however, was not detected in three VC-dechlorinating enrichment cultures derived from Michigan river sediments, where the presence of VC-respiring populations has been established through hydrogen consumption threshold measurements (19), and Dehalococcoides 16S rRNA gene sequences were detected (K. M. Ritalahti, R. Krajmalnik-Brown, and F. E. Löffler, session C1, Abstr. 6th Int. Symp. In Situ and On-Site Bioremediation, 2001). Similarly, the ethene-producing Victoria culture containing Dehalococcoides sp. strain VS (6) did not contain bvcA (Fig. 2C). These findings suggest that a diversity of VC RDase genes exists, and additional sequences must be obtained before development of a comprehensive suite of nucleic acid-based tools to target all VC RDase genes becomes feasible. Nevertheless, nucleic acid-based tools targeting bvcA will complement 16S rRNA gene-based approaches to monitor sites undergoing enhanced treatment where BAV1-type populations are already present (17) or inocula containing BAV1-type populations (e.g., Bio-Dechlor Inoculum or KB-1) are used for bioaugmen-tation.
Nucleotide sequence accession numbers.
The coding sequences of the putative RDase genes and putative B gene fragments were deposited in GenBank under accession numbers AY553222 to AY553228. GenBank accession number AY563562 was assigned to the complete sequence of the putative VC reductive dehalogenase gene bvcA.
ADDENDUM IN PROOF
Müller et al. (25a) characterized the VcrA dehalogenase from Dehalococcoides sp. strain VS, present in the ethene-producing Victoria enrichment culture. The vcrA-targeted primers described by Müller et al. failed to yield amplicons when DNA from Dehalococcoides sp. strain BAV1 was used as the template. This finding further supports our conclusion that multiple VC RDases exist.
Acknowledgments
This work was supported by a National Science Foundation CAREER award to F.E.L., by the Strategic Environmental Research and Development Program, and by Regenesis Bioremediation Products.
We thank J. He for growing cultures of Dehalococcoides sp. strain BAV1, Y. Sung for providing ethene-producing enrichment cultures from the Minerva and Hydrite sites, B. Lynch for operating the ABI sequencer, and B. Amos for helpful discussions. E. Edwards, S. Zinder, and A. Spormann are acknowledged for providing genomic DNA of KB-1, strain 195, and the Victoria culture, respectively.
REFERENCES
- 1.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 2.Allen, M. R., A. Braithwaite, and C. C. Hills. 1997. Trace organic compounds in landfill gas at seven UK waste disposal sites. Environ. Sci. Technol. 31:1054-1061. [Google Scholar]
- 3.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 4.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Görisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 5.Coulston, F., and A. C. Kolbye, Jr. 1994. Vinyl chloride and polyvinyl chloride. Regul. Toxicol. Pharmacol. 19:344-346. [Google Scholar]
- 6.Cupples, A. M., A. M. Spormann, and P. L. McCarty. 2003. Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors as determined by competitive PCR. Appl. Environ. Microbiol. 69:953-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duhamel, M., S. D. Wehr, L. Yu, H. Rizvi, D. Seepersad, S. Dworatzek, E. E. Cox, and E. A. Edwards. 2002. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis-dichloroethene and vinyl chloride. Water Res. 36:4193-4202. [DOI] [PubMed] [Google Scholar]
- 8.Ellis, D. E., E. J. Lutz, J. M. Odom, R. J. Buchanan, C. L. Bartlett, M. D. Lee, M. R. Harkness, and K. A. Deweerd. 2000. Bioaugmentation for accelerated in situ anaerobic bioremediation. Environ. Sci. Technol. 34:2254-2260. [Google Scholar]
- 9.Fennell, D. E., A. B. Carroll, J. M. Gossett, and S. H. Zinder. 2001. Assessment of indigenous reductive dechlorinating potential at a TCE-contaminated site using microcosms, polymerase chain reaction analysis, and site data. Environ. Sci. Technol. 35:1830-1839. [DOI] [PubMed] [Google Scholar]
- 10.He, J., K. M. Ritalahti, K.-L. Yang, S. S. Koenigsberg, and F. E. Löffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 11.He, J. Z., K. M. Ritalahti, M. R. Aiello, and F. E. Löffler. 2003. Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species. Appl. Environ. Microbiol. 69:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrickson, E. R., J. A. Payne, R. M. Young, M. G. Starr, M. P. Perry, S. Fahnestock, D. E. Ellis, and R. C. Ebersole. 2002. Molecular analysis of Dehalococcoides16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 68:485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hölscher, T., R. Krajmalnik-Brown, K. M. Ritalahti, F. V. Wintzingerode, H. Görisch, F. E. Löffler, and L. Adrian. 2004. Multiple non-identical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70:5290-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa, J., and K. Hotta. 1999. Frameplot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 15.Keppler, F., R. Borchers, J. Pracht, S. Rheinberger, and H. F. Schöler. 2002. Natural formation of vinyl chloride in the terrestrial environment. Environ. Sci. Technol. 36:2479-2483. [DOI] [PubMed] [Google Scholar]
- 16.Kielhorn, J., C. Melber, U. Wahnschaffe, A. Aitio, and I. Mangelsdorf. 2000. Vinyl chloride: still a cause for concern. Environ. Health Perspect. 108:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lendvay, J. M., F. E. Löffler, M. Dollhopf, M. R. Aiello, G. Daniels, B. Z. Fathepure, M. Gebhard, R. Heine, R. Helton, J. Shi, R. Krajmalnik-Brown, C. L. Major, Jr., M. J. Barcelona, E. Petrovskis, J. M. Tiedje, and P. Adriaens. 2003. Bioreactive barriers: bioaugmentation and biostimulation for chlorinated solvent remediation. Environ. Sci. Technol. 37:1422-1431. [Google Scholar]
- 18.Löffler, F. E., Q. Sun, J. Li, and J. M. Tiedje. 2000. 16S rRNA gene-based detection of tetrachloroethene-dechlorinating Desulfuromonas and Dehalococcoides species. Appl. Environ. Microbiol. 66:1369-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Löffler, F. E., J. M. Tiedje, and R. A. Sanford. 1999. Fraction of electrons consumed in electron acceptor reduction (Fe) and hydrogen threshold as indicators of halorespiratory physiology. Appl. Environ. Microbiol. 65:4049-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maillard, J., W. Schumacher, F. Vazquez, C. Regeard, W. R. Hagen, and C. Holliger. 2003. Characterization of the corronoid iron-sulfur protein tetrachloroethene reductive dehalogenase of Dehalobacter restrictus. Appl. Environ. Microbiol. 69:4628-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Major, D. W., M. L. McMaster, E. E. Cox, E. A. Edwards, S. M. Dworatzek, E. R. Hendrickson, M. G. Starr, J. A. Payne, and L. W. Buonamici. 2002. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ. Sci. Technol. 36:5106-5116. [DOI] [PubMed] [Google Scholar]
- 23.Maymó-Gatell, X., Y.-T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 24.Maymó-Gatell, X., I. Nijenhuis, and S. H. Zinder. 2001. Reductive dechlorination of cis-dichloroethene and vinyl chloride by “Dehalococcoides ethenogenes.” Environ. Sci. Technol. 35:516-521. [DOI] [PubMed] [Google Scholar]
- 25.Mohn, W. W., and J. M. Tiedje. 1992. Microbial reductive dehalogenation. Microbiol. Rev. 56:482-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Müller, J. A., B. M. Rosner, G. von Abendroth, G. Meshulam-Simon, P. L. McCarty, and A. M. Spormann. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70:4880-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann, A., G. Wohlfarth, and G. Diekert. 1998. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J. Bacteriol. 180:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okeke, B. C., Y. C. Chang, M. Hatsu, T. Suzuki, and K. Takamizawa. 2001. Purification, cloning, and sequencing of an enzyme mediating the reductive dechlorination of tetrachloroethylene (PCE) from Clostridium bifermentans DPH-1. Can. J. Microbiol. 47:448-456. [PubMed] [Google Scholar]
- 28.Regeard, C., J. Maillard, and C. Holliger. 2004. Development of degenerate and specific PCR primers for the detection and isolation of known and putative chloroethene reductive dehalogenase genes. J. Microbiol. Methods 56:107-118. [DOI] [PubMed] [Google Scholar]
- 29.Roberts, A. L., L. A. Totten, W. A. Arnold, D. R. Burris, and T. J. Campbell. 1996. Reductive elimination of chlorinated ethylenes by zero valent metals. Environ. Sci. Technol. 30:2654-2659. [Google Scholar]
- 30.Smidt, H., A. D. L. Akkermans, J. van der Oost, and W. M. de Vos. 2000. Halorespiring bacteria—molecular characterization and detection. Enzyme Microb. Technol. 27:812-820. [DOI] [PubMed] [Google Scholar]
- 31.Suyama, A., M. Yamashita, S. Yoshino, and K. Furukawa. 2002. Molecular characterization of the PceA reductive dehalogenase of Desulfitobacterium sp. strain Y51. J. Bacteriol. 184:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL-X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Pas, B. A., H. Smidt, W. R. Hagen, J. van der Oost, G. Schraa, A. J. M. Stams, and W. M. de Vos. 1999. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J. Biol. Chem. 274:20287-20292. [DOI] [PubMed] [Google Scholar]
- 34.van Pee, K. H., and S. Unversucht. 2003. Biological dehalogenation and halogenation reactions. Chemosphere 52:299-312. [DOI] [PubMed] [Google Scholar]
- 35.Villemur, R., M. Saucier, A. Gauthier, and R. Beaudet. 2002. Occurrence of several genes encoding putative reductive dehalogenases in Desulfitobacterium hafniense/frappieri and Dehalococcoides ethenogenes. Can. J. Microbiol. 48:697-706. [DOI] [PubMed] [Google Scholar]
- 36.Vogel, T. M., C. S. Criddle, and P. L. McCarty. 1987. Transformations of halogenated aliphatic-compounds. Environ. Sci. Technol. 21:722-736. [DOI] [PubMed] [Google Scholar]
- 37.Zhou, J., M. E. Davey, J. B. Figueras, E. Rivkina, D. Gilichinsky, and J. M. Tiedje. 1997. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil. Microbiology 143:3913-3919. [DOI] [PubMed] [Google Scholar]