Abstract
Black band disease (BBD) is a virulent polymicrobial disease primarily affecting massive-framework-building species of scleractinian corals. While it has been well established that the BBD bacterial mat is dominated by a cyanobacterium, the quantitative composition of the BBD bacterial mat community has not described previously. Terminal-restriction fragment length polymorphism (T-RFLP) analysis was used to characterize the infectious bacterial community of the bacterial mat causing BBD. These analyses revealed that the bacterial composition of the BBD mat does not vary between different coral species but does vary when different species of cyanobacteria are dominant within the mat. On the basis of the results of a new method developed to identify organisms detected by T-RFLP analysis, our data show that besides the cyanobacterium, five species of the division Firmicutes, two species of the Cytophaga-Flexibacter-Bacteroides (CFB) group, and one species of δ-proteobacteria are also consistently abundant within the infectious mat. Of these dominant taxa, six were consistently detected in healthy corals. However, four of the six were found in much higher numbers in BBD mats than in healthy corals. One species of the CFB group and one species of Firmicutes were not always associated with the bacterial communities present in healthy corals. Of the eight dominant bacteria identified, two species were previously found in clone libraries obtained from BBD samples; however, these were not previously recognized as important. Furthermore, despite having been described as an important component of the pathogenetic mat, a Beggiatoa species was not detected in any of the samples analyzed. These results will permit the dominant BBD bacteria to be targeted for isolation and culturing experiments aimed at deciphering the disease etiology.
Coral reefs are among the most biologically diverse ecosystems on earth and harbor a large number of unique marine taxa. However, over the last few decades, 27% of coral reefs have been destroyed worldwide, and in places such as Belize, up to 75% of the coral reef habitat has been lost (38). The destruction of coral reef ecosystems is a complex phenomenon and can be attributed to a combination of factors (16, 19, 30). One of the leading causes of reef degradation has been a dramatic increase in reef coral mortality due to disease (30). While historically the study of coral diseases and their role in the ecology of reef ecosystems has been minimal, with increases in both the number and severity of coral diseases, this topic has rapidly gained scientific attention (26, 30). However, while the number of newly described coral diseases has steadily increased, in most cases their etiology has remained unknown (26).
Black band disease (BBD) is one of the most widespread and destructive coral diseases. This is largely due to its preferential infection of the massive-framework-building corals that serve as ecological cornerstones of the reef ecosystem (11). Antonius first described BBD in the 1970s as a black bacterial mat that migrates in a top-down manner across the surface of the coral, destroying the tissue and leaving behind bare skeleton (5, 15). After the tissue has been stripped from the coral surface, algae rapidly colonize the naked skeleton, preventing any subsequent recovery from the infection. Black band disease has been extremely devastating to certain massive corals, suggesting that certain species of coral are more immune to BBD infection than others (3).
BBD is a polymicrobial disease presumably caused by a consortium of microorganisms rather than a single pathogen (26, 28). Early descriptions of the disease, based solely on optical or electron microscopy, reported that the BBD mat consortium was dominated by a filamentous, nonheterocystous, cyanobacterium first classified as Phormidium corallyticum (31, 35). Further optical microscopy studies revealed that there were other organisms accompanying P. corallyticum in the BBD mat: the motile sulfide-oxidizing bacterium Beggiatoa, the sulfate-reducing bacterium Desulfovibrio, numerous heterotrophic bacteria, and marine fungi (10, 15, 25).
Molecular techniques have now shown that the composition of the bacterial community living in the infectious mat is significantly more complex than was initially recognized. As many as 64 different species of bacteria have been identified living in the BBD mat (9, 14). These methods have confirmed the presence of Desulfovibrio and related species in the infectious community but have not been able to verify the presence of Beggiatoa, one of the organisms previously described as a major component of the BBD bacterial mat (9, 14). On the basis of the known 16S rRNA gene sequences, the primers used in these studies should not have had any problem amplifying sequences from Beggiatoa spp. (9).
Molecular analysis of BBD has given new insights into the nature of the cyanobacteria present in the BBD mat. Results using specific primers revealed that the dominant cyanobacteria within BBD mat samples belong to at least three different taxa, despite producing similar patterns and symptoms of disease (13). However, since these species of cyanobacteria have not been formally classified, the name P. corallyticum refers to any of the different filamentous cyanobacteria present in the BBD mat. While there exists a general consensus as to the importance of P. corallyticum during disease development, the reasons to consider it the primary pathogen remain largely circumstantial (9, 28).
To understand the process of infection and to elucidate the possible environmental factors involved in its development, it is first necessary to identify those organisms that play a crucial role in the disease. The aim of the present study is to characterize the composition of the dominant and potentially important bacteria present in the BBD bacterial mat. Terminal-restriction fragment length polymorphism (T-RFLP) analysis was used to identify the most abundant bacteria present in BBD mats. Once identified, the most-probable-number PCR (MPN-PCR) technique was used to quantify the abundance of those species.
Results show that the species of coral being infected does not affect the composition of the infectious bacterial mat. Instead, differences in the mat composition appear to be linked to the species of cyanobacteria dominant in the infection. Quantification of the most abundant organisms present in the infectious mat shows that there are three major groups of bacteria that accompany the cyanobacterium: (i) members of the division Firmicutes (five species), (ii) Cytophaga-Flexibacter-Bacteroides (CFB) group (two species), and (iii) δ-proteobacteria (one species). Of the eight species identified using specific primers, two were not consistently detected in healthy corals, making them ideal candidates for future studies on the etiology of the disease. Other species, while consistently detected, were present at much lower numbers than those found during infection. The combined results of T-RFLP and quantification suggest that the presence of the filamentous cyanobacterium is the driving force that controls the final composition of the BBD mat.
MATERIALS AND METHODS
Fieldwork and sample collection.
BBD mat samples were collected from infected corals at three different locations along the southwestern coast of Curaçao, Netherlands Antilles, in the Caribbean Sea, and at two different locations on the northern coast of New Britain, Papua New Guinea, in the Indo-Pacific Sea. The three sites on Curaçao included a sea aquarium, a water plant, and Playa Kalki (13). At each of these three locations, both healthy and BBD-infected colonies of Montastrea annularis and Diploria strigosa were collected. On New Britain, Papua New Guinea, BBD mat samples of infected colonies of Porites lutea were collected off the northern coast at Father's Reef (13). All samples were collected from a water depth between 4 and 5 m. Small 4-cm2 sections of mat and coral skeleton were individually collected using a small chisel and placed in a sterile disposable 50-ml polypropylene centrifuge tube while using standard scuba techniques to access the reefs. Portions of BBD mats that could be physically peeled off the coral surface using forceps were placed in their own sterile disposable 15-ml polypropylene centrifuge tube. Immediately upon return to shore, the seawater within each tube was decanted, and coral samples were immersed in 80% ethanol for molecular analyses. Live D. strigosa specimens were also collected in Curaçao, transported to the lab in Illinois, and kept alive in an aquarium. One of these colonies developed BBD and was processed as described above.
DNA extraction.
DNA extraction was performed as described previously (14). Bead beating and freeze-thaw cycling protocols were used to extract community genomic DNA from the cells collected from the crushed coral slurries. The same procedure was performed in parallel using ultrapure water as a negative control and 50 μl of an Escherichia coli culture that had been allowed to grow overnight as a positive control.
PCR and T-RFLP analyses.
Specific primers to amplify 16S rRNA genes from cyanobacteria were used (24) to characterize the cyanobacterium infecting the specimen of D. strigosa that developed BBD spontaneously in the aquarium. The amplified products were cloned and sequenced as described below. The cyanobacteria infecting corals sampled from the environment had been characterized previously (13).
T-RFLP analysis was performed by the method of Liu et al. (20). Universal bacterial oligonucleotide primers used in the PCR amplifications were obtained from Integrated DNA Technologies (Coralville, Iowa): forward primer Univ9F (5′-GAGTTTGATYMTGGCTC) with a 5′ carboxyfluorescine (FAM) label and reverse primer Univ1509R (5′-GYTACCTTGTTACGACTT). Reaction mixtures included the following: 10 μl of TaqMaster buffer (an agent which improves thermostability and processivity of the polymerase), 5 μl of Taq reaction buffer containing 15 mM MgCl2 (Eppendorf, Westbury, N.Y.), 4 μl of a mixture of deoxynucleoside triphosphates (2.5 mM concentration of each deoxynucleoside triphosphate) (Gibco/BRL, Rockville, Md.), 1 μl each of forward and reverse primers (200 ng each), 5 to 30 μl of the sample preparation, and water to bring the total volume to 49.5 μl. An initial denaturation or hot start of 5 min at 95°C was followed by the addition of 0.5 μl (2 U) of MasterTaq polymerase (Eppendorf). The hot start was followed by 30 or 35 cycles of the following incubation pattern: 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min. A final extension step of 5 min at 72°C concluded the reaction.
PCR products were combined to a final volume of 300 μl, purified using the Wizard PCR prep kit (Promega, Madison, Wis.), and eluted with 30 μl of water. Clean DNA (10 μl) was aliquoted into each of three tubes, and restriction digestions were performed with RsaI, HhaI, and MspI. The final volume of each digest was 20 μl. At least 300 ng of total DNA per digestion was used. Tubes were first incubated overnight at 37°C, covered with aluminum foil to prevent photobleaching of the FAM label, and kept at −20°C.
Prior to loading on a gel, 1 μl of sample was added to a loading buffer consisting of 1.25 μl of deionized formamide, 0.25 μl of blue loading dye, and 0.3 μl of size standard (tetrachloro-6-carboxyfluorescine [TAMRA] 2500). The samples were then mixed and denatured at 95°C for 3 min prior to loading onto a 5% Long Ranger (proprietary acrylamide formula from BioWhittaker) and 7 M urea gel. Analyses were run on a 377-XL sequencer (Applied Biosystems, Foster City, Calif.) with a run time of 5 h. The data were analyzed using the ABI GeneScan software. T-RFLP profiles were analyzed using the TAP T-RFLP program at the Ribosomal Database Project II website (http://rdp.cme.msu.edu/html/analyses.html).
Cloning and sequencing.
To identify the most abundant peaks from the T-RFLP analysis, digested products were purified by electrophoresis in a 6% polyacrylamide gel under native conditions in a SE 600 series vertical slab gel unit from Hoefer Scientific Instruments (San Francisco, Calif.). One lane in each gel contained the 100-bp DNA size standard ladder available from Invitrogen (Carlsbad, Calif.). The DNA bands that were selected had sizes that corresponded approximately to the results obtained from the T-RFLP analysis (error margin, 50 bp).
To elute the DNA from the acrylamide, 200 μl of crush and soak solution (0.5 M ammonium acetate, 0.1% sodium dodecyl sulfate, 0.1 mM EDTA) was added to the excised band, and the acrylamide was crushed and incubated overnight at 37°C. On the following day, the solid acrylamide was concentrated via centrifugation for 10 min at 16,000 × g. The supernatant was transferred to a clean tube, and another 200 μl of crush and soak solution was added to the solid acrylamide pellet. Following centrifugation, the recovered supernatant was pooled with the initial fraction. DNA was then precipitated and recovered. The DNA fragments obtained from HhaI and MspI digestions were blunted using T4 DNA polymerase (Invitrogen) or the large Klenow polymerase subunit (Invitrogen), following the manufacturer's instructions.
In order to clone the product into pGEM-T vector (Promega), the DNA was incubated for 10 min at 72°C in the presence of Taq polymerase and dATP (2.5 mM). The gel-purified PCR product was then cloned into pGEM-T and transformed into competent DH5αMCR E. coli cells using the manufacturer's instructions and standard techniques (32). Clones were checked for the presence of the insert by PCR using the primers M13 (−21) and T7 (−26). A RFLP analysis of the products was performed to select for clones that presented identical patterns. PCR products were digested with MspI and HinP1 enzymes and analyzed in a 1.6% Metaphor agarose gel. Only clones with the correct sizes and present in more than one RFLP analysis were selected for sequence analysis. These clones were transferred to petri dishes containing Luria broth agar supplemented with 100 μg of ampicillin (Roche Molecular Biochemicals, Indianapolis, Ind.) per ml and incubated overnight at 37°C.
Inoculation, culturing, template preparation, and sequencing were performed by the High Throughput Laboratory of the University of Illinois W. M. Keck Center for Comparative and Functional Genomics. The petri cultures were used to inoculate 2-ml 96-well culture blocks containing Circle Grow medium (Bio 101 Inc., Vista, Calif.) supplemented with ampicillin. Plasmid template DNA was purified from the cultures using an automated system and the QIAwell 96 Turbo prep BioRobot kit (QIAGEN, Valencia, Calif.). The first round of sequencing was completed using the T7 (−26) primer and Big Dye Terminator chemistry (version 2.0) from Applied Biosystems. Sequencing was performed on an ABI 3700 capillary sequencer and then processed in the Bioinformatics Unit of the W. M. Keck Center.
MPN-PCR and dot blot analyses.
MPN-PCR quantification was performed as follows. First, eight primer sets specific for the different bacterial species identified were created (Table 1). Additionally, a primer pair specific for the 16S rRNA gene of cyanobacterium CD1C11 (GenBank accession number AY038527) (13) was also used for the MPN-PCR experiments. Primer sequence specificity was tested using Probe Match at the Ribosomal Database Project II website (http://rdp.cme.msu.edu/html/index.html). The primer pairs were tested for the ability to amplify the 16S rRNA gene from the different clones isolated during this project. No cross-reactivity was detected. To assess the sensitivity of PCR amplification, we performed experiments using different concentrations of E. coli and universal primers for the 16S rRNA gene. Successful amplification occurred with as little as 10 to 50 cells. E. coli has seven copies of the 16S rRNA gene in its genome (17); therefore, the sensitivity for the number of DNA molecules of 16S rRNA ranges from 70 to 350 copies of DNA. For the MPN-PCR analysis, the sensitivity was determined to be 350 copies of 16S rRNA DNA.
TABLE 1.
Primers used to amplify the specific 16s rRNA sequences for MNP-PCR analysis
| Clone | Directiona | Primer sequence |
|---|---|---|
| CD22E1 | Fwd | 5′-CCCGTAGGAGTCTGGTCCGT-3′ |
| Rev | 5′-AGAGGAAGGTCCCCCACACT-3′ | |
| CD22B1 | Fwd | 5′-TTGGAAGCGCAACCCTTG-3′ |
| Rev | 5′-GAGTGCCCAGCATTACCTGC-3′ | |
| CD22E6 | Fwd | 5′-ACTGGGACTGGCTTTTTGGG-3′ |
| Rev | 5′-ACGGGCGGCTAAGGAGTAAT-3′ | |
| R3-M4 | Fwd | 5′-GGAGGATCCGAGCGTTATCC-3′ |
| Rev | 5′-CGCCTACGCACCCTTTAAAC-3′ | |
| CD22B8 | Fwd | 5′-GGATTCGCTAGTAATCGCGC-3′ |
| Rev | 5′-CCGGGAACGTATTCACCG-3′ | |
| CD22E5 | Fwd | 5′-GGGTGAGTAACGCGTGGGTA-3′ |
| Rev | 5′-TCGATGTGTTATCCCCCTGC-3′ | |
| CD22D4 | Fwd | 5′-TCTGACCGTTTCTGTAATGGAAAC-3′ |
| Rev | 5′-CACCTGTCACTTCTGCTCCG-3′ | |
| CD22D5 | Fwd | 5′-CTAACTCCGTGCCAGCAGC-3′ |
| Rev | 5′-CGATTAACGCTCGCACCCT-3′ | |
| CD1C11 | Fwd | 5′-CTGTAGGTGGCCAGCT-3′ |
| Rev | 5′-TTCCCTTCGCAGGTTCGCTGC-3′ |
Fwd, forward; Rev, reverse.
Dot blot analysis was used as a sensitive and rapid means to process a large number of samples. Serial dilutions (1× to 10−4× concentration) of chromosomal DNA from BBD mat samples containing cyanobacterium CD1C11 and three healthy coral samples were prepared in triplicate. The nine specific primers for the different species and cyanobacteria (Table 1) were used to amplify DNA (target DNA) from the dilution series. These PCR products were applied to a nylon membrane (GeneScreen; NEN Life Sciences Products, Boston, Mass.) using a Minifold I sample filtration manifold (Schleicher & Schuell, Keene, N.H.) and bound using a Spectrolinker XL-1500 UV-cross-linker (Spectronics Corp., Westbury, N.Y.).
The probes were synthesized by PCR amplifying 16S rRNA genes from BBD mat samples with the universal bacterial oligonucleotide primers. The probes used for the dot blot analyses were always obtained from a different BBD mat sample than that used to create the target DNA. Probes were labeled and detected using the Renaissance random primer fluorescein labeling kit (Perkin-Elmer Life Sciences, Boston, Mass.) following the manufacturer's instructions. MPN-PCR data were converted to number of 16S rRNA genes using standard MPN tables (37). Concentrations of bacteria present in healthy and BBD coral samples (Table 1) were compared using the Mann-Whitney U test statistic and the SPSS 10.0 statistical package (SPSS Inc., Chicago, Ill.).
Sequence and phylogenetic analyses.
The sequences obtained were compared with the GenBank database using the Basic Local Alignment Search Tool (BLAST) network service (2). Consensus sequences were analyzed using CHIMERA_CHECK version 2.7 at the Ribosomal Database Project II website (http://rdp.cme.msu.edu/html/index.html) (21).
Sequences were aligned using the Clustal X program, and phylogenetic analysis and trees were obtained using the program PAUP* version 4.0b10 (34). Trees were constructed by parsimony and by bootstrapping 10,000 trees from resampled data.
Nucleotide sequence accession numbers.
The sequences of the partial gene fragments identified in this work are deposited in GenBank under accession numbers AY497293 through AY497300.
RESULTS
Characterization of the bacterial communities in different samples of BBD mats.
T-RFLP was used to identify the most abundant organisms present in the BBD bacterial mat. A total of 12 BBD mat samples were analyzed: 2 samples were from infected P. lutea (New Britain), 1 was from M. annularis (Curaçao), and the other 9 were from D. strigosa (Curaçao). An additional sample from D. strigosa living in an aquarium at the University of Illinois was also analyzed.
In order to check the reliability of T-RFLP in our samples, one sample from D. strigosa was analyzed five times, and another three samples from D. strigosa were analyzed in duplicate. Additionally, the sample from M. annularis and one of the samples from P. lutea were also analyzed in duplicate. All repetitions for a given sample provided an identical pattern.
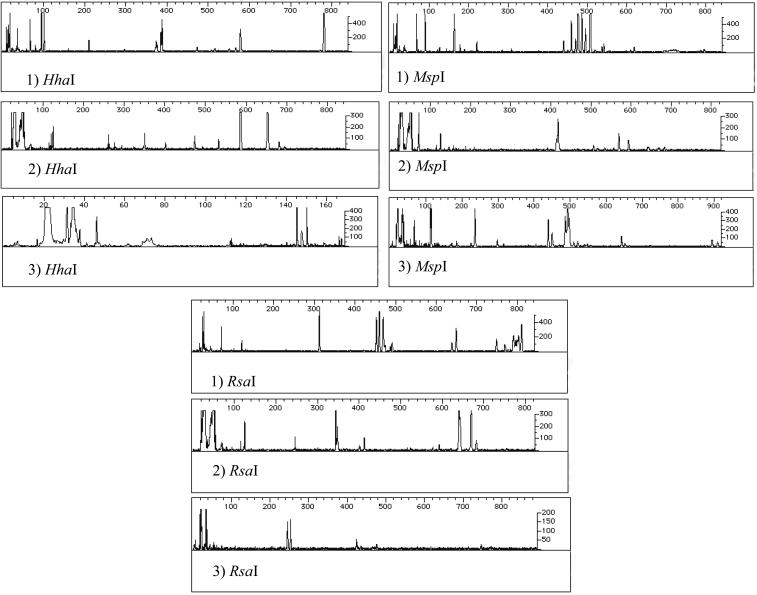
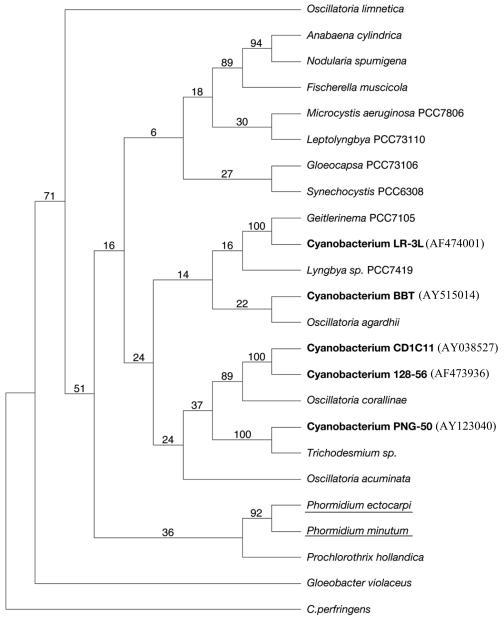
T-RFLP profiles for all BBD samples were the same except for two cases (Fig. 1). The distinct profiles corresponded to a sample of P. lutea taken in New Britain, Papua New Guinea, and the sample of D. strigosa that spontaneously developed BBD in the aquarium. In both cases, the BBD mat was inhabited by a unique species of cyanobacteria. The sample of P. lutea was infected with a species of cyanobacteria previously identified as cyanobacterium PNG-50 (13). The sample in the aquarium that developed BBD was infected by the cyanobacterium BBT. Although different species of cyanobacteria are associated with BBD, all of them are phylogenetically related to marine nonheterocystous filamentous cyanobacteria but not to marine Phormidium species (Fig. 2).
FIG. 1.
T-RFLP analysis of the bacterial communities (see text for details). Profiles: 1) Representative result of 12 samples that contained cyanobacterium CD1C11; 2) representative result of 2 samples that contained cyanobacterium PNG-50; 3) representative result of 2 samples that contained cyanobacterium BBT.
FIG. 2.
Phylogenetic consensus tree based on parsimony analysis of 16S rRNA gene sequences of BBD cyanobacteria (bold type) and other representative cyanobacteria. Clone CD1C11 and cyanobacteria PNG-50, LR-L3, and 128-56 were previously analyzed (13). The numbers at the nodes are the bootstrap values based on a total of 10,000 replicate resamplings. GenBank accession numbers are shown in parentheses. Marine Phormidium spp. are underlined. C. perfringens, Clostridium perfringens.
When cyanobacterial sequence CD1C11 was detected in the BBD mat, the resulting T-RFLP profile was identical regardless of the species of coral infected. Furthermore, comparison of T-RFLP profiles generated from healthy coral samples shows that only one peak is shared between BBD samples and healthy corals (18).
Identification of the predominant bacteria in the BBD mat.
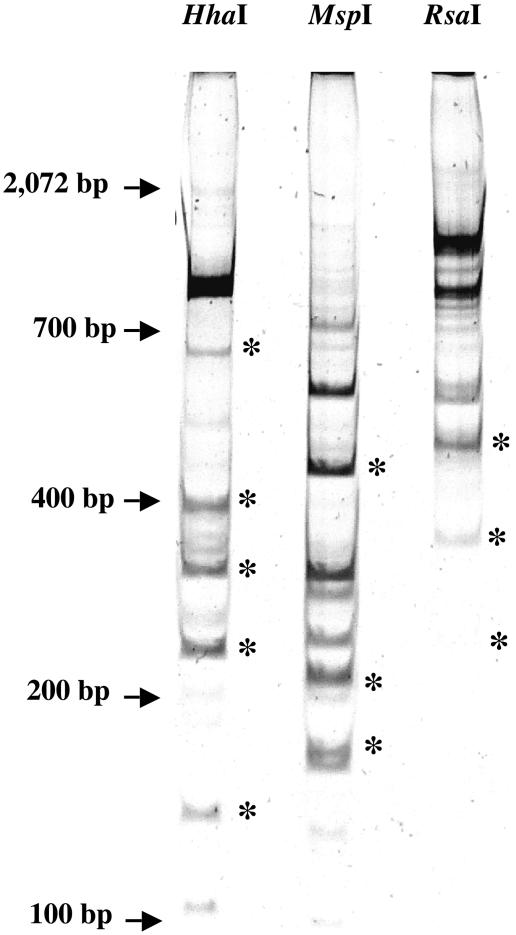
To identify the microorganisms corresponding to the T-RFLP profiles, the patterns were analyzed using the TAP T-RFLP program (21). Unfortunately, no exact matches were found for any of the profiles derived from BBD samples. To overcome this problem, a polyacrylamide gel was used to separate and identify DNA fragments corresponding in size to the peaks obtained from the T-RFLP analysis (Fig. 3). Bands with sizes closely matching those of the peaks identified by T-RFLP were excised, cloned, and sequenced. In all cases, at least four clones from the same band were sequenced. On several occasions, a band derived from one restriction enzyme digest was identical to sequences derived from one or both of the other enzymes. The recurrence of a sequence is a further indication that the bands selected correspond to the most abundant bacteria present in our samples. Eight different sequences were identified (Table 2). No chimeric sequences were detected, and E-values and scores in all cases were lower than E−103.
FIG. 3.
Polyacrylamide gel showing the patterns obtained after digestion with the three different restriction enzymes used for T-RFLP analysis. Bands with sizes observed in all T-RFLP profiles for a given restriction enzyme (*) were excised, cloned, and sequenced. The locations of DNA size standards (not visible) are indicated by arrows to the left of the gel.
TABLE 2.
Best matches of the different sequences obtained from the clone libraries based on the T-RFLP results
| Clone | Length of the band (bp) | GenBank accession no. | Best match (BLASTN) | % Identity/E-value | Division |
|---|---|---|---|---|---|
| CD22E1 | 449 | AY497293 | Uncultured CFB group bacterium partial 16S rRNA gene, clone P. palm C/A 42 | 87/E−103 | CFB |
| CD22B1 | 469 | AY497294 | Uncultured Firmicutes clone CD4B11 | 98/E−138 | Firmicutes |
| CD22E6 | 481 | AY497295 | Uncultured Firmicutes clone CD5D10 | 99/0.0 | Firmicutes |
| R3-M4 | 527 | AY497296 | Cytophaga fermentans | 91/0.0 | CFB |
| CD22B8 | 402 | AY497297 | Uncultured Firmicutes 128-3-6 | 99/E−170 | Firmicutes |
| CD22E5 | 559 | AY497298 | Uncultured Firmicutes clone CD5C11 | 98/0.0 | Firmicutes |
| CD22D4 | 607 | AY497299 | Uncultured bacterium p-1921-s962-3 | 90/0.0 | Firmicutes |
| CD22D5 | 245 | AY497300 | Uncultured δ-proteobacterium 128-9-6 | 99/E−130 | δ-Proteobacteria |
Quantitative analysis of the bacteria present in the BBD mat.
To confirm which of the eight species represented a numerically large fraction of the bacterial mat, the MPN-PCR method was used to quantify the abundance of the species presented in Table 3. This method has been successfully used to quantify microorganisms in different environmental samples (1, 12, 22). To process a larger number of samples simultaneously, the presence of PCR product was checked by DNA dot blot analysis (Table 3). The eight different sequences were detected in all of the samples analyzed. Of these, clones CD22E1, CD22E6, CD22B8, CD22E5, CD22D4, and CD22D5 were also detected in all healthy coral tissue samples; however, clones CD22E1, CD22B1, R3-M4, and CD22B8 were detected at significantly lower concentrations. Furthermore, CD22B1 and R3-M4 were not always detected in the healthy coral tissue (Table 3). Although clones CD22E5 and CD22E6 appeared to be present at higher concentrations in BBD mat samples than in healthy coral samples, due to the large variation in the BBD mat samples, the differences did not prove statistically significant. Clones CD22D4 and CD22D5 were consistently detected at the same level in BBD mat and healthy tissue samples (Table 3).
TABLE 3.
Frequencies of the different bacteria identified as present in high numbers in BBD matsa
| Primer or clone | Division | No. of bacteriab
|
No. of positive results in healthy coral samples (n = 3) | |
|---|---|---|---|---|
| BBD mat (n = 4) | Healthy coral samples (n = 3) | |||
| Universal primers | 100 | 100 | 3 | |
| Clones | ||||
| CD1C11 | 44.40 ± 41.92c | <0.0001 ± 0 | 0 | |
| CD22E1 | CFB | 22.126 ± 20.56c | 0.75 ± 0.36 | 3 |
| CD22B1 | Firmicutes | 6.51 ± 6.27c | 0.26 ± 0.22 | 2 |
| CD22E6 | Firmicutes | 16.84 ± 28.72 | 0.75 ± 0.36 | 3 |
| R3-M4 | CFB | 14.21 ± 8.69c | 0.64 ± 0.55 | 2 |
| CD22B8 | Firmicutes | 17.89 ± 2.21c | 0.75 ± 0.36 | 3 |
| CD22E5 | Firmicutes | 14.42 ± 11.96 | 2.22 ± 1.51 | 3 |
| CD22D4 | Firmicutes | 8.35 ± 7.67 | 8.32 ± 9.45 | 3 |
| CD22D5 | δ-Proteobacteria | 9.05 ± 4.54 | 9.70 ± 8.40 | 3 |
Results were obtained by MPN-PCR and refer to the total number of bacteria obtained using universal primers.
Average (percent respect to the total DNA) ± standard deviation.
Significantly different (P < 0.05) from the value for healthy coral sample as determined by the Mann-Whitney U test.
Microscopy suggests that filamentous cyanobacteria constitute the majority of the BBD biomass (13, 14). However, the use of cyanobacterium-specific primers suggests that the DNA from this group represents less than half of the total DNA present in the samples (Table 3). Three major groups of bacteria besides cyanobacteria are present at high numbers in the infecting BBD mat: CFB, Firmicutes, and δ-proteobacteria. Clone CD22E1, one of the CFB division sequences that was detected in low numbers in healthy coral tissues, was the next most abundant bacterium after the cyanobacteria in the BBD mat. The δ-proteobacterium Desulfovibrio sp. is consistently present in important numbers during infection (9.05%). Nevertheless, it is also present at the same level (9.7%) in the healthy coral samples analyzed (Table 3). This species was previously identified as a part of the BBD mat (7, 28). Three additional species were present in high numbers in BBD mats and at low numbers in healthy corals. CD22B1, a member of the division Firmicutes, was present at relatively low numbers (6.5%) compared with the other species analyzed, and it was detected in only two of three samples. Clone R3-M4 represented 14.21% of the population in the BBD-infected corals but only 0.64% in the healthy corals. Moreover, it was completely absent in one of the healthy coral samples analyzed (Table 3).
DISCUSSION
A filamentous cyanobacterium classified as P. corallyticum, along with Desulfovibrio sp. and Beggiatoa sp., have been proposed as the primary pathogens for BBD (27, 31, 33, 35). However, the evidence supporting this hypothesis is purely circumstantial (9). In that regard, other bacteria have been proposed as possible initiators of the disease (28). This study has identified the most abundant and potentially most important organisms of BBD. These findings will advance future studies of the mechanisms of BBD pathogenesis and roles of the different bacteria in the disease.
Recent work has indicated that healthy corals have a distinct bacterial community associated with them. The composition of that community is dependent upon the species of coral and is not merely a reflection of the bacterial community present in the surrounding water (18, 29). Furthermore, clone libraries obtained from BBD mat samples show that there is a complex and specific bacterial community associated with the pathogenic mats that is distinct from the communities growing on healthy corals (9, 14). On the basis of these clone libraries, it seems that the composition of the bacterial community undergoes a switch from a healthy to diseased profile (9). The relative proportion of clones representing members of the Firmicutes and CFB groups increases in the BBD mat (9, 14). Other groups disappear when healthy corals become diseased (γ- and β-proteobacteria), and finally, there are groups not present in healthy corals that make their appearance in diseased corals (ɛ-proteobacteria) (9, 14). Due to the complexity of the BBD mat, it is difficult to assess which bacteria play an active role in the development of the disease.
After analyzing all of the bands selected from the T-RFLP results, eight different sequences belonging to three different divisions were determined to be important in the BBD mat. The results show that one species of δ-proteobacteria, two species of CFB group, and five species of Firmicutes were the most abundant bacteria present in the infectious mat. Cooney et al. (9) previously identified two of these species in BBD mats. However, an α-proteobacterium that had been described as an important part of the infectious mat (9) has not been detected in the present study.
The first descriptions of the bacterial composition of BBD mats referred to the presence of two major groups of bacteria besides the filamentous cyanobacterium: a sulfate-reducing bacterium presumed to be Desulfovibrio (15) and a sulfide-oxidizing bacterium interpreted to be Beggiatoa (10). There are reports proposing that these two microorganisms play an active role in the destruction of coral tissue (27, 28), including reported oscillations in sulfide and oxygen levels in the BBD mat due to the action of Beggiatoa (7, 28). However, the presence of Beggiatoa has been suggested only on the basis of microscopy (10, 15). In the present study, Beggiatoa was not detected as a member of the infectious bacterial mat. Previous results obtained by different groups also failed to identify Beggiatoa as a part of the BBD mat (9, 14), and as mentioned previously, the primers used in these studies should have recognized the 16S rRNA genes from Beggiatoa spp. (9). A lingering possibility is that Beggiatoa exists at cell counts below PCR detection limits. If Beggiatoa is not present, the oxidation of sulfide in the BBD mat would need to be explained by the activity of other organisms. Previous work consistently detected the presence of an ɛ-proteobacterium in the BBD samples, although it probably represents a small fraction of the total community (9, 14). These species of ɛ-proteobacterium were not present in healthy corals. Marine ɛ-proteobacteria are capable of oxidizing sulfide into sulfate (39), which could explain the observations made by other researchers regarding the sulfide/sulfate profiles observed in BBD and attributed to Beggiatoa (7, 28).
The sulfate-reducing bacterium Desulfovibrio was identified both optically and by using 16S rRNA-targeted oligonucleotide probes (4, 15, 33). The species of δ-proteobacteria we have identified as an important part of the BBD mat is nearly identical to δ-proteobacteria clone 128-9-6, which has high homology with Desulfovibrio species (9). Nonetheless, the same species of δ-proteobacteria was also present in all the healthy coral samples analyzed and at levels comparable to those observed during infection.
One member of the CFB group, clone R3-M4, was not consistently found in the healthy coral samples, although it represented a large fraction of DNA isolated from the BBD mat (14.2%). This species has homology with Cytophaga fermentans, and similar sequences were previously identified in BBD clone libraries (14). Cooney et al. have also found that there is a Cytophaga species present in BBD but not in healthy coral samples (9). The species identified by those researchers seems to be different from the species we have identified. The CFB group includes well-known pathogens that infect a variety of organisms inhabiting many different environments. For example, the CFB group includes species that are responsible for a number of diseases in freshwater and saltwater fish (6, 8, 23, 36). The other CFB group species identified as important in BBD (clone CD22E1) makes up an important fraction of the bacterial community, up to 22.12%. This clone was present only at low levels on healthy corals (0.75%), indicating that BBD disease is somehow advantageous for its growth.
Five species of the division Firmicutes were identified; each species constituted an important fraction of the infectious mat. Four of five species of Firmicutes present in healthy coral samples increased their relative number during infection. Clone CD22B1 has homology with clone CD4B11 (GenBank accession number AY038526), an uncultured firmicute previously identified in BBD samples, but not in healthy corals using clone libraries (16). This clone was present in only two of three healthy coral samples analyzed. However, it may not be important for the progression of the disease, since it is found in widely variable proportions (from 0.50 to 15.33%) during infection.
Clone CD22B8 has homology with clone 128-3-6 (GenBank accession number AF473927), an uncultured member of the Firmicutes that was previously identified in BBD samples by other researchers (10). Although present in all healthy coral samples, there was a 24-fold increase in its relative proportion from healthy to diseased communities.
Finally, the cyanobacteria present in the BBD-infected coral samples are completely absent in the healthy coral samples analyzed. This further confirms the importance of the cyanobacteria in the development of the disease.
The complex community of microorganisms present in BBD mats has made it difficult to understand the onset and progression of the disease. The strategy of comparative T-RFLP analysis, cloning, and sequencing used in this study has identified a subset of bacterial species likely to play important roles in the etiology of BBD. Having identified these species, classical methods of culture may be employed to determine how each may contribute to the disease. This same strategy could be applied in the study of other diseases or to analyze microbial activity in complex systems.
Acknowledgments
We thank the Office of Navel Research (ONR-N00014-00-1-0609) for support of this research.
The conclusions of this study are those of the authors and do not necessarily reflect those of the funding agency.
REFERENCES
- 1.Alam, M. J., K. I. Tomochika, S. I. Miyoshi, and S. Shinoda. 2002. Environmental investigation of potentially pathogenic Vibrio parahaemolyticus in the Seto-Inland Sea, Japan. FEMS Microbiol. Lett. 208:83-87. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Antonius, A. 1981. The “band” diseases in coral reefs, p. 7-14. In E. D. Gomez et al. (ed.), The reef and man. Proceedings of the Fourth International Coral Reef Symposium, vol. 2. Marine Sciences Center, University of the Philippines, Quezon City. [Google Scholar]
- 4.Antonius, A. 1977. Coral mortality in reefs: a problem for science and management. In D. L. Taylor (ed.), Geology. Proceedings of the Third International Coral Reef Symposium, vol. 2. Rosenstiel School of Marine and Atmospheric Science, Miami, Fla.
- 5.Antonius, A. 1973. New observations on coral destruction in reefs, p. 3. Proceedings of the 10th Meeting of the Association of Island Marine Laboratories, vol. 10. Caribbean University of Puerto Rico, Mayaguez.
- 6.Bader, J. A., C. A. Shoemaker, and P. H. Klesius. 2003. Rapid detection of columnaris disease in channel catfish (Ictalurus punctatus) with a new species-specific 16-S rRNA gene-based PCR primer for Flavobacterium columnare. J. Microbiol. Methods 52:209-220. [DOI] [PubMed] [Google Scholar]
- 7.Carlton, R. G., and L. L. Richardson. 1995. Oxygen and sulfide dynamics in a horizontally migrating cyanobacterial mat: black band disease of corals. FEMS Microbiol. Ecol. 18:155-162. [Google Scholar]
- 8.Cipriano, R. C., L. A. Ford, and J. D. Teska. 1995. Association of Cytophaga psychrophila with mortality among eyed eggs of Atlantic salmon (Salmo salar). J. Wildlife Dis. 31:166-171. [DOI] [PubMed] [Google Scholar]
- 9.Cooney, R. P., O. Pantos, M. D. Le Tissier, M. R. Barer, A. G. O'Donnell, and J. C. Bythell. 2002. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 4:401-413. [DOI] [PubMed] [Google Scholar]
- 10.Ducklow, H. W., and R. Mitchell. 1979. Observations on naturally and artificially diseased tropical corals: a scanning electron microscopy study. Microb. Ecol. 5:215-223. [DOI] [PubMed] [Google Scholar]
- 11.Edmunds, P. J. 1991. Extent and effect of black band disease on a Caribbean reef. Coral Reefs 10:161-165. [Google Scholar]
- 12.Fredslund, L., F. Ekelund, C. S. Jacobsen, and K. Johnsen. 2001. Development and application of a most-probable-number-PCR assay to quantify flagellate populations in soil samples. Appl. Environ. Microbiol. 67:1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frias-Lopez, J., G. T. Bonheyo, Q. Jin, and B. W. Fouke. 2003. Cyanobacteria associated with coral black band disease in Caribbean and Indo-Pacific reefs. Appl. Environ. Microbiol. 69:2409-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frias-Lopez, J., A. L. Zerkle, G. T. Bonheyo, and B. W. Fouke. 2002. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 68:2214-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrett, P., and H. Ducklow. 1975. Coral diseases in Bermuda. Nature 253:349-350. [Google Scholar]
- 16.Hughes, T. P., A. H. Baird, D. R. Bellwood, M. Card, S. R. Connolly, C. Folke, R. Grosberg, O. Hoegh-Guldberg, J. B. Jackson, J. Kleypas, J. M. Lough, P. Marshall, M. Nystrom, S. R. Palumbi, J. M. Pandolfi, B. Rosen, and J. Roughgarden. 2003. Climate change, human impacts, and the resilience of coral reefs. Science 301:929-933. [DOI] [PubMed] [Google Scholar]
- 17.Keener, J., and M. Nomura. 1996. Regulation of ribosome synthesis, p. 1417-1431. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 18.Klaus, J. S., J. Frias-Lopez, G. T. Bonheyo, J. M. Heikoop, and B. W. Fouke. Bacterial communities inhabiting the healthy tissues of two Caribbean reef corals: interspecific and spatial variation. Coral Reefs, in press.
- 19.Knowlton, N. 2001. The future of coral reefs. Proc. Natl. Acad. Sci. USA 98:5419-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantynen, V., S. Niemela, S. Kaijalainen, T. Pirhonen, and K. Lindstrom. 1997. MPN-PCR-quantification method for staphylococcal enterotoxin c1 gene from fresh cheese. Int. J. Food Microbiol. 36:135-143. [DOI] [PubMed] [Google Scholar]
- 23.Nomura, S. 1997. Recent knowledge on fish pathogenic bacteria Aeromonas salmonicida, Listonella anguillara and Cytophaga columnaris, and their virulence factors. Nippon Saikingaku Zasshi 52:393-416. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 24.Pichel, F., and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos-Flores, T. 1983. Lower marine fungus associated with black line disease in star corals (Montastrea annularis). Biol. Bull. 165:429-435. [DOI] [PubMed] [Google Scholar]
- 26.Richardson, L. L. 1998. Coral disease: what is really known? Trends Ecol. Evol. 13:438-443. [DOI] [PubMed] [Google Scholar]
- 27.Richardson, L. L. 1996. Horizontal and vertical migration patterns of Phormidium corallyticum and Beggiatoa spp. associated with black-band disease of corals. Microb. Ecol. 32:323-335. [DOI] [PubMed] [Google Scholar]
- 28.Richardson, L. L., K. G. Kuta, S. Schnell, and R. G. Carlton. 1997. Ecology of the black band disease microbial consortium, p. 597-600. Proceedings of the Eighth International Coral Reef Symposium, vol. 1. Smithsonian Tropical Research Institute, Balboa, Panama.
- 29.Rohwer, F., V. Segritan, F. Azam, and N. Knowlton. 2002. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243:1-10. [Google Scholar]
- 30.Rosenberg, E., and Y. Ben-Haim. 2002. Microbial diseases of corals and global warming. Environ. Microbiol. 4:318-326. [DOI] [PubMed] [Google Scholar]
- 31.Rützler, K., and D. L. Santavy. 1983. The black band disease of Atlantic reef corals. I. Description of the cyanophyte pathogen. Mar. Ecol. 4:301-319. [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Schnell, S., B. Assmus, and L. L. Richardson. 1996. Role of sulfate-reducing bacteria in the black band disease of corals. Abstr. Annual Meeting of the VAAM (Vereinigung fuer Allgemeine und Angewandte Mikrobiologie) and GBCH (Gesellschaft fuer Biologische Chemie), abstr.
- 34.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 35.Taylor, D. 1983. The black band disease of Atlantic reef corals. II. Isolation, cultivation, and growth of Phormidium corallyticum. Pubblicazioni della stazione zoologica di Napoli I. Marine Ecol. 4:320-328. [Google Scholar]
- 36.Toncheva-Panova, T., and J. Ivanova. 2000. Influence of physiological factors on the lysis effect of Cytophaga on the red microalga Rhodella reticulata. J. Appl. Microbiol. 88:358-363. [DOI] [PubMed] [Google Scholar]
- 37.U.S. Department of Agriculture. 2003. Microbiology laboratory guidebook. Most probable number tables. Appendix 2.02. Office of Public Health and Science, Food Safety and Inspection Service, U.S. Department of Agriculture, Atlanta, Ga.
- 38.Wilkinson, E. B. C. (ed.). 2003. Status of coral reefs of the world: 2002. Australian Institute of Marine Science, Cape Ferguson, Townsville,Queensland, Australia.
- 39.Wirsen, C. O., S. M. Sievert, C. M. Cavanaugh, S. J. Molyneaux, A. Ahmad, L. T. Taylor, E. F. DeLong, and C. D. Taylor. 2002. Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]