Abstract
We have developed a new method based on the sublimation of adenine from Escherichia coli to estimate bacterial cell counts in natural samples. To demonstrate this technique, several types of natural samples, including beach sand, seawater, deep-sea sediment, and two soil samples from the Atacama Desert, were heated to a temperature of 500°C for several seconds under reduced pressure. The sublimate was collected on a cold finger, and the amount of adenine released from the samples was then determined by high-performance liquid chromatography with UV absorbance detection. Based on the total amount of adenine recovered from DNA and RNA in these samples, we estimated bacterial cell counts ranging from ∼105 to 109 E. coli cell equivalents per gram. For most of these samples, the sublimation-based cell counts were in agreement with total bacterial counts obtained by traditional DAPI (4,6-diamidino-2-phenylindole) staining.
Direct counting methods using DNA-specific fluorochromes, such as 3,6-bis(dimethylamino)acridinium chloride (acridine orange [AO]) and DAPI (4,6-diamidino-2-phenylindole), have traditionally been used to count total microbial cells in aquatic samples (11, 19). DAPI specifically binds to double-stranded DNA, emitting a blue fluorescence when excited by 365-nm UV light, while AO-stained single-stranded DNA and RNA emit an orange-red fluorescence with an excitation maximum of approximately 470 nm, enabling nucleoid-containing cells (NuCC) to be identified (13). DAPI has been rapidly replacing AO as the bacterial stain of choice for a wide range of sample types (13) and is particularly useful for quantifying the total number of nonviable and viable but nonculturable bacteria in natural samples. However, DAPI does not stain bacteria with intact cell membranes that do not contain a visible nucleoid region (non-NuCC) and is less specific for DNA than previously thought (13, 24). Moreover, the enumeration of bacteria in soils and sediments using this method can be difficult due to high background fluorescence and nonspecific staining (13). It is also important to note that any nonstainable organic compounds present in the soil and sediment derived from the degradation of living bacterial cells, such as nucleobases or amino acids, would not be detected by these staining methods.
Previous studies investigated a method to isolate both amino acids and nucleobases from Escherichia coli and other natural samples using a sublimation pyrolysis technique (8-10). In these experiments, the samples were heated at 450 to 500°C for at least 30 s under reduced pressure. Based on numerous tests with pure amino acid and nucleobase standards as well as natural samples, it was found that the maximum sublimation yields were obtained under these heating conditions (8, 9). After sublimation was complete, the residue on the end of the cold finger was dissolved in water and analyzed for amino acids and nucleobases using high-performance liquid chromatography (HPLC).
Although amino acids are the single most abundant compounds in E. coli cells, comprising 55% of the total dry cell weight (18), it was found that most of the amino acids originally present in the bacterial cells (>98%) did not undergo sublimation and were destroyed during heating (10). Even though nucleic acids are less abundant than amino acids, accounting for only 24% of the dry E. coli cell weight (18), purines and pyrimidines are much more resistant to thermal degradation than protein-bound amino acids and readily sublimate directly from native E. coli DNA and RNA when the cells are heated (9). A previous study found that the recovery of adenine from DNA and RNA in E. coli after sublimation (∼90 to 100% of the theoretical amount) was much higher than that of the other nucleobases present in E. coli (9); therefore, adenine was used as a proxy for estimating the bacterial nucleic acid content in natural samples.
In order to use the sublimation recovery of adenine from E. coli to estimate bacterial cell equivalents in natural samples, we have made several assumptions: (i) the efficiency of the sublimation of adenine from E. coli (∼10%) in the serpentine soil analogue is similar to that in the natural biofilm-type environments represented by the samples tested in this study, (ii) E. coli is representative of the types and genome sizes of the bacteria present in the natural samples, and (iii) all of the adenine sublimed from the samples was derived from bacterial DNA and RNA. Each of these assumptions will be critically evaluated in Results and Discussion.
In the present study we describe a new counting method, based on the sublimation of adenine from E. coli, to estimate total bacterial abundances in natural samples. In this technique, total cell counts measured in E. coli cell equivalents (ECE) can be determined by a measurement of the amount of adenine that is sublimed from intact or degraded bacterial DNA and RNA in the samples after heating. In contrast to the fluorescent staining methods that count only cells containing discrete bundles of intact DNA, the sublimation method also accounts for intact or degraded nucleic acids from dead cells. Therefore, the sublimation method of counting could be particularly useful for estimating total bacterial cell abundances in samples containing mostly dead cells and degraded organic matter. It has recently been shown that dead cells represent the largest percentage (∼70%) of total bacterial cells (dead plus living cells) in coastal marine sediments (15).
MATERIALS AND METHODS
E. coli sample preparation and cell count determination.
All glassware used in the analyses was annealed at 500°C overnight. A crushed serpentine (hydrated magnesium silicate) sample that was previously used as a martian soil analogue material (9) was heated at 500°C for 3 h to remove any organic contaminants and then inoculated with E. coli bacteria by the following procedure. E. coli (strain MG1655) cells were grown in glass tubes with 500 mg of serpentine by shaking at 250 rpm overnight in 10 ml of Luria-Bertani (LB) medium (23) at 37°C in a water bath. In addition, E. coli cells were grown in LB medium inside glass tubes that did not contain serpentine. After overnight growth, a 0.5-ml aliquot of the LB medium was diluted 1:10 in culture medium, transferred into a quartz cuvette (1-cm path length), and the optical density at 460 nm (OD460) was then measured using an HP 8452A diode array spectrophotometer. The E. coli cell concentration in the medium was calculated from the OD reading using an extinction coefficient of 108 cells per OD460 absorbance unit (18).
The remaining LB growth medium was centrifuged for 10 min at 5,000 rpm in a Falcon plastic tube to pellet the cells. The medium was decanted from the tube, the inoculated serpentine sample was resuspended in 1.5 ml of potassium-phosphate-buffered saline (KPBS) and centrifuged at 6,000 rpm for 2 min, and the supernatant was removed. The KPBS washing procedure was repeated three more times in order to completely remove the LB medium from the sample. After rinsing, a thin bacterial film coating the top of the serpentine sample was homogenized by mixing with a sterile spatula. The inoculated serpentine sample was transferred into a capped Eppendorf vial and stored at 4°C. A crushed serpentine control blank that had not been inoculated with E. coli cells was also submitted to the LB medium treatment and the KPBS washing and mixing procedure described above. Individual cell pellets generated after the growth and centrifugation of different volumes of LB medium were washed in KPBS and then weighed separately.
Natural samples and cell enumeration using the DAPI staining method.
Several natural samples containing a wide range of bacterial cell concentrations were used in this study. A Nile Delta seafloor sediment from a core collected in 1964 by the German research vessel Meteor at a water depth of 120 m (6) was dried under a vacuum and weighed. This sample was originally taken from the top 0 to 15 cm of the core and was stored at the GEOMAR Research Center for Marine Geosciences (Kiel, Germany) at a temperature of 10 to 15°C from 1989 to 1999 (J. Gruetzner, personal communication). For the last 5 years, a dry sample from the core has been kept at room temperature at the Scripps Institution of Oceanography. In addition, a sample of wet beach sand (∼50 m above the high-tide mark) and a sample of seawater from the surf zone collected in January of 2004 from La Jolla Shores in San Diego, Calif., were also investigated. Two soil samples from an extremely arid region of the Atacama Desert in northern Chile (17) were also analyzed. A sample of fine surface particles and a subsurface (1 cm deep) sample were collected in May and October of 2003, respectively, from the Flat Top Hill site (24°S, 70°W) approximately 100 miles south of the Yungay region (F. Grunthaner, personal communication).
The seawater, sand, soil, and sediment bacteria were stained in solution with DAPI and enumerated using a standard epifluorescence microscopy technique (19). Approximately 1 ml of KPBS was combined with 1 g of each of the solid samples or 1 ml of seawater, and 100 μl of 2% borate-buffered formalin was then added to the mixture. The samples were then sonicated three times for 1 min at 50% power (Fisher Scientific Sonic Dismembrator 50) and centrifuged at 3,000 rpm for 1 min, and the supernatant was removed. Five milliliters of KPBS was added to the supernatant, and the entire solution was filtered through a 0.2-μm-pore-size, 25-mm-diameter black polycarbonate membrane filter (Nuclepore) to collect the bacteria from the sample solution. A 20-μl aliquot of DAPI stain (1 mg/ml) was added to a clean microscope slide, and then the drop was covered with the sample membrane filter and a 25-mm-diameter square coverslip. Cell colonies were counted under UV by using a fluorescence microscope (Olympus BX51, 100× magnification) with a DAPI fluorescence emission filter. The average cell count was determined from 10 separate field counts, and the cell concentration (number of cells per sample) was then inferred from the filter and field areas.
Sublimation heating experiments.
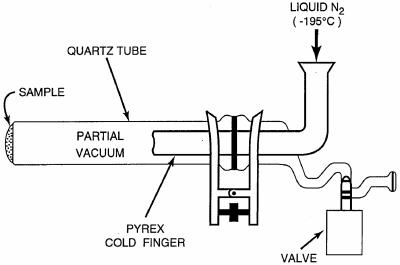
A portion of the inoculated serpentine sample (190 mg) was transferred to a quartz glass sublimation apparatus (SA) (Fig. 1), sealed in air at 0.5 torr, and heated in a tube furnace (Lindberg/Blue M Mini-Mite) set at 1,100°C. Three separate E. coli cell pellets (P1, P2, and P3) weighing 1.3, 6.2, and 7.5 mg, respectively, were also sublimed under the same conditions (Fig. 2). For the other samples, 1 g each of the Nile Delta and Atacama samples, 5 g of beach sand, and 20 ml of seawater were evaporated to dryness and heated separately. A cold finger, attached to the sublimation tube, was kept at −195°C with liquid nitrogen throughout the experiments. After heating for 30 s, the apparatus was removed from the furnace, and the pressure inside the SA was brought up to 1 atm. According to thermocouple measurements of the temperature inside the apparatus, the samples were heated to 500°C for at least 30 s during the experiment. The experimental conditions (i.e., temperature, internal pressure, and duration of heating) used in this study have previously been optimized to achieve maximum sublimation yields of amino acids and nucleobases from natural samples (8, 9).
FIG. 1.
The SA used in the heating experiments. The custom-made SA built at the University of California at San Diego was designed specifically for the Lindberg Blue tube furnace and consists of a quartz tube (2.5 by 31 cm) and a Pyrex glass cold finger (1.6 by 15.5 cm), which are sealed together under a vacuum with a clamp and an O-ring. The total cost of the SA was approximately $400. Several vacuum sublimator designs are also available commercially and can be purchased online from Safety Emporium.
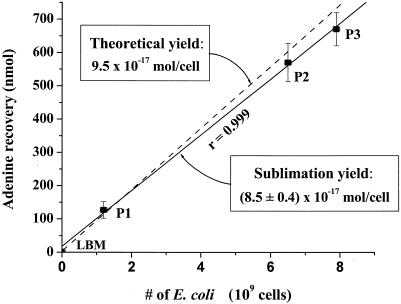
FIG. 2.
The calibration data for the sublimation recovery of adenine from E. coli (solid line). LB medium (LBM) that did not contain E. coli and three separate E. coli pellets (P1, P2, and P3) were heated to 500°C for 30 s in the sublimation apparatus. The total number of E. coli cells was calculated from the mass of each cell pellet given an average wet weight for a single E. coli cell of 9.5 × 10−13 g (18). An average sublimation yield of (8.5 ± 0.4) × 10−17 mol of adenine per cell from E. coli was calculated from the slope of the best-fit line. The uncertainties in the adenine recoveries were based on the standard deviation of the average value of three separate measurements. An error in the sublimation yield of ∼5% was determined from the uncertainty in the slope of the best-fit line. The theoretical yield is also shown for comparison (dashed line).
After sublimation of the E. coli samples was carried out, a yellow residue coated the end of the cold finger. This colored coating was also observed, although to a lesser extent, after sublimation of the Nile Delta sediment, beach sand, and seawater samples. We did not observe any yellow coloration of the cold finger for the Atacama Desert soils or the control blank that did not contain bacterial cells. The residue on the cold finger was carefully rinsed off in 1 ml of double-distilled water, and the water extract was then analyzed directly by HPLC separation and UV absorbance detection. Purines and pyrimidines were separated isocratically using 0.1 M sodium phosphate buffer, pH 4.8, at a flow rate of 1 ml/min on a YMC ODS-AQ reverse-phase column (4.6 by 150 mm) at 25°C using a Beckman 110B pump. Peaks were detected at 260 nm using a Kratos Spectroflow 757 UV and visible-light detector. Peaks were identified by comparison of the retention times with those of a standard run carried out in parallel. The entire sublimation extraction and analysis method can be carried out in less than 3 h, in contrast to traditional acid extraction techniques, which require more than 1 day (9).
RESULTS AND DISCUSSION
E. coli cell counts and sublimation calibration results.
After blank correction using growth medium that did not contain E. coli cells, we measured the OD460 of the E. coli growth medium as 0.65. From the OD reading, we calculate 6.5 × 109 E. coli cells in 10 ml of LB growth medium with a 5% measurement error. We recognize that physiological variation may cause aggregation, changes in cell size, or capsule formation, leading to small differences in the conversion between OD and total cell counts. Therefore, the total number of E. coli cells determined from the OD reading was independently confirmed by a measurement of the mass of a solid E. coli pellet (P2) (Fig. 2) generated by overnight growth and centrifugation of a volume of LB medium identical to that used to inoculate the serpentine. Assuming that the E. coli cells were homogenously mixed into the 0.5-g crushed serpentine sample, a concentration of 1.3 × 1010 cells/g (±5%) for the serpentine is inferred. This value is in close agreement with the cell concentration of 1.1 × 1010 cells/g that was previously determined from the total amount of amino acids present in a soil analogue inoculated with E. coli after hydrolysis (10).
After sublimation of the inoculated serpentine sample, high concentrations (31 to 125 nmol/g) of several nucleobases, including adenine, cytosine, thymine, and uracil, were detected in a water extract of the cold finger (Table 1). Guanine did not sublime from the cells at this temperature. In a previous sublimation experiment with λ DNA, it was demonstrated that at a temperature of 150°C, melting and fragmentation of the DNA occur and the glycosidic bonds that attach the nucleobases to the DNA begin to break, facilitating the sublimation of adenine, cytosine, and thymine directly from the λ DNA (9). Therefore, the nucleobases detected after sublimation of the inoculated serpentine sample and cell pellets were probably derived from native E. coli DNA and RNA. The sublimation process for nucleobases occurs rapidly and to completion when E. coli is heated in the SA at temperatures above 450°C for at least 30 s. We were unable to detect any nucleobases in the UV absorbance chromatogram of the control blank, which indicates that the nucleobases recovered from the inoculated serpentine and cell pellets were entirely derived from the E. coli cells and were not associated with any remnants of the growth medium used for inoculation.
TABLE 1.
Summary of nucleobase recovery after sublimation of several natural samplesa
| Nucleo- base | Recovery (nmol/g) from:
|
|||||
|---|---|---|---|---|---|---|
| Serpentine- E. coli | Sand | Sea- water | Nile Delta sediment | Atacama soilb
|
||
| Surface | Subsurface | |||||
| Adenine | 125 | 0.2 | 0.005 | 12.7 | <0.05 | 0.04 |
| Guaninec | ND | ND | ND | ND | ND | ND |
| Cytosine | 42 | 1.1 | 0.002 | 11.2 | <0.05 | 0.59 |
| Thymine | 31 | 1.0 | 0.010 | 3.0 | <0.05 | 0.13 |
| Uracil | 68 | 0.8 | 0.005 | 16.8 | <0.05 | 0.05 |
The total recoveries of several nucleobases (nmol/g) from water extracts of a cold finger after sublimation are shown. All reported values were corrected using the procedural blank. The uncertainty in these measurements was ±5% based on repeat sublimation experiments of the sand and seawater that gave consistent results within the measurement error.
The upper limits for the recoveries from the Atacama soil were based on the detection limits for these nucleobases obtained using HPLC and UV absorbance.
Guanine did not sublime at these temperatures. ND, not determined.
From the data in Fig. 2, we calculate an average sublimation recovery of adenine from the E. coli cell pellets of (8.5 ± 0.4) × 10−17 mol per cell. These data are reasonably close to the theoretical yield of adenine in E. coli and demonstrate that the sublimation process for adenine from E. coli is linear over the range of cell concentrations tested (Fig. 2). For the inoculated serpentine sample we measured an adenine concentration of 125 nmol per gram (Table 1), which is equivalent to 9.6 × 10−18 mol of adenine per cell based on the measured number of E. coli bacteria in the sample. This sublimation yield is approximately 10% of the adenine recovered directly from solid E. coli pellets sublimed alone (8.5 × 10−17 mol/cell), indicating that the mineral matrix inhibits the sublimation of adenine from the cells. Previous experiments have shown that the sublimation yield of a pure adenine standard is substantially reduced in the presence of serpentine as well as in other media, such as seawater and humic acid, with recoveries ranging from 1 to 10% (7). These recoveries are similar to the adenine yield obtained from the inoculated serpentine sample. Because the data for sublimation from the E. coli cell pellets do not take into account potential matrix effects from reactive mineral surfaces, salts, and larger amounts of organic matter, the recovery of adenine from the inoculated serpentine sample (9.6 × 10−18 mol/cell), rather than that from the E. coli pellets, was used to estimate cell counts in the natural samples. Using the higher theoretical yield of adenine from E. coli to calculate bacterial abundances would result in lower overall cell counts for the same sublimed adenine yield; therefore, the bacterial concentrations reported for the natural samples in Table 2 must be considered to be upper limits. Additional studies will be required to critically evaluate the efficiency of adenine sublimation from E. coli in a variety of different porous media.
TABLE 2.
Bacterial cell counts of the natural samples
| Sample origin | Characterization | Bacterial abundance
|
|
|---|---|---|---|
| Sublimation (106 ECE/g)a | DAPI (106 cells/g)b | ||
| La Jolla | Seawater (surface) | 0.52 ± 0.04 | 1.3 ± 0.2 |
| La Jolla | Beach sand (upper 5 cm) | 21 ± 2 | 6.3 ± 1.1 |
| Nile Delta | Deep-sea core (upper 15 cm) | 1,300 ± 90 | 1.9 ± 0.3 |
| Atacama | Surface fines | <5 | 0.7 ± 0.1 |
| Atacama | Subsurface (1 cm deep) | 4.4 ± 0.3 | 9.6 ± 1.6 |
Bacterial counts in ECE per gram for the soil and sediment samples were calculated by dividing the sublimation recovery of adenine from Table 1 by an adenine yield of (9.6 ± 1.4) × 10−18 mol per cell from the inoculated serpentine sample. Propagated errors in the cell counts were based on the uncertainty of the adenine yields from the sample and E. coli sublimation experiments and ranged from ±7 to 10%.
For the DAPI cell counts (cells/g) we assumed an average uncertainty of ±17% based on measurements made in a previous study (12).
The wide variety of microbes and large genetic diversity of bacteria in soils, seawater, and marine sediments (3, 12, 22) should be taken into consideration when using sublimation to estimate total bacterial counts in natural samples. Although the genome size of E. coli, ∼4 Mbp of DNA (18), is similar to the average size of prokaryotic genomes (0.6 to 10 Mbp) as well as several common marine and soil bacteria (5), we recognize that by using E. coli as a reference we only obtain total bacterial counts for cells with genome sizes similar to that of E. coli. For example, a bacterium with a smaller genome size, such as a mycoplasma (∼0.6 to 0.8 Mbp [5]), would be at least five times more abundant than E. coli for the same sublimed adenine yield. Variable RNA contents in bacterial cells could also influence bacterial cell counts extrapolated from adenine levels using the sublimation method. In addition, the amount of adenine in a given cell can vary depending on its growth stage and rate, which could also impact bacterial cell count extrapolations.
Another possible source of adenine in E. coli is free ATP. However, the concentration of ATP in E. coli is much lower than that found in DNA and RNA, accounting for only a small fraction (∼0.1%) of the adenine derived from nucleic acids (18). In addition to bacteria, there are other potential sources of adenine in the marine and soil samples analyzed in this study, including phytoplankton, zooplankton, algae, plants, and other debris that would contribute to our sublimation-based bacterial counts. Viruses are also abundant in marine seawater, soil, and sediment samples with concentrations ranging from 106 to 109 free particles per ml (2, 20, 25). Since viral genome sizes range from 103 to 105 bp (4), a significant contribution of adenine from viral DNA and RNA could have been present in the sublimed extracts of the natural samples analyzed in this study. Given these additional sources of adenine, our sublimation-based cell counts, expressed as E. coli cell equivalents, likely overestimate the total number of bacteria present in the natural samples. On the other hand, using the DAPI method, many viruses in these samples would not have been counted, since single-stranded viral DNA and RNA are not stained by the DAPI fluorochrome, which is specific for double-stranded DNA (19).
Comparison of bacterial cell counts in marine samples using sublimation and DAPI.
The sublimation results for the natural samples are shown in Fig. 3 and Table 1. Several nucleobases, including adenine, cytosine, thymine, and uracil, were detected on the cold finger after sublimation of the sand, seawater, and Nile Delta sediment samples. The distribution of nucleobases in these sublimed extracts was similar to that of the purines and pyrimidines identified after sublimation of the inoculated serpentine sample containing E. coli. We did not detect any peaks with retention times similar to those of a nucleobase standard in the HPLC chromatograms of the sublimed control serpentine blank that did not contain bacteria (Fig. 3). The concentration of adenine in the sublimed extracts from these marine samples ranged from 0.005 to ∼13 nmol/g. For the seawater, we calculated a cell count of 0.5 × 106 ECE/g based on the average sublimation of adenine from two separate samples (Table 2). This cell concentration agrees reasonably well with the DAPI cell count of 1.3 × 106 cells/g measured in the same sample. Both of these values are well within the range of total microbial cell counts of 0.2 × 106 to 1.6 × 106 cells/g obtained from seawater using whole-cell fluorescent in situ hybridization (16). Our results suggest that the presence of viruses does not bias the total bacterial counts in seawater since the sublimation cell count was significantly lower and not higher than the DAPI count, which does not account for viruses. It is also possible that some adenine from dissolved cyclic AMP (cAMP) present in seawater could contribute to sublimation-based cell counts of the marine samples. However, given a maximum free cAMP concentration of ∼35 pM for seawater (1), the contribution of adenine from cAMP in seawater would translate into a bacterial cell concentration of only 4,000 cells/g. This value is within the uncertainty of the sublimation-based cell count of the seawater and is well below the detection limit for this method.
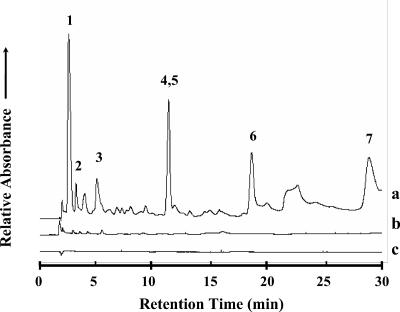
FIG. 3.
The 0- to 30-min region of the reverse-phase HPLC chromatograms of UV absorbance spectra (λ = 260 nm) from sublimed extracts of the (a) Nile Delta deep-sea sediment sample, (b) Atacama Desert subsurface particles, and (c) serpentine blank made after heating the samples in the SA at ∼500°C for 30 s. UV absorbance measurements for the sublimed sand, seawater, and Atacama Desert fine surface particles were also carried out but are not shown here. Peak identifications: (1) HCl front, (2) cytosine, (3) uracil, (4, 5) guanine/hypoxanthine, (6) thymine, and (7) adenine.
The concentrations of sublimed adenine from the beach sand (0.2 nmol/g) and Nile Delta deep-sea sediment (12.7 nmol/g) were higher than that from the seawater sample (0.005 nmol/g), which is not surprising since marine sediments have much higher total bacterial concentrations per gram than seawater. For the Nile Delta sample we calculated a cell concentration of ∼1.3 × 109 ECE/g based on sublimation. This value is within the range (0.1 × 109 to 5 × 109 cells/g) reported for coastal marine sediments (14, 15, 21). An estimate for the cell count based on the total hydrolyzable amino acid abundance (10) of the Nile Delta sediment sample (20.3 ppm) also yields a bacterial concentration of 0.2 × 109 ECE/g, which is consistent with marine sediments. However, the DAPI staining of the Nile Delta sample gave a cell concentration of only 1.9 × 106 cells/g, less than 0.2% of the ECE value obtained by sublimation (Table 2), suggesting that there is a much higher percentage of organic matter associated with dead cells than with intact NuCC in the Nile Delta sample.
In most marine sediments, living bacterial cells (i.e., those displaying an intact membrane) account for roughly 25 to 30% of total bacterial counts, while dead cells represent the most abundant fraction (15). Among the living bacteria, nucleoid-containing cells (i.e., those that actively grow) represent only 4% of total bacterial counts (15). Since the DAPI method only stains NuCC, these total bacterial counts do not take into account dead non-NuCC and any detrital organic matter associated with them. Moreover, since the Nile Delta sediment sample analyzed in the present study was from a dried-out core collected nearly 40 years ago and probably contains mostly dead cells and/or organic matter derived from the long-term degradation of cells, it is not surprising that the DAPI total cell counts were considerably lower than the values obtained by sublimation, which takes into account all organic matter from both dead and living cells. Because the sublimation count of 1.3 × 109 ECE/g for the Nile Delta sample is consistent with DAPI counts for fresh deep-sea sediment cores (15), the sublimation count for the Nile Delta sample may represent the total bacterial population present in this sample at the time of collection. However, adenine that was not derived from nucleic acids in the sediment (e.g., deposition of dead organic matter from the Mediterranean Sea surface waters by gravitational settling) would have added to the bacterially derived adenine content of the Nile Delta sediment sample over time. Therefore, the sublimation-based count must be considered to be an upper limit for the total number of cells present in this sample at the time of collection.
For the beach sand, which was collected within days of analysis, a total bacterial count of 21 × 106 ECE/g was determined based on sublimation (Table 2). This result was confirmed by a repeat sublimation experiment of the sand. The beach sand count was significantly lower than that of the Nile Delta sediment, which is in accordance with the microbial load being much lower in coarse-grained sand than in mud cores, such as the Nile Delta sample (14). With the DAPI method a bacterial count of roughly 6.3 × 106 cells/g was measured for the sand (Table 2), which is consistent with previous estimates for the percentage of living cells (∼30%) in total bacterial counts of marine sediments (15).
Bacterial cell count estimates of the Atacama Desert soil.
Trace levels of nucleobases (0.04 to 0.6 nmol/g) were identified in the sublimed extract of the Atacama subsurface soil sample (Table 1). Based on the amount of adenine sublimed from this sample (0.04 nmol/g), we estimate a total bacterial concentration of 4.4 × 106 ECE/g of soil. We were unable to identify any adenine in the sublimed extract of the Atacama fine surface particles above the level ∼0.05 nmol per gram after heating (Fig. 3; Table 1); therefore, only an upper limit for the number of cells in this sample (<5 × 106 ECE/g) could be obtained by the sublimation method (Table 2). In order to detect adenine released from Atacama surface samples by using this method, a sample size larger than what was available for this study would be required.
Using the DAPI staining method, we measured bacterial counts for the surface and subsurface Atacama samples of ∼0.7 × 106 cells/g and 9.6 × 106 cells/g, respectively. These values compare reasonably well to the sublimation cell count estimates (Table 2). The DAPI counts for the Atacama soils are much higher than the total counts of viable, culturable heterotrophic bacteria (<10 to 104 CFU/g) previously measured by serial dilution plating (17), which could indicate that soil samples from the Atacama Desert contain mostly nonculturable bacteria that are not detected by dilution plating. The extremely dry conditions in the Atacama Desert may inhibit biological productivity in the soils and enhance the survival of photochemically produced oxidants that would destroy organic material (17). Although our sublimation count estimates for the Atacama soils were very near the detection limit of the analytical technique, these results confirm that the subsurface sample contains higher organic content and a higher bacterial cell concentration than fine surface particles. This may be due to the existence of a concentration gradient of soil oxidants corresponding to depth.
Conclusions.
These results demonstrate the feasibility of using sublimation to estimate total bacterial counts in natural samples. Since sublimation-based cell enumeration accounts for all organic matter associated with dead and living cells, this method is particularly useful for determining total cell equivalents in samples that contain only low levels of viable nucleoid-containing cells. In addition, the greater simplicity and robustness of the sublimation technique compared to the DAPI staining method make this approach particularly attractive for use by spacecraft instrumentation.
NASA is currently planning to send a rover to Mars in 2009 in order to assess whether or not organic compounds, especially those that might be associated with life, are present in martian surface samples. The sublimation method for determining the adenine content in martian samples is one approach that should be considered. However, the extrapolation to evidence of extinct or extant life on Mars using this approach may not be warranted, since adenine of chemical origin might also behave similarly in the sublimation procedure. Nonetheless, based on our analyses of the Atacama Desert soil samples, several million bacterial cells per gram of martian soil should be detectable using this technique. Since the Atacama Desert soils contain only traces of organic compounds and extremely low levels of viable bacteria, these samples serve as valuable martian analogue material for the testing of instruments designed for future martian life detection missions.
Acknowledgments
We thank D. H. Bartlett for generously providing the E. coli cells and L. Palekar and A. Purdy for assistance with cell staining. We are also grateful to F. Grunthaner and his JPL research team for providing the Atacama Desert soil samples analyzed in this study. We appreciate the helpful comments of J. Dworkin and three anonymous reviewers.
This research was supported by a grant from the NASA Specialized Center for Research and Training in Exobiology at the University of California at San Diego.
REFERENCES
- 1.Ammerman, J. W., and F. Azam. 1981. Dissolved cAMP in the sea and uptake of cAMP by marine bacteria. Mar. Ecol. Prog. Ser. 5:85-89. [Google Scholar]
- 2.Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heidal. 1989. High abundance of viruses found in aquatic environments. Nature (London) 340:467-468. [DOI] [PubMed] [Google Scholar]
- 3.Bernard, L., H. Schäfer, F. Joux, C. Courties, G. Muyzer, and P. Lebaron. 2000. Genetic diversity of total, active and culturable marine bacteria in coastal seawater. Aquatic Microb. Ecol. 23:1-11. [Google Scholar]
- 4.Cann, A. J. 2001. Principles of molecular virology. Academic Press, San Diego, Calif.
- 5.Cole, S., and I. Saint-Girons. 1999. Bacterial genomes—all shapes and sizes, p. 35-62. In R. Charlebois (ed.), Organization of the prokaryotic genome. ASM Press, Washington, D.C.
- 6.Einsele, G., and F. Werner (ed.). 1968. Meteor scientific research, vol. C, p. 21-42. German Science Foundation (Gebrueder Borntraeger), Berlin, Germany. [Google Scholar]
- 7.Glavin, D. P. 2001. Survival of prebiotic organic compounds during exogenous delivery: implications for the origin of life on Earth and potentially on Mars. Ph.D. dissertation. University of California at San Diego, San Diego.
- 8.Glavin, D. P., and J. L. Bada. 1998. Isolation of amino acids from natural samples using sublimation. Anal. Chem. 70:3119-3122. [DOI] [PubMed] [Google Scholar]
- 9.Glavin, D. P., M. Schubert, and J. L. Bada. 2002. Direct isolation of purines and pyrimidines from nucleic acids using sublimation. Anal. Chem. 74:6408-6412. [DOI] [PubMed] [Google Scholar]
- 10.Glavin, D. P., M. Schubert, O. Botta, G. Kminek, and J. L. Bada. 2001. Detecting pyrolysis products from bacteria on Mars. Earth Planet. Sci. Lett. 185:1-5. [Google Scholar]
- 11.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. The use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kepner, R. L., Jr., and J. R. Pratt. 1994. Use of fluorochromes for direct enumeration of total bacteria in environmental samples: past and present. Microbiol. Rev. 58:603-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llobet-Brossa, E., R. Rosselló-Mora, and R. Amann. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luna, G. M., M. Manini, and R. Danovaro. 2002. Large fraction of dead and inactive bacteria in coastal marine sediments: comparison of protocols for determination and ecological significance. Appl. Environ. Microbiol. 68:3509-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maruyama, A., and M. Sunamura. 2000. Simultaneous direct counting of total and specific microbial cells in seawater, using a deep-sea microbe as target. Appl. Environ. Microbiol. 66:2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navarro-González, R., F. A. Rainey, P. Molina, D. R. Bagaley, B. J. Hollen, J. de la Rosa, A. M. Small, R. C. Quinn, F. J. Grunthaner, L. Cáceras, B. Gomez-Silva, and C. P. McKay. 2003. Mars-like soils in the Atacama Desert, Chile, and the dry limit of microbial life. Science 302:1018-1021. [DOI] [PubMed] [Google Scholar]
- 18.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Inc., Sunderland, Mass.
- 19.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 20.Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature (London) 343:60-62. [Google Scholar]
- 21.Proctor, L. M., and A. C. Souza. 2001. Method for enumeration of 5-cyano-2,3-ditoyl tetrazolium chloride (CTC)-active cells and cell-specific CTC activity of benthic bacteria in riverine, estuarine and coastal sediments. J. Microbiol. Methods 43:213-222. [DOI] [PubMed] [Google Scholar]
- 22.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Schallenberg, M., J. Kalff, and J. B. Rasmussen. 1989. Solutions to problems in enumerating sediment bacteria by direct counts. Appl. Environ. Microbiol. 55:1214-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sobsey, M. D., C. H. Dean, M. E. Knuckles, and R. A. Wagner. 1980. Interactions and survival of enteric viruses in soil materials. Appl. Environ. Microbiol. 40:92-101. [DOI] [PMC free article] [PubMed] [Google Scholar]