Abstract
The methods commonly used for norovirus (NV) detection are based on reverse transcription-PCR (RT-PCR) followed by confirmation of the amplified sequence. To increase sensitivity, an RT-booster PCR was developed. The proposed method showed an increase in sensitivity at least 2 log units for all the NV strains tested compared with the standard RT-PCR method. Higher sensitivity was confirmed in tests on experimentally and naturally contaminated shellfish.
Noroviruses (NVs) represent the most important cause of viral gastroenteritis outbreaks worldwide (11, 14, 15). Food- and waterborne transmission of NVs is common (5), and different epidemiological studies have implicated NVs in outbreaks linked to shellfish consumption (2). In many outbreaks of shellfish-transmitted diseases, however, the causative agent has not been identified due to the lack of a sensitive method of analysis of shellfish samples (12). In fact, as the levels of enteric viruses in mussels are normally low, high-sensitivity detection methods are usually needed.
Different strategies of reverse transcription-PCR (RT-PCR) to enable the molecular detection of the largest number of NV strains possible have been proposed (3). An effective approach to improve the chances of successful amplification consists of associating primers selected in a mostly conserved region (e.g., the RNA-dependent RNA polymerase [RdRp] gene region) with the use of low annealing temperatures (as low as 37°C) (18, 19). This strategy proved to be a promising one, as evidenced by the detection of most of the strains commonly spread in the countries of the European Union, which was confirmed in an international collaborative study (20).
To increase the sensitivity of RT-PCR detection and still retain a useful amplicon length for sequencing, an RT-booster PCR based on a double amplification performed with the same primer pair was developed (6, 16, 17).
Strains.
Strain 99-23 372 (GGII, Hu/NLV/Hawaii virus/1971/US) was used to develop and optimize RT-booster PCR. The method was subsequently tested on two additional GGII strains (patient 5 of Turin outbreak in 2002, Hu/NLV/Melksham/1994/UK, and strain 98 39 449, Hu/NLV/Hawaii virus/1971/US) and a GGI strain (patient 41, La Spezia outbreak in 2002, Hu/NLV/Valetta/1995/Malta).
Sample preparation.
Viral suspensions were prepared as follows. Ten percent stool suspensions in phosphate-buffered saline (Oxoid, Basingstoke, England) from patients implicated in NV outbreaks were clarified by centrifugation at 3,000 rpm for 20 min in an Eppendorf Microfuge. One hundred microliters of the viral suspension of strain 99-23 372 and its 10-fold dilutions (from 10−1 to 10−3) were used to experimentally contaminate 900 μl of shellfish extract (final dilution of virus in shellfish ranged from 10−1 to 10−4) obtained from samples of Mytilus galloprovincialis prepared as elsewhere described (8). For the experiments on naturally contaminated shellfish, 75 g of whole shellfish homogenate from 16 samples (Mytilus galloprovincialis and Tapes philippinarum) collected from local markets was processed as described by De Medici et al. (8) and analyzed together with a positive control (shellfish extract contaminated with strain 99-23 372) and negative controls (uninoculated samples).
RNA extraction.
Both for viral suspensions and experimentally contaminated shellfish, RNA extraction was performed with Nucleospin RNAII (Macherey-Nagel, Düren, Germany) according to manufacturer instructions, and nucleic acid was eluted in a 100-μl volume. For naturally contaminated samples 300 μl of shellfish extract, equivalent to 5 g of shellfish homogenate, was extracted as described by Afzal and Minor (1) and Croci et al. (7). The RNA pellet was resuspended in 10 μl of deionized triply distilled water containing 1 U of RNase inhibitor/μl.
Primers.
A broadly reactive primer set (JV12 and JV13; synthesized by M-Medical Genenco, Milan, Italy; Table 1), amplifying a 326-bp region of the RdRp gene (18), was chosen for RT-booster PCR and single-round RT-PCR (19).
TABLE 1.
Sequences of RT-PCR and RT-booster PCR oligonucleotide primers and probes
| Primer or probe set | Sequence (5′-3′) |
|---|---|
| Primers | |
| JV12 | ATA CCA CTA TGA TGC AGA TTA |
| JV13 | TCA TCA TCA CCA TAG AAA GAG |
| 5′ end biotin-labeled probes | |
| UK-3 | GTC CCC TGA CAT CAT ACA GGC T |
| JV-5 | CTC ACC AGA GGT TGT CCA AGC |
| GG-I | ATG GAY GTT GGY GAY TAT GT |
| GG-II | GAA YTC CAT CRC CCA YTG |
Optimization of RT-booster PCR.
Tests were performed to define the primer concentrations in the first PCR (1, 10, and 100 nM and 1 μM) and in the second PCR (tenfold higher than the ones used for the first PCR), annealing times (1 min 30 s or 4 min) and number of cycles (20, 30, or 40) in the first PCR, and volume of the first PCR product transferred to the second PCR.
RT-booster PCR and RT-PCR.
RT-booster PCR was performed with the Access RT-PCR system (Promega, Madison, Wis.). RT was performed with the following mixture: 1× AMV/Tfl buffer, 200 mM deoxynucleoside triphosphates (dNTPs), 1 mM MgSO4, 5 U of avian myeloblastosis virus reverse transcriptase, 4 μl of 5 μM primer JV13, 5 μl of RNA, and PCR grade water to a final volume of 45 μl. The RT reaction was carried out at 48°C for 60 min (PTC-200 Thermalcycler; MJ Research, Watertown, Mass.) and was terminated with a 5-min step at 99°C. The first PCR was performed by adding 1 μl of Tfl DNA polymerase (5 U/μl), 1 μl of 5 μM primer JV13, 1 μl of 5 μM primer JV12, and 2 μl of PCR grade water directly to the RT vessels. Samples were then denatured for 2 min at 94°C and subjected to 20 cycles of 94°C for 1 min, 37°C for 4 min, and 68°C for 2 min. A final extension at 68°C for 7 min was then performed. For the second PCR, 5 μl of the first PCR product was added to the following mixture: 1× AMV/Tfl buffer (Promega), 200 mM dNTPs, 1 mM MgSO4, 5 U of Tfl polymerase, 1 μM primers JV13 and JV12, and PCR grade water to a final volume of 50 μl. Thermalcycler conditions were as follows: 94°C for 2 min, followed by 40 cycles at 94°C for 1 min, 37°C for 1 min 30 s, and 68°C for 2 min and a final extension at 68°C for 7 min.
A single-round RT-PCR, performed as described by Vinje and Koopmans (19), was used as a comparative method. Both single-round RT-PCR and RT-booster PCR were repeated three times on RNA extracted from viral suspensions and experimentally contaminated shellfish.
Electrophoresis and Southern blotting.
PCR products were run in 2% agarose gel (Kodak, New Haven, Conn.) in the presence of ethidium bromide and photographed by Chemi Doc (Bio-Rad Laboratories, Hercules, Calif.). The resulting amplicon (326 bp) was confirmed by Southern blotting; hybridization was performed as described by Bergmans et al. (4) with the probes reported in Table 1 (J. Vinje, personal communication) at a temperature of 42°C.
Data and conclusions.
This paper describes the development of an RT-booster PCR for the detection of NVs both in shellfish and clinical samples. The data obtained showed that the highest sensitivity was reached when the first PCR was performed with a primer concentration of 0.1 μM, while the detection sensitivity decreased at higher (1 μM) and lower concentrations (10 and 1 nM). The use of a longer annealing time in the first PCR appeared to be slightly more effective in enhancing the overall assay sensitivity than increasing the number of cycles in the first PCR. In fact, a 4-min annealing time instead of 1 min 30 s resulted occasionally in an increase in sensitivity, while an increase in the number of cycles from 20 to 40 didn't produce any change in sensitivity. The use of smaller volumes of the first PCR product in the second PCR resulted in a decrease in the formation of primer dimers, without any evident loss of sensitivity (data not shown). In the conditions defined the RT-booster PCR was able to detect the NV up to the second decimal dilution beyond the last dilution detected by the single-round RT-PCR (Fig. 1). The results for the different strains tested consistently showed an increase in sensitivity: for the GGII strains the increase was at least two decimal dilutions, while, in our conditions (no detection in single-round PCR), the increase in sensitivity for the GGI strain, although perceptible as more than two decimal dilutions, could not be clearly defined (Table 2).
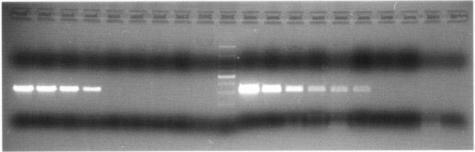
FIG. 1.
Comparison of RT-PCR and RT-booster PCR on RNA from a viral suspension. Lanes 1 to 8 (from left), 10-fold dilutions (from 10−1 to 10−8) of viral RNA (patient 5 from the Turin outbreak) subjected to RT-PCR; lane 9, negative control for RT-PCR; lane 10, marker (100 bp); lanes 11 to 18, same RNA dilutions as in lanes 1 to 8 subjected to RT-booster PCR; lanes 19 and 20, negative controls for the first and second PCRs of RT-booster PCR.
TABLE 2.
End point titration of four fecal samples containing NVs and tested by single-round RT-PCR and RT-booster PCR
| Sample | GG | Strain | Origin | Yr | RT-PCR titer (10−n)a
|
|
|---|---|---|---|---|---|---|
| Single round | Booster | |||||
| 99-23 372 | II | Hu/NLV/Hawaii virus/1971/US | The Netherlands | 1999 | 2b | 4b |
| 98 39 449 | II | Hu/NLV/Hawaii virus/1971/US | United Kingdom | 1998 | 0b | 2/3c |
| Patient 5 Torino | II | Hu/NLV/Melksham/1994/UK | Italy | 2002 | 4b | 6b |
| Patient 41 La Spezia | I | Hu/NLV/Valetta/1995/Malta | Italy | 2002 | NDbd | 2b |
Highest dilution of fecal sample which gave an amplicon of the correct size visible in ethidium bromide-stained agarose gel.
Results obtained in three experiments out of three.
Two experiments out of three showed positive results up to the second dilution; one experiment showed a positive result up to the third dilution.
ND, not detectable.
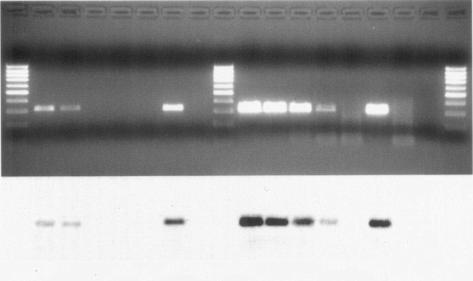
The higher sensitivity of booster amplification was confirmed with experimentally contaminated shellfish, and the results were consistent with those obtained with RNA from viral suspensions; RT-booster PCR, in fact, was able to detect NV up to the fourth decimal dilution, while standard RT-PCR detected NV up to the second decimal dilution in all three experiments done (Fig. 2).
FIG. 2.
Comparison of RT-PCR and RT-booster PCR on experimentally contaminated shellfish (electrophoretic gel and Southern blot). Lanes 1, 9, and 18 (from left), marker (100 bp); lanes 2 to 5, 10-fold dilutions (from 10−1 to 10−4) of NV (strain 99-23 372) in shellfish extract subjected to RT-PCR; lanes 10 to 13, 10-fold dilutions (from 10−1 to 10−4) of NV (strain 99-23 372) in shellfish extract subjected to RT-booster PCR; lanes 6 and 14, RT-PCR and RT-booster PCR on uninoculated shellfish extract; lanes 7 and 15, RT-PCR and RT-booster PCR positive controls; lane 8, RT-PCR negative control; lanes 16 and 17, RT-booster PCR negative controls for the first and second PCRs.
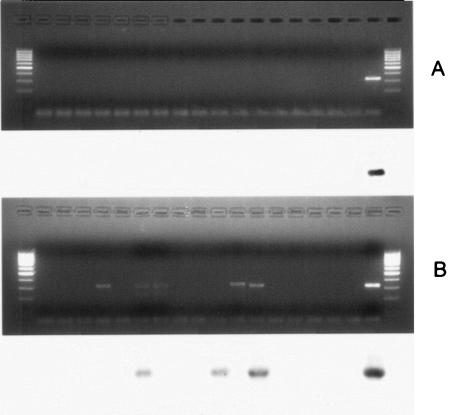
On naturally contaminated shellfish RT-booster PCR followed by Southern blotting detected the presence of NVs (confirmed by sequencing GGII NV strains) in 3 out of 16 examined samples, while standard RT-PCR gave negative results on all samples (Fig. 3). RT-booster PCR gave positive results for one sample in only one of the two experiments (Fig. 3, lane 7), while it gave positive results for two samples in both experiments (Fig. 3, lanes 11 and 13).
FIG. 3.
Comparison of RT-PCR and RT-booster PCR on naturally contaminated shellfish (electrophoretic gels and Southern blots. (A) RT-PCR; (B) RT-booster PCR. Lanes 1 and 20 (from left), marker (100 bp); lanes 2 to 17, shellfish samples; lane 18, negative control; lane 19, positive control.
As opposed to other published methods that use RT-(semi)nested-PCR-based techniques, the method proposed does not reduce the length of the amplicon produced in the first amplification. This strategy allows the sequencing of a long section of the RdRp gene, a region reported as being a significant one for epidemiological research (9, 13). In addition, the sequencing of this region or the use of specific probes (19) allows the confirmation of the amplicon, excluding false-positive reactions that can be developed when low-stringency PCR conditions are used, as in this protocol, to detect a target with high genomic variability (10). The work carried out confirms what was described by Ruano et al. (16): when the target is present in a very low number of copies, the amplification reaction is hampered by the synthesis of primer dimers, with consequent reduction in the target yield. To overcome these problems, the primers must be diluted in such a way as not to exceed the target concentration. Differently from what was proposed by Ruano et al. (16), a longer annealing time only occasionally seemed to raise the PCR detection limit, possibly due to differences in a variety of other PCR parameters (e.g., low annealing temperature). Finally, it was demonstrated that, in such a double amplification, it is not necessary to use a high number of cycles in the first PCR in order to raise the method's detection limit. This allows the sensitivity of the method to be raised without increasing the time necessary to complete the analysis. Indeed, 3 h more than the time required for a single-round RT-PCR is enough to complete the RT-booster PCR, thus making this method suitable for the investigation and control of public health problems arising from the consumption of shellfish.
Acknowledgments
We appreciate the support of M. P. G. Koopmans, F. Ruggeri, and M. N. Losio in kindly providing the strains used in this publication.
This work was supported by Sixth Framework of the European Commission project 506359, “Health promoting, safe seafood of high eating quality in a consumer driven fork to farm concept.”
REFERENCES
- 1.Afzal, M. A., and P. D. Minor. 1994. Instant RNA isolation from virus-infected tissue culture fluid for the polymerase chain reaction. Vaccine 12:976-977. [DOI] [PubMed] [Google Scholar]
- 2.Ang, L. H. 1998. An outbreak of viral gastroenteritis associated with eating raw oysters. Commun. Dis. Publ. Health 1:38-40. [PubMed] [Google Scholar]
- 3.Atmar, R. L., and M. K. Estes. 2001. Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses. Clin. Microbiol. Rev. 14:15-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmans, A. M., J. W. Groothedde, J. F. Schellekens, J. D. van Embden, J. M. Ossewaarde, and L. M. Schouls. 1995. Etiology of cat scratch disease: comparison of polymerase chain reaction detection of Bartonella (formerly Rochalimaea) and Afipia felis DNA with serology and skin tests. J. Infect. Dis. 171:916-923. [DOI] [PubMed] [Google Scholar]
- 5.Boccia, D., A. E. Tozzi, B. Cotter, C. Rizzo, T. Russo, G. Buttinelli, A. Caprioli, M. L. Marziano, and F. M. Ruggeri. 2002. Waterborne outbreak of Norwalk-like virus gastroenteritis at a tourist resort, Italy. Emerg. Infect. Dis. 8:563-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, N. D., E. D. McGruder, H. L. Neibergs, R. W. Behle, D. E. Wallis, and B. M. Hargis. 1994. Detection of Salmonella enteritidis in feces from poultry using booster polymerase chain reaction and oligonucleotide primers specific for all members of the genus Salmonella. Poult. Sci. 73:354-357. [DOI] [PubMed] [Google Scholar]
- 7.Croci, L., D. De Medici, G. Morace, A. Fiore, C. Scalfaro, F. Beneduce, and L. Toti. 1999. Detection of hepatitis A virus in shellfish by nested reverse transcription-PCR. Int. J. Food Microbiol. 48:67-71. [DOI] [PubMed] [Google Scholar]
- 8.De Medici, D., L. Croci, S. Di Pasquale, A. Fiore, and L. Toti. 2001. Detecting the presence of infectious hepatitis A virus in molluscs positive to RT-nested-PCR. Lett. Appl. Microbiol. 33:362-366. [DOI] [PubMed] [Google Scholar]
- 9.Green, S. M., K. E. Dingle, P. R. Lambden, E. O. Caul, C. R. Ashley, and I. N. Clarke. 1994. Human enteric Caliciviridae: a new prevalent small round-structured virus group defined by RNA-dependent RNA polymerase and capsid diversity. J. Gen. Virol. 75:3-1888. [DOI] [PubMed] [Google Scholar]
- 10.Honma, S., S. Nakata, K. Kinoshita-Numata, K. Kogawa, and S. Chiba. 2000. Evaluation of nine sets of PCR primers in the RNA dependent RNA polymerase region for detection and differentiation of members of the family Caliciviridae, Norwalk virus and Sapporo virus. Microbiol. Immunol. 44:411-419. [DOI] [PubMed] [Google Scholar]
- 11.Koopmans, M., C. H. von Bonsdorff, J. Vinje, D. de Medici, and S. Monroe. 2002. Foodborne viruses. FEMS Microbiol. Rev. 26:187-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Guyader, F., F. H. Neill, M. K. Estes, S. S. Monroe, T. Ando, and R. L. Atmar. 1996. Detection and analysis of a small round-structured virus strain in oysters implicated in an outbreak of acute gastroenteritis. Appl. Environ. Microbiol. 62:4268-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lew, J. F., A. Z. Kapikian, J. Valdesuso, and K. Y. Green. 1994. Molecular characterization of Hawaii virus and other Norwalk-like viruses: evidence for genetic polymorphism among human caliciviruses. J. Infect. Dis. 170:535-542. [DOI] [PubMed] [Google Scholar]
- 14.Lopman, B. A., D. W. Brown, and M. Koopmans. 2002. Human caliciviruses in Europe. J. Clin. Virol. 24:137-160. [DOI] [PubMed] [Google Scholar]
- 15.Marks, P. J., I. B. Vipond, D. Carlisle, D. Deakin, R. E. Fey, and E. O. Caul. 2000. Evidence for airborne transmission of Norwalk-like virus (NLV) in a hotel restaurant. Epidemiol. Infect. 124:481-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruano, G., W. Fenton, and K. K. Kidd. 1989. Biphasic amplification of very dilute DNA samples via ‘booster’ PCR. Nucleic Acids Res. 17:5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saulnier, P., and A. Andremont. 1992. Detection of genes in feces by booster polymerase chain reaction. J. Clin. Microbiol. 30:2080-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vinje, J., and M. P. Koopmans. 1996. Molecular detection and epidemiology of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J. Infect. Dis. 174:610-615. [DOI] [PubMed] [Google Scholar]
- 19.Vinje, J., and M. P. Koopmans. 2000. Simultaneous detection and genotyping of Norwalk-like viruses by oligonucleotide array in a reverse line blot hybridization format. J. Clin. Microbiol. 38:2595-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vinje, J., H. Vennema, L. Maunula, C. H. von Bonsdorff, M. Hoehne, E. Schreier, A. Richards, J. Green, D. Brown, S. S. Beard, S. S. Monroe, E. de Bruin, L. Svensson, and M. P. Koopmans. 2003. International collaborative study to compare reverse transcriptase PCR assays for detection and genotyping of noroviruses. J. Clin. Microbiol. 41:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]