Abstract
The colonic microbiota mediates many cellular and molecular events in the host that are important to health. These processes can be affected in the elderly, because in some individuals, the composition and metabolic activities of the microbiota change with age. Detailed characterizations of the major groups of fecal bacteria in healthy young adults, in healthy elderly people, and in hospitalized elderly patients receiving antibiotics were made in this study, together with measurements of their metabolic activities, by analysis of fecal organic acid and ammonia concentrations. The results showed that total anaerobe numbers remained relatively constant in old people; however, individual bacterial genera changed markedly with age. Reductions in numbers of bacteroides and bifidobacteria in both elderly groups were accompanied by reduced species diversity. Bifidobacterial populations in particular showed marked variations in the dominant species, with Bifidobacterium angulatum and Bifidobacterium adolescentis being frequently isolated from the elderly and Bifidobacterium longum, Bifidobacterium catenulatum, Bifidobacterium boum, and Bifidobacterium infantis being detected only from the healthy young volunteers. Reductions in amylolytic activities of bacterial isolates in healthy elderly subjects and reduced short-chain fatty acid concentrations supported these findings, since bifidobacteria and bacteroides are important saccharolytic groups in the colon. Conversely, higher numbers of proteolytic bacteria were observed with feces samples from the antibiotic-treated elderly group, which were also associated with increased proteolytic species diversity (fusobacteria, clostridia, and propionibacteria). Other differences in the intestinal ecosystem in elderly subjects were observed, with alterations in the dominant clostridial species in combination with greater numbers of facultative anaerobes.
Bacterial succession in the infant large bowel has been well documented both by culture (6, 52) and, more recently, by using molecular methods (10, 16, 54), but there have been relatively few studies dealing with changes in the colonic microbiota during the aging process. We therefore know little about the composition of bacterial populations or their metabolic activities in elderly people. Advances in medical science and improved living standards have led to an increased life expectancy in western societies, where people over 60 currently constitute one-fifth of the population, and this number is estimated to rise to one-third by the year 2030 (36).
Good nutrition is important in maintaining gastrointestinal function. Malnutrition is one of the main factors responsible for reduced immune responses in old people (28), and the development of preventative nutritional strategies to promote healthy aging is desirable. Increased thresholds for taste and smell (8, 55), resulting in preferences for bland foods, coupled with swallowing difficulties (5) and masticatory dysfunction caused by loss of teeth and muscle bulk (25, 43) can result in nutritionally imbalanced diets among the elderly. In the stomach, hypochlorhydria due to atrophic gastritis is associated with lowered absorption of calcium, ferric iron, and vitamin B12 (47), reducing micronutrient intake. Furthermore, decreased intestinal motility, resulting in fecal impaction and constipation, is a major problem in the elderly (4, 26, 57).
Physiological changes in the gastrointestinal tract, as well as modifications in diet and host immune system activity, will inevitably affect the composition and metabolism of the gut microbiota. Early studies of the elderly indicated the presence of increased levels of enterobacteria and fungi together with reduced levels of anaerobic lactobacilli (15). Further investigations, in Japan (39, 40, 41), suggested that there were decreased levels of bifidobacteria and increased levels of clostridia, lactobacilli, streptococci, and enterobacteria in older people, changes which also occur in the intestinal microbiota in aging dogs (2). Recent bacteriological studies and 16S rRNA abundance measurements have confirmed these changes in anaerobe populations and the increase in enterobacteria in older people (22). These changes might result in increased putrefaction in the colon and a greater susceptibility to diseases, such as gastroenteritis or Clostridium difficile infection (3, 23, 41).
There is increasing evidence for a general reduction in species diversity in many bacterial groups in older people (13, 23). Changes in bifidobacteria are of particular interest, and a wide range of bifidobacterial species are found in infants and young adults. However, it has been reported that species diversity in the elderly population is reduced to one or two dominant organisms, namely Bifidobacterium adolescentis and Bifidobacterium longum (13, 17, 23, 40).
The aim of this study was to investigate the intestinal microbiota in three different human age groups—healthy young adults, healthy elderly volunteers, and hospitalized elderly patients receiving antibiotics—with particular reference to alterations in species diversity. Fermentation products in feces, such as short-chain fatty acids (SCFA) and ammonia, were quantitated to investigate how aging and antibiotic treatment affected bacterial metabolic activities in the gut. Understanding the changes that occur in older people will facilitate the development of nutritional and therapeutic strategies to reduce or reverse these processes and maintain a healthy colon.
MATERIALS AND METHODS
Volunteer recruitment.
Inclusion criteria for the study for the healthy young (aged 19 to 35 years) and the healthy elderly (aged 65 years or more) volunteers were as follows. The volunteers were recruited as free-living individuals from the local community and were in general good health. Volunteers were excluded if they had received antibiotics in the 8 weeks prior to the start of the study. Current treatment involving the use of steroids, immunosuppressants, acetylsalicylic acid, lactulose, or other stimulants or laxatives was not permitted. Individuals who were medically unstable, terminally ill, or suffering from dysphagia or severe dementia were also excluded. Criteria were the same for hospitalized patients as for the healthy elderly volunteers except for the fact that they were all receiving antibiotics. These subjects were inpatients at Royal Victoria Hospital, Dundee, United Kingdom. Ethical permission for these studies was obtained from the Tayside Medical Research Ethics Committee.
Fecal material.
For bacteriology studies, fresh stools were obtained from 12 healthy young adults aged 19 to 35 years (three males and nine females), 6 healthy elderly people aged 67 to 75 years (six females), and 10 elderly hospitalized patients receiving antibiotics, aged 73 to 101 years (two females and eight males). All samples were weighed and were processed under anaerobic conditions within 1 h of defecation to minimize loss of cell viability. Fecal slurries (10% [wt/vol]) were homogenized in a stomacher bag by using anaerobic 0.1 M phosphate-buffered saline (pH 6.8) and then passed through a 500-μm mesh sieve to remove large particulate matter. For measurements of bacterial fermentation products, stools were obtained from 14 healthy young people, 70 healthy elderly volunteers, and 9 antibiotic-treated elderly people, a group which included the subjects used for bacteriological analysis. Fecal slurries were prepared as described above, and aliquots (5 ml) were frozen and stored at −20°C for analysis of fecal organic acids and ammonia concentrations.
Isolation and enumeration of bacteria from feces.
One milliliter of fecal slurry was vortex mixed with sterile anaerobic half-strength peptone water (9 ml) to form a 1 in 10 dilution series (10−2 to 10−9). Samples (0.1 ml) from the tubes were spread onto prereduced duplicate agar plates, as outlined below. Anaerobes were isolated by using Wilkins-Chalgren agar. To observe proteolytic and amylolytic activities of bacteria after isolation, the medium was supplemented with casein (10 g liter−1) or starch (10 g liter−1). Zones of casein hydrolysis were observed directly as areas of opacity on the plates or as cleared areas after treatment with dilute HCl. Starch hydrolysis was observed by the addition of iodine. Neutral red (8 mg liter−1) was also added to some of the agar plates to enhance the differentiation of bacterial colonies. Wilkins-Chalgren agar with gram-negative antibiotic supplements was used to isolate Bacteroides spp., Prevotella spp., and other gram-negative anaerobes. Beerens agar (1) was used to isolate bifidobacteria, while MRS agar was employed in the isolation of lactobacilli and lactococci. Clostridia were isolated by using perfringens agar with the manufacturer's antibiotic supplements, reinforced clostridial agar with added novobiocin and colistin (0.008 g liter−1 each) and azide blood base agar. These media were also supplemented with casein to visualize proteolysis. Plates were incubated at 37°C in an anaerobic chamber (Don Whitley Scientific, Shipley, Yorkshire, England) containing an atmosphere of CO2, H2, and N2 (10:10:80) for 48 to 72 h. Facultative anaerobes were cultured by using nutrient agar (for total counts), azide blood agar base with casein (for enterococci), and MacConkey agar number 2 (for enterobacteria) and were incubated aerobically at 37°C for 48 h.
Identification of bacteria.
Bacteria were identified based on their cellular fatty acid profiles. Fatty acid methyl esters (FAME) were extracted from bacterial pellets obtained from approximately 40 ml of culture in anaerobic peptone yeast extract broth (21) supplemented with glucose (10 g liter−1) for anaerobic isolates or from 40 mg of bacteria grown on BBL trypticase soy broth agar (Becton Dickinson Ltd., Oxford, United Kingdom) for aerobic isolates, by saponification, methylation, and extraction, as described previously (38). The FAME were separated by using a model 5898A microbial identification system (Microbial ID, Inc., Newark, Del.), which consisted of a Hewlett-Packard model 6890 gas chromatograph fitted with a 5% phenyl-methyl silicone capillary column (0.2 mm by 25 m), a flame ionization detector, a Hewlett-Packard model 7637A automatic sampler, and a Hewlett-Packard Vectra XM computer (Palo Alto, Calif.). Gas chromatography parameters were as follows: carrier gas, ultra-high-purity hydrogen; column head pressure, 60 kPa; injection volume, 2 μl; column split ratio, 100:1; septum purge, 5 ml min−1; column temperature, 170 to 270°C; injection port temperature, 300°C; and detector temperature, 300°C. Peaks were automatically integrated, and fatty acid names and percentages were calculated, with numerical analysis done by using the standard MIS Library Generation software (Microbial ID, Inc.). Bacterial isolates were identified by comparing FAME profiles to those of known cultures in the MIS aerobe and anaerobic standard libraries. The system was calibrated by using a standard Microbial Identification System FAME calibration mix before each run and validated by using the type strains Stenotrophomonas maltophilia ATCC 13637, Bacteroides fragilis ATCC 25285, and Clostridium perfringens ATCC 13124. Yeasts were identified by using a API 20 C AUX biochemical identification system (bioMérieux, Basingstoke, Hampshire, England).
Chemical analyses.
Ammonia levels were determined by using the phenol-hypochlorite method (51). Standards containing known concentrations of ammonium chloride were prepared. All samples were tested in triplicate. Ammonia levels were analyzed by using a microplate reader (model 450; Bio-Rad, Hercules, Calif.) with single wavelength absorbance (630 nm). SCFA, lactate, and succinate from fecal supernatants were analyzed by using methods described previously (21, 30). SCFA were first acidified with 50 mM H2SO4 to convert them into free fatty acids, which were subsequently extracted into ether. SCFA were separated by gas chromatography by using an Agilent Technologies model 6890N gas chromatograph, following split injection (40:1) with an HP-INNOwax column (model 19091N-133; cross-linked polyethylene glycol; 30 m by 0.25 mm by 0.25 μm; Agilent Technologies). Injector and detector temperatures were 250 and 300°C, respectively. The initial column temperature (120°C) was held for 1 min, thereafter increasing in stages of 10°C per min until reaching 265°C, which was maintained for 2 min. Flow rates of the helium carrier gas, H2, air, and N2 (the makeup gas) were set at 1.8 ml min−1, 40 ml min−1, 450 ml min−1, and 45 ml min−1, respectively. Lactate and succinate were measured by using the same analytical system and operating conditions, after methylation of samples and extraction into chloroform (21). All samples were quantitated by comparisons of sample peak heights with those of authentic standards and internal standards by using Hewlett-Packard Integrated Chemstation software.
Chemicals.
All bacteriological culture media and selective antibiotic supplements were obtained from Oxoid (Basingstoke, Hampshire, England). Unless otherwise stated, all chemicals were purchased from the Sigma Chemical Co. (Poole, Dorset, England).
Statistics.
Statistical analyses were done by using the SPSS version 9.0 statistics package (SPSS, Chicago, Ill.). The independent-sample t test was applied to compare bacterial counts from the healthy young and healthy elderly groups and from the healthy elderly and antibiotic-treated elderly groups. The Mann-Whitney U test was used to compare fecal SCFA and ammonia levels from the healthy young and healthy elderly subjects and from the healthy elderly and antibiotic-treated elderly groups.
RESULTS
Stool weight.
Mean fecal weights were 107 g (± 22.2), 82.7 g (± 17.4), and 70.3 g (± 11.5) for the healthy young subjects (HY), healthy elderly subjects (HE), and antibiotic-treated elderly subjects (AE), respectively.
Bacteriology of different subject groups.
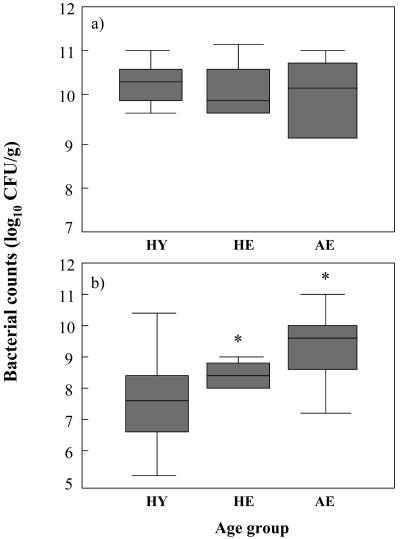
The results shown in Fig. 1A demonstrate that while total anaerobe numbers were similar for all three study groups, the range of counts was noticeably increased for AE. Total facultative anaerobes (Fig. 1B) were generally lower in HY than in HE. However, a significant increase (P < 0.05) in levels of facultative anaerobes was found for AE.
FIG. 1.
Box plot showing mean fecal bacterial counts from individuals in different age groups: a total anaerobe counts, b total facultative anaerobe counts. Results are expressed as mean log10 CFU per gram of feces ± interquartile range. Error bars represent standard errors of the mean. Healthy young (HY), n = 12; healthy elderly (HE), n = 6; antibiotic-treated elderly (AE), n = 10; *, significant difference (P < 0.05) between HE and AE.
Bacteroides and prevotellas.
Table 1 shows counts of bacteroides and prevotellas for each subject group, including details of the different species isolated. Marked reductions in total bacteroides occurred in both HE and AE, in comparison to HY, with lowered species diversity in the elderly subjects. The predominant Bacteroides spp. in samples from HY were B. ovatus and B. thetaiotaomicron, which had the highest prevalence and were present in the greatest numbers. Other common species, such as B. vulgatus, B. distasonis, B. fragilis, B. uniformis, and B. caccae, were detected in similar numbers but in fewer volunteers. Reductions in total bacteroides in HE and AE were accompanied by decreased species diversity. The principal bacteroides in HE was Bacteroides sp. strain FO, whereas B. ovatus was the predominant species in AE. A similar pattern was observed with prevotellas. Prevotella tannerae was the dominant species in HY and HE, although the organism was found in lower numbers in the latter group and was supplanted by Prevotella loescheii in AE. A general reduction in levels of prevotella was observed for both elderly groups, especially the group receiving antibiotics.
TABLE 1.
Comparison of bacteroides and prevotella species counts for different age groups
| Organism | HY
|
HE
|
AE
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of isolates | Meana | Rangeb | No. of isolates | Mean | Range | No. of isolates | Mean | Range | |
| B. ovatus | 7 | 5.8 ± 1.4 | 8.8-10.0 | 1 | 1.8 ± 1.8 | NA | 6 | 5.5 ± 1.5 | 7.5-10.9 |
| B. thetaiotaomicron | 7 | 5.2 ± 1.3 | 7.4-10.1 | 1 | 1.7 ± 1.7 | NA | 3 | 2.6 ± 1.3 | 8.0-9.2 |
| Bacteroides sp. strain F1 | 1 | 0.8 ± 0.8 | NA | 1 | 1.8 ± 1.8 | NA | NF | NF | |
| B. eggerthii | NF | NF | 1 | 1.6 ± 1.6 | NA | NF | NF | ||
| B. caccae | 5 | 3.6 ± 1.3 | 7.0-10.0 | 1 | 1.5 ± 1.5 | NA | 1 | 0.8 ± 0.8 | NA |
| Bacteroides sp. strain FO | 2 | 1.5 ± 1.0 | 8.3-10.0 | 2 | 3.1 ± 1.9 | 9.0-9.3 | NF | NF | |
| B. vulgatus | 6 | 4.6 ± 1.4 | 8.9-9.6 | NF | NF | 1 | 0.9 ± 0.9 | NA | |
| B. coagulans | 1 | 0.6 ± 0.6 | NA | NF | NF | NF | NF | ||
| B. uniformis | 4 | 3.2 ± 1.4 | 8.8-10.0 | NF | NF | NF | NF | ||
| B. tectus | 1 | 0.8 ± 0.8 | NA | NF | NF | NF | NF | ||
| B. fragilis | 3 | 2.3 ± 1.2 | 8.3-10.3 | NF | NF | 2 | 1.8 ± 1.2 | 8.0-9.5 | |
| B. forsythus | NF | NF | NF | NF | 1 | 1.0 ± 1.0 | NA | ||
| B. distasonis | 4 | 3.1 ± 1.3 | 8.9-9.9 | NF | NF | 1 | 0.8 ± 0.8 | NA | |
| Total bacteroides | 41 | 9.9 ± 0.1 | 9.0-10.7 (12)c | 7 | 6.5 ± 2.1 | 9.0-11.2 (4) | 15 | 6.7 ± 1.5 | 8.5-10.9 (7) |
| P. tannerae | 6 | 4.6 ± 1.4 | 8.4-10.0 | 2 | 3.1 ± 2.0 | 9.3-9.5 | 1 | 0.9 ± 0.9 | NA |
| P. loeschii | 4 | 3.2 ± 1.4 | 8.9-10.0 | 2 | 2.9 ± 1.9 | 8.7-8.9 | 1 | 1.1 ± 1.1 | NA |
| Prevotella sp. strain D68 | NF | NF | 1 | 1.3 ± 1.3 | NA | NF | NF | ||
| P. melaninogenica | 1 | 0.6 ± 0.6 | NA | NF | NF | 1 | 0.8 ± 0.8 | NA | |
| P. enoeea | 1 | 0.8 ± 0.8 | NA | NF | NF | NF | NF | ||
| Prevotella sp. strain D88 | 1 | 0.7 ± 0.7 | NA | NF | NF | 1 | 0.9 ± 0.9 | NA | |
| P. ruminicola | 1 | 0.8 ± 0.8 | NA | NF | NF | 1 | 0.9 ± 0.9 | NA | |
| Prevotella sp. strain D64 | 2 | 1.3 ± 0.9 | 8.0 | NF | NF | NF | NF | ||
| Total prevotellas | 16 | 7.1 ± 1.3 | 8.0-10.3 (9) | 5 | 6.1 ± 1.9 | 8.7-9.5 (4) | 5 | 3.7 ± 1.5 | 8.0-10.8 (4) |
Results are expressed as the population mean log10 CFU (gram wet weight of feces)−1 ± standard error of the mean for each group. NA, not applicable; NF, not found. Samples were taken from 12 HY, 6 HE, and 10 AE.
Results show the range log10 CFU (gram wet weight of feces)−1 of subjects in which the bacteria were isolated.
Values in parentheses are the number of subjects positive for carriage of the bacteria.
Bifidobacteria and lactic acid bacteria.
Although there was a decline in total bifidobacterial counts from HY to HE (Table 2), the reduction was not significant. Bifidobacterium angulatum was the predominant species in both groups. A small decrease in the numbers of bifidobacterial species was found for HE. In comparison, total counts for AE were significantly lower (P < 0.05) than those for HE, with many patients having no bifidobacteria at all. Consequently, overall species diversity was low in these individuals. Bifidobacterium catenulatum, Bifidobacterium boum, and Bifidobacterium infantis were isolated from feces samples from HY but were not detected for either elderly group, whereas Bifidobacterium coryneforme was only present in samples from AE.
TABLE 2.
Comparison of bifidobacteria and lactic acid bacteria species for different age groupsa
| Organism | HY
|
HE
|
AE
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of isolates | Mean | Range | No. of isolates | Mean | Range | No. of isolates | Mean | Range | |
| Bifidobacterium adolescentis | 10 | 7.3 ± 1.0 | 6.7-10.0 | 2 | 3.2 ± 2.0 | 9.5-9.8 | 2 | 1.6 ± 1.6 | 7.9-8.3 |
| Bifidobacterium angulatum | 11 | 8.3 ± 0.8 | 8.4-10.0 | 5 | 7.4 ± 1.5 | 7.9-9.3 | 2 | 1.7 ± 1.2 | 8.4-8.9 |
| Bifidobacterium pseudocatenulatum | 1 | 0.8 ± 0.8 | NA | 1 | 1.5 ± 1.5 | NA | NF | ||
| Bifidobacterium bifidum | 2 | 1.4 ± 1.0 | 7.9-9.0 | 2 | 3.2 ± 2.0 | 9.0-10.0 | 1 | 1.0 ± 1.0 | NA |
| Bifidobacterium breve | 3 | 2.2 ± 1.1 | 8.3-9.3 | 2 | 3.1 ± 1.9 | 8.6-9.7 | NF | ||
| Bifidobacterium longum | 5 | 3.5 ± 1.3 | 7.7-9.0 | 3 | 4.6 ± 2.0 | 8.3-9.8 | 2 | 1.3 ± 0.9 | 6.0-7.1 |
| Bifidobacterium dentium | 3 | 2.0 ± 1.1 | 6.7-9.0 | 1 | 1.6 ± 1.6 | NA | 1 | 0.8 ± 0.8 | NA |
| Bifidobacterium sp. strain DO5 | NF | NF | 1 | 1.6 ± 1.6 | NA | NF | NF | ||
| Bifidobacterium suis | NF | NF | 1 | 1.2 ± 1.2 | NA | NF | NF | ||
| Bifidobacterium catenulatum | 7 | 4.8 ± 1.2 | 7.0-9.8 | NF | NF | NF | NF | ||
| Bifidobacterium boum | 1 | 0.6 ± 0.6 | NA | NF | NF | NF | NF | ||
| Bifidobacterium coryneforme | NF | NF | NF | 1 | 0.9 ± 0.9 | NA | |||
| Bifidobacterium infantis | 1 | 0.7 ± 0.7 | NA | NF | NF | NF | |||
| Total bifidobacteria | 44 | 9.5 ± 0.2 | 8.5-10.3 (12) | 18 | 8.1 ± 1.6b | 9.2-10.5 (5) | 9 | 3.3 ± 1.4b | 6.0-9.6 (4) |
| L.c rhamnosus | 6 | 0.4 ± 0.4 | NA | 3 | 3.7 ± 1.6 | 6.7-8.0 | NF | NF | |
| L. paracasei subsp. paracasei | 6 | 3.4 ± 1.2 | 3.9-10.4 | 1 | 1.1 ± 1.1 | NA | 4 | 3.6 ± 1.5 | 7.5-10.7 |
| L. paracasei subsp. tolerans | NF | NF | 1 | 0.9 ± 0.9 | NA | NF | NF | ||
| L. breve | NF | NF | 1 | 1.1 ± 1.1 | NA | NF | NF | ||
| L. plantarum | 1 | 0.5 ± 0.5 | NA | 1 | 1.2 ± 1.2 | NA | NF | NF | |
| L. oris | NF | NF | 1 | 1.5 ± 1.5 | NA | NF | NF | ||
| Lactobacillus strain D10 | 2 | 1.6 ± 1.1 | 9.2-9.5 | NF | NF | 1 | 0.9 ± 0.9 | NA | |
| L. coryneformis subsp. coryneformis | 1 | 0.5 ± 0.5 | NA | NF | NF | 2 | 1.7 ± 1.1 | 8.2-8.5 | |
| L. coryneformis subsp. torquens | 1 | 0.5 ± 0.5 | NA | NF | NF | 1 | 0.7 ± 0.7 | NA | |
| L. delbruekii subsp. lactis | 2 | 0.8 ± 0.6 | 3.3-6.3 | NF | NF | 1 | 1.1 ± 1.1 | NA | |
| L. cateneforme | 1 | 0.6 ± 0.6 | NA | NF | NF | NF | NF | ||
| L. buchneri | 1 | 0.5 ± 0.5 | NA | NF | NF | 1 | 0.7 ± 0.7 | NA | |
| L. sharpeae | 1 | 0.3 ± 0.3 | NA | NF | NF | NF | NA | ||
| L. curvatus | NF | NF | NF | NF | 2 | 1.7 ± 1.1 | 7.6-9.0 | ||
| L. animalis | NF | NF | NF | NF | 1 | 0.9 ± 0.9 | NA | ||
| L. helveticus | NF | NF | NF | NF | 2 | 1.6 ± 1.1 | 6.7-9.3 | ||
| L. reuteri | NF | NF | NF | NF | 1 | 0.9 ± 0.9 | NA | ||
| L. acidophilus | 1 | 0.6 ± 0.6 | NA | NF | NF | 1 | 0.9 ± 0.9 | NA | |
| Total lactobacilli | 23 | 6.3 ± 1.0 | 4.2-10.4 (10) | 8 | 4.1 ± 1.8 | 7.7-9.0 (3) | 17 | 6.5 ± 1.5 | 7.9-10.8 (7) |
| Lactococcus lactis | NF | NF | 2 | 2.4 ± 1.5 | 6.7-7.9 | NF | NF | ||
| Total lactococci | NF | NF | 2 | 2.4 ± 1.5 | 6.7-7.9 (2) | NF | NF | ||
Total lactobacilli numbers in samples from HE were lower than in those from HY; however, the highest counts occurred in samples from AE, with almost a 100-fold difference compared to counts for HE. Samples from AE also had greater species diversity, and many of these organisms were not found in HE or HY. Large variations in total lactobacilli counts were evident for all three groups, particularly HE and AE.
Clostridia, fusobacteria, and propionibacteria.
Counts of these bacteria are shown in Table 3. Clostridial diversity was markedly higher in AE patients compared to the other two groups. Conversely, the lowest numbers were found in samples from HE, with an associated reduction in species diversity. A large number of different species were observed in samples within each cohort, particularly HY and AE, with no particular organism predominating in any group. Nevertheless, many of the clostridia, such as C. difficile, in samples from AE were not detected in samples from the other volunteers. A comparatively limited number of fusobacteria and propionibacteria were isolated in this study. Fusobacteria were not detected at all in stools of HY; however, they were present in low numbers in samples from AE and HE. The HE had the highest counts, but AE had slightly higher species diversity. Propionibacteria counts were low, particularly in HY and HE, where this genus was detected in one individual only. The increased frequency of isolation of propionibacteria in AE was accompanied by an increase in the total numbers of propionibacteria for that group relative to other groups.
TABLE 3.
Comparison of clostridia, fusobacteria, and propionibacteria for different age groupsa
| Organism | HY
|
HE
|
AE
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of isolates | Mean | Range | No. of isolates | Mean | Range | No. of isolates | Mean | Range | |
| C. sardiniensis | NF | NF | 1 | 1.0 ± 1.0 | NA | NF | |||
| C. malenominatum | 2 | 1.1 ± 0.7 | 5.6-7.4 | 1 | 1.3 ± 1.3 | NA | 2 | 1.6 ± 1.1 | 7.3-8.6 |
| C. inocuum | 2 | 1.1 ± 0.7 | 5.3-7.4 | 2 | 2.5 ± 1.6 | 7.3-7.7 | NF | NF | |
| C. cocleatum | NF | NF | 2 | 2.8 ± 1.8 | 8.0-8.9 | 1 | 0.8 ± 0.8 | NA | |
| C. bifermentans | 2 | 1.5 ± 1.0 | 7.5-10.0 | 1 | 1.6 ± 1.6 | NA | 4 | 3.3 ± 1.4 | 7.4-9.0 |
| C. borati | 1 | 0.5 ± 0.5 | NA | NF | NF | NF | NF | ||
| C. sordellii | 2 | 1.3 ± 0.8 | 7.0-8.0 | NF | NF | 2 | 1.5 ± 1.0 | 6.6-8.8 | |
| C. ramosum | 2 | 1.0 ± 0.7 | 5.1-7.0 | NF | NF | 1 | 0.9 ± 0.9 | NA | |
| C. inoculum | 1 | 0.5 ± 0.5 | NA | NF | NF | NF | NF | ||
| C. perfringens | 2 | 1.2 ± 0.8 | 7.1-7.3 | NF | NF | 1 | 0.7 ± 0.7 | NA | |
| C. sporogenes | 1 | 0.6 ± 0.6 | NA | NF | NF | 1 | 0.8 ± 0.8 | NA | |
| C. sphenoides | 1 | 0.5 ± 0.5 | NA | NF | NF | NF | NF | ||
| C. clostridiiforme | 1 | 0.6 ± 0.6 | NA | NF | NF | 3 | 2.2 ± 1.1 | 6.0-8.3 | |
| C. butyricum | NF | NF | NF | NF | 1 | 0.5 ± 0.5 | NA | ||
| C. sporosphaerides | NF | NF | NF | NF | 1 | 0.7 ± 0.7 | NA | ||
| C. difficile | NF | NF | NF | NF | 1 | 0.6 ± 0.6 | NA | ||
| C. novyi type A | NF | NF | NF | NF | 1 | 0.8 ± 0.8 | NA | ||
| C. tertium | NF | NF | NF | NF | 1 | 0.8 ± 0.8 | NA | ||
| C. indolis | NF | NF | NF | NF | 1 | 0.7 ± 0.7 | NA | ||
| Total clostridia | 17 | 5.6 ± 1.0 | 5.3-10.0 (9) | 7 | 5.3 ± 1.7 | 6.0-9.6 (4) | 21 | 7.6 ± 0.9 | 7.7-9.5 (9) |
| Fusobacterium prausnitziib | NF | NF | 1 | 1.2 ± 1.2 | NA | 1 | 0.9 ± 0.9 | NA | |
| Fusobacterium alocis | NF | NF | 1 | 1.4 ± 1.4 | NA | 1 | 0.9 ± 0.9 | NA | |
| Fusobacterium russii | NF | NF | NF | 2 | 1.6 ± 1.1 | 7.3-9.0 | |||
| Total fusobacteria | NF | NF | 2 | 2.6 ± 1.6 (2) | 7.2-8.3 | 4 | 1.7 ± 1.1 | 7.3-9.0 (2) | |
| Propionibacterium acnes | 1 | 0.7 ± 0.7 | NA | 1 | 1.0 ± 1.0 | NA | 4 | 3.7 ± 1.5 | 8.0-10.4 |
| Propionibacterium jensenii | NF | NF | 1 | 1.3 ± 1.3 | NA | NF | NF | ||
| Total propionibacteria | 1 | 0.7 ± 0.7 (1) | NA | 2 | 1.3 ± 1.3 (1) | NA | 4 | 3.7 ± 1.5 | 8.0 ± 10.4 (4) |
Eubacteria and anaerobic cocci.
Fecal eubacterial levels were similar for the different study groups, though greater numbers were present in AE and HE than in HY (Table 4). Although the HE had the highest total eubacterial counts, samples from those individuals had the lowest species diversity, with only two different organisms, Eubacterium rectale and Eubacterium cylindroides, being detected. Eubacterium aerofaciens and Eubacterium rectale were the most prevalent species in HY, whereas Eubacterium limosum predominated in AE. Anaerobic gram-positive and gram-negative cocci were present in low numbers in HY, HE, and AE.
TABLE 4.
Comparison of eubacteria and anaerobic cocci species for different age groupsa
| Organism | HY
|
HE
|
AE
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of isolates | Mean | Range | No. of isolates | Mean | Range | No. of isolates | Mean | Range | |
| Eubacterium aerofaciens | 4 | 2.8 ± 1.2 | 8.3-8.6 | NF | NF | 1 | 0.6 ± 0.6 | NA | |
| Eubacterium dolchium | 1 | 0.8 ± 0.8 | NA | NF | NF | NF | NF | ||
| Eubacterium sp. strain D13 | 1 | 0.8 ± 0.8 | NA | NF | NF | NF | NF | ||
| Eubacterium eligens | 1 | 0.7 ± 0.7 | NA | NF | NF | 1 | 0.9 ± 0.9 | NA | |
| Eubacterium rectale | 3 | 2.0 ± 1.0 | 6.5-8.9 | 1 | 1.5 ± 1.5 | NA | 1 | 0.7 ± 0.7 | NA |
| Eubacterium timidium | 2 | 1.4 ± 0.9 | 7.3-9.3 | NF | NF | NF | NF | ||
| Eubacterium bioforme | 1 | 0.8 ± 0.8 | NA | NF | NF | 1 | 0.7 ± 0.7 | NA | |
| Eubacterium plauti | 1 | 0.6 ± 0.6 | NA | NF | NF | NF | NF | ||
| Eubacterium age | 1 | 0.7 ± 0.7 | NA | NF | NF | NF | NF | ||
| Eubacterium lentum | NF | NF | NF | NF | 1 | 0.9 ± 0.9 | NA | ||
| Eubacterium ventriosum | NF | NF | NF | NF | 1 | 0.8 ± 0.8 | NA | ||
| Eubacterium cylindroides | NF | NF | 3 | 3.5 ± 1.6 | 5.8-7.9 | 1 | 1.0 ± 1.0 | NA | |
| Eubacterium brachyi | NF | NF | NF | NF | 1 | 0.9 ± 0.9 | NA | ||
| Eubacterium limosum | NF | NF | NF | NF | 2 | 1.9 ± 1.3 | 9.2-10.0 | ||
| Eubacterium ramulus | NF | NF | NF | NF | NF | NF | |||
| Total eubacteria | 15 | 5.3 ± 1.3 | 8.4-9.5 (7) | 4 | 6.5 ± 1.4 | 5.8-9.3 (5) | 10 | 5.9 ± 1.4 | 7.0-10.0 (6) |
| Peptopstreptococcus sp. strain BM CFA 2 | NF | NF | 1 | 1.5 ± 1.5 | NA | NF | NF | ||
| Peptopstreptococcus productis strain IIB | NF | NF | NF | NF | 1 | 0.9 ± 0.9 | NA | ||
| Veillonella atypical | NF | NF | 1 | 1.2 ± 1.2 | NA | NF | NF | ||
| Peptococcus niger | NF | NF | 1 | 1.2 ± 1.2 | NA | NF | NF | ||
| Ruminococcus obeum | 1 | 0.8 ± 0.8 | NA | NF | NF | NF | NF | ||
| Total anaerobic cocci | 1 | 0.8 ± 0.8 | NA | 3 | 3.8 ± 1.7 | 7.0-9.0 (3) | 1 | 0.9 ± 0.9 | NA |
See footnotes to Table 1.
Aerobes and facultative anaerobes.
The most predominant member of the Enterobacteriaceae detected in all three groups of volunteers was Escherichia coli (Table 5). This organism was the only enterobacterium isolated from samples from HE and had the largest numbers and prevalence in that group; samples from HY and AE showed similar prevalences, but there were reduced counts in samples from HY. Various other enterobacteria, including Providencia spp. and Proteus spp., were found in HY and AE.
TABLE 5.
Comparison of aerobes and facultatively anaerobic species for different age groupsa
| Organism | HY
|
HE
|
AE
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of isolates | Mean | Range | No. of isolates | Mean | Range | No. of isolates | Mean | Range | |
| Escherichia coli | 8 | 4.3 ± 1.0 | 3.3-7.9 | 6 | 7.3 ± 0.4 | 6.0-8.7 | 7 | 5.6 ± 1.2 | 6.6-8.8 |
| Klebsiella pneumoniae | 1 | 0.4 ± 0.4 | NA | NF | NF | 1 | 0.7 ± 0.7 | NA | |
| Klebsiella ascorbata | 1 | 0.5 ± 0.5 | NA | NF | NF | NF | NF | ||
| Serratia fonticola | 2 | 0.9 ± 0.6 | 5.5-5.8 | NF | NF | NF | NF | ||
| Morganella morganii | 2 | 1.0 ± 0.7 | 5.2-6.9 | NF | NF | 1 | 0.6 ± 0.6 | NA | |
| Proteus vulgatis | NF | NF | NF | NF | 1 | 0.8 ± 0.8 | NA | ||
| Proteus mirabilis | NF | NF | NF | NF | 1 | 0.8 ± 0.8 | NA | ||
| Providencia stuartii | NF | NF | NF | NF | 1 | 0.8 ± 0.8 | NA | ||
| Total enterobacteria | 14 | 5.8 ± 0.6 | 3.3-7.9 (11) | 6 | 7.3 ± 0.4 | 6.0-8.7 (6) | 12 | 5.7 ± 1.3 | 6.6-8.9 (7) |
| E. durans | 4 | 2.7 ± 1.2 | 6.0-10.0 | NF | NF | 5 | 4.9 ± 1.6 | 7.9-11.0 | |
| E. faecalis | 7 | 3.8 ± 1.0 | 5.0-8.5 | NF | NF | 8 | 6.5 ± 1.1 | 6.6-9.3 | |
| E. faecium | 2 | 1.2 ± 0.8 | 7.0-7.4 | NF | NF | 4 | 3.0 ± 1.2 | 6.3-8.1 | |
| E. avium | 1 | 0.8 ± 0.8 | NA | NF | NF | 3 | 2.6 ± 1.3 | 7.8-9.4 | |
| E. pseudoavium | NF | NF | NF | NF | 1 | 0.8 ± 0.8 | NA | ||
| E. solitarius | NF | NF | NF | NF | 2 | 1.8 ± 1.2 | 8.5-9.3 | ||
| E. hirae | NF | NF | NF | NF | 2 | 1.7 ± 1.1 | 8.2-8.3 | ||
| Total enterococci | 14 | 6.5 ± 0.9 | 5.0-10.1 (9) | NF | NF | 25 | 9.1 ± 0.4 | 6.8-11.1 (10) | |
| Streptococcus salvarius | NF | NF | 2 | 2.8 ± 1.8 | 8.0-9.0 | NF | NF | ||
| Streptococcus parasanguis | NF | NF | 2 | 2.2 ± 1.4 | 5.7-7.3 | NF | NF | ||
| Streptococcus sp. strain M7 | NF | NF | 1 | 1.2 ± 1.2 | NA | NF | NF | ||
| Streptococcus intermedius | NF | NF | 1 | 1.2 ± 1.2 | NA | NF | NF | ||
| Streptococcus mutans | 2 | 1.2 ± 0.8 | 6.5-8.2 | NF | NF | NF | NF | ||
| Streptococcus plantarum | NF | NF | NF | NF | 2 | 1.8 ± 1.2 | 8.4-9.1 | ||
| Streptococcus sp. strain SO3 | NF | NF | NF | NF | 1 | 1.0 ± 1.0 | NA | ||
| Total streptococci | 2 | 1.2 ± 0.8 | 6.5-8.2 (2) | 6 | 2.8 ± 1.8 | 8.0-9.0 (2) | 3 | 2.7 ± 1.4 | 8.4-9.6 (3) |
| Staphylococcus epidermidis | 3 | 2.1 ± 1.1 | 7.5-10.0 | 2 | 2.6 ± 1.6 | 7.5-7.8 | 4 | 2.8 ± 1.2 | 6.7-7.5 |
| Staphylococcus warneri | 2 | 1.4 ± 1.0 | 6.8-9.9 | 1 | 1.3 ± 1.3 | NA | 2 | 2.1 ± 1.4 | 10.0-10.5 |
| Staphylococcus simulans | NF | NF | 1 | 1.2 ± 1.2 | NA | NF | NF | ||
| Staphylococcus aureus | 1 | 0.7 ± 0.7 | NA | NF | NF | NF | NF | ||
| Staphylococcus haemolyticus | NF | NF | NF | NF | 1 | 0.7 ± 0.7 | NA | ||
| Staphylococcus kloosii | NF | NF | NF | NF | 1 | 0.7 ± 0.7 | NA | ||
| Total staphylococci | 6 | 2.2 ± 1.2 | 7.5-10.3 (3) | 4 | 5.1 ± 1.6 | 7.0-8.0 (4) | 8 | 4.9 ± 1.4 | 7.0-10.5 (6) |
| Candida albicans | 2 | 1.2 ± 0.8 | 7.0-7.3 | 3 | 3.6 ± 1.6 | 6.0-8.5 | 2 | 1.4 ± 0.9 | 7.0-10.5 |
| Pseudomonas aeruginosa | NF | NF | NF | NF | 1 | 0.6 ± 0.6 | NA | ||
See footnotes to Table 1.
Proportionally high numbers of enterococci were observed in samples from AE, with seven different species being detected. The HY had lower total numbers and reduced prevalences of these bacteria, although for both HY and AE, the individual counts were spread over a large range. E. faecalis was the predominant species in both HY and AE; however, E. durans had the greatest range, with some individuals having extremely high numbers in their feces. Enterococci were not isolated from any samples from the HE.
Streptococci were relatively uncommon, especially for HY, although the numbers of species observed was higher in samples from HE. Levels of staphylococci were also high for both elderly groups, compared to HY, with only a small increase in species diversity and a similar range of individual counts between the groups. Candida albicans occurred mainly in samples from HE, where it was detected more frequently than in samples from the other groups. HY had the lowest numbers and prevalence of Candida albicans. Pseudomonas aeruginosa was isolated from one AE fecal donor and was not detected in samples from HY or HE.
Microbiological species diversity.
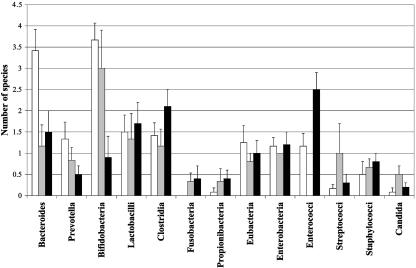
A summary of the changes in species diversity for each bacterial group detected in feces samples from HY, HE, and AE is shown in Fig. 2. A marked reduction in the numbers of species in both elderly populations was observed for many anaerobic bacterial groups, such as bacteroides, prevotella, and bifidobacteria. Lactobacilli and clostridial species diversity were decreased for HE compared to levels for the other groups, with samples from AE demonstrating the greatest species diversity in both bacterial genera. Similar changes occurred in eubacteria and enterobacteria populations. The large increase in enterococci in samples from AE can be clearly seen in Fig. 2, with an increase in streptococci and yeasts being evident for HE.
FIG. 2.
Summary of bacterial species diversity in feces from individuals in different age groups. Results are expressed as mean values ± standard errors of the mean. Healthy young (white), healthy elderly (grey), and antibiotic-treated elderly (black).
Proteolytic and amylolytic activities.
The abilities of fecal isolates to hydrolyze casein were determined by observing hydrolysis zones around bacterial colonies. The abundances of proteolytic species were calculated as percentages of the total numbers of bacteria detected, and the results showed that 15.8, 4.6, and 19.4% of all isolates from HY, HE, and AE, respectively, were proteolytic. Amylolytic activities were visualized similarly on starch plates, where hydrolysis was observed for 14.3, 3.6, and 9.0% of isolates from HY, HE, and AE, respectively.
Bacterial fermentation products in feces.
Median fecal ammonia levels were highest in samples from HY and lowest in those from AE. Fecal ammonia was significantly lower (P < 0.05) for AE than for HE. The amounts of fermentation acids in samples from HY and HE were generally similar (Table 6), with only small reductions in fecal acetate, butyrate, and valerate, and total SCFA for HE. However, the production levels of these fermentation products were altered for AE, with significant decreases in propionate, butyrate, isobutyrate, and valerate being observed in comparison to levels for HE. The reduction in total SCFA between HE and AE was found to be approaching significance. Concentrations of fecal lactate and succinate were also determined. The results demonstrated the presence of succinate in samples from all three subject groups, with the highest concentrations occurring for AE and the lowest for HE (Table 6). Lactate was not detected in any of the fecal samples.
TABLE 6.
Comparison of fecal organic acid concentrations for different age groupsa
| Study group | Acetate
|
Propionate
|
Butyrate
|
Isobutyrate
|
Isovalerate
|
Valerate
|
Isocaproate
|
Caproate
|
Total SCFA
|
Lactate
|
Succinate
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | 25 | 75 | Median | 25 | 75 | Median | 25 | 75 | Median | 25 | 75 | Median | 25 | 75 | Median | 25 | 75 | Median | 25 | 75 | Median | 25 | 75 | Median | 25 | 75 | Median | 25 | 75 | Median | 25 | 75 | |
| HY | 29.6 | 16.9 | 36.0 | 11.4 | 6.4 | 19.9 | 11.0 | 5.2 | 16.7 | 0.5 | 0.3 | 1.1 | 1.3 | 0.8 | 2.1 | 1.2 | 0.8 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.4 | 81.2 | 43.2 | 87.1 | 0.0 | 0.0 | 0.0 | 3.2 | 1.2 | 4.8 |
| HE | 25.5 | 7.9 | 47.6 | 12.7 | 7.1 | 19.8 | 9.2 | 4.2 | 15.4 | 0.8 | 0.5 | 1.4 | 1.3 | 0.9 | 2.0 | 1.1 | 0.6 | 2.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.4 | 67.4 | 33.9 | 117.1 | 0.0 | 0.0 | 0.0 | 0.7 | 0.5 | 1.3 |
| AE | 23.2 | 19.9 | 25.0 | 8.7b | 5.5 | 10.4 | 3.7b | 0.8 | 5.4 | 0.3b | 0.0 | 0.9 | 0.9 | 0.0 | 1.0 | 0.0b | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 36.5c | 34.3 | 42.3 | 0.0 | 0.0 | 0.0 | 7.7 | 4.9 | 9.5 |
Results are expressed as the median (mmol kg−1) ± interquartile range. 25, 25th percentile; 75, 75th percentile. Samples were taken from 14 HY, 70 HE, and 9 AE.
Significant difference between HE and AE.
Approaching significance.
DISCUSSION
A number of molecular techniques are available for studying intestinal bacteria such as dot blot hybridizations of rRNA gene sequences (22, 49), denaturing gradient gel electrophoresis (18, 27), real-time PCR (12, 34), and fluorescent in situ hybridization analysis (33, 46). However, all of these methods have limitations, ranging from lack of sensitivity and limited probe availability (dot blot hybridizations, fluorescent in situ hybridization analysis), to being nonquantitative (denaturing gradient gel electrophoresis). Culturing bacteria from the fecal material has the drawback that some species cannot be readily recovered from intestinal samples; however, this problem can be circumvented, to some extent, by using a variety of growth media and culturing conditions which are sensitive and quantitative and capable of detecting a large range of species. The present investigation is the largest study undertaken to investigate the microbiota in elderly people in such detail. In total, bacteriological analyses were performed on samples from 28 individuals, and chemical analyses of feces were performed for 93 subjects.
Bacteroides populations in the HY were found to be consistent, though levels of these bacteria decreased with the ages of the subjects. A large variety of species was detected, with an average of just over three different species per person. The most dominant of these species was B. ovatus, closely followed by B. thetaiotaomicron, B. vulgatus, and B. caccae (Table 1). Other studies have reported similar results (23, 37). However, the previously observed increase in bacteroides diversity in elderly people living in the Cambridge area in southeast England (23) was not repeated in this study, where marked reductions in numbers of different species occurred in samples from both HE and AE. Since the analytical methods used to investigate these organisms were similar in the two investigations, the reduction might result from nutritional, lifestyle, genetic, or environmental differences for healthy elderly people living in two different geographic locations. In the present study, HY and AE were of mixed genders, while due to the inability to recruit older males, the HE group was composed entirely of females; they were postmenopausal, however, which would eliminate many hormonal differences.
Bacteroides are nutritionally versatile and are able to utilize a wide variety of different polymerized carbon sources; consequently, they are believed to be responsible for the majority of polysaccharide digestion occurring in the colon (32, 48). Changes in such a nutritionally important subpopulation in the bowel may have effects on the host through changes in SCFA production (30) as well as for other bacteria in the gut that depend nutritionally on a complex cross-feeding network in the ecosystem (14). Similar reductions in prevotella numbers and species diversity were also observed in the elderly, particularly in the AE, but the ecological significance of these organisms in the gut is unclear.
The appearance of fusobacteria exclusively in elderly people may indicate a trend towards putrefaction in the large bowel, principally in patients undergoing antibiotic therapy (Table 3). These bacteria mainly ferment amino acids, resulting in the production of several toxic metabolic end products, such as ammonia and indoles (20, 21). The increase in proteolytic species, such as fusobacteria, propionibacteria, and clostridia, in the AE coincided with the marked increase in proteolytic potential observed for these individuals.
The genus Clostridium comprises a heterogeneous group of microorganisms with highly diverse nutritional requirements and habitats. A marked increase in total numbers of these bacteria was observed for the AE and was accompanied by a distinct increase in species diversity (Table 3). An average of 2.1 different species per person were detected, almost double that for the HY and HE, with the predominant Clostridium species including C. bifermentans, C. clostridioforme, C. sordellii, and C. malenominatum. The pathogen C. difficile was found in the sample from one patient, demonstrating one of the side effects of antibiotic therapy in the hospital environment. Unlike many other clostridia, C. malenominatum and C. sporosphaeroides utilize pyruvate and lactate (19, 21), and their increased numbers and prominence in AE patients may have been related to increased lactobacilli (Table 2), as suggested by Hopkins and Macfarlane (23). However, the increase in clostridial numbers in AE patients differs from the results of Ljungberg et al. (29), where a marked reduction in levels of these bacteria was found following administration of ciprofloxacin to young and elderly volunteers.
Eubacteria have complex nutritional requirements, and their increase in total numbers in the HE group may have health consequences due to the greater potential for bile acid transformations, creating potentially harmful metabolites in the gut. Additionally, cell material from Eubacterium aerofaciens has been reported to induce arthritis in rats inoculated intraperitoneally with bacterial cell wall components (50). If this effect occurs in vivo, these bacteria could contribute to the increase in arthritic conditions seen in elderly people. Eubacteria have been reported to be second only to bacteroides in numbers isolated from the large intestine (11), but results obtained in the present study do not corroborate this finding (Table 4), possibly due to the fastidious nature of some members of this genus.
The beneficial properties of lactobacilli have long been recognized, and an increase in their numbers and species diversity with increased subject age and use of antibiotic therapy has been reported in earlier studies (23, 26). This increase also occurred with fecal lactobacilli in the AE in the present investigation. The abilities of lactobacilli to persist through antibiotic treatment may accentuate their value in the colonic microbiota, particularly against antibiotic-associated diarrhea. Conversely, the presence of numerous plasmids, transposons, and insertion sequences in various lactobacilli (56) might be less beneficial and could potentially provide a mechanism for the spread of antibiotic resistance in the gut.
Elderly people had reduced levels of bifidobacteria (Table 2), which has been reported in earlier studies (2, 13, 22, 40, 41). Other studies have indicated that microbiotas in healthy young adults contain on average three to four different bifidobacterial species (35), which was also found here. However, in HE and AE, the average number of species was reduced (to 3 and 0.9, respectively). Bifidobacterium adolescentis is generally considered to be the dominant species in the bifidobacterial community among adults (13, 40), although in this study, Bifidobacterium angulatum and Bifidobacterium longum were more prevalent. Bifidobacterium catenulatum, Bifidobacterium boum, and Bifidobacterium infantis were not detected in either of the elderly groups, with Bifidobacterium pseudocatenulatum and Bifidobacterium breve also being absent in AE patients. Many of these species, however, have been reported to be more common in children (13, 40). Changes in the adhesive abilities of bifidobacteria have been put forward as a reason for reductions in these bacteria in the aging intestinal ecosystem (17) or for alterations in the chemical composition and structure of intestinal mucus with age (44). In view of the immunomodulatory and potentially protective properties of many species in this genus, these changes could result in a reduced functionality and immune responsiveness in the gut and an increased susceptibility to gastrointestinal infections.
Further evidence to support changes in bacterial communities in the aging gut was the marked rise in numbers of facultative anaerobes (Fig. 1B), with significant increases (P < 0.05) demonstrated in samples from AE. Levels of enterobacteria, streptococci, staphylococci, and yeasts all increased, particularly for HE, although enterococci, which were not isolated from samples from a single HE volunteer, were found in highest numbers in samples from AE. These findings might be linked to increased serum antibodies to commensal gut microorganisms, such as Escherichia coli and E. faecalis, as reported by Percival et al. (45). Specific changes in levels of Enterobacteriaceae family members were most noticeable (Table 5), with Escherichia coli being the only species found in samples from HE. In contrast, the variety of species detected in samples from AE was much greater, with Proteus spp. and Providencia spp. being isolated. These bacteria have been associated with gastrointestinal illness (42), with the latter expressing invasive properties in vitro (24). The presence of the strict aerobe Pseudomonas aeruginosa and the increase in Candida albicans in AE indicated that there was a marked shift in environmental conditions in the gut ecosystem following antibiotic treatment. The proliferation of enterococci in this group may also be due to the depletion of other organisms following therapy, resulting in reduced competition for nutrients and space.
Changes in metabolic functionality and fermentative activities in the colonic microflora were assessed by analysis of SCFA, lactate, succinate, and ammonia in feces. An overall reduction in fecal excretion of SCFA occurred with increased age, particularly in association with antibiotic treatment (Table 6), which correlated with the bacteriological changes. Since the vast majority (ca. 95 to 99%) of SCFA and other fermentation products formed by intestinal bacteria are absorbed from the gut or are metabolized further by the microbiota (7), small changes in fecal excretion indicate large differences in production in the gut. The reductions in acetate found in samples from healthy elderly volunteers and antibiotic-treated elderly patients may be associated with the decline in bacteroides and bifidobacteria, although this SCFA is the principal fermentation product formed by most gut microorganisms. Furthermore, levels of branched-chain fatty acids, which are products of branched-chain amino acid fermentation, decreased for AE. HE had marked reductions in fecal succinate (Table 6), which could be associated with the decline in bacteroides numbers. As described above, changes in levels of some clostridial species, particularly following antibiotic therapy, may be due to the increased production of lactate by lactobacilli, where it is an important electron sink product. Utilization of this product by gut anaerobes may thus explain the absence of fecal lactate.
The increase in proteolytic species in AE patients might predispose them towards a corresponding increase in putrefactive fermentation products, however, as was observed with branched-chain fatty acids. The reverse was found for levels of ammonia, which decreased from HY to HE to AE. The reduction in fecal weight with increased age provided a good indication that physiological changes were occurring in the gastrointestinal tracts of elderly people. Low fecal weights have been linked to slow intestinal transit times, reduced excretion of bacterial cell mass (53), and increased levels of protein breakdown and amino acid fermentation, resulting in the formation of toxic bacterial metabolites (31). From the bacteriological measurements made in this investigation and studies of bacterial metabolites, it is clear that in terms of size and complexity, the metabolic potential of the colonic microbiota in older people is much reduced.
Acknowledgments
This work was funded by the Medical Research Council and The Health Foundation.
REFERENCES
- 1.Beerens, H. 1990. An elective and selective isolation medium for Bifidobacterium spp. Lett. Appl. Microbiol. 11:155-157. [Google Scholar]
- 2.Benno, Y., H. Nakao, K. Uchida, and T. Mitsuoka. 1992. Impact of the advances in age on the gastrointestinal microflora of beagle dogs. J. Vet. Med. Sci. 54:703-706. [DOI] [PubMed] [Google Scholar]
- 3.Brandt, L. A., K. A. Kosche, D. A. Greenwald, and D. Berkman. 1999. Clostridium difficile-associated diarrhea in the elderly. Am. J. Gastroenterol. 94:3263-3266. [DOI] [PubMed] [Google Scholar]
- 4.Brocklehurst, J. C. 1972. Bowel management in the neurologically disabled. The problems of old age. Proc. R. Soc. Med. 65:66-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castell, D. O. 1988. Eating and swallowing disorders in the elderly. Pract. Gastroenterol. 12:32-43. [Google Scholar]
- 6.Cooperstock, M. S., and A. J. Zedd. 1983. Intestinal flora of infants, p. 78-93. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, New York, N.Y.
- 7.Cummings, J. H. 1981. Short chain fatty acids in the human colon. Gut 22:763-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doty, R. L., P. Sharman, S. L. Applebaum, R. Giberson, L. Siksorski, and L. Rosenbery. 1984. Smell identification ability: changes with age. Science 22:1441-1443. [DOI] [PubMed] [Google Scholar]
- 9.Duncan, S. H., G. L. Hold, H. J. Harmsen, C. S. Stewart, and H. J. Flint. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. E vol. Microbiol. 52:2141-2146. [DOI] [PubMed] [Google Scholar]
- 10.Favier, C. F., E. E. Vaughan, V. M. de Vos, and A. D. L. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finegold, S. M., V. L. Sutter, and G. E. Mathisen. 1983. Normal indigenous intestinal microflora, p. 3-31. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, New York, N.Y.
- 12.Fite, A., G. T. Macfarlane, J. H. Cummings, M. J. Hopkins, S. C. Kong, E. Furrie, and S. Macfarlane. 2004. Identification and quantitation of mucosal and faecal desulphovibrios using real-time PCR. Gut 53:523-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gavini, F., C. Cayuela, J.-M. Antoine, C. Lecoq, B. Lefebvre, J.-M. Membre, and C. Neut. 2001. Differences in the spatial distribution of bifidobacterial and enterobacterial species in human faecal microflora of three different (children, adults, elderly) age groups. Microb. Ecol. Health Dis. 13:40-45. [Google Scholar]
- 14.Gibson, G. R., J. H. Cummings, and G. T. Macfarlane. 1988. Use of a three-stage continuous culture system to study the effect of mucin on dissimilatory sulfate reduction and methanogenesis by mixed populations of human gut bacteria. Appl. Environ. Microbiol. 54:2750-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbach, S. L., L. Nahas, P. I. Lerner, and L. Weinstein. 1967. Studies of intestinal microflora. I. Effects of diet, age, and periodic sampling on numbers of fecal microorganisms in man. Gastroenterology 53:845-855. [PubMed] [Google Scholar]
- 16.Harmsen, H. J., A. C. Wildeboer-Veloo, G. C. Raangs, A. A. Wagendorp, N. Klijn, J. G. Bindels, and G. W. Welling. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61-67. [DOI] [PubMed] [Google Scholar]
- 17.He, F., A. C. Ouwehand, E. Isolauri, M. Hosoda, Y. Benno, and S. Salminen. 2001. Differences in composition and mucosal adhesion of bifidobacteria isolated from healthy adults and healthy seniors. Curr. Microbiol. 43:351-354. [DOI] [PubMed] [Google Scholar]
- 18.Heilig, H. G. H. J., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. L. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hippe, H., J. R. Andreesen, and G. Gottschalk. 1992. The genus Clostridium—nonmedical, p. 1800-1866. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. Springer-Verlag, New York, N.Y.
- 20.Holdeman, L. V., and L. H. Moore. 1974. Genus I. Bacteroides (Castellani and Chalmers, 1919), p. 385-404. In R. E. Buchanan and N. E. Gibbons (ed.), Bergey's manual of determinative bacteriology. Williams & Wilkins, Baltimore, Md.
- 21.Holdeman, L. V., E. P. Cato, and W. E. C. Moore. 1977. Anaerobe laboratory manual. 4th ed. Blacksburg, Virginia Polytechnic Institute and State University, Blacksburg, Va.
- 22.Hopkins, M. J., R. Sharp, and G. T. Macfarlane. 2001. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut 48:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hopkins, M. J., and G. T. Macfarlane. 2002. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J. Med. Microbiol. 51:448-454. [DOI] [PubMed] [Google Scholar]
- 24.Janda, J. M., S. L. Abbott, D. Woodward, and S. Khashe. 1998. Invasion of Hep-2 and other eukaryotic cell lines by Providenciae; further evidence supporting the role of Providencia alcalifaciens in bacterial gastroenteritis. Curr. Microbiol. 37:159-165. [DOI] [PubMed] [Google Scholar]
- 25.Karlsson, S., M. Persson, and G. E. Carlsson. 1991. Mandibular movement and velocity in relation to state of dentition and age. J. Oral Rehabil. 18:1-8. [DOI] [PubMed] [Google Scholar]
- 26.Kleessen, B., B. Sykura, H. J. Zunft, and M. Blaut. 1997. Effects of inulin and lactose on fecal microflora, microbial activity, and bowel habit in elderly constipated persons. Am. J. Clin. Nutr. 65:1397-1402. [DOI] [PubMed] [Google Scholar]
- 27.Konstantinov, S. R., N. Fitzsimons, E. E. Vaughan, and D. L. Akkermans. 2002. From composition to functionality of the intestinal microbial communities, 59-84. In G. W. Tannock (ed.), Probiotics and prebiotics: where are we going. Caister Academic Press, Wymondham, United Kingdom.
- 28.Lesourd, B. M., C. Laisney, R. Slavatore, S. Meaume, and R. Moulias. 1994. Decreased maturation of T-cell populations in the healthy elderly: influence of nutritional factors on the appearance of double negative CD4-CD8-CD2+ cells. Arch. Gerontol. Geriatr. 4:S149-S154. [DOI] [PubMed] [Google Scholar]
- 29.Ljungberg, B., I. Nilsson-Ehle, C. Edlund, and C. E. Nord. 1990. Influence of ciprofloxacin on the colonic microflora in young and elderly volunteers: no impact of the altered drug absorption. Scand. J. Infect. Dis. 22:205-208. [DOI] [PubMed] [Google Scholar]
- 30.Macfarlane, G. T., and H. N. Englyst. 1986. Starch utilization by the human large intestinal microflora. J. Appl. Bacteriol. 60:195-201. [DOI] [PubMed] [Google Scholar]
- 31.Macfarlane, G. T., J. H. Cummings, S. Macfarlane, and G. R. Gibson. 1989. Influence of retention time on degradation of pancreatic enzymes by human colonic bacteria grown in a 3-stage continuous culture system. J. Appl. Bacteriol. 67:520-527. [PubMed] [Google Scholar]
- 32.Macfarlane, G. T., and G. R. Gibson. 1991. Co-utilization of polymerized carbon sources by Bacteroides ovatus grown in a two-stage continuous culture system. Appl. Environ. Microbiol. 57:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macfarlane, S., and Macfarlane, G. T. 2003. Bacterial growth on mucosal surfaces and biofilms in the large bowel, p. 262-286. In M. Wilson and D. Devine (ed.), Medical implications of biofilms. Cambridge University Press, Cambridge, United Kingdom.
- 34.Malinen, E., A. Kassinen, T. Rinttila, and A. Palva. 2003. Comparison of real-time PCR with SYBR Green I or 5′-nuclease assays and dot-blot hybridization with rDNA-targeted oligonucleotide probes in quantification of selected bacteria. Microbiology 149:269-277. [DOI] [PubMed] [Google Scholar]
- 35.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMurdo, M. E. T. 2000. A healthy old age: realistic or futile goal? BMJ 321:1149-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meijer-Severs, G. J., and E. van Santen. 1986. Variations in the anaerobic faecal flora of ten healthy human volunteers with special reference to the Bacteroides fragilis-group and Clostridium difficile. Zntbl. Bakteriol. Mikrobiol. Hyg. Ser. A 261:43-52. [DOI] [PubMed] [Google Scholar]
- 38.Microbial ID, Inc. 1992. Microbial identification system operational manual. Microbial ID, Inc., Newark, Del.
- 39.Mitsuoka, T., and K. Hayakawa. 1972. The fecal flora of man. I. Communication: the composition of the fecal flora of different age groups. Zntbl. Bakteriol. Mikrobiol. Hyg. 223:333-342. [PubMed] [Google Scholar]
- 40.Mitsuoka, T., K. Hayakawa, and N. Kimura. 1974. The faecal flora of man. II. The composition of bifidobacterium flora of different age groups (author's transl.). Zntbl. Bakteriol. Abt. 1 Orig. A 226:469-478. [PubMed] [Google Scholar]
- 41.Mitsuoka, T. 1992. Intestinal flora and aging. Nutr. Rev. 50:438-446. [DOI] [PubMed] [Google Scholar]
- 42.Müller, H. E. 1986. Occurrence and pathogenic role of Morganella-Proteus-Providencia group bacteria in human feces. J. Clin. Microbiol. 23:404-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newton, J. P., R. Yemm, R. W. Abel, and S. Menhinick. 1993. Changes in human jaw muscles with age and dental state. Gerontology 10:16-22. [DOI] [PubMed] [Google Scholar]
- 44.Ouwehand, A. C., E. Isolauri, P. V. Kirjavainen, and S. J. Salminen. 1999. Adhesion of four Bifidobacterium strains to human intestinal mucus from subjects in different age groups. FEMS Microbiol. Lett. 172:61-64. [DOI] [PubMed] [Google Scholar]
- 45.Percival, R. S., P. D. Marsh, and S. J. Challacombe. 1996. Serum antibodies to commensal oral and gut bacteria vary with age. FEMS Immunol. Med. Microbiol. 15:35-42. [DOI] [PubMed] [Google Scholar]
- 46.Rigotteir-Gois, L., V. Rochet, N. Garrec, A. Suau, and J. Dore. 2003. Enumeration of Bacteroides species in human feces by fluorescent in situ hybridization combined with flow cytometry using 16S rRNA probes. Syst. Appl. Microbiol. 26:110-118. [DOI] [PubMed] [Google Scholar]
- 47.Russell, R. M. 1992. Changes in gastrointestinal function attributed to aging. Am. J. Clin. Nutr. 55:1203S-1207S. [DOI] [PubMed] [Google Scholar]
- 48.Salyers, A. A. 1984. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 38:293-313. [DOI] [PubMed] [Google Scholar]
- 49.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Dore. 2000. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66:2263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Severijnen, A. J., R. van Kleef, M. P. Hazenburg, and J. van de Merwe. 1989. Cell wall fragments from major residents of the human intestinal flora induce chronic arthritis in rats. J. Rheumatol. 16:1061-1068. [PubMed] [Google Scholar]
- 51.Solorzano, L. 1969. Determination of ammonia in natural waters by the phenolhypochlorite method. Limnol. Oceanogr. 14:799-801. [Google Scholar]
- 52.Stark, P. L., and A. Lee. 1982. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J. Med. Microbiol. 15:189-203. [DOI] [PubMed] [Google Scholar]
- 53.Stephen, A. M., H. S. Wiggins, and J. H. Cummings. 1987. Effect of changing transit time on colonic microbial metabolism in man. Gut 28:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiffenbach, J. M., B. J. Baum, and R. Burghauser. 1982. Taste thresholds: quality specific variation with human aging. J. Gerontol. 37:372-377. [DOI] [PubMed] [Google Scholar]
- 56.Wells, J. M., and C. Allison. 1995. Molecular genetics of intestinal anaerobes, p. 25-60. In G. R. Gibson and G. T. Macfarlane (ed.), Human colonic bacteria: role in nutrition, physiology and pathology. CRC Press, Boca Raton, Fla.
- 57.Yagamata, A. 1965. Histopathological studies of the colon in relation to age. Jpn. J. Gastroenterol. 62:229-234. [Google Scholar]