Abstract
Biosensor technology has a great potential to meet the need for sensitive and nearly real-time microbial detection from foods. An antibody-based fiber-optic biosensor to detect low levels of Listeria monocytogenes cells following an enrichment step was developed. The principle of the sensor is a sandwich immunoassay where a rabbit polyclonal antibody was first immobilized on polystyrene fiber waveguides through a biotin-streptavidin reaction to capture Listeria cells on the fiber. Capture of cells on the fibers was confirmed by scanning electron microscopy. A cyanine 5-labeled murine monoclonal antibody, C11E9, was used to generate a specific fluorescent signal, which was acquired by launching a 635-nm laser light from an Analyte 2000 and collected by a photodetector at 670 to 710 nm. This immunosensor was specific for L. monocytogenes and showed a significantly higher signal strength than for other Listeria species or other microorganisms, including Escherichia coli, Enterococcus faecalis, Salmonella enterica, Lactobacillus plantarum, Carnobacterium gallinarum, Hafnia alvei, Corynebacterium glutamicum, Enterobacter aerogenes, Pseudomonas aeruginosa, and Serratia marcescens, in pure or in mixed-culture setup. Fiber-optic results could be obtained within 2.5 h of sampling. The sensitivity threshold was about 4.3 × 103 CFU/ml for a pure culture of L. monocytogenes grown at 37°C. When L. monocytogenes was mixed with lactic acid bacteria or grown at 10°C with 3.5% NaCl, the detection threshold was 4.1 × 104 or 2.8 × 107 CFU/ml, respectively. In less than 24 h, this method could detect L. monocytogenes in hot dog or bologna naturally contaminated or artificially inoculated with 10 to 1,000 CFU/g after enrichment in buffered Listeria enrichment broth.
Listeria monocytogenes is a gram-positive, rod-shaped food-borne pathogen that causes listeriosis particularly in immunocompromised populations and abortion in pregnant women (9, 27, 32). The recent well-publicized outbreaks and food recalls due to L. monocytogenes (9, 23, 31, 39) have increased the need for more rapid, sensitive, and specific methods for detection of this bacterium in foods.
Conventional methods for the detection and identification of bacteria in food are greatly restricted by prolonged assay times (up to 7 days), requiring initial enrichment for detection of pathogens that are initially present in low numbers. Immunological assays with antibodies provide specific, reproducible, and reliable detection of bacteria, viruses, or toxins. Even though antibody-based detection may greatly reduce the assay time compared to traditional culture techniques, it still lacks the ability to detect biomolecules in real time. Biosensors use a combination of biological receptors and physical or chemical transducers, which represent a new and unique technology with great potential to meet the need for the rapid detection of low levels of biomolecules (5, 14). Fiber-optic biosensors exploit the measurement of fluorescent light excited by an evanescent wave generated by a laser to quantitatively detect biomolecules immobilized on the fiber surface (1, 24, 26). A portable sensor (Analyte 2000; Research International, Woodinville, Wash.) has been developed by using the above principle. The assay principle is based on a sandwich immunoassay, using a capture antibody, immobilized onto the optical fibers, and a cyanine 5 (Cy5)-labeled antibody for detection (24). The Analyte 2000 uses a 635-nm laser diode as an excitation light that is launched into the proximal end of an optical fiber. The Cy5 fluorescent molecules within several hundred nanometers of the fiber are excited by an evanescent field, and a portion of their emission energy reemits into the fiber. A photodiode allows for quantization of the collected emission light at wavelengths of 670 to 710 nm (1). This assay has been used to detect 2,4,6,-trinitrotoluene and hexahydro-1,3,5-trinitro-1,3,5-triazine (2), staphylococcal enterotoxin B, (36), Clostridium botulinum toxin (29), polymyxin B (15), Salmonella enterica serovar Typhimurium (40), Escherichia coli O157:H7 (7, 8), and PCR products of Listeria spp. (35). Recently, Tims et al. (37) used this assay to detect pure cultures of L. monocytogenes, reported a detection limit of 4.1 × 108 CFU/ml, and concluded that the quality of antibodies is key in improving sensitivity. The feasibility of their assay to differentially detect L. monocytogenes in the presence of other Listeria spp. or other common food-contaminating microorganisms was not tested. Also, no food samples were tested in their study (37).
Sensitivity and specificity of antibodies are critical for immunodetection of biomolecules in antibody-based biosensors. In our project, we attempted to increase the sensitivity and specificity of the fiber-optic biosensor by using two different antibodies. A polyclonal antibody (PAb) developed against whole cells of L. monocytogenes in the rabbit was used as a capture antibody, while a Cy5-labeled murine monoclonal antibody (MAb), C11E9 (4), which reacts with L. monocytogenes and some selected strains of Listeria innocua (4, 20), was used for detection. Sensitivity and specificity of the sensor were further evaluated by testing with other microorganisms in a mixed-culture setup. Finally, the ability of this sensor to detect L. monocytogenes cells from artificially inoculated (10 to 1,000 CFU/g) and naturally contaminated ready-to-eat (RTE) hot dog or bologna samples following enrichment was tested.
MATERIALS AND METHODS
Bacteria.
Listeria monocytogenes strain V7, a milk isolate (4), Listeria ivanovii ATCC 19119, Listeria grayi ATCC 19120, Listeria seeligeri SE 31, Listeria welshimeri ATCC 35897, L. innocua strains F4248 and LA-1, Citrobacter freundii ATCC 3624, E. coli ATCC 52739, Enterococcus faecalis ATCC 344, Salmonella enterica serovar Typhimurium, Salmonella enterica serovar Enteritidis, Bacillus cereus 4AC, Lactobacillus plantarum NCDO 955, Hafnia alvei, Corynebacterium glutamicum ATCC 31834, Enterobacter aerogenes, Pseudomonas aeruginosa ATCC 10145, Carnobacterium gallinarum ATCC 49517, and Serratia marcescens ATCC 14756 from our collection were maintained on brain heart infusion (BHI) agar (1.5%) slants (Difco Laboratories) at 25°C for the duration of this study. All fresh cultures for experiments were obtained by inoculating a loop of slant cultures into BHI broth and incubating them at 37°C for 16 h. In some cases, bacteria were adjusted to approximately the same concentration by using a spectrophotometer (Beckman-Coulter, Fullerton, Calif.) (12).
Preparation of polyclonal (capture) and monoclonal (detection) antibodies.
Anti-Listeria PAb was used as a capture antibody and was prepared according to the method described by Harlow and Lane (13). Briefly, freshly cultured L. monocytogenes cells (final concentration, approximately 1 × 109 CFU/ml) were inactivated at 70°C for 15 min and were subcutaneously injected at 2-week intervals for 3 months into a New Zealand White rabbit (Harlan Sprague-Dawley, Indianapolis, Ind.). The rabbit was maintained in the Purdue Biology Animal Facility (West Lafayette, Ind.). The serum from the third bleeding, showing very high antibody titer (1:10,000), was partially purified by ammonium sulfate precipitation (13). A Protein A column (ActaPrime; Pharmacia-Amersham, Uppsala, Sweden) was used for affinity purification of antibodies, and the final concentration was adjusted to 2.0 mg/ml. The reaction characteristics of this antibody to different bacterial cultures were determined by an indirect enzyme-linked immunosorbent assay (ELISA) (12, 20).
MAb C11E9 was used as a detection antibody. Frozen, stored hybridoma cell line C11E9 (4) was grown in cell culture medium in a cultivation chamber of CELLine 1000 (Integra Biosciences, East Dundee, Ill.) as previously described (12). The antibody from the supernatant was partially purified by ammonium sulfate precipitation (13), followed by the use of a Protein G column (ActaPrime) for affinity purification, and the final concentration was adjusted to 1.4 mg/ml.
Antibody labeling.
An antibody labeling kit (Cy5-Ab labeling kit; Amersham Biosciences, Piscataway, N.J.) was used for labeling MAb C11E9 according to the manufacturer's instructions and the suggestions of Kada et al. (18). Briefly, purified antibody was first ion exchanged from 0.1 M glycine-HCl (pH 2.7) to 0.1 M carbonate-bicarbonate buffer (pH 9.3) by using a desalting column (Amersham Biosciences). Second, 2 ml of antibody (1 mg/ml) was added to a dye vial wrapped with aluminum foil and incubated at room temperature for 30 min with mixing approximately every 10 min. Then, free dye was removed by a gel filtration column provided by the labeling kit. The final concentration of labeled antibody, measured by a DU-640 spectrophotometer (Beckman-Coulter), was 0.7 mg/ml, and the final molecular ratio of dye to antibody was estimated to be 2:1.
A long-chain biotin (EZ-Link NHS-LC-Biotin; Pierce, Rockford, Ill.) was used for biotinylation of the polyclonal capture antibody according to the manufacturer's instructions. One milligram of biotin was dissolved in 1 ml of dimethyl sulfoxide, and 75 μl of this solution was added to 1 mg of the antibody in 1 ml of carbonate-bicarbonate solution (5.7 g of NaHCO3, 3.4 g of Na2CO3 in 1 liter of water, pH 9.3). The solution was then incubated in an ice bucket for 2 h. Free biotin was removed by column chromatography (PD-10; Amersham Biosciences), and the final biotinylated antibody was estimated to be 0.7 mg/ml. Cy5- and biotin-labeled antibodies were stored in phosphate-buffered saline (PBS) containing bovine serum albumin (1 mg/ml; Sigma) at 4°C until used.
Western blotting.
L. monocytogenes cells grown in BHI broth at 37°C for 16 to 18 h were harvested by centrifugation (6,000 × g, 10 min) and washed three times with 20 mM PBS, pH 7.4, and cell concentration was adjusted to an A595 of 0.93. The washed cell pellets were resuspended with sample solvent buffer containing sodium dodecyl sulfate buffer (5%), β-mercaptoethanol (0.5%), and Tris (1.5%), pH 6.8, and incubated at 37°C for 60 min (4). After centrifugation (10,000 × g, 10 min), the cellular protein extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (7.5% polyacrylamide), transferred to Immobilon-P membranes (Millipore, Bedford, Mass.) and immunoprobed with MAb C11E9 (1:400) or anti-Listeria PAb (1:1,000). Membranes were developed with horseradish peroxidase-conjugated goat anti-mouse (for MAb C11E9) or anti-rabbit (for PAb) antibody (1:20,000) utilizing diamino benzidine tetrahydrochloride (Sigma) containing H2O2 as a substrate (6).
Determination of optimal concentration of detection antibody required for detection of L. monocytogenes.
L. monocytogenes V7 cells, with serial dilutions from 109 CFU/ml to 102 CFU/ml in 1 ml of PBS, were centrifuged, and the supernatants were discarded. The pellets of each concentration were incubated with 200 μl of 500-, 200-, 100-, or 50-μg/ml Cy5-labeled C11E9 for 1 h at 37°C. Then the cells were centrifuged, washed with PBS three times, and resuspended in 200 μl of PBS. Finally, cell suspensions were added into a 96-well black microtiter plate (Costar, Corning, N.Y.) and measured by a spectrofluorometer (Molecular Devices, Sunnyvale, Calif.) at excitation and emission wavelengths of 650 and 667 nm, respectively.
Fiber preparation.
The fibers were prepared according to the method described by Tims et al. (37) with modifications. The polystyrene fibers (4 cm in length and 0.78 mm in diameter) (Research International) were precleaned with 50% isopropanol and air-dried under a biosafety cabinet with laminar flow of air. Fibers were inserted into glass capillary tubes (VWR, Willard, Ohio) to form a reaction chamber and were incubated overnight at 4°C with 80 μl of 0.1-mg/ml streptavidin (Promega, Madison, Wis.). The fibers were rinsed with PBS containing 0.05% Triton X-100 (PBS-Triton) and then incubated with 80 μl of biotinylated capture antibody (0.1 mg/ml) at room temperature for 1 h in capillary tubes.
Blocking and background reading.
Fibers were first rinsed with PBS-Triton and then incubated with 80 μl of 1-mg/ml biotinylated bovine serum albumin (Pierce) in capillary tubes at room temperature for 1 h to block nonspecific binding sites. PBS-Triton was used to rinse fibers before inserting them into the waveguide holder that was prefabricated with proximal and distal outlets. The waveguide holder was injected with 200 μl of PBS and then connected to the Analyte 2000 equipped with a 635-nm laser light source (Research International) for a final reading taken at a wavelength of 670 to 710 nm. This reading value, recorded in picoamperes (pA), was considered to be the background for each fiber.
Fiber-optic assays.
Eighty microliters of bacteria in PBS was injected into capillary tubes and incubated with fibers at room temperature for 1 h. After rinses with PBS-Triton, fibers with captured bacteria were placed in a waveguide holder. Briefly, 200 μl of Cy5-labeled C11E9 (100 μg/ml) was injected into the holder through the proximal outlet, with the distal outlet blocked by a clip. Then, the fiber was inserted into the holder and incubated with the detection antibody for 252 s (on-site signals were read every 28 s for 10 readings). Consequently, 1 ml of PBS-Triton was injected into the holder by a syringe through the proximal outlet to wash out unbound antibody. Again, readings were taken every 28 s, and the values at the end of 252 s (10 readings) were considered final. The changes in signal due to the binding of bacteria were calculated as the final value minus the background reading. For each concentration of L. monocytogenes or each of the bacterial species, three to four fibers were used to generate average values and standard deviations.
Sensitivity and specificity of the biosensor.
To determine the sensitivity (detection limit) of this sensor, fresh culture of L. monocytogenes was washed and serially diluted (from 4.3 × 109 to 4.3 × 102 CFU/ml) in sterile PBS, pH 7.4. Eighty microliters of each dilution was incubated with fibers separately, and signals were acquired as described above. The minimum concentration with a signal significantly (P < 0.05) greater than the signal for a negative control (0 CFU/ml) was considered to be the limit of detection. The ability of the biosensor to detect stressed L. monocytogenes cells (2.8 × 109 CFU/ml) grown at low temperature (10°C) in the presence of NaCl (3.5%) for 48 h was also determined. The cells were serially diluted (from 2.8 × 107 to 2.8 × 105 CFU/ml) in PBS, and signals were acquired as described above.
The specificity of the biosensor to discriminate L. monocytogenes from closely related Listeria species (L. ivanovii, L. grayi, L. seeligeri, L. welshimeri, and L. innocua) or competitive microflora (Lactobacillus plantarum, Carnobacterium gallinarum, Serratia marcescens, H. alvei, Enterobacter aerogenes, P. aeruginosa, and Corynebacterium glutamicum) and other bacterial cultures (E. coli, Enterococcus faecalis, and S. enterica serovar Typhimurium) was tested. Cultures were grown in BHI separately, washed with PBS, and adjusted to uniform concentrations (approximately 4 × 109 CFU/ml), and 80 μl of each culture was tested separately.
To determine the fiber-optic signal response for L. monocytogenes cells in the presence of other bacteria, two mixed-culture conditions were used. In the first one, L. monocytogenes (1.0 × 109 CFU/ml) was mixed in equal proportions with E. coli, Enterococcus faecalis, and S. enterica serovar Typhimurium. For the control without L. monocytogenes, an equivalent volume of PBS was added. In the second one, L. monocytogenes (about 7.8 × 108 CFU/ml) was mixed with equal concentrations of other Listeria spp. listed above. In the control, L. monocytogenes or L. innocua was omitted.
The performance of the fiber-optic immunosensor in the presence of competitive microflora was evaluated. L. monocytogenes cells at 4.1 × 105, 4.1 × 104, and 4.1 × 103 CFU/ml were mixed separately with a constant number of Lactobacillus plantarum cells (2.6 × 107 CFU/ml) and analyzed. In addition, the same culture mixture was added to hot dog samples, enriched in buffered Listeria enrichment broth (BLEB) for 20 h, and analyzed. The fibers with PBS or L. monocytogenes cells only (4.1 × 109 CFU/ml) were used as controls.
Scanning electron microscopy.
Binding patterns of L. monocytogenes to fibers coated with capture antibody (PAb) were analyzed by scanning electron microscopy (SEM). No detection antibody was used in this experiment. The fibers were prepared and blocked as described above and incubated with L. monocytogenes for 1 h. The control fibers were devoid of the capture antibody. The fibers were then washed in three changes of buffered 2% gluteraldehyde (0.025 M KH2PO4, 0.025 M Na2HPO4, pH 7.2) for 15 min per change, rinsed quickly with buffer, immersed in buffered osmium tetrachloride (1%) for 30 min, rinsed briefly with distilled water, dehydrated in a graded series of ethanol exchanges until reaching 100%, dried in a critical-point dryer (Ladd Research Industries, Burlington, Vt.), mounted on metal stubs with silver paste, and coated with a layer of gold (Hummer I Sputter Coater). Finally, three portions, the top, the middle, and the bottom, of the fibers were examined under an SEM (JSM 840; Jeol USA Inc., Peabody, Mass.) at 5 kV.
The number of cells captured on the surface of cylindrical fibers was calculated with two formulae. First, the surface area was calculated as At = π × d × l, where At is the surface area, d is the diameter, and l is the length. Second, the cells in SEM images (Ni) were counted, and image area (Ai) was determined. Finally, the total number of cells (Nt) captured by the fibers was estimated by Nt = Ni × At/Ai. Two fibers were used to generate average values. These calculated values were confirmed by the plate-counting method on BHI agar plates, where the captured cells were released by vigorous vortexing for 10 min in Eppendorf tubes.
Selection of suitable selective enrichment broth for testing meat samples with fiber-optic sensor.
Several selective enrichment broths commonly used for Listeria detection were examined for their suitability for use with the fiber-optic immunosensor application. This selection was done by assaying reaction patterns of viable L. monocytogenes cells grown in different selective enrichment broths for 20 h at 37°C with the capture antibody (anti-L. monocytogenes rabbit PAb) and the detection antibody (murine MAb C11E9) separately in an ELISA format as described previously (12, 28). The following selective enrichment broths were purchased from Becton Dickinson (Sparks, Md.) or Oxoid (Ogdensburg, N.Y.): University of Vermont medium (UVM), Fraser broth (FB), Listeria repair broth (LRB), and BLEB. All media were prepared according to the manufacturer's directions or as described previously (33). In the immunoassay, L. monocytogenes cells grown in BLEB gave overall the highest reactions with both antibodies; therefore, BLEB was a logical choice for food sample analyses using the fiber-optic sensor.
Detection of L. monocytogenes from artificially inoculated or naturally contaminated hot dog and bologna.
Packages of hot dog and bologna were purchased from area grocery stores, and 25 g of each was aseptically transferred into a stomacher bag with a filter lining (Seward, Cincinnati, Ohio). L. monocytogenes cells were artificially inoculated into the selected meat samples, with final concentrations of 101, 102, and 103 cells/g, and allowed to stand for 10 min. Meats with or without inoculation were suspended in 100 ml of BLEB (Oxoid) in stomacher bags and were massaged by hand for 2 min and incubated at 37°C for 20 h. One-milliliter aliquots of the samples were withdrawn, the cells were harvested by centrifugation (6,000 × g for 5 min) and washing (three times), and the cells were finally resuspended in 1 ml of PBS. Eighty microliters of each sample was used for fiber-optic detection. Three fibers were used to generate average values and standard deviations. After fiber-optic analysis, fibers with adsorbed cells were cut, and the thin tips were transferred to Eppendorf tubes with 1 ml of PBS. The cells captured on the fibers were released into PBS by vigorous vortexing for 10 min and plated on Oxford agar plates. Bacterial counts in enriched samples were determined by serial dilution and plating on Oxford and BHI agar plates. In parallel, samples were analyzed by the U.S. Department of Agriculture Food Safety and Inspection Service (USDA FSIS) method (25). Colonies on Oxford plates recovered from the enrichment broth or the fibers were confirmed by CAMP test and ribotyping (16).
Statistical analysis.
Data were presented as means ± standard deviations. Statistical analysis of the results was done using a Duncan multiple comparison test or a Tukey’s t test. Differences were significant if P was <0.05.
RESULTS
Reaction characteristics of anti-Listeria PAb.
A limited number of cultures were tested by ELISA (A490). As expected, the anti-Listeria PAb showed strong reactions with all species of Listeria tested, including L. monocytogenes strains V7 (2.95) and Scott A (1.66), L. innocua strains LA-1 (2.74) and F4248 (1.49), L. welshimeri (0.98), L. ivanovii (2.20), L. seeligeri (1.71), and L. grayi (2.79). Among the non-Listeria cultures, antibody positively reacted with Citrobacter freundii (1.99) but showed weak or no reactions (<0.5) with S. enterica serovar Enteritidis, S. enterica serovar Typhimurium, E. coli, and B. cereus.
Determination of optimal concentration of detection antibody.
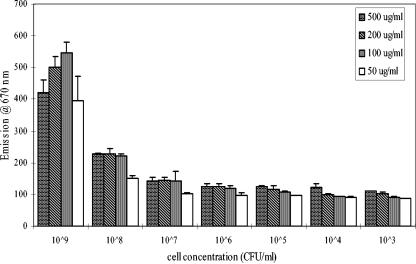
Concentrations of cells or detection antibodies (Cy5-conjugated C11E9) needed to show optimum reaction were determined in a spectrofluorometer, and the strongest signal was seen when 109 cells/ml was mixed with 100 μg of Cy5-conjugated C11E9/ml. When the cell concentrations decreased, the signal strengths decreased logarithmically (Fig. 1). When antibody concentrations increased, the signal strengths also decreased because of fluorescent quenching (21). Based on this experiment, Cy5-conjugated C11E9 was used at 100 μg/ml.
FIG. 1.
Determination of optimal concentration of detection antibody, Cy5-conjugated C11E9, and L. monocytogenes cells required in a 96-well microtiter plate using a spectrofluorometer. Measurement was done at excitation and emission wavelengths of 650 and 670 nm, respectively.
Western blotting reaction patterns of two antibodies.
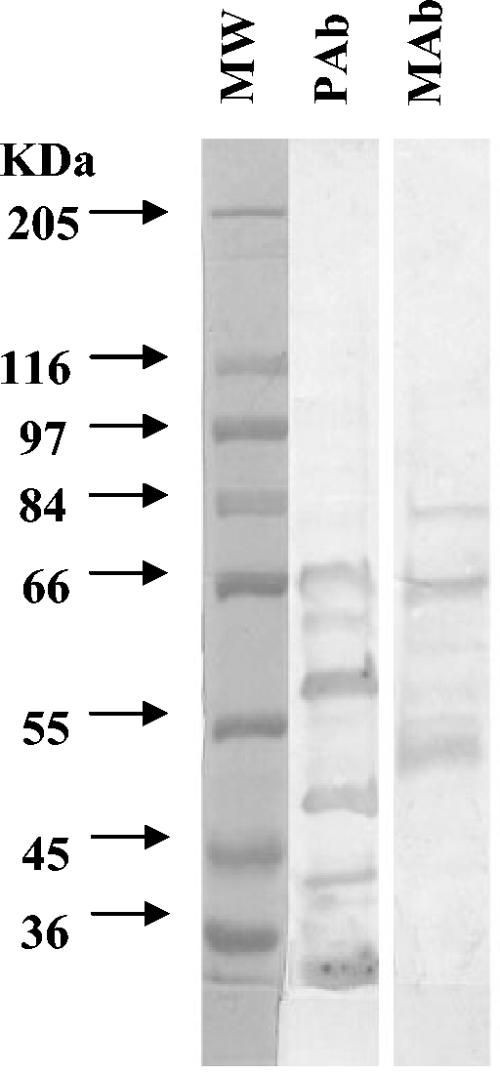
Western blotting showed that the major proteins recognized by anti-Listeria PAb were about 68, 62, 58, 50, 43, and 30 kDa, while the expected bands for MAb C11E9 were 76, 66, and 52 kDa (4). Molecular weights of the bands for each antibody were apparently different from each other; therefore, it is anticipated that there will be less competition for epitopes between capture and detection antibodies on the waveguide (Fig. 2).
FIG. 2.
Reaction patterns of antibodies with surface protein extract of L. monocytogenes with Western blotting. Lane 1, molecular weight standard (MW); lane 2, L. monocytogenes protein extracts reacted with anti-Listeria rabbit PAb; and lane 3, mouse MAb C11E9.
Sensitivity and specificity of the biosensor.
An L. monocytogenes concentration of 4.3 × 109 CFU/ml gave the strongest signal (1,700 pA), while the signal strength decreased logarithmically when the cell concentration decreased (Table 1). The lowest cell concentration that gave a positive signal compared to a background control (no bacteria) was 4.3 × 103 CFU/ml and was considered to be the detection limit for this sensor. The strength of the signal obtained for this cell concentration was 230 pA, which was significantly higher than that of the signal emitted by a control (89 pA) or by a cell concentration of 4.3 × 102 CFU/ml (93 pA).
TABLE 1.
Sensitivity of the immunosensor using a serial dilution of L. monocytogenes suspended in PBS (pH 7.2) grown in optimal (37°C, 0.5% NaCl) or stressful (10°C, 3.5% NaCl) environments
| Species or control | Density (CFU/ml) | Preculture | Mean signal strength (pA)a ± SD | Resultsb |
|---|---|---|---|---|
| L. monocytogenes V7 | 4.3 × 109 | 37°C, 0.5% NaCl | 1,649 ± 218 | Positive |
| 4.3 × 108 | 649 ± 114 | Positive | ||
| 4.3 × 107 | 446 ± 112 | Positive | ||
| 4.3 × 106 | 419 ± 108 | Positive | ||
| 4.3 × 105 | 381 ± 28 | Positive | ||
| 4.3 × 104 | 309 ± 21 | Positive | ||
| 4.3 × 103 | 230 ± 10 | Positive | ||
| 4.3 × 102 | 93 ± 24 | Negative | ||
| Control (PBS) | 0 | 89 ± 14 | Negative | |
| L. monocytogenes V7 | 2.8 × 107 | 10°C, 3.5% NaCl | 748 ± 140 | Positive |
| 2.8 × 106 | 348 ± 35 | Negative | ||
| 2.8 × 105 | 229 ± 135 | Negative | ||
| Control (PBS) | 0 | 332 ± 52 | Negative |
Signal strengths are based on the average of results from three independent measurements (fibers) from a representative experiment.
Results were positive when the values were significantly greater than that of the control without bacteria (P < 0.05).
When cells were subjected to a stress environment of cold (10°C) and salt (3.5% NaCl) concurrently, the signal output was severely diminished. Signals emitted by cells at bacterial concentrations of 2.8 × 105 CFU/ml (299 pA) and 2.8 × 106 CFU/ml (348 pA) were not significantly stronger than that of the control (332 pA). A positive signal was obtained only from cell concentrations of 2.8 × 107 CFU/ml (738 pA), thus indicating that bacterial cells subjected to stress can severely affect the result (Table 1). When the sensor was evaluated for its reaction with other Listeria spp. at approximate concentrations of 4 × 109 CFU/ml each, L. monocytogenes showed a significantly (P < 0.05) stronger signal (1,728 pA) than L. ivanovii (346 pA), L. grayi (204 pA), L. seeligeri (381 pA), L. welshimeri (533 pA), or L. innocua (720 pA) (Table 2).
TABLE 2.
Specificity of the immunosensor for L. monocytogenes when compared with Listeria spp. and other microorganismsa
| Species or control | Density (CFU/ml) | Mean signal strength (pA) ± SDb |
|---|---|---|
| L. monocytogenes V7 | 4.0 × 109 | 1,728 ± 325 A |
| L. ivanovii ATCC 19119 | 4.0 × 109 | 346 ± 85 D |
| L. grayi ATCC 19120 | 4.0 × 109 | 203 ± 9 D |
| L. seeligeri SE 31 | 4.0 × 109 | 381 ± 186 BCD |
| L. welshimeri ATCC 35897 | 4.0 × 109 | 533 ± 57 BC |
| L. innocua F4248 | 4.0 × 109 | 720 ± 11 B |
| Control (PBS) | 0 | 114 ± 87 D |
| L. monocytogenes V7 | ∼109 | 2,179 ± 366 A |
| S. enterica serovar Typhimurium | ∼109 | 553 ± 77 B |
| E. coli ATCC 52739 | ∼109 | 374 ± 39 BC |
| Enterococcus faecalis ATCC 344 | ∼109 | 110 ± 41 CD |
| Lactobacillus plantarum NCDO 955 | ∼109 | 310 ± 82 BCD |
| Serratia marcescens ATCC 14756 | ∼109 | 147 ± 82 CD |
| H. alvei | ∼109 | 73 ± 51 D |
| Enterobacter aerogenes | ∼109 | 141 ± 28 CD |
| P. aeruginosa ATCC 10145 | ∼109 | 327 ± 44 BCD |
| Corynebacterium glutamicum ATCC 31834 | ∼109 | 281 ± 58 BCD |
| Carnobacterium gallinarum ATCC 49517 | ∼109 | 342 ± 78 BCD |
| Control (PBS) | 0 | 347 ± 12 BCD |
Bacterial cultures were precultured at 37°C and with 0.5% NaCl.
Mean signal strengths were based on the average of results from three independent measurements (fibers) from a representative experiment. Results with different letters (A, B, C, and D) are significantly (P < 0.05) different from each other.
Evaluation of the sensor with other common food contaminants indicated that L. monocytogenes (∼109 CFU/ml) generated a signal of 1,636 pA, which was significantly (P < 0.05) stronger than that of the equivalent concentrations of S. enterica serovar Typhimurium (375 pA), E. coli (110 pA), Enterococcus faecalis (553 pA), Lactobacillus plantarum (310 pA), Serratia marcescens (147 pA), H. alvei (73 pA), Enterobacter aerogenes (141 pA), P. aeruginosa (327 pA), Carnobacterium gallinarum (342 pA), and Corynebacterium glutamicum (281 pA) (Table 2).
In a mixed-culture setup, the signal for L. monocytogenes with other Listeria spp. was 475.5 pA, while that for L. monocytogenes alone was 456 pA (Table 3). Mixed cultures without L. monocytogenes or L. monocytogenes and L. innocua had a significantly (P < 0.05) lower-strength signal than L. monocytogenes-containing mixed culture. The bacterial concentrations for each were about 7.8 × 108 CFU/ml, and the signal for L. monocytogenes alone was slightly lower in strength than the expected value for this concentration observed in Table 1. When L. monocytogenes was mixed with a high concentration of Lactobacillus plantarum, the signal for L. monocytogenes at 4.1 × 103 CFU/ml was 316 pA, equivalent to that of a negative control (PBS). Signals from L. monocytogenes at 4.1 × 104 CFU/ml (883 pA) and 4.1 × 105 CFU/ml (1,041 pA) were significantly stronger than that of the control (Table 3), which suggests that the presence of competitive microflora could decrease the detection limit by 1 log. The signal strength from the mixed cultures containing L. monocytogenes, E. coli, Enterococcus faecalis, and S. enterica serovar Typhimurium was 1,009 pA, while that from cultures containing L. monocytogenes alone was 995 pA (Table 3). When L. monocytogenes was absent from the mixture, the signal strength was significantly decreased and was similar to that of the negative control (PBS only). These studies suggest that a specific signal for L. monocytogenes could be acquired even in the presence of other Listeria spp. or common food contaminants.
TABLE 3.
Fiber-optic signal acquisition for L. monocytogenes in mixture in PBS, pH 7.2a
| L. monocytogenes density (CFU/ml) | Added organism, density (CFU/ml) | Mean signal strength (pA)b ± SD | Resultsc |
|---|---|---|---|
| 7.8 × 108 | 476 ± 204 | Positive | |
| 7.8 × 108 | All other Listeria spp., ∼3 × 109 | 456 ± 30 | Positive |
| 0 (control) | 104 ± 6 | Negative | |
| 0 | All other Listeria spp., ∼3 × 109 | 165 ± 45 | Negative |
| 0 | All other Listeria spp., except L. innocua, ∼3 × 109 | 198 ± 37 | Negative |
| 1.0 × 109 | 995 ± 77 | Positive | |
| 1.0 × 109 | E. coli, Enterococcus faecalis, and serovar Typhimurium, ∼3 × 109 | 1,009 ± 142 | Positive |
| 0 | 304 ± 33 | Negative | |
| 0 | E. coli, Enterococcus faecalis, and serovar Typhimurium, 1.0 × 109 | 298 ± 68 | Negative |
| 4.1 × 109 | 1,844 ± 123 | Positive | |
| 4.1 × 103 | L. plantarum, 2.6 × 107 | 316 ± 55 | Negative |
| 4.1 × 104 | L. plantarum, 2.6 × 107 | 883 ± 205 | Positive |
| 4.1 × 105 | L. plantarum, 2.6 × 107 | 1,041 ± 187 | Positive |
| 0 | 294 ± 111 | Negative |
Cells were precultured at 37°C and with 0.5% NaCl.
Signal strengths are based on the average of results from three independent measurements (fibers) from a representative experiment.
Results were positive when the values were significantly greater than that of the control without bacteria (P < 0.05) for a given experiment.
SEM analysis of fibers.
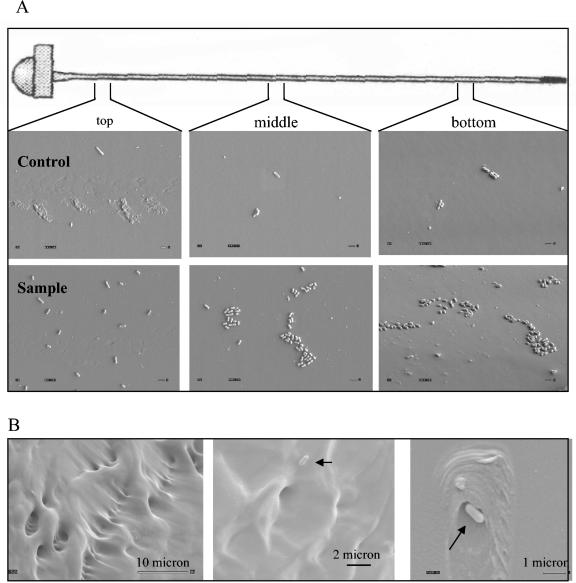
SEM images taken from three sections, i.e., the top, the middle, and the bottom of the fibers, showed that the number of cells captured by the fibers decreased from the top to the bottom (Fig. 3A). Cell distributions on fibers appeared to be in clusters. The average number of cells captured by fibers was estimated to be 7.6 × 106 cells/fiber, and the recovery rate was about 10% from 8 × 107 bacteria present in 80 μl of solution. The number of cells bound to fibers without capture antibody was calculated to be about 4.6 × 105 CFU/fiber, and the recovery rate was about 0.6%. When cells were released from fibers by vortexing, the average count from two fibers was 8.6 × 106 CFU/fiber, with a 10% recovery rate, and the count from control fibers was 2.8 × 105 CFU/fiber, with about 0.4% recovery. These values were comparable to the estimated values.
FIG. 3.
SEM analyses of fibers with captured L. monocytogenes cells. (A) L. monocytogenes cell binding was examined in three arbitrary sections (top, middle, and bottom) of the fiber. The top row (control) is without capture antibody, and the bottom row (sample) is with rabbit polyclonal capture antibody. Magnification, ×4,000. Bar, 1.0 μm. (B) Fibers showing scratches and manufacturing defects. Magnifications for each panel are presented in the figure.
The numbers of cells captured on the surface of the fibers decreased from the top to the bottom. This result could be due to the concentration gradient when cells were incubated with fibers in a vertical tube without agitation for a long time (1 h).
Close inspection of the fibers also revealed dents, scratches, and manufacturing defects with attached bacterial cells (Fig. 3B). These defects are thought to mask the acquisition of signals from bacteria and may contribute to fiber-to-fiber variation in signal output.
BLEB as a suitable enrichment broth for fiber-optic assay.
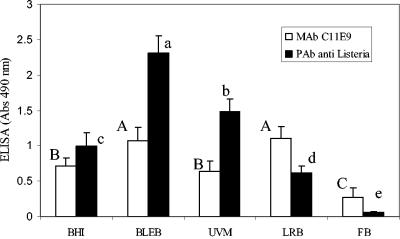
ELISA data (A490) showed that the reaction of anti-Listeria PAb to L. monocytogenes cells (2.30) grown in BLEB was significantly (P < 0.05) higher than that of cells grown in UVM (1.5), LRB (0.62), or FB (0.05) (Fig. 4). The reaction of MAb C11E9 to L. monocytogenes cells grown in BLEB (1.0) was similar to the reaction to cells grown in LRB (1.10) but significantly (P < 0.05) higher than that to cells grown in UVM (0.64) or FB (0.3). BHI broth was used as a nonselective medium control, and the reaction to cells grown in it was lower than that to cells grown in BLEB for both antibodies.
FIG. 4.
Comparison of reactions of MAb C11E9 and anti-Listeria PAb to L. monocytogenes grown in BHI broth, BLEB, UVM, LRB, and FB in an ELISA. Cultures were obtained after 20 h of growth and centrifuged to remove media, and cell concentrations were adjusted to an A595 of 0.37 with a spectrophotometer. Bars marked with different capital letters (A, B, or C) for MAb C11E9 and different lowercase letters (a, b, c, d, or e) for anti-Listeria PAb are significantly different, with P values of <0.05.
Testing of food samples.
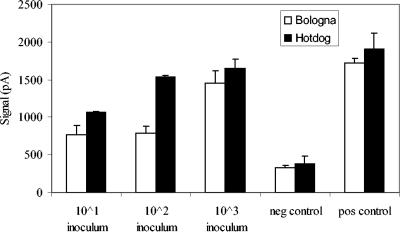
Artificially contaminated hot dogs with initial inocula of L. monocytogenes at concentrations of approximately 101, 102, and 103 CFU/g followed by a 20-h enrichment gave fiber-optic signals of 1,064, 1,540, and 1,652 pA, respectively. The final cell concentrations after enrichment were enumerated to be 3.8 × 108, 2.3 × 109, and 4.2 × 109 CFU/ml, respectively (Fig. 5). These values were significantly (P < 0.05) greater than those from samples without inoculation. Likewise, spiked bolognas with initial inocula of 101, 102, and 103 CFU/ml gave fiber-optic signals of 764, 790, and 1,452 pA, respectively, and the final cell concentrations after enrichment were estimated to be 2.3 × 108, 6.8 × 108, and 3.9 × 109 CFU/ml (Fig. 5). No background Listeria spp. were found in either hot dog or bologna samples. The cells captured on the fibers were randomly picked from Oxford plates and were positive by CAMP test, identifying them as L. monocytogenes.
FIG. 5.
Fiber-optic signal acquisition for L. monocytogenes artificially inoculated into hot dog or bologna at 101, 102, or 103 CFU/ml. The meat samples were enriched for 20 h in BLEB before fiber-optic analysis. The negative (neg) control contained only PBS, and the positive (pos) control contained pure culture of L. monocytogenes.
Analyses of 11 hot dog or bologna samples purchased from area grocery stores (six stores) with the fiber-optic sensor revealed only one hot dog sample to be positive for L. monocytogenes, with an average signal of 1,400 pA. The isolate obtained from this positive fiber formed a black colony on an Oxford agar plate, gave a positive CAMP reaction, and was identified as L. monocytogenes with a ribotype pattern of DUP 1093. The presence of L. monocytogenes in this sample was further confirmed by the USDA FSIS method. The average signals for negative meat samples ranged from 338 to 549 pA, and none of the samples contained any L. monocytogenes or other Listeria species, as analyzed by the USDA FSIS method and CAMP testing.
DISCUSSION
L. monocytogenes accounted for the greatest number of food product recalls (9, 31, 39) and several recent outbreaks (10, 11, 23, 31). Therefore, rapid and sensitive methods are required to detect this pathogen in food. Conventional methods are time-consuming. Several rapid methods, such as PCR (17) and biosensors based on their transducer properties (21), are sensitive and specific but often expensive, sophisticated, and nonportable, and thus with limited usefulness. Furthermore, the efficacy of those biosensors to detect pathogens from real-world food samples has not been thoroughly investigated (5, 14).
Antibody-based fiber-optic biosensors could be one of the best methods to meet the need for a rapid, sensitive, and portable microbial detection system. A previous report showed that E. coli O157:H7 could be detected at levels as low as 30 CFU/ml with a fiber-optic sensor (8). Recently, Liu et al. (22) developed a chemiluminescent fiber-optic biosensor that could detect about 100 CFU of E. coli O157:H7/ml from ground beef, chicken carcass, and lettuce samples within 1.5 h, but preseparation with immunomagnetic beads was required. Zhou et al. (40) used a compact fiber-optic evanescent system that could detect 104 CFU of S. enterica serovar Typhimurium/ml. Using a fiber-optic system, Tims et al. (37) reported the detection limit for L. monocytogenes to be ∼108 CFU/ml; however, this study did not evaluate the performance of the sensor with other Listeria spp. or other microorganisms or with food samples.
In this study, our goal was to develop a fiber-optic immunosensor and optimize conditions that would be able to detect viable L. monocytogenes cells from ready-to-eat meat samples in less than 24 h from the point of food sampling. To develop a sensitive immunosensor, the quality and specificity of the antibodies are important. We developed a PAb in rabbit against whole cells of L. monocytogenes, which was used as a capture antibody on the fiber waveguide. Subsequent detection was accomplished by using MAb C11E9 (4, 20). This combination improved the sensitivity of the fiber-optic biosensor to 4.3 × 103 CFU of L. monocytogenes/ml. In contrast, when MAb C11E9 was used for both capture and detection, the sensitivity was very poor and results were extremely variable (data not shown). Overall, the strength of the fiber-optic signals decreased logarithmically when L. monocytogenes cells were serially diluted. A similar reaction pattern was observed when antibody binding (detection antibody; MAb C11E9) was examined by immunofluorescence assay in a 96-well microtiter plate, and the detection limit appeared to be 107 to 108 CFU/ml (Fig. 1). The detection limit for L. monocytogenes by sandwich or indirect ELISA methods could vary from 106 to 108 CFU/ml (3, 30, 34, 38). A similar detection limit was reported for the fiber-optic sensor (37) or surface plasmon resonance sensor (19, 20) for L. monocytogenes. In comparison, the sensitivity of the present fiber-optic sensor is severalfold higher.
When a low concentration of L. monocytogenes cells was mixed with a high concentration of competitive microflora, the sensitivity of the sensor decreased and the detection limit increased by 1 log. This result implied that the presence of microflora might interfere with the binding of antibodies to L. monocytogenes, thus affecting the detection sensitivity. When cells were grown in a stress environment with a combination of cold and salt, the sensitivity of the sensor decreased by 4 log. This result was expected, since cold (4°C) or salt (1.5 to 10.5% NaCl) treatments displayed diminished reactions with MAb C11E9 (12). The introduction of an enrichment step prior to detection by immunosensor would eliminate such problems.
The specificity of this fiber-optic biosensor was tested in the presence of E. coli, Enterococcus faecalis, S. enterica serovar Typhimurium, different Listeria species, and common bacterial contaminants, which may be present in a food product. Taken together, these data indicated that the fiber-optic sensor was specific for L. monocytogenes. However, the detection antibody (C11E9) used here is known to show cross-reactions with certain strains (23%) of L. innocua (20). It is possible that the fiber-optic sensor in its present configuration may pick up a signal from certain L. innocua strains. Therefore, a more specific L. monocytogenes antibody would be able to eliminate this problem.
The fiber-optic sensor sometimes showed fiber-to-fiber variation in signal strength, which was counted as background and subtracted from the final reading. This background reading varied from 300 pA to 500 pA (data not shown). SEM analyses of surface topography of some of the fibers revealed scratches, dents, and manufacturing defects that possibly contributed to fiber-to-fiber variation in signal acquisition. Besides, the ratios of biotin or Cy5 to antibodies were changed batch-to-batch when antibodies were labeled (data not shown). The in-batch variation could also contribute to signal variation. The standardization of this procedure needs to be improved in future studies.
In this study, we had set as our goal that fiber-optic sensor-based detection be completed in less than 24 h from the point of food sampling. To achieve that goal, we needed a suitable enrichment broth that would support sufficient antigen expression for capture and detection antibodies to generate strong signals in the fiber-optic sensor. Several enrichment broths (UVM, FB, LRB, and BLEB) were evaluated. Certain selective enrichment broths were reported previously to affect antibody-based detection of L. monocytogenes. Nannapaneni et al. (28) indicated that Listeria enrichment broth supported antigen expression for MAb EM-7G1 (6), while UVM and FB suppressed the expression. In this study, we also observed a similar trend, where cells cultured in BLEB had the overall highest level of reaction, followed by those cultured in UVM, LRB, and FB. Cells from FB showed the lowest level of reaction, and incidentally, this broth is used in the USDA FSIS method (25) for detection of L. monocytogenes. Based on this data, we used BLEB as an enrichment broth for the fiber-optic sensor application.
Since Listeria is problematic in ready-to-eat meats, artificially inoculated hot dogs and bolognas were tested after 20 h of enrichment in BLEB. Results showed that L. monocytogenes cells could be detected directly from the enrichment broth even with low levels of inoculations (10 to 1,000 cells/g). The enriched samples were centrifuged and washed to remove food particles, since excessive food particles gave increased background noise (data not shown).
In an attempt to validate the performance of the fiber-optic sensor to detect L. monocytogenes from food samples, we tested 11 RTE meat samples and found only one to be positive. The sell-by date for this particular positive hot dog sample had expired. Although a limited number of samples were tested, this study indicates that the fiber-optic sensor can detect L. monocytogenes from enriched RTE meat samples with initial low levels of inoculation (contamination) in under 24 h. Our future goal is to determine the earliest time, preferably in a single working day (8 h), in which the assay could be completed with an initial contamination level of 1 to 100 CFU/g of RTE products. This achievement would require optimization of both enrichment conditions and the fiber-optic assay method.
Acknowledgments
This research was supported through a cooperative agreement with the Agricultural Research Service of the United States Department of Agriculture project number 1935-42000-035 and the Center for Food Safety and Engineering at Purdue University.
We thank Kwang-pyo Kim, Ziad Jaradat, Jennifer Wampler, Chunping Huang, Felipe Camacho, and Angela Valadez for their technical assistance.
REFERENCES
- 1.Anderson, G. P., K. A. Breslin, and F. S. Ligler. 1996. Assay development for a portable fiberoptic biosensor. ASAIO J. 42:942-946. [DOI] [PubMed] [Google Scholar]
- 2.Bakaltcheva, I. B., F. S. Ligler, C. H. Patterson, and L. C. Shriver-Lake. 1999. Multi-analyte explosive detection using a fiber optic biosensor. Anal. Chim. Acta 399:13-20. [Google Scholar]
- 3.Beumer, R. R., and E. Brinkman. 1989. Detection of Listeria spp. with a monoclonal antibody-based enzyme-linked immunosorbent assay (ELISA). Food Microbiol. 6:171-177. [Google Scholar]
- 4.Bhunia, A. K., P. H. Ball, A. T. Faud, B. W. Kurz, J. W. Emerson, and M. G. Johnson. 1991. Development and characterization of a monoclonal antibody specific for Listeria monocytogenes and Listeria innocua. Infect. Immun. 59:3176-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhunia, A. K., and A. Lathrop. 2003. Pathogen detection, foodborne, p. 320-323. In D. Blumel and A. Rappaport (ed.), McGraw-Hill 2003 yearbook of science and technology. McGraw-Hill Professional, New York, N.Y.
- 6.Bhunia, A. K., and M. G. Johnson. 1992. Monoclonal antibody specific for Listeria monocytogenes associated with a 66-kilodalton cell surface antigen. Appl. Environ. Microbiol. 58:1924-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeMarco, D. R., and D. V. Lim. 2002. Detection of Escherichia coli O157:H7 in 10- and 25-gram ground beef samples with an evanescent-wave biosensor with silica and polystyrene waveguides. J. Food Prot. 65:596-602. [DOI] [PubMed] [Google Scholar]
- 8.DeMarco, D. R., E. W. Saaski, D. A. McCrae, and D. V. Lim. 1999. Rapid detection of Escherichia coli O157:H7 in ground beef using a fiber-optic biosensor. J. Food Prot. 62:711-716. [DOI] [PubMed] [Google Scholar]
- 9.Donnelly, C. W. 2001. Listeria monocytogenes: a continuing challenge. Nutr. Rev. 59:183-194. [DOI] [PubMed] [Google Scholar]
- 10.Dorozynski, A. 2000. Seven die in French Listeria outbreak. Br. Med. J. 320:601. [PMC free article] [PubMed] [Google Scholar]
- 11.Frye, D. M., R. Zweig, J. Sturgeon, M. Tormey, M. LeCavalier, I. Lee, L. Lawani, and L. Mascola. 2002. An outbreak of febrile gastroenteritis associated with delicatessen meat contaminated with Listeria monocytogenes. Clin. Infect. Dis. 35:943-949. [DOI] [PubMed] [Google Scholar]
- 12.Geng, T., K. P. Kim, R. Gomez, D. M. Sherman, R. Bashir, R. M. R. Ladisch, and A. K. Bhunia. 2003. Expression of cellular antigens of Listeria monocytogenes that react with monoclonal antibodies C11E9 and EM-7G1 under acid-, salt- or temperature-induced stress environments. J. Appl. Microbiol. 95:762-772. [DOI] [PubMed] [Google Scholar]
- 13.Harlow, E., and D. Lane. 1988. Antibodies, a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 14.Ivnitski, D., I. Abdel-Hamid, P. Atanasov, and E. Wilkins. 1999. Biosensors for detection of pathogenic bacteria. Biosens. Bioelectron. 14:599-624. [DOI] [PubMed] [Google Scholar]
- 15.James, E. A., K. Schmeltzer, and F. S. Ligler. 1996. Detection of endotoxin using an evanescent wave fiber-optic biosensor. Appl. Biochem. Biotechnol. 60:189-202. [DOI] [PubMed] [Google Scholar]
- 16.Jaradat, Z. W., G. E. Schutze, and A. K. Bhunia. 2002. Genetic homogeneity among Listeria monocytogenes strains from infected patients and meat products from two geographic locations determined by phenotyping, ribotyping and PCR analysis of virulence genes. Int. J. Food Microbiol. 76:1-10. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, M. G., D. K. Winters, and R. F. Wang. 1997. PCR and nucleic acid amplification methods, p. 183-205. In M. L. Tortorello and S. M. Gendel (ed.), Food microbiological analysis. Marcel Dekker, Inc., New York, N.Y.
- 18.Kada, G., G. Schuetz, H. G. Knaus, G. S. Harms, C. Hahn, and H. J. Gruber. 2000. Optimization of probe labeling with cy3 and cy5 for single molecule fluorescence microscopy. Biophys. J. 78:250a. [Google Scholar]
- 19.Kubova, V., E. Brynda, L. Karasova, J. Skvor, J. Homola, J. Dostalek, P. Tobiska, and J. Rosicky. 2001. Detection of foodborne pathogens using surface plasmon resonance biosensors. Sensors Actuators B 74:100-105. [Google Scholar]
- 20.Lathrop, A. A., Z. W. Jaradat, T. Haley, and A. K. Bhunia. 2003. Characterization and application of a Listeria monocytogenes reactive monoclonal antibody C11E9 in a resonant mirror biosensor. J. Immunol. Methods 281:119-128. [DOI] [PubMed] [Google Scholar]
- 21.Leonard, P., S. Hearty, J. Brennan, L. Dunne, J. Quinn, T. Chakraborty, and R. O'Kennedy. 2003. Advances in biosensors for detection of pathogens in food and water. Enzyme Microb. Technol. 32:3-13. [Google Scholar]
- 22.Liu, Y. C., J. M. Ye, and Y. B. Li. 2003. Rapid detection of Escherichia coli O157:H7 inoculated in ground beef, chicken carcass, and lettuce samples with an immunomagnetic chemiluminescence fiber-optic biosensor. J. Food Prot. 66:512-517. [DOI] [PubMed] [Google Scholar]
- 23.MacDonald, P. D., R. E. Whitwam, J. D. Boggs, J. W. Reardon, R. R. Saah, M. Beatty, J. Sobel, L. M. Graves, S. B. Hunter, and J. N. MacCormack. 2001. Outbreak of Listeria-associated birth complications linked with homemade Mexican-style cheese, North Carolina, 2000. Clin. Infect. Dis. 33:868. [Google Scholar]
- 24.Marazuela, M. D., and M. C. Moreno-Bondi. 2002. Fiber-optic biosensors—an overview. Anal. Bioanal. Chem. 372:664-682. [DOI] [PubMed] [Google Scholar]
- 25.McClain, D., and W. H. Lee. 1989. FSIS method for the isolation and identification of Listeria monocytogenes from processed meat and poultry products. Laboratory communication no. 57. U.S. Department of Agriculture, FSIS, Microbiology Division, Beltsville, Md.
- 26.Mehrvar, M., C. Bis, J. M. Scharer, M. Moo-Young, and J. H. Luong. 2000. Fiber-optic biosensors—trends and advances. Anal. Sci. 16:677-692. [Google Scholar]
- 27.Menudier, A., C. Bosiraud, and J. A. Nicolas. 1991. Virulence of Listeria monocytogenes serovars and Listeria spp. in experimental-infection of mice. J. Food Prot. 54:917-921. [DOI] [PubMed] [Google Scholar]
- 28.Nannapaneni, R., R. Story, A. K. Bhunia, and M. G. Johnson. 1998. Unstable expression and thermal instability of a species-specific cell surface epitope associated with a 66-kilodalton antigen recognized by monoclonal antibody EM-7G1 within serotypes of Listeria monocytogenes grown in nonselective and selective broths. Appl. Environ. Microbiol. 64:3070-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogert, R. A., J. E. Brown, B. R. Singh, L. C. Shriver-Lake, and F. S. Ligler. 1992. Detection of Clostridium botulinum toxin A using a fiber optic-based biosensor. Anal. Biochem. 205:306-312. [DOI] [PubMed] [Google Scholar]
- 30.Oladepo, D. K., A. A. G. Candlish, and W. H. Stimson. 1992. Detection of Listeria monocytogenes using polyclonal antibody. Lett. Appl. Microbiol. 14:26-29. [Google Scholar]
- 31.Olsen, S. J., M. C. Evans, S. Hunter, V. Reddy, L. Kornstein, W. R. Mackenzie, K. Lane, S. A. Bidol, G. Stoltman, D. M. Frye, I. Lee, S. Hurd, D. Gerber, T. F. Jones, T. N. Laporte, W. Dewitt, L. M. Graves, M. Wiedmann, D. J. Schoonmaker-Bopp, A. J. Huang, C. Vincent, A. Bugenhagen, J. Corby, E. R. Carloni, M. Holcomb, R. Woron, S. M. Zansky, G. Dowdle, F. Smith, S. Ahrabi-Fard, A. R. Ong, N. Tucker, N. A. Hynes, and P. Mead. 2001. Multistate outbreak of Listeria monocytogenes infections linked to deli turkey meat. Clin. Infect. Dis. 33:1237. [Google Scholar]
- 32.Schlech, W. F., III. 2000. Foodborne listeriosis. Clin. Infect. Dis. 31:770-775. [DOI] [PubMed] [Google Scholar]
- 33.Silk, T. M., T. M. T. Roth, and C. W. Donnelly. 2002. Comparison of growth kinetics for healthy and heat-injured Listeria monocytogenes in eight enrichment broths. J. Food Prot. 65:1333-1337. [DOI] [PubMed] [Google Scholar]
- 34.Siragusa, G. R., and M. G. Johnson. 1990. Monoclonal antibody specific for Listeria monocytogenes, Listeria innocua, and Listeria welshimeri. Appl. Environ. Microbiol. 56:1897-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strachan, N. J. C., and D. I. Gray. 1995. A rapid general method for the identification of PCR products using a fiber-optic biosensor and its application to the detection of Listeria. Lett. Appl. Microbiol. 21:5-9. [DOI] [PubMed] [Google Scholar]
- 36.Tempelman, L. A., K. D. King, G. P. Anderson, and F. S. Ligler. 1996. Quantitating staphylococcal enterotoxin B in diverse media using a portable fiber-optic biosensor. Anal. Biochem. 233:50-57. [DOI] [PubMed] [Google Scholar]
- 37.Tims, T. B., S. S. Dickey, D. R. Demarco, and D. V. Lim. 2001. Detection of low levels of Listeria monocytogenes within 20 hours using an evanescent wave biosensor. Am. Clin. Lab. 20:28-29. [PubMed] [Google Scholar]
- 38.Torensma, R., M. J. C. Visser, C. J. M. Aarsman, M. J. J. G. Poppelier, A. D. C. Fluit, and J. Verhoef. 1993. Monoclonal antibodies that react with live Listeria spp. Appl. Environ. Microbiol. 59:2713-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong, S., D. Street, S. I. Delgado, and K. C. Klontz. 2000. Recalls of foods and cosmetics due to microbial contamination reported to the US Food and Drug Administration. J. Food Prot. 63:1113-1116. [DOI] [PubMed] [Google Scholar]
- 40.Zhou, C. H., P. Pivarnik, S. Auger, A. Rand, and S. Letcher. 1997. A compact fiber-optic immunosensor for Salmonella based on evanescent wave excitation. Sensors Actuators B 42:169-175. [Google Scholar]