Abstract
Human enteric viruses can contaminate municipal drinking-water wells, but few studies have examined the routes by which viruses enter these wells. In the present study, the objective was to monitor the municipal wells of La Crosse, Wisconsin, for enteric viruses and determine whether the amount of Mississippi River water infiltrating the wells was related to the frequency of virus detection. From March 2001 to February 2002, one river water site and four wells predicted by hydrogeological modeling to have variable degrees of surface water contributions were sampled monthly for enteric viruses, microbial indicators of sanitary quality, and oxygen and hydrogen isotopes. 18O/16O and 2H/1H ratios were used to determine the level of surface water contributions. All samples were collected prior to chlorination at the wellhead. By reverse transcription-PCR (RT-PCR), 24 of 48 municipal well water samples (50%) were positive for enteric viruses, including enteroviruses, rotavirus, hepatitis A virus (HAV), and noroviruses. Of 12 river water samples, 10 (83%) were virus positive by RT-PCR. Viable enteroviruses were not detected by cell culture in the well samples, although three well samples were positive for culturable HAV. Enteroviruses detected in the wells by RT-PCR were identified as several serotypes of echoviruses and group A and group B coxsackieviruses. None of the well water samples was positive for indicators of sanitary quality, namely male-specific and somatic coliphages, total coliform bacteria, Escherichia coli, and fecal enterococci. Contrary to expectations, viruses were found in all wells regardless of the level of surface water contributions. This result suggests that there were other unidentified sources, in addition to surface water, responsible for the contamination.
Human enteric viruses occur in groundwater (2, 5, 19) and are thought to be responsible for a significant proportion of infectious gastroenteritis related to groundwater consumption. Between 1971 and 1996, there were 642 outbreaks of waterborne disease in the United States, of which 371 (58%) were associated with groundwater sources. In 9% of the outbreaks, the etiologic agent was identified as a virus (56). In the most recent compilation of waterborne disease outbreak data for the years 1999 and 2000, 26 of 37 infectious disease outbreaks were attributed to groundwater, and a virus was identified in four of these (35). The etiologic agent could not be identified in roughly half of groundwater-related outbreaks, but these outbreaks were presumably viral in origin because of the absence of bacterial and protozoan pathogens (12, 56).
Several studies have systematically monitored groundwater sources of drinking water for the occurrence of enteric viruses. In a nationwide study, samples from 448 groundwater sites in 35 states were analyzed by reverse transcription-PCR (RT-PCR) for enteroviruses, rotavirus, hepatitis A virus (HAV), and noroviruses (formerly called Norwalk-like viruses). Among all sites, 141 (31.5%) were positive for at least one virus, and 68 (15.2%), 62 (13.8%), and 31 (6.9%) sites were positive for enteroviruses, rotavirus, and HAV, respectively (1). A subset of the sites in this study, 317, was tested for noroviruses, and 3 (0.9%) were found to be positive (1). Borchardt et al. (5) tested 50 private household wells located throughout Wisconsin for enteric viruses. Four wells (8%) were positive by RT-PCR. Three wells were positive for HAV. Another well was positive for rotavirus and norovirus in one sample and an enterovirus in another sample. Fout et al. (19) analyzed 321 samples from 29 groundwater sites by RT-PCR and found that 50 samples (16%) and 21 sites (72%) were virus positive. Other unpublished studies report virus contamination in community and noncommunity wells (56). These previous studies have primarily focused on estimating the prevalence of virus contamination. An ideal next step would be to model groundwater flow near potential fecal sources and develop the ability to estimate the likelihood of virus contamination in a particular well.
Wells located in sand and gravel aquifers are more likely to be virus contaminated than those in other aquifer types (1). The pore sizes of sand and gravel sediments are too large in diameter to impede virus transport by physical straining. Electrostatic adsorption to the sand and gravel grains is the primary virus removal mechanism, which, depending on the virus-specific isoelectric point, may be too weak to significantly attenuate transport (61). Wells pumping from alluvial sand and gravel aquifers are often in proximity to surface water that may be contaminated with human fecal wastes and enteric viruses. If the pumping rate is great enough, the well will induce infiltration of surface water, carrying viruses towards the well. For regulatory purposes, a drinking-water well is classified as under the influence of surface water if a microscopic particulate analysis (MPA) shows algae, rotifers, Giardia, and other organisms typically found in surface water to be present in the well water (54). The MPA method, however, may underestimate the importance of surface water inputs transporting viruses into wells because the MPA indicator organisms are 100 to 1,000 times larger than viruses and more likely to be removed from the infiltrating surface water by physical straining.
Stable hydrogen and oxygen isotope ratios can be valuable tools for investigating surface water infiltration into groundwater systems (7, 37). These isotopes are ideal conservative tracers of water sources because they are part of the water molecule itself. Stable isotope ratios of water are conserved in aquifers at low temperature, but the water becomes isotopically fractionated on the surface at less than 100% humidity (20). Because the vapor pressure of H216O is greater than that of H218O, the residual liquid is characterized by a higher H218O content after evaporation. Protium (1H) and deuterium (2H) fractionate to a greater extent than oxygen due to their larger percent relative mass difference. Thus, characteristic 18O/16O and 2H/1H ratios can provide fingerprints for water sources. These approaches have only recently been applied to drinking-water supplies (10, 38).
Many communities in the United States rely on groundwater that is under the influence of surface water. The City of La Crosse, Wisconsin, is located adjacent to the Mississippi River, but its drinking water source is the alluvial sand-gravel aquifer beneath the city. The U.S. Geological Survey and the Wisconsin Geological and Natural History Survey recently conducted a Source Water Area Protection project for La Crosse County to identify quantitatively the zone of contribution for each municipal groundwater supply in the county, from which a three-dimensional model was completed (29). Simulations using the model suggested that some wells in the City of La Crosse receive large amounts of surface water, some receive only small amounts, and others do not receive any. The Mississippi River likely contains human enteric viruses. Therefore, we conjectured that virus occurrence in the La Crosse wells may be related to surface water contributions.
The objectives of this study were to test for enteric viruses in wells that had various predicted amounts of surface water, quantify the amount of surface water in the discharge of these wells using water isotope analysis, and then relate the amount of surface water to the presence and frequency of virus detection. This understanding could be transferable to other communities with similar hydrogeologic characteristics, thus enabling identification of wells vulnerable to virus contamination without costly virus sampling of all wells in a pumping system.
MATERIALS AND METHODS
Sampling design.
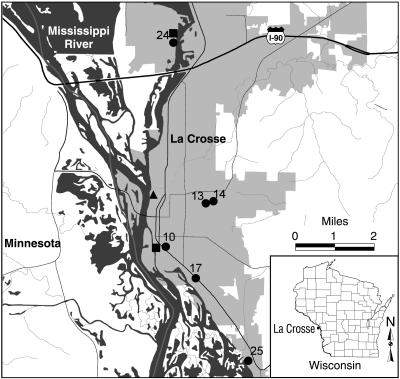
Four drinking-water wells and one surface water site in La Crosse, Wisconsin, were sampled monthly for 1 year from March 2001 to February 2002 (Fig. 1). La Crosse, with a population of 51,818 as of the 2000 census, is located in southwestern Wisconsin on the Mississippi River. The surface water site was at a park located within the city limits. The wells selected for sampling included one well predicted to have high surface water contributions (well 24), one well predicted to have low surface water contributions (well 14), and two wells with intermediate contributions (wells 10 and 25). Due to maintenance issues, wells 10 and 14 were shut down during one sampling period, at which time the samples were drawn from wells in the same proximity, wells 17 and 13, respectively (Fig. 1). All samples from municipal wells were collected at the wellhead, prior to chlorination, from a standard tapered coliform bacteria-sampling tap. River water samples were collected from a levee with a centrifugal pump submerged 0.5 to 1.0 m. In addition to these primary sampling locations, two piezometers were installed in November 2001. One piezometer was located between the river and well 24, and the other was located between the river and well 10 (Fig. 1). In December 2001, samples were collected from both piezometers by using a battery-powered peristaltic pump.
FIG. 1.
Map of La Crosse, Wisconsin, indicating surface water (▴), piezometer (▪), and municipal well (•) sampling sites. The number next to the symbol is the well number.
Analysis of indicators of water sanitary quality.
Indicator samples were collected aseptically before the virus filtration apparatus was attached to the tap. Four liters of well water was collected into a sterile polypropylene container and transported to the laboratory on ice on the day of collection. All indicator analyses were set up the same day or the next day, in which case the samples were stored overnight at 4°C.
Somatic and male-specific coliphages were enumerated with the single agar layer procedure described by Environmental Protection Agency (EPA) method 1602 (57). The analysis volume for each coliphage type was 100 ml. Samples were analyzed as a batch (i.e., one river and four well samples each analysis period). Each batch included a positive and a negative control to test for method efficacy, and each sample included matrix controls to test for interferences. Matrix controls were performed by seeding 80 PFU of somatic coliphage φX174 (ATCC 13706-B1; American Type Culture Collection, Manassas, Va.) and 200 PFU of male-specific coliphage MS2 (ATCC 15597-B1) into separate 100-ml volumes of the sample and performing the assay following the same procedure as for the unknowns. Percent recovery for the matrix controls was calculated by R = 100 × (Nsp − Nusp)/T, where R is the percent recovery, Nsp is the number of PFU in the spiked sample, Nusp is the number of PFU in the unspiked sample, and T is the number of coliphage spiked. In addition, the recovery efficiency of the method was measured with each sample batch by testing two 100-ml volumes of sterile reagent-grade water, one that had been seeded with 80 PFU of φX174 and one that had been seeded with 200 PFU of MS2.
Total coliforms and Escherichia coli bacteria were detected with the Colilert and Colisure chromogenic substrate assays (IDEXX Laboratories, Westbrook, Maine). Both assays were performed in parallel for each sample, and if either assay was positive, a sample was classified as indicator positive. Fecal enterococci were tested with the Enterolert chromogenic substrate assay (IDEXX Laboratories). The test volume for all bacterial indicators was 100 ml. Results were scored as present or absent.
Water isotope analysis.
Water isotope samples were collected unfiltered immediately prior to virus sampling in clean, dry, 20-ml glass vials and sealed with a polyseal cap. Paraffin film was used to secure the cap after sample collection to prevent evaporation that can alter 18O/16O and 2H/1H ratios. Water isotopes were analyzed at the U.S. Geological Survey Stable Isotope Laboratory, Reston, Va. Oxygen isotope ratios were measured using CO2-H2O equilibration (18). Stable hydrogen isotope ratios were determined by H2-H2O equilibration (11). Oxygen and hydrogen isotopic results are reported per mill relative to Vienna Standard Mean Ocean Water and normalized on scales so that the oxygen and hydrogen isotopic values of Standard Light Antarctic Precipitation were −55.5‰ and −428‰, respectively (8). Analytical error (2σ) was estimated at ±0.2‰ and ±2.0‰ for δ18O and δ2H, respectively (http://isotopes.usgs.gov/Quality.htm).
Virus sampling.
Viruses were sampled by filtering a target water volume of 1,500 liters through a 1-MDS filter (CUNO, Meriden, Conn.) according to standard methods (4). A sediment prefilter (McMaster and Carr, Los Angeles, Calif.) was used for the river site. All samples were collected aseptically. Filter housings were autoclaved, and tubing, gauges, centrifugal pump, and other nonautoclavable components of the virus filtration apparatus were disinfected with 0.52% chlorine for 30 min followed by rinsing with 0.05% Na2S2O3 in sterile H2O for 30 min (58). Water temperature and pH were measured prior to virus sampling. Filters were transported to the laboratory on ice and processed within 2 days after collection.
Virus elution and flocculation.
Viruses were eluted by slowly passing 1 liter of 1.5% beef extract (wt/vol) with 0.05 M glycine (pH 9.5) through the filter twice. The prefilter from the river sample was eluted in series with the 1-MDS filter. For the first 45 samples (March 2001 to November 2001), the eluate was flocculated by acidification, where the beef extract was adjusted to pH 3.5, stirred for 30 min at room temperature, and centrifuged at 2,500 × g for 15 min at 4°C. The pellet was resuspended in 30 ml of sterile 0.15 M Na2HPO4 (pH 9.25) and centrifuged again at 6,000 × g for 10 min at 4°C, and the supernatant was adjusted to pH 7.25 ± 0.25. For the last 17 samples (December 2001 to February 2002), the eluate was flocculated with polyethylene glycol (PEG). The reason for switching flocculation methods was that during the sample collection period, we validated in our laboratory (26) that the PEG method yielded higher virus recovery efficiency in a smaller final concentrated sample volume (FCSV) than acidification (23, 36, 45). Flocculation with PEG was accomplished by adjusting the beef extract pH to 7.5 ± 0.25 with 1 N HCl, adding NaCl (0.2 M final concentration) and 8% PEG 8,000 (wt/vol) (Sigma Chemical Co., St. Louis, Mo.), stirring this mixture for 16 h at 4°C, and centrifuging at 4,200 × g for 45 min at 4°C. The pellet was resuspended in 3 to 6 ml of sterile 0.15 M Na2HPO4 (pH 9.25), aliquoted, and frozen immediately at −80°C. The final concentrated samples that had been flocculated by acidification (March 2001 to November 2001) were subsequently flocculated again by the PEG method so that (to the extent possible) samples collected from different periods were concentrated to a similar degree. To accomplish this, 24 ml of final concentrated sample from acidification was treated with PEG as described previously and with the amounts of reagents adjusted accordingly. The remaining portion (approximately 6 ml) of the acidified FCSV was archived. All final concentrated samples were stored at −80°C.
A method recovery control was performed approximately every 3 months during the study period according to the quality control procedure for virus monitoring that is described in the ICR Microbial Laboratory Manual (55). Two hundred PFU of poliovirus (vaccine strain Sabin 3) was seeded into 40 liters of dechlorinated tap water, which was then filtered through a sterile 1-MDS filter, eluted into 1 liter of beef extract as described above, and flocculated by either acidification (three times) or PEG (one time). The seeded virus captured in the FCSV was cultured on buffalo green monkey kidney (BGMK) cells, and the quantity of virus was determined by the most probable number assay (55). The mean recovery for flocculation by acidification was 79% (n = 3; range, 33 to 130%). Recovery was 137% for the single control when PEG was used as the flocculation agent.
Viral RNA extraction and concentration.
Prior to RNA extraction, all work surfaces and pipettors were scrubbed with RNase ZAP (Sigma), 10% bleach, and 70% ethanol to remove nucleases and contaminants. RNA extraction and purification were performed as described by Borchardt et al. (5). RNA extract (750 μl) was applied to a sterile Sephadex G-100 column (Sigma) and eluted with four successive volumes (750 μl) of Tris-EDTA. Of the four eluate fractions, fraction 3, the fraction found previously to contain the highest concentration of viral RNA (5), was concentrated with a sterile, nuclease-free Centricon YM-100 ultrafilter (Millipore, Bedford, Mass.). The ultrafilter was sterilized with 70% ethanol and treated with 0.2% diethyl pyrocarbonate-treated water at 37°C for 2 h prior to use. The entire fraction 3 volume was placed on the filter and centrifuged at 2,000 × g for 12 min at 4°C. RNA storage solution (300 μl) (Ambion, Inc., Austin, Tex.) was pipetted onto the filter, which was then inverted and placed on a new, autoclave-sterilized cupule and centrifuged (1,000 × g for 2 min at 4°C). The fraction 3 RNA concentrate was stored at −80°C.
RT-PCR.
A two-step RT-PCR was performed to detect five groups of viruses: enteroviruses, rotavirus, HAV, and norovirus genogroups 1 and 2 (G1 and G2). The RT and PCR procedures and the primers used were identical to those described by Borchardt et al. (5). The rotavirus forward primer reported by Borchardt et al. (5) incorrectly includes an extra base. The correct sequence is 5′-TTGCCACCAATTCAGAATAC-3′. The RT-PCR amplicons were resolved by electrophoresis in 1.6% agarose gels containing ethidium bromide, followed by UV light illumination (Gel-Doc system; Bio-Rad Laboratories, Hercules, Calif.).
Controls for extraction, RT-PCR, and inhibition.
The negative extraction control consisted of final concentrate from 1 liter of beef extract that had been flocculated by PEG. An aliquot of this final concentrate (500 μl) was extracted each day a batch of samples was extracted, reserving the control for last in the sequence of extractions so that if contamination occurred it would be more likely to be detected. The negative RT-PCR control consisted of the same reaction components as the samples, except that 50 μl of water was substituted for the fraction 3 RNA concentrate. The positive RT-PCR controls consisted of the five target viruses seeded into 1 liter of beef extract and flocculated by PEG. The quantities of viruses added to the 1 liter of beef extract were 3 × 104 PFU of poliovirus Sabin 3, 4 × 104 fluorescent foci group A rotavirus, 2.5 × 108 PFU of formalin-inactivated HAV, a 10−3 dilution of G1 norovirus, and a 10−1 dilution of G2 norovirus, with both genogroups diluted from freon-extracted human stool. To minimize the possibility of contamination, the final concentrates of the virus-positive controls were extracted on different dates than those of the field samples. Moreover, the RT-PCRs were batched by virus, with only one positive control per batch. Negative and positive controls were processed and analyzed in the same fashion as the field samples. Each sample was tested for RT-PCR inhibition by seeding 50 μl of fraction 3 RNA concentrate with a Norwalk virus RNA internal standard (46), as described by Borchardt et al. (5).
Southern hybridization.
All gels were vacuum blotted, and Southern hybridization was performed following the methods and using the oligoprobes described by Borchardt et al. (5). A sample was scored as virus positive when the oligoprobe hybridized to the membrane at the location of the amplicon specific to the virus group tested. If the gel was positive for the amplicon but did not hybridize, the sample was scored as virus negative. Samples scored as positive were reanalyzed, beginning with a new RNA extract from the FCSV to confirm the results.
RT-PCR method detection limit.
To estimate the limit of the RT-PCR and Southern hybridization method for detecting low levels of virus present in groundwater samples, we seeded approximately 250 PFU of poliovirus (Sabin 3) into 1 liter beef of extract. The poliovirus was enumerated each time it was seeded by the standard agar overlay plaque assay with BGMK cells. The plaque assay was performed in duplicate using both 1:10 and 1:2 serial dilutions (i.e., two independent serial dilutions of the poliovirus stock) so that more than one dilution yielded countable plaques. The method detection limit was estimated on three separate occasions, twice where the beef extract was flocculated by acidification and once where flocculation was by the PEG method. After flocculation, the final concentrated samples were diluted serially twofold (1:2 to 1:512) with unseeded final concentrated sample as the diluent, and then each dilution was carried through the remainder of the procedure to the final hybridization step.
Culturable enteroviruses and HAV.
All well water and river samples were analyzed for enteroviruses by cell culture in BGMK, rhabdomyosarcoma, and Caco-2 cell lines. The cell culture methods were the same as those described by Borchardt et al. (5), with several modifications. (i) All cell cultures were rinsed with 15 ml of sterile ELAH (0.5% lactalbumin hydrolysate in Earle's balanced salt solution) to remove any residual fetal bovine serum (FBS) before sample inoculation (59). (ii) The volume of final concentrated sample inoculated into each culture in a 175-cm2 flask was 0.13 to 0.37 ml amended with Earle's balanced salt solution to 5.0 ml/flask to facilitate even dispersion during the virus adsorption step. The combined inoculation volume for the six flasks (two flasks per cell line) represented approximately 30% of the original water sample volume (range, 24 to 34%). (iii) Controls included a negative control inoculated with 0.15 M Na2HPO4 and four positive controls per cell line: poliovirus (Sabin 3), echovirus 30, coxsackievirus A6, and coxsackievirus B4. The latter three viruses were human isolates obtained from fecal specimens submitted to a clinical reference laboratory. All cultures were monitored for 14 days for viral cytopathic effect and then frozen and aliquoted into new cultures, which were monitored for another 14 days to confirm the first passage results.
Four samples positive for HAV by RT-PCR were analyzed by cell culture to determine HAV infectivity. Fetal rhesus monkey kidney cells (FRhK-4; ATCC CRL-1688) were grown to confluent monolayers in 75-cm2 tissue culture flasks with Eagle's minimal essential medium with Earle's salt, HEPES buffer, penicillin-streptomycin-amphotericin B (Fungizone) solution, 0.1 mM nonessential amino acids (Cambrex, Walkersville, Md.), and 10% FBS. Cell cultures were rinsed with 15 ml of ELAH, and 0.4 to 0.5 ml of filter-sterilized final concentrated sample was inoculated into each of four flasks per sample. The combined inoculation volume for the four flasks represented 33% of the original water sample volume. Inoculated cultures were rocked for 120 min at room temperature, and 50 ml of Eagle's minimal essential medium with 2% FBS was added to each flask. All flasks were incubated at 37°C. Of the four flasks per sample, two were passaged weekly, while the other two were passaged every 2 weeks. Cultures were passaged by trypsinizing the cell monolayer, pelleting the cells (700 × g for 5 min), washing once with sterile phosphate-buffered saline, lysing by bead vortexing, and transferring 1 ml of cell lysate into a new FRhK-4 cell culture in a 75-cm2 flask. Samples were passaged for 6 weeks. After each passage, trypsinized and washed FRhK-4 cells were stained with fluorescein isothiocyanate-conjugated anti-HAV monoclonal antibody (Chemicon, Temecula, Calif.), counterstained with Evans Blue, and examined with an epifluorescence microscope. The negative control (0.15 M Na2HPO4) and positive control (HAV strain HM175/18f from ATCC) were cultured and passaged simultaneously with the samples. Immunofluorescent positive cultures were further confirmed to contain HAV by testing trypsinized cells for HAV by RT-PCR and Southern hybridization.
Enterovirus identification by sequencing.
The method of Ishiko et al. (30) for determining the serotype of an enterovirus from its nucleotide sequence was used for those samples found to be enterovirus positive by RT-PCR with panenterovirus primers (15). Primers MD91 and OL68-1 (30) were used in the two-step RT-PCR described above to produce a ∼750-bp amplicon encoding the entire viral protein 4 and portions of the 5′-nontranslated region and viral protein 2. Amplicon was extracted and purified from gels with a QIAquick gel extraction kit (QIAGEN, Valencia, Calif.) and cloned with the pGEM-T Easy Vector system (Promega, Madison, Wis.). DNA sequencing was performed by the dideoxy chain-termination method with an ABI Prism Big Dye Terminator cycle sequencing kit (Applied Biosystems) on the ABI model 3100 automated sequencer. Sequences were determined in both directions by using standard pUC/M13 forward and reverse primers. Consensus sequences were analyzed with Lasergene software (DNAstar, Madison, Wis.), and enterovirus identity was determined by using BLAST (3) at the National Center for Biotechnology Information website.
RESULTS
Characteristics of study wells and water samples.
The six municipal wells sampled in the present study were conventionally drilled wells in active service (Table 1). All the wells were constructed in an alluvial sand-gravel aquifer. The physical characteristics of the water samples are reported in Table 2. Seven samples from well 24 had a pH of ≥8.00, and these were adjusted to pH 7.00 to 7.50 with 0.1 N HCl during sample filtration to facilitate virus adsorption. Only one river sample had a pH of >8.0.
TABLE 1.
Construction and pumping data for La Crosse, Wisconsin, municipal well sites
| Well no. | Yr constructed | Well depth (m) | Well casing depth (m) | Static water level (m) | Avg GPMa over study periodb | Study periodb total gallons pumped (106) |
|---|---|---|---|---|---|---|
| 10 | 1936 | 45.7 | 15.2 | 11.9 | 224 | 117 |
| 13 | 1953 | 46.0 | 30.8 | 13.1 | 1,135 | 598 |
| 14 | 1952 | 44.2 | 29.0 | 14.0 | 813 | 429 |
| 17 | 1955 | 48.8 | 33.5 | 7.8 | 789 | 416 |
| 24 | 1980 | 32.9 | 21.6 | 7.5 | 726 | 382 |
| 25 | 1984 | 30.2 | 19.2 | 3.0 | 828 | 437 |
Gallons pumped per minute.
March 2001 to February 2002.
TABLE 2.
Physical characteristics of La Crosse municipal wells and Mississippi River samples
| Measurement | Well (n = 48)
|
River (n = 12)
|
||||
|---|---|---|---|---|---|---|
| pH | Temp (°C) | Sample vol (liters) | pH | Temp (°C) | Sample vol (liters) | |
| Minimum | 7.3 | 9.8 | 1,484 | 7.7 | 2.0 | 223 |
| Maximum | 8.4 | 15.0 | 2,752 | 8.1 | 26.1 | 681 |
| Mean | 7.6 | 12.7 | 1,611 | 7.9 | 12.5 | 431 |
| Median | 7.5 | 12.8 | 1,526 | 7.9 | 11.0 | 445 |
All of the well water samples for viruses met or exceeded the 1,500-liter target sample volume, but the river samples clogged the prefilter after several hundred liters (Table 2). The fraction of the sample original volume analyzed by RT-PCR for viruses averaged 2.6% (median, 1.7%; range, 0.4 to 5.0%). These fractions corresponded to an average sample volume analyzed of 36.0 liters (median, 25.2 liters; range, 1.0 to 137.6 liters).
Virus occurrence as determined by RT-PCR.
Of the 62 water samples collected during the present study, 36 (58%) contained at least one enteric virus as detected by RT-PCR and confirmed by Southern hybridization (Table 3). Fifty percent (24 of 48) of the well water samples were positive for at least one virus group. Eleven out of the 48 well water samples (23%) were positive for two or more virus groups. Viruses detected in the wells included enteroviruses (20 samples [42%]), rotavirus (10 samples [21%]), HAV (4 samples [8%]), and norovirus genogroup 1 (3 samples [6%]). Among the river samples, 83% (10 of 12) were positive for at least one virus, including enteroviruses, rotavirus, and norovirus genogroup 1 (Table 3). HAV was not detected in the river, and norovirus genogroup 2 was not detected in any of the samples. Both piezometer samples were virus positive, one for an enterovirus and the other for rotavirus.
TABLE 3.
Virus occurrence by site as determined by RT-PCR and Southern hybridization
| Sample site | No. of samples | No. of virus-positive samples | Positive samples by virus type
|
||||
|---|---|---|---|---|---|---|---|
| Enteroviruses | Rotavirus | Hepatitis A virus | Noroviruses
|
||||
| G1 | G2 | ||||||
| River | 12 | 10 | 5 | 8 | 0 | 1 | 0 |
| Well 13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Well 14 | 11 | 5 | 4 | 1 | 1 | 1 | 0 |
| Well 17 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| Well 10 | 11 | 8 | 6 | 4 | 2 | 1 | 0 |
| Well 24 | 12 | 4 | 4 | 3 | 1 | 0 | 0 |
| Well 25 | 12 | 6 | 5 | 2 | 0 | 1 | 0 |
RT-PCR inhibition.
RT-PCR was inhibited in 10 samples, as indicated by the absence of amplicon from the Norwalk virus internal standard. River samples were inhibited at the highest frequency. Inhibition did not appear to be seasonally related. River samples were inhibited in April, June, August, September, October, and December. The well samples were inhibited in May, July, January, and March. Inhibition was alleviated in all 10 samples by diluting the fraction 3 RNA concentrate by 1:10 in nuclease-free water. Of the 10 diluted samples, five were subsequently determined to be rotavirus positive, and the remaining five samples were negative for all viruses tested (see reference 26 for additional discussion of inhibition).
RT-PCR method detection limit.
The method detection limits estimated twice with the acidification flocculation method were 0.005 PFU and 0.004 PFU of poliovirus per liter of water sample. When the PEG flocculation method was used, the method detection limit was 0.006 PFU liter−1. These numbers were calculated assuming that viruses were recovered from the 1-MDS filter in beef extract eluate with 79% efficiency (see Materials and Methods) and that the water sample volume was 1,500 liters.
Surface water infiltration into municipal wells.
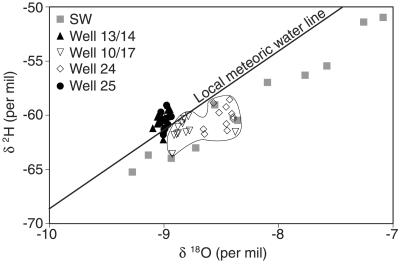
Only wells 10 and 24 contained appreciable amounts of surface water, as indicated by water isotope 18O/16O and 2H/1H ratios that were substantially on the evaporative side (right side) of the local meteoric water line (LMWL) (Fig. 2). The LMWL was constructed from δ18O and δ2H previously published data collected from a U.S. Geological Survey monitoring site located approximately 16 miles north of La Crosse at the Black River at Galesville, Wis. (9). The isotopic compositions of the surface water collected in the present study lie to the right of the LMWL, reflecting the evaporative effect of a large Mississippi River impoundment located a few miles upstream from the sample site (Fig. 1). The majority of samples from wells 13 or 14 and 25 were on the left side of the LMWL, indicating that surface water was not entering these wells. The one or two samples from these wells that were slightly on the evaporative side of the LMWL were within the natural variability of unevaporated sources, given that the LMWL is considered more of a flattened ellipse than a discrete, sharp line (28).
FIG. 2.
Surface water (SW) content in municipal well samples as determined by 18O/16O and 2H/1H ratios.
Association of surface water infiltration with virus occurrence.
The relationship between virus occurrence and the amount of surface water infiltrating a well could not be determined because nearly all wells were virus positive regardless of surface water amount. Wells 13 or 14 and 25, which had no detectable surface water, were positive for viruses in 5 of 12 and 6 of 12 samples, respectively. Well 24, which had an appreciable amount of surface water, was virus positive in 4 of 12 samples. Well 10 had an intermediate level of surface water contributions, but it had the greatest proportion of virus-positive samples, 8 out of 11.
Virus occurrence compared to clinical seasonality.
The temporal correspondence between virus occurrence in the wells and the seasonal pattern of clinical infections typically observed in Wisconsin was equivocal and did not provide any insights into potential fecal sources, transport routes, or virus travel times (Table 4). HAV and rotavirus were found in the wells at approximately the same time the viruses are found in patients, the winter months (27, 32), although rotavirus was also found in the wells in the autumn before the usual clinical season. Enteroviruses, whose infections usually occur in the late summer and autumn (39), were found in the La Crosse wells in nearly every monthly sample. Noroviruses occur clinically year round (33), but they were found in the wells in only 1 month. The temporal correspondence was not any clearer when the wells were subgrouped into wells with high, intermediate, and low surface water contributions.
TABLE 4.
Seasonality of virus occurrence in La Crosse municipal wells, compared to virus clinical seasonality
| Virus | 2001a
|
2002
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | A | M | J | J | A | S | O | N | D | J | F | |
| Enteroviruses | − | − | + | + | + | + | + | + | + | + | + | |
| Rotavirus | − | − | − | − | − | + | + | − | + | + | + | + |
| Hepatitis A virus | − | − | − | − | − | − | − | − | + | + | + | − |
| Noroviruses | − | + | − | − | − | − | − | − | − | − | − | − |
+, at least one sample taken from the wells that month was positive for the given virus. The leftmost column is March 2001, and columns proceed chronologically to February 2002. Boldface symbols indicate months when the given virus is commonly detected in clinical cases in Wisconsin.
Indicators of water sanitary quality.
The coliphage matrix controls did not show any evidence of interferences from the river or well water samples. The mean matrix recovery of male-specific coliphage was 95% (n = 62; median, 94%; range, 74 to 153%), whereas the matrix recovery of somatic coliphage had a mean of 96% (n = 62; median, 95%; range, 49 to 174%). The lower end of the recovery range for both phage types fell within the quality control acceptance criteria of EPA method 1602 (57). The mean method recovery control for male-specific coliphage was 90% (n = 13; median, 90%; range, 78 to 104%), and the mean method recovery for the somatic coliphage was 95% (n = 13; median, 95%; range, 74 to 118%).
None of the 50 well and piezometer samples was positive for any of the indicators. All 12 river samples were positive for total coliforms and somatic coliphages, and 11 river samples were positive for E. coli and fecal enterococci. The mean concentration of somatic coliphages in the river samples was 71 PFU 100 ml−1 (n = 12; median, 64 PFU 100 ml−1; range, 4 to 141 PFU 100 ml−1). Male-specific coliphages were identified in only 4 out of 12 river samples, with a mean concentration of 2 PFU 100 ml−1 (n = 12; median, 0; range, 0 to 17).
Culturable enteroviruses and HAV.
Of the 48 well water samples cultured for enteroviruses, none was positive. In contrast, 9 out of 12 river samples, the 5 samples that were positive by RT-PCR and 4 additional samples, were culture positive. After sequencing, these four samples were found to be bovine enteroviruses, which would not have amplified with the human enterovirus primers used in the RT-PCR. Of the four samples positive for HAV by RT-PCR, three samples were positive for culturable HAV. These three samples were taken from wells 10, 14, and 24.
Identity of enteroviruses detected by RT-PCR.
Enteroviruses from all 25 river and well samples positive for enteroviruses by RT-PCR were successfully sequenced. Thirteen enterovirus serotypes were identified, including coxsackievirus A and B groups and echoviruses (Table 5).
TABLE 5.
Identity of enteroviruses detected by RT-PCR in well and river samples
| Enterovirus type | No. of positive samples | Sample source | BLAST searcha
|
|
|---|---|---|---|---|
| % Identity | E-score | |||
| Coxsackie B3 | 2 | Well | 97-100 | 0.0 |
| Coxsackie B5 | 2 | Well, river | 88-91 | 1e−173, 0.0 |
| Coxsackie B4 | 5 | Well | 93-96 | 0.0 |
| Coxsackie A3 | 3 | Well, river | 96-100 | 0.0 |
| Coxsackie A11 | 1 | Well | 100 | 0.0 |
| Coxsackie A16 | 1 | Well | 97 | 0.0 |
| Echo 9 | 1 | Well | 86 | 1e−164 |
| Echo 18 | 4 | Well | 94-97 | 1e−93, 0.0 |
| Echo 19 | 1 | Well | 100 | 1e−166 |
| Polio 1 | 2 | Well | 99 | 0.0 |
| Polio 3 | 1 | Well | 98 | 0.0 |
| Echo 30 | 1 | River | 98 | 0.0 |
| Coxsackie B1 | 1 | River | 93 | 0.0 |
The ranges in percent identities and E-scores are reported when two or more samples were positive for the identified enterovirus type.
DISCUSSION
Association of surface water infiltration with virus occurrence.
The primary objective of this study was to relate the amount of Mississippi River water infiltrating La Crosse, Wisconsin, municipal drinking water wells with the detection frequency of human enteric viruses in these wells. Such a relationship would be useful for predicting those wells most vulnerable to virus contamination, indicating where additional treatment might be necessary to produce safe drinking water. Four wells were selected for study that were predicted to have high, intermediate, and low surface water contributions based on previous hydrogeological modeling (29). Indeed, based on 18O/16O and 2H/1H ratio measurements in the present study, the hydrogeological model was correct except for well 25, which did not receive appreciable river inputs due to local hydrogeological heterogeneity that enhanced the well's capture of water not associated with the river (6). Nonetheless, the four study wells did vary substantially in their levels of surface water contributions. The isotope ratios for these wells represent the extremes of the distribution of surface water contributions among all 13 operating La Crosse drinking water wells sampled for isotopes during the study period (29a), and we expected the level of virus contamination to correspond. Instead, all the wells, regardless of the level of surface water contribution, had about the same frequency of virus detection. Wells 25 and 13 or 14, without any surface water inputs, were virus positive for 5 and 6 monthly samples out of 12, respectively, and well 24, with high surface water inputs, was virus positive for 4 out of 12 months. Well 10 or 17 had an intermediate level of river water, but their virus occurrence rates were the highest. Therefore, it was not possible to relate surface water contributions to virus contamination in the wells, and viruses found in the wells that had little or no surface water must have been derived from other sources.
The most likely other source of fecal contamination was municipal sanitary sewer lines. Other potential fecal sources, such as manure, septage, sludge land application sites, wastewater lagoons, wastewater irrigation sites, landfills, and injection wells are located outside the La Crosse city limits. There are a few septic systems at the edges of the city limits but far outside the capture zones of the study wells. Moreover, the enterovirus primers used in the present study are specific to human serotypes, suggesting that the fecal source was human. Except for well 24, the study wells were located in residential areas underlain with a network of sanitary sewers. Sewer pipes, particularly older pipes, are leaky (34), and given the sandy alluvial outwash on which La Crosse is built, fecal viruses could be transported readily to the capture zones of the wells. Sewer lines are typically laid at a depth of 8 feet (2.4 m) in La Crosse, only a short distance from the water table (Table 1), and some lines are laid at 30 feet (9.1 m) (M. Johnson, personal communication). Interestingly, well 10, which had the highest number of virus-contaminated samples, is located only 203 m from a wastewater lift station where the sewer line is pressurized and located within the capture zone of well 10 (6). Moreover, well 14, which had no evidence of surface water contributions, was positive for infectious HAV, suggesting that the fecal contamination source must have been sufficiently close so that the virus was not completely inactivated during transport.
Bank transport of surface water viruses into municipal wells.
Unlike the other wells, well 24 was in a nonresidential area, with the nearest sanitary sewer more than 640 m from the wellhead, far outside the capture zone of the well (6). The source of virus contamination for this well was likely infiltrating surface water from the river, which was heavily contaminated with enteric viruses and fecal indicator organisms. Recall that the isotopic composition of well 24 water demonstrated the contribution of nearby surface water (Fig. 2), indicating that the well was capturing river water through the riverbank. Based on the water isotope time series and the seasonal variation in the isotopic signature, it was estimated that the river water traveled to well 24 in 2 to 3 months during the spring flood of 2001, and under nonflood conditions the travel time to well 24 was 6 to 9 months (29a). The distance between well 24 and the river is 68 m. Surface water may have also contributed some viruses into well 10, which according to the isotopic signature was receiving some surface water, albeit less than well 24. Well 10 is located further from the river, and it took greater than 1 year for surface water to reach the well (29a). These travel time estimates were corroborated by a local numerical groundwater flow model (6) and also by analyzing inflections in the continuously recorded temperature signatures in the river and wells (29a).
At least for well 24, the travel time was sufficiently short that viruses moving with the surface water could reach the well in an infectious state. Enteric viruses can survive in the subsurface environment for 6 to 9 months, depending on the virus type, temperature, moisture content, and other abiotic and biotic factors (14, 62, 63). Adsorption is the primary virus removal mechanism, but for coarse sediments, such as the alluvial sand and gravel comprising the aquifer for La Crosse, virus transport is expected to have little attenuation because there is less surface area for adsorption and fewer virus-to-sediment particle interactions than with more finely textured sediments like clay (16, 21).
In the case of water traveling from riverbank to well 24, however, transport through the alluvial sediment appears to have removed most of the microbial contaminants. The river samples were frequently positive for coliform bacteria, E. coli, fecal enterococci, coliphages, and culturable enteroviruses, but all of the well 24 samples were negative for these microbes. One well 24 sample was positive for culturable HAV, consistent with the finding that surface water travel times were within the survival times of viruses, and four samples were virus positive by the RT-PCR method. Thus, we conclude that the riverbank removed most but not all of the microbial contaminants in the river. It is important to note that well 24 was pumped regularly during the study period, which should have improved the consistency of measurements of any surface water effects (48). Riverbank filtration in The Netherlands has been reported to provide a 4 log reduction in enterovirus and reovirus concentrations in drinking-water wells located 25 to 30 m from the rivers (52). During a slug test for virus transport in an alluvial sand-gravel aquifer in Montana, 1011 PFU of poliovirus injected into the groundwater was almost completely attenuated after 19.4 m (13). In the same study, the bacteriophages MS2, PRD-1 and φX174 were not nearly as attenuated as the poliovirus and were readily transported 40 m. The effectiveness of riverbank filtration to remove viruses is likely to vary substantially by virus type and the properties (grain size, degree of sorting, and organic matter content) of the aquifer.
Virus incidence in La Crosse municipal wells.
Previous studies of virus occurrence in municipal drinking-water wells in the United States have sampled from multiple municipalities located in one or more states (56). The largest study of this type investigated 448 wells in 35 states and found that 141 wells (31.5%) were positive by RT-PCR for an enterovirus, rotavirus, HAV, or norovirus (1). In contrast, the present study focused on one municipality and found that among 48 samples from six drinking-water wells, 24 samples (50%) and five wells (83%) were positive by RT-PCR for the same groups of viruses. This virus incidence rate for well water samples is higher than those from previously reported studies probably because the La Crosse wells are located in a highly permeable sand-gravel aquifer with a shallow water table, and the virus incidence rate on a per well basis is higher probably because the wells were sampled monthly, increasing the odds of finding virus contamination if it was present. It is important to note that the La Crosse wells were all properly constructed, with a minimum casing depth of 15 m, appropriately grouted, and capped with a sanitary seal.
Spring flood of 2001.
During the second month of virus sampling in April 2001, heavy rainfall events during snowmelt resulted in the Mississippi River cresting at 16.41 ft at La Crosse, the third highest crest on record, and the most severe flood in the area since 1965 (National Weather Service Forecast Office, La Crosse, Wisc., The 2001 Mississippi River flood [http://www.crh.noaa.gov/arx/flood2001.html]; U.S. Geological Survey, Mississippi River flood: April 2001 [http://www.umesc.usgs.gov/flood_2001/flood.html]). Compared to the other months of virus sampling, the number of virus detections did not increase in April during the flood, nor shortly thereafter in May or June. However, with the large infiltration of low-ionic-strength rainfall, viruses may have been desorbed from subsurface sediments, transported to the wells, and detected at a later date. In a year when the spring flood is less severe, the incidence of viruses in La Crosse municipal wells receiving appreciable contributions of surface water may be lower than what was observed during the present study.
Microbial indicators of virus contamination.
A secondary objective of this study was to relate microbial indicators of water sanitary quality with the occurrence of enteric viruses. One goal among drinking-water microbiologists is to find an easily detected indicator organism that is highly correlated with virus occurrence so that the expense and technical expertise involved with waterborne virus testing can be avoided. The 50 well and piezometer samples in the present study were all negative for total coliforms, E. coli, fecal enterococci, somatic coliphages, and male-specific coliphages. In contrast, 26 of the 50 samples were positive by RT-PCR for enteric viruses. By this comparison, the indicators were clearly not associated with virus occurrence. Other studies have reported a lack of association between virus occurrence and microbial indicators of sanitary quality (1, 5, 22, 25, 31). On the other hand, all 50 samples were negative for culturable enteroviruses, so by this virus detection method the negative predictive value of the indicators for enterovirus occurrence was 100%. These kinds of comparisons are difficult to interpret because the detection methods and sample volumes analyzed for the indicators and pathogens are so different, which calls into question the value of looking for such an association. Ultimately, the association of interest is a drinking-water indicator that is highly predictive of disease outcome, not pathogen occurrence, as has been shown for the value of fecal enterococci and E. coli for predicting acute gastroenteritis among recreational bathers in marine waters and freshwater, respectively (60).
Identity of enteroviruses.
A variety of group B coxsackieviruses, group A coxsackieviruses, and echoviruses were identified in the La Crosse municipal wells. These viruses can cause acute gastroenteritis and febrile illness, as well as chronic severe illnesses, such as pleurodynia, myocarditis, and meningitis (39). It is unknown whether these illnesses are attributable to waterborne enteroviruses, and for this reason they are listed on the EPA Drinking Water Contaminant Candidate List as viruses requiring future study (http://www.epa.gov/safewater/ccl/cclfs.html). Two viruses, coxsackievirus A3 and coxsackievirus B5, were common both to the river and to the municipal wells, specifically wells 10 and 14. The coxsackievirus B5 found in well 10 could have originated in the river, but it is unlikely given the >1-year travel time and opportunity for viral RNA degradation. The coxsackievirus A3 in well 14 could not have originated from the river, as the well does not contain surface water. The diversity of enteroviruses found in the wells over a 1-year period suggests that the fecal source was derived from a large number of people, as would be expected if the fecal source was municipal sanitary sewers.
Public health implications.
Although this study reports widespread presence of viral RNA in La Crosse municipal wells, the public health implications of these findings are unknown. The viruses were detected by the RT-PCR method, which does not discriminate between infectious and inactivated virions. The 24 RT-PCR positive detections in the wells may all have been due to inactivated viruses. This contention is supported by the culturable enterovirus results, where all 48 well samples were analyzed for infectious enteroviruses in three cell lines, representing 30% of the original sample volume, and yet none of the samples was positive. On the other hand, three well samples were positive for infectious HAV. Hepatitis A infection is a federal and state reportable disease, and when the records from the state department of health were checked, no hepatitis outbreaks had been reported in La Crosse during the study period. Only one hepatitis A case occurred in La Crosse during the study, in April 2001, but there was no evidence to link this case with water consumption (J. Kazmierczak, personal communication).
Another possible explanation for the negative enterovirus cell culture results is that the ambient virus concentrations were below the detection limit. If this were the case, the RT-PCR-positive wells could still present some risk to public health. The RT-PCR method is 10- to 1,000-fold more sensitive than cell culture (24, 42). In the present study, the RT-PCR method detection limit was 0.004 to 0.006 PFU of polioviruses per liter of water sample. Other studies have reported similar detection limits (42, 44). This level of sensitivity is within the realm of public health significance. Regli et al. (43) estimated that virus concentrations in drinking water needed to be less than 1.9 × 10−3 liter−1 to 2.22 × 10−7 liter−1, depending on the virus type, to adequately ensure that the annual risk of infection would be less than 1 person per 10,000. The U.S. EPA conducted a risk analysis for the proposed Groundwater Rule based on a virus concentration of 0.36 PFU 100 liter−1 (i.e., 0.0036 PFU liter−1) for properly constructed wells and 29 PFU 100 liter−1 (i.e., 0.29 PFU liter−1) for improperly constructed wells. The analysis suggested that there are 98,000 viral illnesses annually in the U.S. due to consumption of groundwater from public water systems that do not disinfect (56).
The City of La Crosse does disinfect, and the virus-positive results, both by RT-PCR and cell culture methods, apply to samples taken before chlorination. The city doses gaseous chlorine at a rate of 1 mg liter−1 at each wellhead and maintains an average free chlorine residual of 0.4 mg liter−1 in the distribution system (M. Johnson, personal communication). This residual may be adequate for inactivating viruses to a level less than their infectious dose, but that inference is difficult to make because the literature is inconclusive about what constitutes an effective chlorine concentration for inactivating waterborne viruses. In one study, for example, poliovirus seeded into drinking water with a 0.4 mg liter−1 free chlorine residual at 4°C or 20°C was not inactivated even after 1,000 min (40), whereas in another study, the virus concentration of poliovirus seeded into demand-free water at pH 6 and 5°C and treated with 1.0 mg liter−1 of free chlorine was reduced 3 log in 10 min (49). These kinds of discrepancies between studies exist because chlorination efficacy depends on the chlorine demand of the water, virus strain (17, 41), pH and temperature (17), ionic composition (47), and whether the viruses are dispersed or aggregated with organic material (51).
The U.S. EPA has recommended free chlorine Ct values (free-chlorine concentration multiplied by the time the pathogen is exposed to this concentration) for disinfecting drinking water from a surface water source. These Ct values were derived from laboratory studies of HAV seeded into chlorine demand-free water (50), which were further increased by a factor of 3 to improve the margin of safety (53). At pH 7 and a temperature of 5°C, the EPA recommends a Ct of 8 mg-min liter−1. If we assume that this Ct value is applicable to La Crosse groundwater, then for effective disinfection the contact time from the wellhead to the drinking-water consumer needs to be between 8 and 20 min. For most points of the La Crosse distribution system, 8 mg-min liter−1 is probably achieved, except perhaps for those homes located next to the wellheads, where the water transit time from pump house to tap could be less than the required contact time.
The vulnerability of the La Crosse municipal wells to virus contamination is probably not unique. Nearly one-third of the groundwater withdrawn in the United States is pumped from alluvial sand and gravel aquifers (1). Municipalities situated like the City of La Crosse on an alluvial floodplain adjacent to a river probably also have some virus transport into their drinking-water wells from surface water infiltration. If these municipalities have shallow wells located near sanitary sewers, it is also likely that the alluvial sand and gravel is having little effect in attenuating virus transport from leaking sewers to the wells. However, detecting viruses in untreated groundwater is not prima facie evidence for inferring the risk of disease transmission. To adequately estimate disease risk, it will be necessary to conduct an epidemiological study where virus occurrence in drinking-water wells, preferably measured quantitatively, is related to a defined health end point.
Acknowledgments
We gratefully acknowledge the capable laboratory assistance of Phil Bertz (enterovirus and HAV cultures), Lynn Ivacic (HAV culture), and Susan Spencer (coliphage assays and enterovirus sequencing). Linda Weis, Steve Offer, and Alice Stargardt assisted with manuscript preparation. Carla Schofield provided data management services. Mary Estes, Baylor College of Medicine, kindly provided the Norwalk virus internal standard. We also sincerely thank Mark Johnson, Utilities Manager, and Thomas Berendes, Water Superintendent, City of La Crosse for their cooperation with this study.
This study was funded by grants from the Wisconsin Department of Natural Resources and by the U.S. Geological Survey Water Resources Cooperative Program.
REFERENCES
- 1.Abbaszadegan, M., M. LeChevallier, and C. Gerba. 2003. Occurrence of viruses in US groundwaters. J. Am. Water Works Assoc. 95:107-120. [Google Scholar]
- 2.Abbaszadegan, M., P. Stewart, and M. LeChevallier. 1999. A strategy for detection of viruses in groundwater by PCR. Appl. Environ. Microbiol. 65:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.American Public Health Association. 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, D.C.
- 5.Borchardt, M. A., P. D. Bertz, S. K. Spencer, and D. A. Battigelli. 2003. Incidence of enteric viruses in groundwater from household wells in Wisconsin. Appl. Environ. Microbiol. 69:1172-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapel, D. M., K. R. Bradbury, and R. J. Hunt. 2003. Delineation of 5-year zones of contribution for municipal wells in La Crosse County, Wisconsin: Wisconsin Geological and Natural History Survey open file report 2003-02. Wisconsin Geological and Natural History Survey, Madison, Wis.
- 7.Clark, I. D., and P. Pritz. 1997. Environmental isotopes in hydrogeology. Lewis Publishers, Inc., Boca Raton, Fla.
- 8.Coplen, T. B. 1994. Reporting of stable hydrogen, carbon, and oxygen isotopic abundances. Pure Appl. Chem. 66:273-276. [Google Scholar]
- 9.Coplen, T. B., and C. Kendall. 2000. Stable hydrogen and oxygen isotope ratios for selected sites of the U.S. Geological Survey's NASQAN and Benchmark surface-water networks: U.S. Geological Survey open-file report 00-160. U.S. Geological Survey, Reston, Va. [Online.] http://pubs.water.usgs.gov/ofr-00-160.
- 10.Coplen, T. B., H. L. Herczeg, and C. Barnes. 1999. Isotope engineering—using stable isotopes of the water molecule to solve practical problems, p. 79-110. In P. Cook and A. Herczeg (ed.), Environmental tracers in subsurface hydrology. Kluwer Academic Publishers, Boston, Mass.
- 11.Coplen, T. B., J. D. Wildman, and J. Chen. 1991. Improvements in the gaseous hydrogen-water equilibrium technique for hydrogen isotope ration analysis. Anal. Chem. 63:910-912. [Google Scholar]
- 12.Craun, G. F., P. S. Berger, and R. L. Calderon. 1997. Coliform bacteria and waterborne disease outbreaks. J. Am. Water Works Assoc. 89:96-104. [Google Scholar]
- 13.DeBorde, D. C., W. W. Woessner, Q. T. Kiley, and P. Ball. 1999. Rapid transport of viruses in a floodplain aquifer. Water Res. 33:2229-2238. [Google Scholar]
- 14.DeBorde, D. C., W. W. Woessner, B. Lauerman, and P. Ball. 1998. Virus occurrence and transport in a school septic system and unconfined aquifer. Ground Water 36:825-834. [Google Scholar]
- 15.De Leon, R., C. Sheih, R. S. Baric, and M. D. Sobsey. 1990. Detection of enteroviruses and hepatitis A virus in environmental samples by gene probes and polymerase chain reaction, p. 833-853. In Proceedings of the American Water Works Association Water Quality and Technology Conference. American Water Works Association, Denver, Colo.
- 16.Duboise, S. M., B. E. Moore, and B. P. Sagik. 1976. Poliovirus survival and movement in a sandy forest soil. Appl. Environ. Microbiol. 31:536-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelbrecht, R. S., M. J. Weber, B. L. Salter, and C. A. Schmidt. 1980. Comparative inactivation of viruses by chlorine. Appl. Environ. Microbiol. 40:249-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epstein, S., and T. Mayeda. 1953. Variation of 18O content of water from natural sources. Geochim. Cosmochim. Acta 4:213-244. [Google Scholar]
- 19.Fout, G. S., B. C. Martinson, M. W. Moyer, and D. R. Dahling. 2003. A multiplex reverse transcription-PCR method for detection of human enteric viruses in groundwater. Appl. Environ. Microbiol. 69:3158-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gat, J. R. 1970. Environmental isotope balance of Lake Tiberias, p. 109-127. In Isotope hydrology. International Atomic Energy Agency, Vienna, Austria.
- 21.Gerba, C. P., and G. Bitton. 1984. Groundwater pollution microbiology, p. 65-88. John Wiley & Sons, New York, N.Y.
- 22.Gerba, C. P., S. M. Goyal, R. L. LaBelle, I. Cech, and G. F. Bodgan. 1979. Failure of indicator bacteria to reflect the occurrence of enteroviruses in marine waters. Am. J. Public Health 69:1116-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green, D. H., and G. D. Lewis. 1995. Enzymatic amplification of enteric viruses from wastewaters. Water Sci. Technol. 31:329-336. [Google Scholar]
- 24.Greening, G. E., J. Hewitt, and G. D. Lewis. 2002. Evaluation of integrated cell culture-PCR (C-PCR) for virological analysis of environmental samples. J. Appl. Microbiol. 93:745-750. [DOI] [PubMed] [Google Scholar]
- 25.Griffin, D. W., C. J. Gibson III, E. K. Lipp, K. Riley, J. H. Paul III, and J. B. Rose. 1999. Detection of viral pathogens by reverse transcriptase PCR and of microbial indicators by standard methods in the canals of the Florida Keys. Appl. Environ. Microbiol. 65:4118-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas, N. L. 2003. Susceptibility of La Crosse, Wisconsin, municipal wells to enteric virus contamination from surface water contributions. Masters thesis. University of Wisconsin—La Crosse, La Crosse.
- 27.Hollinger, F. B., and J. R. Ticehurst. 1996. Hepatitis A virus. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 3rd ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 28.Hunt, R. J., T. D. Bullen, D. P. Krabbenhoft, and C. Kendall. 1998. Using stable isotopes of water and strontium to investigate the hydrology of a natural and a constructed wetland. Ground Water 36:434-443. [Google Scholar]
- 29.Hunt, R. J., D. A. Saad, and D. M. Chapel. 2003. Numerical simulation of ground water flow in La Crosse County, Wisconsin and into nearby pools of the Mississippi River. United States Geological Survey water-resources investigations report 03-4154. U.S. Geological Survey, Middleton, Wis.
- 29a.Hunt, R. J., T. B. Coplen, N. L. Haas, D. A. Saad, and M. A. Borchardt. J. Hydrol., in press.
- 30.Ishiko, H., Y. Shimada, M. Yonaha, O. Hashimoto, A. Hayashi, K. Sakae, and N. Takeda. 2002. Molecular diagnosis of human enteroviruses by phylogeny-based classification by use of the VP4 sequence. J. Infect. Dis. 185:744-754. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, S., R. Nobel, and W. Chu. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapikian, A. Z., and R. M. Chanock. 1996. Rotaviruses, p. 1657-1708. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 33.Kapikian, A. Z., M. K. Estes, and R. M. Chanock. 1996. Norwalk group of viruses, p. 783-810. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 34.LeChevallier, M. W., R. W. Gullick, M. R. Karim, M. Friedman, and J. E. Funk. 2003. The potential for health risks from intrusion of contaminants into the distribution system from pressure transients. J. Water Health 1:3-14. [PubMed] [Google Scholar]
- 35.Lee, S. H., D. A. Levy, G. F. Craun, M. J. Beach, and R. L. Calderon. 2002. Surveillance for waterborne-disease outbreaks—United States, 1999-2000. Morb. Mortal. Wkly. Rep. Surveill. Summ. 51:1-47. [PubMed] [Google Scholar]
- 36.Le Guyader, F., E. Dubois, D. Menard, and M. Pommepuy. 1994. Detection of hepatitis A virus, rotavirus, and enterovirus in naturally contaminated shellfish and sediment by reverse transcription-seminested PCR. Appl. Environ. Microbiol. 60:3665-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazor, E. 1997. Applied chemical and isotopic groundwater hydrology, 2nd ed., p. 413. Marcel Dekker Inc., New York, N.Y.
- 38.McCarthy, K. A., W. D. McFarland, J. M. Wilkinson, and L. D. White. 1992. The dynamic relationship between groundwater and the Columbia River: using deuterium and oxygen-18 as tracers. J. Hydrol. 135:1-12. [Google Scholar]
- 39.Melnick, J. L. 1996. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 655-712. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 40.Payment, P. 1999. Poor efficacy of residual chlorine disinfectant in drinking water to inactivate waterborne pathogens in distribution systems. Can. J. Microbiol. 45:709-715. [PubMed] [Google Scholar]
- 41.Payment, P., M. Tremblay, and M. Trudel. 1985. Relative resistance to chlorine of poliovirus and coxsackievirus isolates from environmental sources and drinking water. Appl. Environ. Microbiol. 49:981-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puig, M., J. Jofre, F. Lucena, A. Allard, G. Wadell, and R. Girones. 1994. Detection of adenoviruses and enteroviruses in polluted waters by nested PCR amplification. Appl. Environ. Microbiol. 60:2963-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regli, S., J. B. Rose, C. N. Haas, and C. P. Gerba. 1991. Modeling the risk from Giardia and viruses in drinking water. J. Am. Water Works Assoc. 83:76-84. [Google Scholar]
- 44.Schwab, K. J., R. De Leon, and M. D. Sobsey. 1995. Concentration and purification of beef extract mock eluates from water samples for the detection of enteroviruses, hepatitis A virus, and Norwalk virus by reverse transcription-PCR. Appl. Environ. Microbiol. 61:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwab, K. J., R. De Leon, and M. D. Sobsey. 1996. Immunoaffinity concentration and purification of waterborne enteric viruses for detection by reverse transcriptase PCR. Appl. Environ. Microbiol. 62:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwab, K. J., M. K. Estes, F. H. Neill, and R. L. Atmar. 1997. Use of heat release and an internal RNA standard control in reverse transcription-PCR detection of Norwalk virus from stool samples. J. Clin. Microbiol. 35:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharp, D. G., D. C. Young, R. Floyd, and J. D. Johnson. 1980. Effect of ionic environment on the inactivation of poliovirus in water by chlorine. Appl. Environ. Microbiol. 39:530-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheets, R. A., R. A. Darner, and B. L. Whitteberry. 2002. Lag times of bank filtration at a well field, Cincinnati, Ohio, USA. J. Hydrol. 266:162-174. [Google Scholar]
- 49.Shin, G. A., D. Battigelli, and M. D. Sobsey. 1998. Reduction of Norwalk virus, poliovirus 1, and coliphage MS2 by free chlorine, chlorine dioxide, and ozone disinfection of water. In Proceedings of the Water Quality Technology Conference. American Water Works Association, Denver, Colo.
- 50.Sobsey, M. D., T. Fuji, and P. A. Shields. 1988. Inactivation of hepatitis A virus and model viruses in water by free chlorine and monochloramine. Water Sci. Technol. 20:385-391. [Google Scholar]
- 51.Thurston-Enriquez, J. A., C. N. Haas, J. Jacangelo, and C. P. Gerba. 2003. Chlorine inactivation of adenovirus type 40 and feline calicivirus. Appl. Environ. Microbiol. 69:3979-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tufenkji, N., J. N. Ryan, and M. Elimelech. 2002. The promise of bank filtration. Environ. Sci. Technol. 36:422A-428A. [DOI] [PubMed] [Google Scholar]
- 53.U.S. Environmental Protection Agency. 1989. Guidance manual for compliance with filtration and disinfection requirements for public water systems using surface water sources. Office of Water, U.S. Environmental Protection Agency, Washington, D.C.
- 54.U.S. Environmental Protection Agency. 1992. Consensus method for determining groundwaters under the direct influence of surface water using microscopic particulate analysis (MPA). EPA 910/9-92-029. U.S. Environmental Protection Agency, Washington, D.C.
- 55.U.S. Environmental Protection Agency. 1996. ICR microbial laboratory manual. EPA/600/R-05/178. Office of Research and Development, U.S. Environmental Protection Agency, Washington, D.C. [Online.] http://www.epa.gov/nerlcwww/icrmicro.pdf.
- 56.U.S. Environmental Protection Agency. 2000. Proposed rules. Federal Register. 65:30193-30274. [Online.] http://www.epa.gov/fedrgstr/EPA-WATER/2000/May/Day-10/w10763.htm. [PubMed] [Google Scholar]
- 57.U.S. Environmental Protection Agency. 2001. Method 1602: male-specific (F+) and somatic coliphage in water by single agar layer (SAL) procedure. U.S. Environmental Protection Agency, Washington, D.C. [Online.] http://www.epa.gov/nerlcwww/1602ap01.pdf.
- 58.U.S. Environmental Protection Agency. 2001. USEPA manual of methods for virology. Concentration and processing of waterborne viruses by positive charge 1 MDS cartridge filters and organic flocculation. EPA/600/4-84/013 (N14). Office of Research and Development, U.S. Environmental Protection Agency, Washington, D.C. [Online.] http://www.epa.gov/nerlcwww/chapt14.pdf.
- 59.U.S. Environmental Protection Agency. 2001. USEPA manual of methods for virology. Total culturable virus quantal assay. EPA/600/4-84/013 (N15). Office of Research and Development, U.S. Environmental Protection Agency, Washington, D.C. [Online.] http://www.epa.gov/nerlcwww/chapt15.pdf.
- 60.Wade, T. J., N. Pai, J. N. Eisenberg, and J. M. Colford, Jr. 2003. Do U.S. Environmental Protection Agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environ. Health Perspect. 111:1102-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woessner, W. W., P. N. Ball, D. C. DeBorde, and T. L. Troy. 2001. Viral transport in a sand and gravel aquifer under field pumping conditions. Ground Water 39:886-894. [DOI] [PubMed] [Google Scholar]
- 62.Yates, M. V., C. P. Gerba, and L. M. Kelley. 1985. Virus persistence in groundwater. Appl. Environ. Microbiol. 49:778-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yates, M. V., and S. R. Yates. 1988. Modeling microbial fate in the subsurface environment. Crit. Rev. Environ. Control 17:307-344. [Google Scholar]