Abstract
The complete SfiI and I-CeuI physical maps of four Bacillus subtilis (natto) strains, which were previously isolated as natto (fermented soybean) starters, were constructed to elucidate the genome structure. Not only the similarity in genome size and organization but also the microheterogeneity of the gene context was revealed. No large-scale genome rearrangements among the four strains were indicated by mapping of the genes, including 10 rRNA operons (rrn) and relevant genes required for natto production, to the loci corresponding to those of the B. subtilis strain Marburg 168. However, restriction fragment length polymorphism and the presence or absence of strain-specific DNA sequences, such as the prophages SPβ, skin element, and PBSX, as well as the insertion element IS4Bsu1, could be used to identify one of these strains as a Marburg type and the other three strains as natto types. The genome structure and gene heterogeneity were also consistent with the type of indigenous plasmids harbored by the strains.
A traditional Japanese food, natto (fermented soybean), is made from soybeans fermented by a bacterium classified as Bacillus subtilis (natto). Natto-producing strains, referred to as B. subtilis (natto) in this paper, are classified as closely related strain to the laboratory strain B. subtilis Marburg 168 (10, 29), which is the best-characterized gram-positive bacterium and has about 4,100 protein-encoding genes in a 4,215-kb genome sequence (19). Natto is an ideal food because it is easily made and full of nutrients and it can be stored with the fungicidal antibiotics produced by this bacterium (49). Many studies have attempted to determine the complicated process by which natto is produced, a process which can be divided into several steps, including secretion of proteases on the surface of soybeans, incorporation of digested amino acids, racemization of l-glutamic acid into the d form in the cytoplasm, and synthesis of poly-γ-glutamic acid (γ-PGA), the major constituent of viscous biofilm-like material (2, 8).
Extensive biochemical and molecular genetic studies have been conducted on the genes and enzymes involved in the fermentation of natto (4-7, 13, 47), although the genes required for a natto production bioprocess have been yet to be totally clarified. A limited number of genetically and biochemically characterized gene homologues, including psgABCD (ywsC-ywtABCV in the Marburg 168 counterpart) involved in the synthesis of γ-PGA (6, 47), iep (degQ) involved in the regulation of protease secretion (40), and glr and yrpC involved in the racemization of glutamate (8), are also present in the genome of B. subtilis Marburg 168(1A1). This laboratory strain does not produce capsular PGA, suggesting that highly coordinated regulation of gene expression, as well as physiological conditions during growth on the soybean surface, are required for good natto starters. It was demonstrated by Itaya and Matsui (16) that repeated transformation of a Marburg 168 derivative with genomic DNA from B. subtilis (natto) conferred the ability to produce natto to the recipient. As more DNA sequence from the natto strain was transferred to the recipient Marburg 168 strain, the natto fermentation ability was gradually enhanced in proportion to the amount of natto strain-derived DNA in the recipient. This study highlighted the conclusion that the gene regulation of natto fermentation could function in Marburg 168 if the relevant genes are appropriately transferred to corresponding loci of the genome via homologous recombination and that the involvement of plasmids could clearly be ruled out.
Other genes from natto strains have been studied, including competence development genes (3) and insertion elements (25). Sequences of these genes from B. subtilis (natto) show high levels of similarity to Marburg 168 sequences regardless of involvement in natto production. The analysis of gene functions is hampered by the fact that one-third of the open reading frames of Marburg 168 have not been functionally characterized so far, and the presence of strain-specific genes has been demonstrated in Helicobacter pylori (2), Haemophilus influenzae (12), Chlamydia subspecies (31), Prochlorococcus (33), and Bacillus anthracis (30).
A preliminary analysis of the genome structure of a B. subtilis (natto) strain suggested that it was similar to that of the Marburg strains (16). Our aim is to fully understand the process of fermentation by B. subtilis (natto) from boiled soybeans to natto. Construction of complete physical maps of B. subtilis (natto) strains and mapping of natto-related genes could serve as a starting point for this project and should be helpful in sequencing the entire genome. Thus, I-CeuI and SfiI maps of four B. subtilis (natto) strains were constructed by using pulsed-field gel electrophoresis (14) and SfiI-linking clone-based Southern hybridization. In addition to the similarities in genome size and organization, differences in genome-specific characteristics were found, which provided a method for discriminating variants of natto starter strains.
MATERIALS AND METHODS
Bacterial strains and media.
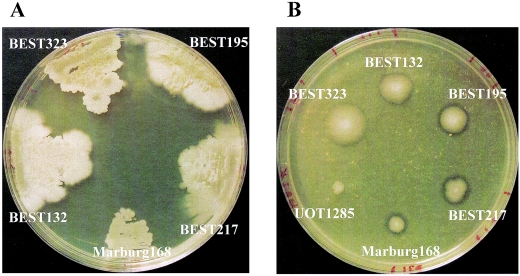
B. subtilis Marburg 168(1A1) was obtained from the Bacillus Genetic Stock Center (Ohio State University, Columbus). The B. subtilis (natto) strain BEST195 is an isolate from Miyagino-based natto (16). BEST217 (= IAM1168) and BEST323 (= IAM1163) were obtained from the Institute of Applied Microbiology, University of Tokyo, and BEST132 (= IFO3335) was obtained from the Institute of Fermentation, Osaka (43). The ability to produce γ-PGA was determined based on the morphology of colonies and the production of mucous material on GSP medium (1.5% [wt/vol] glutamic acid, 3% sucrose, 1.5% Phytone peptone [Becton Dickinson, Cockeysville, Md.], 0.25% KH2PO4, 0.17% Na2HPO4, 0.05% MgCl2, 0.05% NaCl, 100 μg of biotin per liter) plates incubated at 42°C for 18 h (Fig. 1). All of the strains except BEST195 have lost the ability to produce γ-PGA, which is explained below. GSP medium was solidified by addition of 1.5% Bacto Agar (Becton Dickinson). The amount of protease secreted was estimated from the transparent zone that formed around a colony on a Schaeffer sporulation agar plate supplemented with 1% (vol/vol) skim milk (36).
FIG. 1.
Colony morphology and extracellular protease production by B. subtilis (natto) strains. (A) Strains grown on GSP agar plates for 24 h at 42°C. Only BEST195 formed mucoid colonies. (B) Protease production on agar plates supplemented with skim milk incubated for 17 h at 37°C. Strain UOT1285 (trpC2 lys1 nprR2 nprE18 aprEΔ3) (47) was a non-protease-secreting control strain.
Escherichia coli DH5α [F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gryA96 relA1] and TOP10F− [F− φ80dlacZ {lacIq Tn10(Tetr)} mcrA Δ(mrr-hsdRMS-mcrBC) ΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 nupG] were used as the hosts for molecular cloning in the laboratory. E. coli and B. subtilis were grown at 37°C unless indicated otherwise.
Preparation of plasmid.
Typically, a covalently closed circular plasmid DNA (cccDNA) was purified from 100 ml of a culture in Luria-Bertani medium grown overnight at 37°C. The gently treated DNA solution was purified by ultracentrifugation twice in the presence of cesium chloride and ethidium bromide as described previously (43). The DNA was finally dissolved in 50 μl of TE solution (10 mM Tris-HCl [pH 8.0], 1.0 mM EDTA).
Preparation of genomic DNA in an agarose plug and pulsed-field gel electrophoresis.
B. subtilis genomic DNA in solution or in agarose plugs was prepared as described previously (14). Digestion with the SfiI or I-CeuI endonuclease was carried out by using a method described previously (14, 44). The genomic DNA prepared from a mixture of two strains was used to determine the subtle size difference. The running conditions for pulsed-field gel electrophoresis (Bio Craft, Tokyo, Japan) are described below. After electrophoresis, DNAs were transferred onto a nylon membrane for Southern hybridization (37). Digoxigenin-11-dUTP was used to prepare probes for Southern hybridization (Dig-High Prime kit; Roche Diagnostics Co., Penzberg, Germany). Both the cloned DNA in plasmids and PCR products were used to prepare probes. For hybridization and detection we used the manufacturer's protocol.
Preparation of SfiI-linking clones for strain Marburg168 and B. subtilis (natto) strain BEST195.
An SfiI-linking clone including each SfiI site flanked by about 1 kb and was used to order the SfiI restriction fragments of genomic DNA. The linking clones were prepared by the PCR amplification method. The primer sets were designed to amplify approximately 1-kb fragments flanking each side of the SfiI sites of the Marburg 168 genome (http//bacillus.genome.ad.jp). Primer sequences used for the 26 SfiI sites are shown in Table 1. PCR for DNA cloning was done by using the Ex-Taq Hot start version (Takara Bio Co., Kyoto, Japan). The amplified segments were characterized by using several restriction enzymes and were confirmed by sequencing both ends with an ABI3100 sequencer (Applied Biosystems, Foster City, Calif.). The PCR product was cloned into plasmid pCR2.1-TOPO by using a TOPO TA PCR cloning kit (Invitrogen Inc., Carlsbad, Calif.). Plasmid preparation from E. coli was carried out by using a Rapid Plasmid mini system kit (Marligen Bioscience Inc., Ijamsville, Md.). Most restriction enzymes were obtained from Takara Bio Co.; the only exception was I-CeuI, which was obtained from New England Biolabs Inc., Beverly, Mass. Fifteen PCR fragments were cloned into the E. coli pCR2.1-TOPO vector plasmid. Although the remaining 11 PCR fragments were not successfully cloned into the pCR2.1-TOPO vector, these fragments could be used as probes for Southern hybridization. Hence, both cloned and uncloned fragments were designated SfiI-linking clones and used in this study. Amplification from the B. subtilis (natto) strain BEST195 genome was attempted by using the same primer sets.
TABLE 1.
Linking clone primers and linking clone probes used in this studya
| Linking clone | Forward primer | Reverse primer | Linking clone coordinates (bp) | Probe template |
|---|---|---|---|---|
| pAG-168 | 5′-ATGCAGAATAGCCCCTTTTAA-3′ | 5′-CGTCCAGCCGTTAAAATG-3′ | 2127942-2130265 | TOPO construct |
| pBC-168 | 5′-ATAACGGGGAAGATGCG-3′ | 5′-TGCTCACAAGAGGAGAAAGC-3′ | 642935-645127 | TOPO construct |
| pCZ-168 | 5′-CGCGATCAAATTAGCCAGT-3′ | 5′-GCTTCCAAGGCTCCATTC-3′ | 995682-998304 | TOPO construct |
| pDE-168 | 5′-CACTTCATCGGCACTCAC-3′ | 5′-GAGTAGGAAAGAAAACCGCA-3′ | 2833532-2835697 | TOPO construct |
| pEM-168 | 5′-CATCACGATGATAAGGAGGA-3′ | 5′-CCATAAATATGCCTTGCGTT-3′ | 3096907-3099044 | PCR product |
| pFB-168 | 5′-CCCTTGGCACTTAGATTCC-3′ | 5′-AAGCCAAATGTGCCTCTTT-3′ | 205592-207634 | TOPO construct |
| pGK-168 | 5′-ATCTGAACTGACACCGGA-3′ | 5′-GAAGTAGCCGGTTCACAC-3′ | 2325518-2327701 | TOPO construct |
| pHP-168 | 5′-GATCGCAATTGACCCACT-3′ | 5′-GATTGCAGCAATTGTGCC-3′ | 3437833-3439984 | TOPO construct |
| pIY-168 | 5′-AAGGCATTGGCTGGATTG-3′ | 5′-TTCTTCGAGCTCCTGTGT-3′ | 3735053-3737244 | PCR product |
| pJQ-168 | 5′-AACAAACGGCAACTACGG-3′ | 5′-CAAACGGAAAGAGGACGAC-3′ | 3933539-3935831 | TOPO construct |
| pKS-168 | 5′-GTGATCAGCACTCTCGTTC-3′ | 5′-GCTCATTATCGCGGGATT-3′ | 2488261-2490611 | PCR product |
| pLW-168 | 5′-CCAAACGCAATGGGTTCT-3′ | 5′-CAAAAGAATGCCTGTCCGT-3′ | 1222673-1225333 | PCR product |
| pMH-168 | 5′-TTCGGCTTTGTGGAATGG-3′ | 5′-GGTATGCGTTGGCCTATG-3′ | 3241798-3244090 | TOPO construct |
| pNF-168 | 5′-GTGAGTAAATCGGATCGAGG-3′ | 5′-CGTCTGAAGATAGATGGCTG-3′ | 4181779-4184036 | TOPO construct |
| pOT-168 | 5′-AGATCGGGATCAAGCCAC-3′ | 5′-TGAACGATGACACGGTCTT-3′ | 1375293-1377581 | PCR product |
| pPI-168 | 5′-ACTCATTCTCTCCACTTCCA-3′ | 5′-GGTGATGCCTAAAGTGCC-3′ | 3557964-3560071 | PCR product |
| pQX-168 | 5′-TTCAGCCAGTTCTCGTCT-3′ | 5′-AAGAGCAACAAGCGGAAC-3′ | 4028442-4030953 | TOPO construct |
| pRL-168 | 5′-CGAAAATTGGGAGACGGAG-3′ | 5′-GCTTGTGCTGATTATGGGT-3′ | 1079392-1081702 | PCR product |
| pSD-168 | 5′-TTCATACCCTGCATTGCC-3′ | 5′-TGGCACCTTTCAGTGTTC-3′ | 2523743-2525988 | TOPO construct |
| pTA-168 | 5′-CTGCGAGATCGTTTACGG-3′ | 5′-CAAGATTCAGGAATCGGCA-3′ | 1403319-1405511 | TOPO construct |
| pUR-168 | 5′-TTATGCGACATTGGGAGAG-3′ | 5′-CTGAGGTTAAGGCTGCAA-3′ | 1023117-1025305 | PCR product |
| pVJ-168 | 5′-AAGGCGATGAATTAGGCA-3′ | 5′-CGTAAATGCAAAAGGAGACA-3′ | 3762175-3764643 | PCR product |
| pWO-168 | 5′-CATTCCGAATTGATCGCAAG-3′ | 5′-TCTCAACGTCTCTACAGTCA-3′ | 1244278-1246490 | TOPO construct |
| pXN-168 | 5′-GGCAACGGATTTTAATCCTG-3′ | 5′-GCGTAAGGAATTCCGATCT-3′ | 4044997-4047192 | TOPO construct |
| pYV-168 | 5′-CACTACAGCATCGCCAAA-3′ | 5′-TGGAAATGGGTATCCCGT-3′ | 3742524-3744887 | PCR product |
| pZU-168 | 5′-GAATGAACGCAAATGGTGG-3′ | 5′-ATCATGTTCTTTCCGTGCT-3′ | 998957-1001411 | PCR product |
Linking clones or restriction fragments were designated as follows. As the 26 SfiI fragments of the Marburg 168 strain were alphabetically designated from the largest (AS) to the smallest (ZS) (14), a clone that links adjacent SfiI fragments was designated by a combination of the first characters of the SfiI fragments. For example, the combination of AS and GS was designated pAG-168. Similarly, the counterpart that came from BEST195 was designated pAG-195.
RESULTS
Indigenous plasmids.
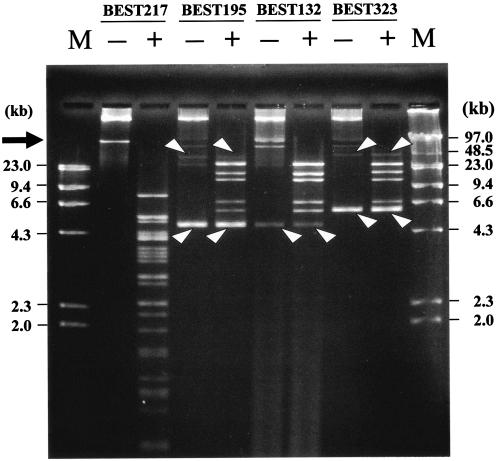
The presence of indigenous plasmids was demonstrated for all of the B. subtilis (natto) strains. Two plasmids, one large plasmid and one small plasmid, were present in BEST132, BEST195, and BEST323. Only one large plasmid was isolated from BEST217 (17, 22, 23, 41, 43). The sequences of the small plasmids from BEST195 (5.8 kb), BEST132 (5.8 kb), and BEST323 (6.6 kb) revealed that these plasmids are variants of the rolling-circle replicating plasmid pTA10015 (5.8 kb) harbored by a B. subtilis (natto) strain (23). The large plasmids from BEST195 and BEST323 are variants of the pLS20 plasmid harbored by BEST132 (approximately 55 kb), which is commonly observed in B. subtilis (natto) (26). Comparisons based on the results of restriction enzyme digestion by, for example, BglII (Fig. 2), revealed that pLS32 (approximately 80 kb) of strain BEST217 is distinct from the pLS20 variant. This is consistent with the difference in the sequences of the region including the replication origin and the mechanism of initiation of DNA replication for pLS20 (22) and pLS32 (42). Constant amounts of plasmids were reproducibly prepared for all plasmids except those of BEST195 and BEST132. No indigenous plasmid was found in BEST195 according to a previous report (16). We observed that the amount of plasmid from BEST195 varied significantly among experiments (data not shown). Because no special selection procedure was employed, the copy number of plasmids of BEST195 might depend on subtle growth conditions. It has been demonstrated that the cognate plasmids of natto strains are not involved in the fermentation process (26). These plasmids had no SfiI site (data not shown). They remained cccDNA after SfiI or I-CeuI digestion and migrated differently from linear double-stranded DNA in the gel under the pulsed-field gel electrophoresis conditions (44). Thus, plasmids were not included in construction of the SfiI and I-CeuI map of these host genomes.
FIG. 2.
Plasmids isolated from B. subtilis (natto) strains. Plasmids from the strains were subjected to pulsed-field gel electrophoresis. The conditions were as follows: 2.3 V/cm, a pulse time of 6 s, and a running time of 15 h. Lanes + contained BglII digests. Bands not digested by BglII indicated by arrowheads are the small plasmids (monomeric and dimeric forms) that migrated as cccDNA because they did not have a BglII site. The arrow indicates the area where large plasmids migrated. Several other bands are probably bands for open circular forms because of susceptibility to BglII digestion. Lanes M contained the HindIII digests of λphage DNA (left) and genomic DNA of λphage (right) as size markers.
Genome size and organization.
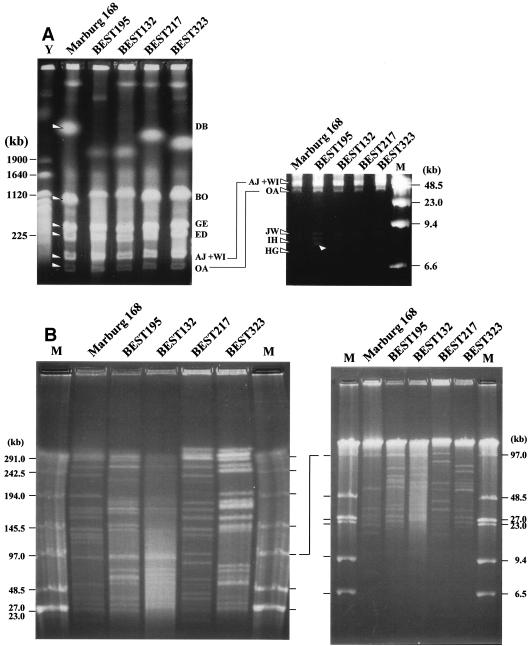
Construction of physical maps by using the endonucleases I-CeuI and SfiI is described below. The former enzyme recognizes and cleaves a 22-base sequence that appears in the rRNA (rrn) operons (19), and therefore the number of I-CeuI fragments is equivalent to the number of rrn operons per genome. B. subtilis Marburg 168 had 10 rrn operons, as revealed by whole-genome sequencing (19) and also by experiments (44). The four B. subtilis (natto) strains all yielded 10 I-CeuI fragments (Fig. 3A). Tandem alignments of three rrn operons (rrnI-rrnH-rrnG) and two rrn operons (rrnJ-rrnW) were conserved among the B. subtilis (natto) strains, as indicated by the three small I-CeuI fragments obtained for all strains (Fig. 3A, right gel). The apparent size difference in the other I-CeuI fragments (Fig. 3A, left gel) was interpreted as an accumulation of small variations in the SfiI fragment sizes. A large-scale inversion, like that detected in the genome of B. subtilis 166 by using I-CeuI digestion (15, 32), seems less likely for the four B. subtilis (natto) strains.
FIG. 3.
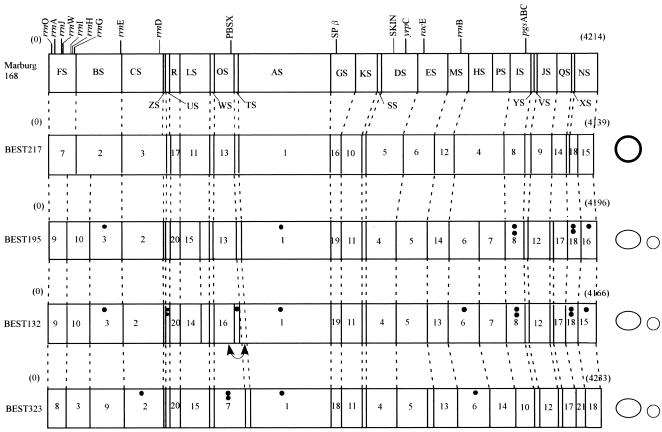
Identification of I-CeuI and SfiI fragments from B. subtilis (natto) strains. (A) I-CeuI digests of genomic DNA of four B. subtilis (natto) strains. I-CeuI digests of the strains indicated were electrophoresed. Lanes Y and M contained yeast genome DNA and lambda phage DNA, respectively, as markers; sizes are indicated on the left and the right. I-CeuI fragments of the Marburg 168 genomic DNA designated in reference 44 are indicated by arrowheads. The fragment-like band in BEST195 indicated by an arrowhead was an artifact because it was not reproduced in separate running conditions. The running conditions for the gel on the left were 3 V/cm, a pulse time of 8 min, and a running time of 44 h. Resolution of the I-CeuI fragments smaller than 150 kb is shown in the gel on the right, for which the running conditions were as follows: 3 V/cm, a pulse time of 3 min, and a running time of 32 h. (B) SfiI digests of four B. subtilis (natto) strains. SfiI fragments of different strains were resolved by pulsed-field gel electrophoresis with the following running conditions: 3 V/cm, a pulse time of 24 s, and a running time of 56 h (left panel) and 3 V/cm, a pulse time of 6 s, and a running time of 24 h (right panel). Resolution of the SfiI restriction fragments smaller than 100 kb is shown in the right panel. Lanes M contained lambda phage DNA as a size marker. (C) Schematic diagram of the SfiI fragments of the four B. subtilis (natto) strains. All the SfiI fragments determined experimentally are included. Sizes are shown in Table 2.
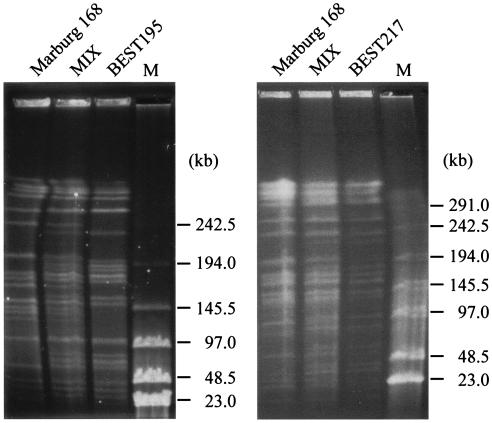
The other endonuclease, SfiI, recognizes and cleaves the sequence GGCCN/NNN/NGGCC. There are 26 SfiI sites in the Marburg 168 genome (14, 19). The patterns of the SfiI fragments of the four B. subtilis (natto) strains are shown in Fig. 3B and C. The number and size of fragments were directly estimated by comparing restriction fragments derived from the culture mixed with Marburg 168, as shown in Fig. 4. A subtle difference in size could be determined by using this method, and estimates of the number and size of SfiI fragments are shown in Table 2. BEST195 and BEST132 yielded 28 SfiI restriction fragments, and BSET323 had 29 SfiI sites, while only 24 fragments were identified for BEST217. The genome sizes of these natto strains are estimated to range from 4,139 to 4,237 kb; these sizes are similar to the size of the 4,215-kb genome of Marburg 168.
FIG. 4.
Direct comparison of strains to measure the subtle size difference in SfiI fragments. SfiI digests of genomic DNA prepared from strains Marburg 168, BEST195, and BEST217 were electrophoresed. A mixture of two strains (MIX) revealed all the SfiI fragments from both strains and the subtle size difference. Details are described in the Results. Lane M contained lambda phage DNA as a size marker. The running conditions for both gels were as follows: 3.3 V/cm, a pulse time of 24 s, and incubation for 48 h at 14°C.
TABLE 2.
SfiI restriction fragments of B. subtilis (natto) strains and their counterparts in the chromosome of B. subtilis Marburg 168
| Marburg 168 SfiI fragment | Coordinates in Marburg 168 genome (kb) | Size (kb) | Size (kb) and designation in:
|
|||
|---|---|---|---|---|---|---|
| BEST217 | BEST132 | BEST195 | BEST323 | |||
| AS | 1404-2129 | 725 | 745 (S1) | 700 (S1) | 680 (S1) | 680 (S1) |
| BS | 206-644 | 437 | 420 (S2) | 310 (S3) + 127 (S10)a | 310 (S3) + 127 (S10)a | 310 (S3) + 127 (S9)a |
| CS | 644-997 | 353 | 340 (S3) (+ZS)b | 353 (S2) | 353 (S2) | 376 (S2) |
| DS | 2525-2835 | 310 | 320 (S5) | 283 (S4) | 283 (S4) | 283 (S4) |
| ES | 2835-3098 | 283 | 283 (S6) | 283 (S5) | 283 (S5) | 56 (24) + 255 (S5)c |
| FS | 4183-206 | 237 | 237 (S8) | 185 (S9) + 50 (S10)a | 185 (S9) + 50 (S10)a | 185 (S9) + 50 (S9)a |
| GS | 2129-2327 | 198 | 64 (S16) | 64 (S19) | 64 (S19) | 64 (S18) |
| HS | 3243-3439 | 196 | 330 (S4) (+PS)b | 235 (S6) | 235 (S6) | 255 (S6) |
| IS | 3559-3736 | 177 | 177 (S8) | 188 (S8) | 188 (S8) | 184 (S10) |
| JS | 3763-3935 | 172 | 172 (S9) | 172 (S12) | 172 (S12) | 147 (S12) |
| KS | 2327-2490 | 163 | 172 (S10) | 177 (S11) | 177 (S11) | 177 (S11) |
| LS | 1081-1226 | 145 | 145 (S11) | 100 (S14) + 39 (S21)c | 100 (S15) + 39 (S21)c | 145 (S15) |
| MS | 3098-3243 | 145 | 145 (S12) | 145 (S13) | 145 (S14) | 145 (S13) |
| NS | 4046-4183 | 137 | 90 (S15) + 42 (S18)c | 100 (S15) | 100 (S16) | 90 (S18) |
| OS | 1245-1376 | 131 | 137 (S13) | 95 (S16) | 147 (S13) | 198 (S7) |
| PS | 3439-3559 | 120 | (S4) (+HS)b | 190 (S7) | 190 (S7) | 150 (S14) |
| QS | 3935-4030 | 95 | 100 (S14) | 80 (S17) + 24 (S26)c | 80 (S17) + 24 (S26)c | 80 (S17) + 24 (S26)c |
| RS | 1024-1081 | 56 | 56 (S17) | 56 (S20) | 56 (S20) | 58 (S20) |
| SS | 2490-2525 | 37 | 37 (S20) | 37 (S22) | 37 (S22) | 37 (S22) |
| TS | 1376-1404 | 28 | 28 (S21) | 28 (S23) | 28 (S23) | 28 (S23) |
| US | 1000-1024 | 24 | 24 (S22) | 26 (S25) | 24 (S25) | 24 (S25) |
| VS | 3744-3763 | 19 | 37 (S19) (+YS)b | 27 (S24) (+YS)b | 27 (S24) (+YS)b | 27 (S24) (+YS)b |
| WS | 1226-1245 | 19 | 19 (S23) | 19 (S27) | 19 (S27) | 19 (S27) |
| XS | 4030-4046 | 16 | 16 (S24) | 70 (S18) (+ part of NS)b | 70 (S18) (+ part of NS)b | 56 (S21) (+ part of NS)b |
| YS | 3736-3744 | 8 | (S19) (+VS)b | (S24) (+VS)b | (S24) (+VS)b | (S24) (+VS)b |
| ZS | 997-1000 | 3 | (S3) (+CS) | 3 (S28) | 3 (S28) | 3 (S28) |
| Total | 0-4214 | 4,214 | 4,139 | 4,166 | 4,196 | 4,233 |
A 127-kb sequence from the BS counterpart and a 50-kb sequence from FS counterpart are combined into a single SfiI fragment in the BEST132, BEST195, and BEST323 strains.
Combined SfiI fragments.
Two novel fragments corresponding to a single counterpart in the Marburg 168 strain.
Alignment of SfiI fragments by using linking clones.
The SfiI-linking clones were amplified from natto strains by using the same Marburg 168 genome sequence-based primer sets and were checked by SfiI digestion (Table 1). Twenty-five linking clones were amplified from the BEST195 strain, and 16 were successfully cloned into the E. coli pCR2.1-TOPO vector. Twenty-three of these clones had the SfiI site and were considered SfiI-linking clones from strain BEST195. Loss of the expected SfiI sites in two clones, pYV-195 (at bp 31743629 of Marburg 168) and pFB-195 (at bp 206607), was due to a point mutation in the SfiI recognition sequence (data not shown). Only one fragment corresponding to pNF-195 (at bp 4182914 of Marburg 168) was not amplified. By using this information, SfiI fragments of natto strains were aligned and ordered based on two principles. First, two bands were aligned next to each other and should have been contiguous if a linking clone probe generated two Southern bands. We found that most fragments fell into this category. If a linking clone generated a single SfiI band in the Southern analysis, it was postulated that the SfiI site was lost in the hybridized fragments, such as pFB-195 and pYV-195. BEST217 also lost two SfiI sites because only a single fragment was hybridized by pHP-168 or pCZ-168. Second, the SfiI fragments were ordered by using the partial digestion method. Obviously, the hybridized bands would have appeared in a ladder form above the Southern band of complete SfiI digests when the probe was used to hybridize the partial SfiI digests. The adjacent fragments were readily predicted by determining the size increase of these hybridized ladder bands. Thus, the physical map of BEST195 was constructed and compared with that of Marburg 168 (Fig. 5). SfiI maps of the other three strains were similarly constructed by using the SfiI-linking clones listed in Table 1 and applying the two basic principles and analyses of SfiI partial digests. The location of the I-CeuI site in the SfiI fragments was determined based on two-dimensional analyses with progressive digestion by two enzymes (44). These SfiI fragments were aligned in the circular genome, and the SfiI and I-CeuI physical maps are both shown in Fig. 5. The SfiI-I-CeuI physical map indicated that there is no large-scale DNA rearrangement in these four strains, except for a small inversion described in the legend to Fig. 5. This is clearly different from the large inversion observed in some closely related strains (11). The inversion suggested by comparative gene orders in the genome of the distantly related organism Bacillus halodurans is interesting in this regard (39).
FIG. 5.
Physical maps of B. subtilis (natto) strains. The map of Marburg 168, linearized at the SfiI site between FS and NS closest to the origin of replication (oriC), is shown at the top. Similar linearized forms of SfiI maps for the four natto strains are shown below the Marburg 168 map. Plasmids that have no SfiI site are shown on the right. Large plasmids are represented by large ovals, and small plasmids are represented by small circles. pL32 from BEST217 is represented by a boldface circle to distinguish it from the pLS20 family plasmid represented by an oval. Plasmid sizes are not to scale. Locations of the 10 rrn operons containing the sites for I-CeuI, prophages, PBSX, SPβ, and skin are shown on the Marburg 168 map. Genes mapped in this study are also indicated on the Marburg 168 map. The number of dots indicates the number of IS determined from the data in Fig. 6. A small inversion was suggested in the BEST132 genome. This inversion, indicated by a double-headed curved arrow, is based on the Southern hybridization pattern for the two adjacent linking clones (pTA-168 and pOT-168), which was different from that of other strains. The order of hybridized bands with these two linking clones for BEST132 was reversed.
Mapping of genes required for natto production.
The number of genes which are known to be required for natto production is limited. During the fermentation process, proteases are secreted to break down soybean proteins into amino acids, including the glutamic acid that is incorporated into γ-PGA (6). γ-PGA, the major component of capsules, is synthesized by the catalysis of the synthases encoded by ywsC-ywtABC (45, 46), and the l-glutamic acid is first processed by the major racemase encoded by glr (racE) and yrpC (4, 8). Fragments that included these genes were amplified from all the strains by using specific primers designed for Marburg 168 (data not shown). The location of these genes in the B. subtilis (natto) genome was determined by Southern hybridization and included in the physical maps (Fig. 5). These genes were mapped at the expected SfiI fragments (Fig. 5).
Phages.
The size variation in SfiI fragments in which prophages PBSX (28 kb), skin element (48 kb), and SPβ (134 kb) reside drew our attention because these phages have been biochemically and genetically characterized in the Marburg 168 strain (19, 48). Absence of SPβ and skin element was predicted based on the significant decrease in the size of the relevant SfiI fragments of the B. subtilis (natto) genome. The locations and sizes of these prophages are also indicated on the physical map (Fig. 5). The insertion of skin element was further examined by amplification of the flanking region of the skin attachment site (data not shown). Only BEST217 had the skin element inserted into the sigK gene (38). The other three strains had an intact sigK gene lacking the prophage. This is consistent with the report that some B. subtilis strains have an intact sigK gene in their genomes (35). However, the prophage SPβ is absent in all the B. subtilis (natto) strains, as confirmed by PCR-mediated amplification and sequencing of the flanking regions of the SPβ attachment site (data not shown). A putative capsular polysaccharide synthesis gene was recognized at the attachment site of BEST195, which is homologous to the ypqP and yodU genes of Marburg 168 interrupted by SPβ. The SPβ inserted putative gene of BEST195 contains the attachment site (a 16-base sequence) for SPβ (21). SPβ may have been lost by BEST195, or SPβ may have never infected the natto strains, including BEST195. PBSX is present in all strains, which was also indicated by amplification of the fragments with the PCR primer sets covering the borders of this temperate phage DNA in the Marburg 168 genome.
Insertion sequence.
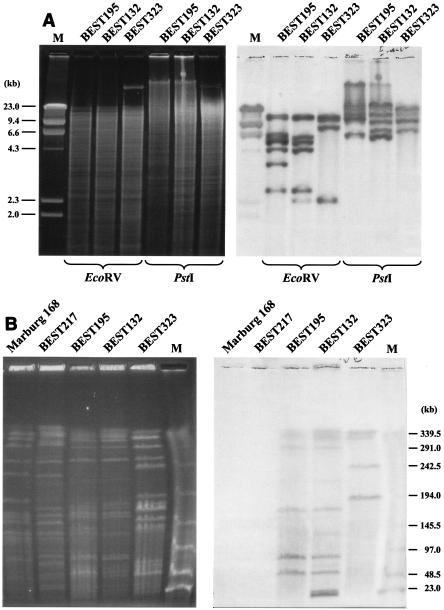
Many B. subtilis (natto) strains harbor various copies of the insertion sequence (IS) IS4Bsu1 at locations that have not been determined yet (25). The IS probe was used to hybridize to EcoRI and PstI digests of genomic DNA of the five strains (Fig. 6A). As IS4Bsu1 has no EcoRV or PstI site, the number of copies per genome is at least equivalent to the number of hybridized bands. The numbers of IS were estimated to be 7, 11, and 5 for BEST132, BEST195, and BEST323, respectively. On the other hand, the same IS4Bsu1 probe identified several SfiI fragments with different levels of hybridization intensity (Fig. 6B). The copy numbers of IS4Bsu1 estimated from the relative intensities of the hybridized SfiI bands are shown in Fig. 5. To our surprise, BEST217 does not have any IS4Bsu1-like IS. As Marburg 168 has no insertion element, the data provided additional evidence that BEST217 could be categorized as a Marburg-like (non-natto) strain. The possibility that other types of IS elements may be present in the four B. subtilis (natto) strains cannot be ruled out.
FIG. 6.
Analysis of IS elements in the B. subtilis (natto) genome. (A) Estimation of the copy number of IS4Bsu1. EcoRV and PstI digests of genomic DNA of three strains were subjected to agarose gel electrophoresis (left gel) and hybridized with the IS4Bsu1 probe (right gel). The number of IS copies per genome is equivalent to the number of hybridized bands because there are no EcoRV and PstI sites in the IS4Bsu1sequence. Lane M contained lambda phage DNA as a size marker. (B) Mapping on SfiI fragments with multiple copies of the IS element. The SfiI digests of genomic DNA of strains were resolved by using a contour-clamped homogeneous electric field with the following running conditions: 3 V/cm, a pulse time of 24 s, and a running time of 56 h (left gel). The blot was hybridized (right gel) with IS4Bsu1 as a probe (25). The intensities of the Southern signal were digitally treated, and number of a particular SfiI fragment was calculated based on the total number per genome derived from the data in panel A. The results are shown in Fig. 5. Lane M contained lambda phage DNA as a size marker.
DISCUSSION
Various methods have been developed for comparative analysis of prokaryotic genomes, including subtractive hybridization (1) and shotgun optical mapping (20). Restriction fragment length polymorphism has been widely used to detect genes or genomic regions responsible for biochemical and physiological differences. The whole genomes of various strains or serotypes of some important pathogenic bacteria have been sequenced, and comparisons have been made by using this most informative top-down approach. For example, a genomic comparison between E. coli K-12 (9) and O157:H7 strains (18, 28) led to identification of K-island and O-island. A similar comparison has been performed for the B. anthracis strains (30). Obviously, large pathogenic islands and other laterally transferred elements can be easily detected in this way. In the present study, we showed that linking-clone-based physical mapping is a powerful and less expensive method for B. subtilis (natto) strains.
The characteristics of the plasmids, phages, and insertion elements investigated in this study were rather strain specific. We therefore propose that strain BEST217 is more closely related to Marburg 168, while BEST132, BEST195 and BEST323 are almost identical except for the small inversion in BEST132 (Fig. 5). Although the number of mapped genes in the physical maps is still limited, it is intriguing that the genes required for natto production are conserved among all the strains. Each of the strains secretes proteases (Fig. 1). However, only BEST195 was able to produce γ-PGA on a GSP medium assay plate (Fig. 1) as well as produce natto from boiled soybeans. A previous study revealed that the Marburg 168 strain, a non-natto producer, was converted to the natto producer BEST3145 as a result of substitution of a sequence that was up to 350 kb long from the BEST195 genome through repeated transformation (16). This result is conceivable given the conserved genome organization of the two strains, including several relevant genes required for natto production mapped in this study. The complicated process of natto production remains to be elucidated. Sequencing the entire genomes of strain BEST195 and the mosaic strain BEST3145 should provide important insights into the biochemical and physiological differences of these strains.
BEST132, BEST217, and BEST323 obtained from stock centers had lost the ability to produce γ-PGA, although they all had a swarming morphology, unlike Marburg 168 (Fig. 1). We suspect that these strains somehow lost the ability to produce γ-PGA during cultivation. This is partially explained by the observation that organisms that did not produce γ-PGA frequently arose during cultivation of BEST195 in media other than GSP medium (Qiu and Itaya, unpublished data). However, the ability to produce γ-PGA was stabilized in BEST3145, whose genome is a mosaic of the Marburg 168 and BEST195 genomes (15, 19). Since BEST3145 has no IS (Itaya, unpublished data), frequent loss of γ-PGA production by BEST195 may be attributed to IS transposition into the relevant genes, such as comP (25).
The Marburg 168 genome has been analyzed based on codon usage and repeat elements and by using hidden Markov models, and a series of laterally transferred elements have been identified (24, 27, 34). To some extent, the computational predictions may be substantiated when some B. subtilis (natto) genome sequences become available. The present results should be helpful for genome sequencing of natto starter strains, and more detailed comparative genome analyses could be conducted, as in the case of B. anthracis (30). The present macro restriction fragment length polymorphism-based classification provides a less expensive analysis for a large number of B. subtilis strains for taxonomic evaluation.
Acknowledgments
We thank K. Matsui and R. Kuniyasu for technical assistance.
REFERENCES
- 1.Agron, P. G., M. Macht, L. Radnedge, E. W. Skowronski, W. Miller, and G. L. Andersen. 2002. Use of subtractive hybridization for comprehensive surveys of prokaryotic genome differences. FEMS Microbiol. Lett. 211:175-182. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Ashikaga, S., H. Nanamiya, Y. Ohashi, and F. Kawamura. 2000. Natural genetic competence in Bacillus subtilis natto OK2. J. Bacteriol. 182:2411-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashiuchi, M., K. Soda, and H. Misono. 1999. Characterization of yrpC gene product of Bacillus subtilis 3336 as glutamate racemase isozyme. Biosci. Biotechnol. Biochem. 63:792-798. [DOI] [PubMed] [Google Scholar]
- 5.Ashiuchi, M., K. Soda, and H. Misono. 1999. A poly-γ-glutamate synthetic system of Bacillus subtilis IFO3336: gene cloning and biochemical analysis of poly-γ-glutamate produced by Escherichia coli clone cells. Biochem. Biophys. Res. Commun. 263:6-12. [DOI] [PubMed] [Google Scholar]
- 6.Ashiuchi, M., C. Nawa, T. Kamei, J. J. Song, S. P. Hong, M. H. Sung, K. Soda, and H. Misono. 2001. Physiological and biochemical characteristics of poly-γ-glutamate synthetase complex of Bacillus subtilis. Eur. J. Biochem. 268:5321-5328. [DOI] [PubMed] [Google Scholar]
- 7.Ashiuchi, M., and H. Misono. 2002. Biochemistry and molecular genetics of poly-γ-glutamate synthesis. Appl. Microbiol. Biotechnol. 59:9-14. [DOI] [PubMed] [Google Scholar]
- 8.Ashiuchi, M., E. Kuwana, K. Komatsu, K. Soda, and H. Misono. 2003. Differences in effects on DNA gyrase activity between two glutamate racemases of Bacillus subtilis, the poly-γ-glutamate synthesis-linking Glr enzyme and the YrpC (MurI) isozyme. FEMS Microbiol. Lett. 223:221-225. [DOI] [PubMed] [Google Scholar]
- 9.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, and G. F. Mayhew. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 10.Burkholder, P. R., and N. H. Giles. 1947. Induced biochemical mutations in Bacillus subtilis. Am. J. Bot. 34:345-348. [PubMed] [Google Scholar]
- 11.Eisen, J. A., J. F. Heidelberg, O. White, and S. L. Salzberg. 2000. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 1:0011.1-0011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 13.Hara, T., A. Aumayr, Y. Fujio, and S. Ueda. 1982. Elimination of plasmid-linked polyglutamate production by Bacillus subtilis (natto) with acridine orange. Appl. Environ. Microbiol. 44:1456-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itaya, M., and T. Tanaka. 1991. Complete physical map of the Bacillus subtilis 168 chromosome constructed by a gene-directed mutagenesis method. J. Mol. Biol. 220:631-648. [DOI] [PubMed] [Google Scholar]
- 15.Itaya, M. 1997. Physical map of the Bacillus subtilis 166 genome: evidence for the inversion of an approximately 1900 kb continuous DNA segment, the translocation of an approximately 100 kb segment and the duplication of a 5 kb segment. Microbiology 143:3723-3732. [DOI] [PubMed] [Google Scholar]
- 16.Itaya, M., and K. Matsui. 1999. Conversion of Bacillus subtilis 168: natto producing Bacillus subtilis with mosaic genomes. Biosci. Biotechnol. Biochem. 63:2034-2037. [DOI] [PubMed] [Google Scholar]
- 17.Koehler, T. M., and C. B. Thorne. 1987. Bacillus subtilis (natto) plasmid pLS20 mediates interspecies plasmid transfer. J. Bacteriol. 169:5271-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudva, I. T., P. S. Evans, N. T. Perna, T. J. Barrett, F. M. Ausubel, F. R. Blattner, and S. B. Calderwood. 2002. Strains of Escherichia coli O157:H7 differ primarily by insertions or deletions, not single-nucleotide polymorphisms. J. Bacteriol. 184:1873-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 20.Lai, Z., J. Jing, C. Aston, V. Clarke, J. Apodaca, E. T. Dimalanta, D. J. Carucci, M. J. Gardner, B. Mishra, and T. S. Anatharaman. 1999. A shotgun optical map of the entire Plasmodium falciparum genome. Nat. Genet. 23:309-313. [DOI] [PubMed] [Google Scholar]
- 21.Lazarevic, V., A. Dusterhoft, B. Soldo, H. Hilbert, C. Mauel, and D. Karamata. 1999. Nucleotide sequence of the Bacillus subtilis temperate bacteriophage SPβc2. Microbiology 145:1055-1067. [DOI] [PubMed] [Google Scholar]
- 22.Meijer, W. J., A. J. de Boer, S. van Tongeren, G. Venema, and S. Bron. 1995. Characterization of the replication region of the Bacillus subtilis plasmid pLS20: a novel type of replicon. Nucleic Acids Res. 23:3214-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meijer, W. J., A. de Jong, G. Bea, A. Wisman, H. Tjalsma, G. Venema, S. Bron, and J. M. van Dijl. 1995. The endogenous Bacillus subtilis (natto) plasmids pTA1015 and pTA1040 contain signal peptidase-encoding genes: identification of a new structural module on cryptic plasmids. Mol. Microbiol. 17:621-631. [DOI] [PubMed] [Google Scholar]
- 24.Moszer, I., E. P. Rocha, and A. Danchin. 1999. Codon usage and lateral gene transfer in Bacillus subtilis. Curr. Opin. Microbiol. 2:524-528. [DOI] [PubMed] [Google Scholar]
- 25.Nagai, T., L. S. Phan Tran, Y. Inatsu, and Y. Itoh. 2000. A new IS4 family insertion sequence, IS4Bsu1, responsible for genetic instability of poly-γ-glutamic acid production in Bacillus subtilis. J. Bacteriol. 182:2387-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagai, T., K. Koguchi, and Y. Itoh. 1997. Chemical analysis of poly-γ-glutamic acid produced by plasmid-free Bacillus subtilis (natto): evidence that plasmids are not involved in poly-γ-glutamic acid production. J. Gen. Appl. Microbiol. 43:139-143. [DOI] [PubMed] [Google Scholar]
- 27.Nicolas, P., L. Bize, F. Muri, M. Hoebeke, F. Rodolphe, S. D. Ehrlich, B. Prum, and P. Bessieres. 2002. Mining Bacillus subtilis chromosome heterogeneities using hidden Markov models. Nucleic Acids Res. 30:1418-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 29.Priest, F. G. 1993. Systematics and ecology of Bacillus, p. 1-16. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 30.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 31.Read, T. D., G. S. Myers, R. C. Brunham, W. C. Nelson, I. T. Paulsen, J. Heidelberg, E. Holtzapple, H. Khouri, N. B. Federova, H. A. Carty, L. A. Umayam, D. H. Haft, J. Peterson, M. J. Beanan, O. White, S. L. Salzberg, R. C. Hsia, G. McClarty, R. G. Rank, P. M. Bavoil, and C. M. Fraser. 2003. Genome sequence of Chlamidophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Regamey, A., V. Lazarevic, P. Hauser, and D. Karamata. 2000. Study of chromosome rearrangements associated with trpE26 mutation of Bacillus subtilis. Mol. Microbiol. 36:1234-1249. [DOI] [PubMed] [Google Scholar]
- 33.Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotype reflects oceanic niche differentiation. Nature 424:1042-1047. [DOI] [PubMed] [Google Scholar]
- 34.Rocha, E. P., A. Danchin, and A. Viari. 1999. Analysis of long repeats in bacterial genomes reveals alternative evolutionary mechanisms in Bacillus subtilis and other competent prokaryotes. Mol. Biol. Evol. 16:1219-1230. [DOI] [PubMed] [Google Scholar]
- 35.Sato, T., and Y. Kobayashi. 1998. The ars operon in the skin element of Bacillus subtilis confers resistance to arsenate and arsenite. J. Bacteriol. 180:1655-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Southern, E. M. 1975. Detection of specific sequences among DNA fragments by gel electrophoresis. J. Mol. Biol. 98:503-519. [DOI] [PubMed] [Google Scholar]
- 38.Stragier, P., B. Kunkel, L. Kroos, and R. Losick. 1989. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science 243:507-512. [DOI] [PubMed] [Google Scholar]
- 39.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka, T., and M. Kawata. 1988. Cloning and characterization of Bacillus subtilis iep, which has positive and negative effects on production of extracellular proteases. J. Bacteriol. 170:3593-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka, T., M. Kuroda, and K. Sakaguchi. 1977. Isolation and characterization of four plasmids from Bacillus subtilis. J. Bacteriol. 129:1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka, T., and M. Ogura. 1998. A novel Bacillus natto plasmid pLS32 capable of replication in Bacillus subtilis. FEBS Lett. 422:243-246. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka, T., and T. Koshikawa. 1977. Isolation and characterization of four types of plasmids from Bacillus subtilis (natto). J. Bacteriol. 131:699-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toda, T., and M. Itaya. 1995. I-CeuI recognition sites in the rrn operon of the Bacillus subtilis 168 chromosome: inherent landmarks for genome analysis. Microbiology 141:1937-1945. [DOI] [PubMed] [Google Scholar]
- 45.Urushibata, Y., S. Tokuyama, and Y. Tahara. 2002. Characterization of the Bacillus subtilis ywsC gene, involved in γ-polyglutamic acid production. J. Bacteriol. 184:337-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urushibata, Y., S. Tokuyama, and Y. Tahara. 2002. Difference in transcription levels of cap genes for γ-polyglutamic acid production between Bacillus subtilis IFO16449 and Marburg 168. J. Biosci. Bioeng. 93:252-254. [DOI] [PubMed] [Google Scholar]
- 47.Yamashita, S., H. Yoshikawa, F. Kawamura, H. Takahashi, T. Yamamoto, Y. Kobayashi, and H. Saito. 1986. The effect of spo mutations on the expression of spo0A- and spo0F-lacZ fusions. Mol. Gen. Genet. 205:28-33. [DOI] [PubMed] [Google Scholar]
- 48.Zahler, S. A. 1993. Temperate bacteriophages. p. 831-842. In A.L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 49.Zuber, P., M. M. Nakano, and M. A. Marahiel. 1993. Peptide antibiotics, p. 897-916. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.