Abstract
Yeasts do not possess an endogenous biochemical pathway for the synthesis of vitamin C. However, incubated with l-galactose, l-galactono-1,4-lactone, or l-gulono-1,4-lactone intermediates from the plant or animal pathway leading to l-ascorbic acid, Saccharomyces cerevisiae and Zygosaccharomyces bailii cells accumulate the vitamin intracellularly. Overexpression of the S. cerevisiae enzymes d-arabinose dehydrogenase and d-arabinono-1,4-lactone oxidase enhances this ability significantly. In fact, the respective recombinant yeast strains even gain the capability to accumulate the vitamin in the culture medium. An even better result is obtainable by expression of the plant enzyme l-galactose dehydrogenase from Arabidopsis thaliana. Budding yeast cells overexpressing the endogenous d-arabinono-1,4-lactone oxidase as well as l-galactose dehydrogenase are capable of producing about 100 mg of l-ascorbic acid liter−1, converting 40% (wt/vol) of the starting compound l-galactose.
Vitamin C or l-ascorbic acid is an important metabolite for most living organisms. In higher organisms, it is indispensable for different physiological functions and thus becomes an essential nutrient for animals (like humans) lacking its biosynthetic pathway. From the variety of cellular functions affected by ascorbate (18, 19), its role in the synthesis of collagen has been clearly established. Beneath the maintenance of metal ions in their reduced form, l-ascorbic acid acts as a scavenger of reactive oxygen species. Hence, an important function is to protect tissues from harmful oxidative products. It is this capacity that attracts attention to this compound and that leads to an increasing demand for l-ascorbic acid as a food additive.
l-Ascorbic acid is conventionally synthesized by a variety of known methods, which are generally variations of the Reichstein process (20, 21). These are mostly relatively complex chemical processes that utilize glucose as a starting material.
Novel biotechnological processes that convert glucose to vitamin C in one step would be desirable. In this respect, yeasts such as Saccharomyces cerevisiae, offer themselves as biocatalysts due to their generally recognized as safe (GRAS) status. However, yeast cells naturally lack the ability to produce l-ascorbic acid. Instead, erythroascorbic acid, a structurally related compound with chemical properties very similar to those of l-ascorbic acid, is the molecule occurring in yeast cells (11).
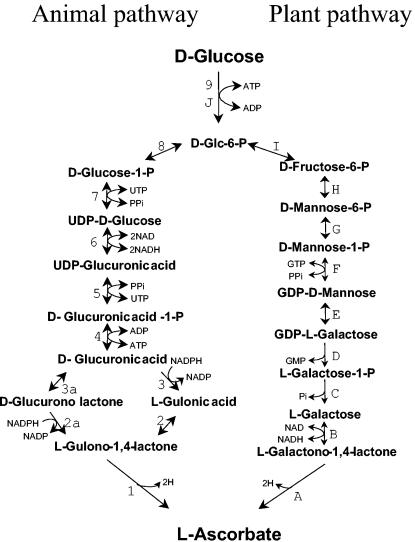
While the synthesis of l-ascorbic acid from d-glucose (Glc) proceeds via glucuronic acid and l-gulono-1,4-lactone (Gul) in animals (1) (Fig. 1), in plants the Glc is epimerized to d-mannose and then to l-galactose, and finally l-galactono-1,4-lactone (Gal) is converted into the vitamin (29) (Fig. 1).
FIG. 1.
Schematic representation of the biosynthetic pathways leading from Glc to l-ascorbic acid in plants or animals. The following enzymes are involved (the corresponding steps [letters or numbers] are in parentheses following Enzyme Commission numbers): l-galactono-1,4-lactone dehydrogenase (EC 1.3.2.3) (A), l-galactose dehydrogenase (B), sugar phosphatase (EC 3.1.3.23; putative) (C), hydrolase (putative) (D), GDP-mannose-3,5-epimerase (EC 5.1.3.18) (E), mannose-1-phosphate guanylyltransferase (EC 2.7.7.22) (F), phosphomannomutase (EC 5.4.2.8) (G), mannose-6-phosphate isomerase (EC 5.3.1.8) (H), glucose-6-phosphate isomerase (EC 5.3.1.9) (I), hexokinase (EC 2.7.1.1), (J), l-gulono-1,4-lactone oxidase (EC 1.1.3.8) (1), aldonolactonase (EC 3.1.1.17) (2), glucurono lactone reductase (EC 1.1.1.20) (2a), d-glucuronate reductase (EC 1.1.1.19) (3), uronolactonase (EC 3.1.1.19) or spontaneous (3a), d-glucurono kinase (EC 2.7.1.43) (4), glucuronate-1-phosphate uridylyltransferase (EC 2.7.7.44) (5) UDP-d-glucose dehydrogenase (EC 1.1.1.22) (6), UTP-glucose-1-phosphate uridylyltransferase (EC 2.7.7.9) (7), phosphoglucomutase (EC 5.4.2.2) (8), and hexokinase (EC 2.7.1.1) (9).
Interestingly, the last two steps of the plant pathway (Fig. 1) show some similarities to the pathway of yeast cells that leads to erythroascorbic acid. d-Arabinose is converted to d-arabinono-1,4-lactone, which is further converted to d-erythroascorbic acid. The structural motifs of the molecules changed during this conversion are identical to the structural motifs changed during the conversion of l-galactose via gal to l-ascorbic acid. The corresponding enzymes have been purified and characterized in more detail (10, 11, 12, 13). It was shown that the arabinono-1,4-lactone oxidases (ALOs) of Candida albicans and S. cerevisiae are able to convert not only l-arabinono-1,4-lactone but also Gal and Gul, among other structurally related lactones, in vitro; for C. albicans, it was shown that upon incubation of the cells with Gal, l-ascorbic acid accumulates in measurable amounts inside the cells (10), as well as in the culture medium (22, 23). The S. cerevisiae ALO1 gene was expressed in Escherichia coli, conferring the ability to convert Gal to l-ascorbic acid and d-arabinono-1,4-lactone to erythroascorbic acid in this organism (15). However, the products are accumulated intracellularly only.
d-Arabinose dehydrogenase (ARA) catalyzes the second-last step in the erythroascorbic acid biosynthetic pathway in yeast cells (12, 13), which resembles the second-last step of l-ascorbic acid biosynthesis in plants (Fig. 1). For this enzyme, the substrate range is also quite broad. It was reported that the respective enzymes from C. albicans and S. cerevisiae are able to convert not only d-arabinose to d-arabinono-1,4-lactone but also l-galactose to Gal in vitro (12, 13). The gene for this enzyme in S. cerevisiae has been cloned, and it has been shown that deletion of the gene leads to a complete absence of erythroascorbic acid in this organism (13). Further, it has been shown for S. cerevisiae that intact cells accumulate l-ascorbic acid intracellularly upon incubation with l-galactose (8), which is most probably due to endogenous ALO and ARA activities.
To our knowledge, there is no report of any overexpression of ARA1 or LGDH (the latter encoding l-galactose dehydrogenase) in yeast cells; despite a variety of groups worldwide that are concerned with the creation of yeast strains overproductive of l-ascorbic acid (9), it has not yet been proven that metabolically engineered yeast cells can in fact sufficiently release the vitamin in the culture medium.
Here, we present the production of vitamin C from metabolically engineered S. cerevisiae as well as from the nonconventional yeast Zygosaccharomyces bailii.
MATERIALS AND METHODS
Yeast strains.
The following yeast strains were used: S. cerevisiae GRF18U (MATα his3 leu2 ura3; NRRL Y-30320) (25) and W303 1B (MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100) (25) and Z. bailii ATCC 60483 (25). In this publication, the term wild type (wt) used to describe a yeast strain refers to the above-mentioned nontransformed strains. From the genotype, it becomes obvious that wt means that no recombinant gene is expressed.
Gene amplification and cloning.
All genes were amplified by PCR. PfuTurbo DNA polymerase (Stratagene catalogue no. 600252) was used on a GeneAmp PCR System 9700 (PE Applied Biosystems, Inc.) Standard conditions used were 400 μM deoxynucleoside triphosphates, 0.5 μM primers, 0.5 mM MgCl2 (in addition to the supplied buffer), and 3.75 U of Pfu per 100 μl of reaction mixture. The program used for amplification of genes encoding l-galactono-1,4-lactone dehydrogenase from Arabidopsis thaliana (AGD), ALO from S. cerevisiae (ALO1), l-galactose dehydrogenase from A. thaliana (LGDH), ARA from S. cerevisiae (ARA1), and l-gulono-1,4-lactone oxidase from Rattus norvegicus (Gulo) was as follows. After 5 min at 94°C, 33 cycles (each cycle consisting of 45 s at 94°C, 30 s at Tx, and 1 min 40 s at 72°C) were carried out, followed by 7 min at 72°C. Tx settings were as follows: for AGD, 53.5°C; for ALO1, 50°C; for LGDH, 56°C; for ARA1, 56°C, and for Gulo, 72°C. Template DNA for AGD and LGDH consisted of 50 ng of the plasmid cDNA library pFL61 from A. thaliana (16) (ATCC 77500); the DNA for Gulo consisted of 0.5 ng of rat liver marathon-ready cDNA library (Clontech 7471-1); for ALO1 and ARA1, we used 50 ng of genomic DNA from S. cerevisiae GRF18U, extracted by a standard method.
Oligonucleotides.
Oligonucleotide pairs were as follows: for AGD, CAAGAAGGCCTAAATGTTCCGTTACGCTCC and ATGGGCCCTTAAGCAGTGGTGGAGACTGGG; for ALO1, TTTCACCATATGTCTACTATCC and AAGGATCCTAGTCGGACAACTC; for LGDH, ATGACGAAAATAGAGCTTCGAGC and TTAGTTCTGATGGATTCCACTTGG; for ARA1, ATGTCTTCTTCAGTAGCCTCAACC and TTAATACTTTAAATTGTCCAAGTTTGGTC; and for Gulo, TGAGGGGTCAGGGTGGTTTGTTTCCA and TGGAATCATGGTCCATGGGTACAAAGGG.
PCR products were blunt-end cloned into the EcoRV site of pSTBlue-1 with the perfectly blunt cloning kit from Novagen, Inc. (catalogue no. 70191-4). Finally, the coding regions were cloned into the S. cerevisiae expression vectors of the pYX series (R&D Systems, Inc.) or the Z. bailii expression vectors pZ3 and pAG26 in the following manner. AGD and ALO1 were subcloned into pYX042 (integrative; LEU2 auxotrophic marker and S. cerevisiae triose phosphate isomerase [TPI] promoter) with EcoRI. LGDH was subcloned into pYX022 (integrative; HIS3 auxotrophic marker and TPI promoter) with EcoRI. ARA1 was subcloned into pYX022 by cutting pSTBlue-ARA SacI, blunt ending, cutting with BamHI, and cloning into pYX022, cutting with EcoRI, blunt ending, and cutting with BamHI. For construction of pAG26TPI-LGDH (autonomous replication sequence-centromeric [ARS-CEN] region; hygromycin resistance marker and TPI promoter), the expression cassette consisting of the TPI promoter, the LGDH gene, and the terminator was cut from pYX022-LGDH with the restriction enzymes AatII and PvuII, blunt ended, cloned into pAG26 (7), cut with ApaI, and blunt ended. The plasmid pZ3 (ARS-CEN region; G418 resistance marker and TPI promoter) was constructed as described by Branduardi et al. (2, 3). In short, the ARS-CEN region was cut from Ycplac33 (5) by cutting it with the restriction enzyme ClaI, blunt ending, further cutting with SpeI, and cloning into pYX022, which had been cut with DraIII, blunt ended, and cut with SpeI. The resulting plasmid was opened with KpnI and blunt ended to receive the G418 resistance cassette, cut from pFA6-kanMX4 (28) with the restriction enzymes SphI/SacI, and blunt ended. ALO1 and Gulo were cloned into pZ3 with the restriction enzyme EcoRI, following blunt ending.
Standard procedures were employed for all cloning purposes (24).
All the plasmids utilized in the experiments described here are listed in Table 1, together with their main properties.
TABLE 1.
Plasmids constructed for the experiments presented in this studya
| Expression vector | Promoter | Heterologous protein | Plasmid property | Selection |
|---|---|---|---|---|
| pL-AGD | Sc TPI | At AGD | INT | Sc LEU2 |
| pH-LGDH | Sc TPI | At LGDH | INT | Sc HIS3 |
| pL-ALO | Sc TPI | Sc ALO | INT | Sc LEU2 |
| pH-ARA | Sc TPI | Sc ARA | INT | Sc HIS3 |
| pL-RGLO | Sc TPI | Rn Gulo | INT | Sc LEU2 |
| pAG26TPI-LGDH | Sc TPI | At LGDH | CEN | Hphr |
| pZ-ALO | Sc TPI | Sc ALO | CEN | Kanr |
| pZ-RGLO | Sc TPI | Rn Gulo | CEN | Kanr |
A complete description of plasmids construction is given in Materials and Methods. Abbreviations: Sc, S. cerevisiae; At, A. thaliana; Rn, R. novergicus; TPI, triose phosphate isomerase; AGD, l-galactono-1,4-lactone dehydrogenase; ALO, d-arabinono-1,4-lactone oxidase; ARA, d-arabinose dehydrogenase; LGDH, l-galactose dehydrogenase; Gulo, l-gulono-1,4-lactone oxidase; LEU2, HIS3, and URA3, gene markers conferring growth to auxotrophic yeast strains in the absence of leucine, histine and uracile, respectively; Kanr and Hphr, cassettes conferring resistance to G418 and hygromycin, respectively; INT and CEN, integrative and centromeric plasmids, respectively.
Yeast transformation and cultivation.
Transformation of S. cerevisiae (6) and Z. bailii (3) yeast cells was done following the standard lithium acetate-single-stranded carrier DNA-polyethylene glycol method. Wild-type and transformed strains were cultivated in shake flasks in minimal medium (0.67% [wt/vol] YNB medium [catalogue no. 919-15; Difco Laboratories, Detroit, Mich.]; 2% [wt/vol] glucose, 50 mg of the appropriate amino acid liter−1 [adenine or uracil, respectively]) by shaking at 30°C (final cellular concentration, 5 × 107 to 10 × 107 ml−1). For cultivation of Z. bailii, the antibiotics G418 and hygromycin were added in a concentration of 200 and 100 μg ml−1, respectively.
The optical density at 660 nm (OD660) of the inoculum was 0.05. For the bioconversion of the different intermediates, cells were directly grown in the presence of the respective intermediate or pregrown for 1 day, and then the respective intermediate was added. For intracellular l-ascorbic acid determination, the cells were recovered by centrifugation at 4,000 rpm for 5 min at 4°C, washed once with cold distilled H2O, and treated as follows. The cells were resuspended in about three times the pellet volume of cold 10% (wt/vol) trichloroacetic acid, vortexed vigorously, and kept on ice for about 20 min, and then the supernatant was cleared from the cell debris by centrifugation.
Determination of l-ascorbic acid.
l-Ascorbic acid concentrations were determined spectrophotometrically following a method adapted from that of Sullivan and Clarke (27): 135 μl of sample was mixed in a cuvette with 40 μl of H3PO4 (85%), 675 μl of α,α′-bipyridyl (0.5%), and 135 μl of FeCl3 (1%). After 10 min at room temperature, the absorbance at 525 nm was measured.
The identity of the l-ascorbic acid was confirmed by high-performance liquid chromatography with a Tracer Extrasil C8 column (5-μm inner diameter, 15 by 0.46 cm, catalogue no. TR-016077; Teknokroma S. Coop. C. Ltda.) with 5 mM cetyltrimethylammonium bromide and 50 mM KH2PO4 in 95/5 H2O/acetonitrile as eluent, a flow rate of 1 ml min−1, and UV detection set at 254 nm, with pure l-ascorbic acid (Aldrich catalogue no. A9290-2) as the standard.
Statistics.
All experiments were carried out at least three times. Here, we report mean values; standard deviations were never >10%.
RESULTS AND DISCUSSION
l-Ascorbic acid production from nontransformed yeasts.
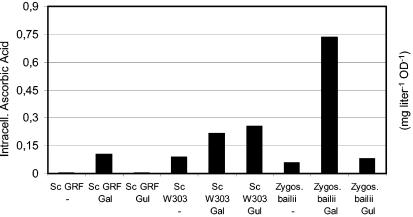
Figure 1 shows the metabolic pathways leading from Glc to l-ascorbic acid in animals and plants (1, 29; Kyoto Encyclopedia of Genes and Genomes, release 27.0 [http://www.genome.ad.jp/kegg/pathway/map/map00053.html]). It has been shown that yeasts do not produce ascorbic acid naturally (10), but it was reported that intact S. cerevisiae cells accumulate l-ascorbic acid intracellularly upon incubation with Gal, the last intermediate of the plant vitamin C biosynthetic pathway, and Gul, the last intermediate of the animal metabolic pathway (8). To address the questions of whether these intermediates can also be converted by another interesting nonconventional yeast and whether the vitamin can eventually be released from the yeast cells, two different S. cerevisiae yeast strains (GRF18U and W303 1B) and a Z. bailii strain were incubated with both of these compounds for 72 h (Fig. 2). In no case was vitamin C detectable in the culture broth (data not shown). Upon incubation of the cells with Gal, all strains accumulated l-ascorbic acid intracellularly in significant amounts. This proves that, like S. cerevisiae, Z. bailii yeast cells are capable of taking up the substrate and that these yeast cells bear an ALO-like activity, which probably retains its broad substrate specificity under in vivo conditions. The S. cerevisiae ALO1 gene is therefore a promising candidate for the construction of a recombinant yeast strain overproductive for l-ascorbic acid. Incubation of the yeast cells with Gul led to a measurable accumulation of vitamin C, but only in S. cerevisiae W303 1B (Fig. 2).
FIG. 2.
Determination of the endogenous ability of yeast strains to convert Gal or Gul to l-ascorbic acid (in milligrams per liter per optical density). Nontransformed yeast cells (S. cerevisiae strains GRF18U and W303 1B and Z. bailii) were grown on mineral medium (2% [wt/vol] glucose, 0.67% [wt/vol] YNB) in the presence of 17.82 g of Gal or Gul liter−1, respectively, for 72 h (initial OD660 = 0.05). −, no substrate was added. While l-ascorbic acid was accumulated within the cell, no l-ascorbic acid could be detected in the culture broth.
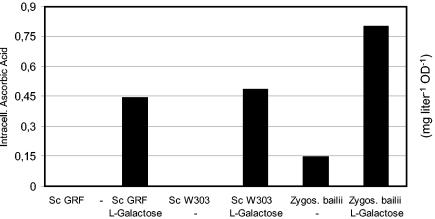
The rare sugar l-galactose is the next intermediate, when the plant biosynthetic pathway is traced backwards. Incubation of S. cerevisiae GRF18U and W303 1B (controls) and Z. bailii with l-galactose for 24 h led to the intracellular accumulation of l-ascorbic acid (Fig. 3), while no vitamin C was detectable in the culture broth (data not shown).
FIG. 3.
Determination of the endogenous ability of yeast cells to convert l-galactose to l-ascorbic acid (measured in milligrams per liter per optical density). Nontransformed S. cerevisiae (GRF18U and W303 1B) and Z. bailii were pregrown overnight on mineral medium (2% [wt/vol] glucose, 0.67% [wt/vol] YNB), starting with an OD660 of 0.05. After 24 h, 250 mg of l-galactose liter−1 was added, and the cultures were kept under standard conditions for another 24 h before the determination of the l-ascorbic acid concentration. All of these strains accumulated l-ascorbic acid intracellularly, while no vitamin C was detectable in the culture broth. −, no substrate was added.
To address the previously mentioned important question of whether the vitamin could be secreted by yeast en route to the biotechnological production of vitamin C, we set out to increase enzymatic activities by converting the above-mentioned substrates.
l-Ascorbic acid production from recombinant yeasts.
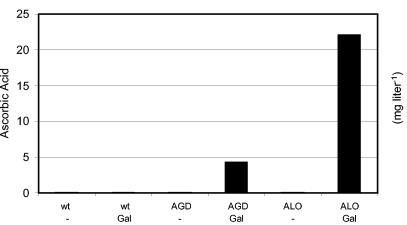
To enhance the natural ability of S. cerevisiae to convert Gal to l-ascorbic acid, we cloned S. cerevisiae ALO1 (encoding ALO) and of A. thaliana AGD (encoding l-galactono-1,4-lactone dehydrogenase) (Table 1) (17). The coding sequences were PCR amplified and cloned into an integrative yeast shuttle vector. The strong and constitutive S. cerevisiae TPI promoter drives the expression of the recombinant genes. The S. cerevisiae strains GRF18U and W303 1B were transformed with the respective plasmids and incubated with Gal for 72 h as described in Material and Methods. Surprisingly, the transformed yeast cells accumulated considerable amounts of vitamin C into the culture broth, as was seen with the transformed S. cerevisiae GRF18U cells (Fig. 4). Interestingly, these transformed cells also accumulated l-ascorbic acid in the culture broth upon incubation with l-galactose (Table 2). This indicates that for the conversion of this rare sugar into the vitamin, the last step is rate limiting.
FIG. 4.
Conversion of Gal to l-ascorbic acid (in milligrams per liter) by transformed S. cerevisiae GRF18U cells. S. cerevisiae GRF18U wt or GRF18U transformed with AGD or ALO1 was grown on mineral medium (2% [wt/vol] glucose, 0.67% [wt/vol] YNB), starting with an OD660 of 0.05 in the presence of 8.91 g of Gal liter−1 for 72 h, when samples were taken and the concentration of l-ascorbic acid in the culture broth was determined. While the untransformed cells did not accumulate l-ascorbic acid in the culture medium, cells transformed with AGD or ALO1 secreted considerable amounts of vitamin C. No ascorbate could be detected in cultures without the addition of Gal (−).
TABLE 2.
Conversion of l-galactose to l-ascorbic acid by transformed yeastsa
| Yeast strain | Gene overexpressed | l-Ascorbic acid accumulation in culture broth (mg liter−1) |
|---|---|---|
| S. cerevisiae GRF18U | Controlb | — |
| ARA1 | — | |
| LGDH | — | |
| ALO1 | 0.9 | |
| AGD | 0.2 | |
| ARA1 + ALO1 | 4.1 | |
| LGDH + AGD1 | 2.2 | |
| LGDH + ALO1 | 13.2 | |
| Z. bailii | Controlb | — |
| ALO1 | 0.8 | |
| LGDH + ALO1 | 2.8 |
Yeasts were pregrown on mineral medium (2% [wt/vol] glucose, 0.67% [wt/vol] YNB) starting from an OD660 of 0.05 overnight. After 24 h of growth, 250 mg of l-galactose liter−1 was added, and the cultures were kept under standard conditions for another 24 h when samples were taken and the l-ascorbic acid concentration in the culture broth was determined.
Control, yeast cells transformed with the empty vectors.
To additionally increase the natural ability of yeast to convert l-galactose into Gal, we cloned S. cerevisiae ARA1 (encoding ARA) and A. thaliana LGDH (encoding l-galactose dehydrogenase) (4, 26). Expression of the ARA1 or LGDH gene alone did not lead to any measurable accumulation of vitamin C in the culture medium upon incubation of the yeast strains with l-galactose. This result is in accordance with the indication that the last step converting l-galactose into l-ascorbic acid is the limiting step in yeast cells. Therefore, an increase in yield of the second-last step alone did not lead to any secretion of the vitamin. In contrast, cotransformation of LGDH with ALO1 or AGD as well as of ALO1 with ARA1 significantly increases the ability of the respective yeast strain to convert l-galactose to vitamin C (Table 2).
The S. cerevisiae ALO1 and A. thaliana LGDH genes were also successfully expressed in Z. bailii (Table 2). Z. bailii was selected for this study because it tolerates high-sugar concentrations, acidic environments, and relatively high temperatures and can survive in the presence of high concentrations of chemical preservatives (3). Because of some of these features, fermentation bioprocesses could be performed at very low pH values, simplifying the downstream process and, at the same time, preventing bacterial contamination. While the cells transformed with vectors conferring only the resistance to the antibiotics did not accumulate any vitamin C in the culture broth, functional expression of the ALO1 gene alone or ALO1 with LGDH led to a significant accumulation of the vitamin in the culture medium. This also proves that Z. bailii, a new nonconventional yeast host recently developed for the production of heterologous proteins by secretion (3), has the ability not only to produce l-ascorbic acid upon introduction of the appropriate metabolic pathway but also to secrete the vitamin.
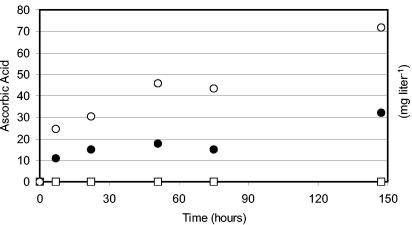
Finally, Fig. 5 shows data from a high-density culture converting l-galactose. The respective yeast strains were grown overnight in standard minimal medium. On the next day, the cells were concentrated 10 times, then 250 mg of l-galactose liter−1 was added, and the cultures were incubated under standard conditions for 6 days. After 6 days, the strain transformed with ALO1 and LGDH accumulated more than 70 mg of l-ascorbic acid per liter of culture medium. A total of 30 mg of vitamin C liter−1 was accumulated intracellularly (data not shown). Taken together, these values correspond to a conversion of about 40% of the added l-galactose to l-ascorbic acid within 6 days. The plant pathway, as shown before, proves to be a passable way for the production of l-ascorbic acid from Glc by metabolically engineered yeasts.
FIG. 5.
Example of a high-cell-density culture of transformed S. cerevisiae yeast cells converting l-galactose to l-ascorbic acid (values expressed as milligrams per liter per optical density). S. cerevisiae GRF18U wt or GRF18U transformed with ALO1 or LGDH and ALO1 was pregrown overnight on mineral medium (2% [wt/vol] glucose and 0.67% [wt/vol] YNB), starting with an OD660 of 0.05. At time zero, the cells were concentrated 10 times, and 250 mg of l-galactose liter−1 was added. The cultures were kept under standard conditions for 6 days. At the indicated times, samples were taken, and the l-ascorbic acid concentration in the culture broth was determined. While the wt (□) cells did not accumulate l-ascorbic acid in the culture medium, cells transformed with ALO1 alone (•) or ALO1 and LGDH (○) secreted considerable amounts of vitamin C.
However, it remains conceivable that other biosynthetic pathways could be employed or a new and unique pathway utilizing appropriate heterologous enzymes could be created. For example, we expressed the rat liver Gulo gene (encoding l-gulono-1,4-lactone oxidase) (Table 1) (14) in S. cerevisiae as well as in Z. bailii. The intracellular production of l-ascorbic acid from Gul was thereby increased 3- and 15-fold for S. cerevisiae and Z. bailii, respectively (data not shown). Secretion of the vitamin remains undetectable so far.
The presented results are highly promising, and the goal of creating yeast cell factories for the easy conversion of a cheap carbon source into l-ascorbic acid appears closer than ever. Corresponding work is ongoing in our laboratory, particularly to fill the metabolic gap between Glc and l-galactose; this is a required step for the development of a commercially acceptable technology leading to vitamin C production. In this respect, it can also be safely assumed that production of vitamin C in grams-per-liter concentrations is attainable.
Acknowledgments
This work was supported by A. E. Staley Manufacturing Company, Decatur, Ill., and it was partially supported by MURST-Cofin 2002 (prot. 2002052349 to D.P.).
REFERENCES
- 1.Banhegyi, G., L. Braun, M. Csala, F. Puskas, and J. Mandl. 1997. Ascorbate metabolism and its regulation in animals. Free Radic. Biol. Med. 23:793-803. [DOI] [PubMed] [Google Scholar]
- 2.Branduardi, P., M. Valli, L. Alberghina, and D. Porro. 2002. Process for expression and secretion of proteins by the non-conventional yeast Zygosaccharomyces bailii. German patent application 102 52 245.6.
- 3.Branduardi, P., M. Valli, L. Brambilla, M. Sauer, L. Alberghina, and D. Porro. 2004. The yeast Zygosaccharomyces bailii: a new host for heterologous protein production, secretion and for metabolic engineering applications. FEMS Yeast Res. 4:493-504. [DOI] [PubMed] [Google Scholar]
- 4.Gatzek, S., G. L. Wheeler, and N. Smirnoff. 2002. Antisense suppression of l-galactose dehydrogenase in Arabidopsis thaliana provides evidence for its role in ascorbate synthesis and reveals light modulated l-galactose synthesis. Plant J. 30:541-553. [DOI] [PubMed] [Google Scholar]
- 5.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 6.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae.. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 8.Hancock, R. D., J. R. Galpin, and R. Viola. 2000. Biosynthesis of l-ascorbic acid (vitamin C) by Saccharomyces cerevisiae. FEMS Microbiol. Lett. 186:245-250. [DOI] [PubMed] [Google Scholar]
- 9.Hancock, R. D., and R. Viola. 2002. Biotechnological approaches for l-ascorbic acid production. Trends Biotechnol. 20:299-305. [DOI] [PubMed] [Google Scholar]
- 10.Huh, W. K., S. T. Kim, K. S. Yang, Y. J. Seok, Y. C. Hah, and S. O. Kang. 1994. Characterization of d-arabinono-1,4-lactone oxidase from Candida albicans ATCC 10231. Eur. J. Biochem. 225:1073-1079. [DOI] [PubMed] [Google Scholar]
- 11.Huh, W. K., B. H. Lee, S. T. Kim, Y. R. Kim, G. E. Rhie, Y. W. Baek, C. S. Hwang, S. J. Lee, and S. O. Kang. 1998. d-Erythroascorbic acid is an important antioxidant molecule in S. cerevisiae. Mol. Microbiol. 30:895-903. [DOI] [PubMed] [Google Scholar]
- 12.Kim, S. T., W. K. Huh, J. Y. Kim, S. W. Hwang, and S. O. Kang. 1996. d-Arabinose dehydrogenase and biosynthesis of erythroascorbic acid in Candida albicans. Biochim. Biophys. Acta 1297:1-8. [DOI] [PubMed] [Google Scholar]
- 13.Kim, S. T., W. K. Huh, B. H. Lee, and S. O. Kang. 1998. d-Arabinose dehydrogenase and its gene from Saccharomyces cerevisiae. Biochim Biophys Acta 1429:29-39. [DOI] [PubMed] [Google Scholar]
- 14.Koshizaka, T., M. Nishikimi, T. Ozawa, and K. Yagi. 1988. Isolation and sequence analysis of a complementary DNA encoding rat liver l-gulono-γ-lactone oxidase, a key enzyme for l-ascorbic acid biosynthesis. J. Biol. Chem. 263:1619-1621. [PubMed] [Google Scholar]
- 15.Lee, B. H., W. K. Huh, S. T. Kim, J. S. Lee, and S. O. Kang. 1999. Bacterial production of d-erythroascorbic acid and l-ascorbic acid through functional expression of Saccharomyces cerevisiae d-arabinono-1,4-lactone oxidase in Escherichia coli. Appl. Environ. Microbiol. 65:4685-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minet, M., M. E. Dufour, and F. Lacroute. 1992. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 2:417-422. [DOI] [PubMed] [Google Scholar]
- 17.Østergaard, J., G. Persiau, M. W. Davey, G. Bauw, and M. Van Montagu. 1997. Isolation of a cDNA coding for l-galactono-γ-lactone dehydrogenase, an enzyme involved in the biosynthesis of ascorbic acid in plants. Purification, characterization, cDNA cloning, and expression in yeast. J. Biol. Chem. 272:30009-30016. [DOI] [PubMed] [Google Scholar]
- 18.Padh, H. 1990. Cellular functions of ascorbic acid. Biochem. Cell Biol. 68:1166-1173. [DOI] [PubMed] [Google Scholar]
- 19.Padh, H. 1991. Vitamin C: newer insights into its biochemical functions. Nutr. Rev. 49:65-70. [DOI] [PubMed] [Google Scholar]
- 20.Reichstein, T. 1941. Process for the manufacture of levoascorbic acid (vitamin C). U.S. patent 2,265,121.
- 21.Reichstein, T. 1942. 2-Keto-levo-gulonic acid and process for the manufacture of same. U.S. patent 2,301,811.
- 22.Roland, J. F., T. Cayle, R. C. Dinwoodie, and D. W. Mehnert. 1986. Fermentation production of ascorbic acid from l-galactonic substrate. U.S. patent 4,595,659.
- 23.Roland, J. F., T. Cayle, R. C. Dinwoodie, and D. W. Mehnert. 1990. Bioconversion production of ascorbic acid with l-galactono-1,4-oxidase. U.S. patent 4,916,068.
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sauer, M., and D. Porro. 2000. Ascorbic acid production from yeast. International patent application WO 02/10425.
- 26.Smirnoff, N., and G. Wheeler. 1999. Plant galactose dehydrogenase. International patent application WO 99/33995.
- 27.Sullivan, M. X., and H. C. N. Clarke. 1955. A highly specific procedure for ascorbic acid. J. Assoc. Off. Agr. Chem. 38:514-518. [Google Scholar]
- 28.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 29.Wheeler, G. L., M. A. Jones, and N. Smirnoff. 1998. The biosynthetic pathway of vitamin C in higher plants. Nature 393:365-369. [DOI] [PubMed] [Google Scholar]