Abstract
Post-translational modifications (PTMs) are important steps in the biosynthesis of proteins. Aside from their integral contributions to protein development, i.e. perform specialized proteolytic cleavage of regulatory subunits, the covalent addition of functional groups of proteins or the degradation of entire proteins, PTMs are also involved in enabling proteins to withstand and recover from temporary environmental stresses (heat shock, microgravity and many others). The literature supports evidence of thousands of recently discovered PTMs, many of which may likely contribute similarly (perhaps, even, interchangeably) to protein stress response. Although there are many PTM actors on the biological stage, our study determines that these PTMs are generally cast into organism-specific, preferential roles. In this work, we study the PTM compositions across the mitochondrial (Mt) and non-Mt proteomes of 11 diverse organisms to illustrate that each organism appears to have a unique list of PTMs, and an equally unique list of PTM-associated residue reaction sites (RSs), where PTMs interact with protein. Despite the present limitation of available PTM data across different species, we apply existing and current protein data to illustrate particular organismal biases. We explore the relative frequencies of observed PTMs, the RSs and general amino-acid compositions of Mt and non-Mt proteomes. We apply these data to create networks and heatmaps to illustrate the evidence of bias. We show that the number of PTMs and RSs appears to grow along with organismal complexity, which may imply that environmental stress could play a role in this bias.
Keywords: PTM bias, reaction site bias, amino acid bias, organism complexity
Introduction
Post-translational modifications
It is extremely likely that all proteins naturally undergo some level of structural and, therefore, functional alteration by post-translational modification (PTMs). Although many thousands of PTMs have been discovered (7308 experimentally identified PTMs and 234 938 putative modifications on 530 264 proteins according to [1]), there are some (i.e. acetylation, glycosylation, phosphorylation, proteolysis, lipidation, methylation, nitrosylation, ubiquitination and others) that commonly interact with proteins at specific reaction sites (RSs). The amino acids are at precise locations of the protein chain, and their interactions with PTMs inspire changes in protein conformations. PTMs have been shown to play prominent roles in protein alteration for destruction [2, 3], general regulation [4, 5] and stress response [6–8]. In this way, PTMs are able to greatly expand the functional diversity of the proteome and disprove the ‘one-gene-one-protein’ hypothesis.
To add functional diversity and adaption to their alternative environments [9], proteins may respond to stresses by a transformation of structure and, hence, function. For instance, protein stress may result from an event or treatment, which leads to protein failure when the protein is forced to sustain its duties under unnatural circumstances, such as environmental stress. Across seemingly all proteins, PTMs offer an extremely rapid solution for withstanding naturally occurring environmental stresses, such as microgravity [10], drought [11], thermic shock [12, 13] and others. Stress responses resulting from the proteins themselves are also conducted by phosphorylation as in the case of initiation and regulation of tumor suppression by the p53 complex [6] and SUMOylation for its response to oxidative stress [7]. Furthermore, by intervening with PTM function, therapies may be created to treat types of cancer [14] or be used to maintain cellular homeostasis [8, 15]. A modification is immediate, as it does not necessitate the resynthesis of a new protein to cope with environment stresses. Once the stress is removed, PTMs are often able to restore the protein to its previous conformation [16].
Biases
PTMs also show evidence of preferential treatment. Discussed in Khoury et al. [1], PTM activities from acetylation, glycosylation and phosphorylation were frequently observed in their data; however, there were many PTMs that were rarely exerted (i.e. flavin adenine dinucleotide, bromination and many others). A PTM bias between related proteins may conveniently be observed using public data such as UniProt [17]. For example, Sir1, also known as the nicotinamide adenine dinucleotide-dependent protein deacetylase sirtuin1, is a regulatory protein found in human and mouse. The Sir1 Human (UniProt: Q96EB6) and Sir1 Mouse (UniProt: Q923E4) had a listing of 19 and 13 observed PTM interactions, respectively. Although there were 16 phosphorylation sites in Sir1 Human and only 10 in Sir1 Mouse, acetylation was observed only in mouse protein. Here, we note that these two similar proteins offer high granularity evidence for the existence of PTM bias between human and mouse.
Research statement
In this article, we extend our original study in [18], which described some of the initial patterns of PTM bias inherent in some of the organisms of the present study. We studied the proteomes of 11 diverse organisms shown in Table 1 to show that each organism has unique PTM biases and an associated RS bias. We present evidence that the number of observed PTMs and RSs by organism appears to increase with organismal complexity. To clearly describe these biases, we use heatmaps and networks, which are built from relative frequency data that we harvested from parsing data available from UniProt. Because mitochondria (Mt) have unique genomes and therefore unique proteomes, we extend our study of protein PTM biases to these organelles to describe their PTM and RS biases by organism. Because Mt are highly conserved across biology, we show that their PTM use is not a conserved entity. Finally, we show that the trends of increasing PTM and RS bias are both observed in Mt and non-Mt with similar degrees of clarity.
Table 1.
Diverse organisms of the study. We noted that glycosylation and phosphorylation were commonly the most frequently occurring PTMs in our data
| Organism | Number of proteins | Top PTMs |
|---|---|---|
| Mustard plant | 3155 | Glycosylation |
| A. thaliana | Phosphorylation | |
| Fungi | 254 | Glycosylation |
| A. nidulans | Methylation | |
| Nematode worm | 572 | Glycosylation |
| C. elegans | Lipidation | |
| Domestic dog | 616 | Glycosylation |
| C. familiaris | Phosphorylation | |
| Zebrafish | 622 | Glycosylation |
| D. rerio | Phosphorylation | |
| Human | 11 884 | Phosphorylation |
| H. sapiens | Glycosylation | |
| House mouse | 10 388 | Phosphorylation |
| M. musculus | Glycosylation | |
| European rabbit | 641 | Glycosylation |
| O. cuniculus | Phosphorylation | |
| Norway rat | 5413 | Glycosylation |
| R. norvegicus | Phosphorylation | |
| Bakers yeast | 3013 | Phosphorylation |
| S. cerevisiae | Glycosylation | |
| African clawed frog | 671 | Glycosylation |
| X. laevis | Phosphorylation |
Methods
For the organisms of Table 1, protein data were downloaded in June 2015 from UniProt, a public protein knowledge base that provides curated data. At the time of our study, our downloaded set was the most currently available. The curated protein records were divided into Mt and non-Mt sets, depending on their origins for each organism. For every protein of each set, the PTM data were assembled—the type and number of PTM as well as their associated RSs, which were often unique to each particular PTM. In Figure 1 (created by http://bioinformatics.psb.ugent.be/webtools/Venn/), we illustrate the counts of PTMs, which were obtained across the organismal Mt and non-Mt sets, taken all together. Because the organization of the Mt genome is highly conserved in insects, as in most other bilateral animals [19, 20], we maintain that the patterns that we were able to find in Mt may likely be extended to other types of organisms as well, although the nuclear proteins may not be similar.
Figure 1.

A comparison between the number of PTMs in our Mt and non-Mt sequence data. Here, we exclude all PTMs that are labeled by UniProt as ‘InterChain’ because of a lack of information available for our study.
We noted that there were often cases where a specific PTM type was given by UniProt that fell into a more general category. For example, N-acetylalanine and N-acetylaspartate are two specific types of acetylation. There were many other cases where specific PTMs (often specifically named because of their associated RSs) could be reduced to more general denominations. To simplify PTM quantifications during our analysis, we followed the PTM conversion documentation available by UniProt to record the general PTM denominations. These frequently occurring PTMs describe evidence of PTM bias even from a high granularity. Specifically, the order of the first and second most frequently observed PTMs (generally, glycosylation and phosphorylation) were not unanimously conserved across the organisms, as noted in Table 1.
Organismal protein samples
In Table 1, we indicate the actual number of proteins analyzed as well as the two most commonly occurring PTMs from the Mt and non-Mt protein sets of each organism. In the second and third columns of Table 2, we display the number of processed Mt and non-Mt UniProt protein records, respectively. In the fourth column, we present the size of the exhaustive list of organism-specific (curated) proteins from UniProt from the time of our study. In this column, there are records containing PTM information, as well as many where PTMs are not discussed. By comparing the numbers of records where PTM information is known with the numbers where it is lacking, it is obvious that much work is yet to be done to complete our knowledge of PTMs.
Table 2.
The table to show the number of protein records by organism available for our work
| Organism | PTM records (Mt) | PTM records (non-Mt) | Total protein records (Mt and non-Mt) | Total NCBI articles |
|---|---|---|---|---|
| A. thaliana | 116 | 3809 | 13 943 | 260 |
| A. nidulans | 4 | 283 | 914 | 21 |
| C. elegans | 16 | 711 | 3537 | 339 |
| C. familiaris | 24 | 613 | 812 | 2 |
| D. rerio | 22 | 611 | 2945 | 8 |
| H. sapiens | 589 | 11 419 | 20 207 | 191 |
| M. musculus | 564 | 10 032 | 16 718 | 21 |
| O. cuniculus | 30 | 661 | 889 | 0 |
| R. norvegicus | 374 | 5115 | 7923 | 7 |
| S. cerevisiae | 212 | 3005 | 7900 | 461 |
| X. laevis | 22 | 692 | 3394 | 67 |
The second and third columns display the number of Mt and non-Mt UniProt protein records, respectively. The forth column describes the exhaustive number of protein records where PTMs are discussed in some of the articles. The fifth column provides an estimation of the number of scientific articles from the literature that may have been sources of PTM information for protein records. These data were furnished by text mining the NCBI body of literature.
In the fifth column, we illustrate an estimation for the number of scientific articles available from the National Center for Biotechnology Information (NCBI), where PTM information may be extracted to populate protein records with PTM information (likely by UniProt and others). To estimate the number of NCBI articles, we applied the text mining analysis implemented in [21, 22], which served to locate all article abstracts containing relevant keywords: protein names, PTM types and other words for syntax.
The organisms were diverse and represented a wide spectrum of biology [23]. We divided the protein data between Mt and non-Mt sets. Unlike the Mt genome, which may be highly conserved across biology, the non-Mt protein is generally more diverse and may be more revealing of natural bias from organism to organism. Proceeding protein by protein for each organism, we determined the types of PTMs, the count of each and their associated RS type. We note that although there are many different kinds of PTMs in nature, we restricted our study only to those PTMs that have been observed to interact with single amino acids (i.e. a length-1 motif) along the protein sequence (a single RS). Figure 2 describes the procedure for capturing the data, which we then used to calculate frequencies (explained in the section on ‘Computing Frequencies’). We note that the data used to calculate these frequencies may have had incomplete references because of the general difficulties of extracting PTM information from physical protein samples in a wet lab. In light of such a limitation, however, we believe that the design of this study is still worthy of providing detailed patterns of PTM bias across the organismal data. Furthermore, as more data become available, our method may again be applied to discover new patterns.
Figure 2.

All Mt and non-Mt proteins were examined in each organism of our study. We recorded the protein type (Mt or non-Mt), the PTMs of the protein and their associated RSs. This information was used to assemble relative frequency data.
Computing frequencies
Because of the common hardships of applying limited computing resources to processing voluminous quantities of data, a statistical analysis is often appropriate [24, 25]. Furthermore, frequency analysis is especially well suited for comparing large data sets and discovery, as it embraces convenient techniques of network analysis to ascertain natural patterns [26, 27]. Here, we discuss the collection of frequency information, which is later used to build networks to discover PTM and RS biases.
We used relative frequencies to determine all PTM occurrence magnitudes for elements that have been observed to interact within the Mt and non-Mt proteomes of an organism. We note that frequency distributions are collected in isolation for each proteome of each organism. This implies that the frequency distribution of any proteome may be compared with any other distribution. All records of proteins were downloaded from UniProt, which were parsed using an in-house program. Across all the organisms of Table 1, we made a tally of the number of PTMs and RSs that had been observed throughout the proteins of each proteome for each organism. Additionally, we also collected the occurrence magnitudes for each non-RS amino acid for the later comparison of RS distributions with ordinary amino acids in each proteome.
We note that PTMs have specific names, which generally imply information about their RSs. In our work, we generalized the PTM names into basic rubrics (i.e. N-acetylalanine, N-acetylaspartate, N-acetylatedlysine and N-acetylcysteine are all kinds of acetylation) because we were also collecting the associated information about RSs. Once all the proteins of an organism were parsed for their PTM, RS and amino-acid tallies, we applied these data to three equations to derive relative frequency information. Using Equation (1), we calculated the PTM frequencies. This equation determined the occurrence magnitude of each unique type of PTM by dividing the number of its counts into the combined number of all observed PTMs by proteome. For example, glycosylation generally appeared many times in a proteome, and our calculation combined all its observations by proteome to create one relative frequency value.
The information concerning PTM and RS interactions was recorded. Similar to how we calculated PTM frequencies, we used Equation (2) to calculate RS frequencies. The relative frequency of a particular RS type was found by dividing its tally into the combined number of all observed RSs by proteome. Visualized in Figure 3, a count of each PTM type was created for each organism, and the PTM frequencies were calculated from this information in each of the Mt and non-Mt protein data sets for each organism. We used this information to populate Table 1. We noted an apparent preference for individual PTMs across the organisms. For instance, although glycosylation and phosphoserine were popular PTMs for many organisms, they do not appear to always achieve the same first and second rankings in the organisms. In addition, we noticed that Caenorhabditis elegans was the only organism of our set that had a high frequency for lipidation and Aspergillus nidulans was the only organism to exhibit methylation.
Figure 3.

An example of how relative frequency information was extracted from protein data. For each organism, all Mt and non-Mt protein records were queried to ascertain their observed PTMs that have been curated by UniProt. The type and count of each PTM, including its associated RS, were recorded to calculate frequencies by Equations (1) and (2). Not shown, the occurrence magnitudes of all amino acids (non-RSs) were also obtained and applied to Equation (3) to determine the general amino-acid compositions of each proteome.
We now discuss the equations. Across each organism j, for a specific element i (i.e. PTM, RS or amino acid), the relative frequency of a particular and its associated RS, , were calculated by Equations (1) and (2), respectively. We note the use of the function, which determines the number of occurrences of the element in the current data set. Across all PTMs of organism j, the relative frequency of a particular may be found by the following:
| (1) |
Across all reactive sites found associated with the PTMs of organism j, the frequency of a particular amino-acid RS, , may be found by the following equation.
| (2) |
The counts of each amino acid of each proteome were also tallied to determine relative frequencies for each organism, j. Akin to simply placing all the protein sequences of a proteome end-to-end to create one sequence, Seq, we determined the amino-acid composition and frequencies using Equation (3).
| (3) |
Building heatmaps and networks
Heatmaps: A heatmap is a color-coded matrix of numerical values, which have been clustered across the top and the side. We used heatmaps to determine the amino-acid compositions across proteomes for comparison with the PTMs biases in their proteomes. Heatmaps are useful in comparing different large sets of data together in terms of their frequency or other numerical information. Our general heatmaps of Figures 4 and 5 were created from the relative frequency data from Equations (1) and (2), respectively, and applied to the method described by [28]. Equation (3) was used to calculate the frequency of occurrence of each amino acid, regardless of also being an RS. These results are shown in Figure 6.
Figure 4.
Mt and non-Mt PTM compositions prepared using Equation (3). High magnitudes of frequency are described by lighter colors. We note that phosphorylation and acetylation were common PTMs across the organisms. We note that all frequency values > 0.18 (threshold) are included here.
Figure 5.
Mt and non-Mt RS compositions prepared using Equation (2). High magnitudes of frequency are described by lighter colors. Unlike the non-Mt heatmap, where nearly all amino acids played a roles as RSs, there were many amino acidss in the Mt proteomes that were never involved with the PTMs.
Figure 6.
Mt and non-Mt amino-acid compositions prepared using Equation (3). High magnitudes of frequency are described by lighter colors. Although all organisms display a common theme of color bands, indicating that their amino-acid composition is similar, we note that related organisms have especially similar patterns of color, indicating that the amino-acid distributions are similar.
Because PTMs (i.e. phosphorylation and others, for example) may interact with several different RSs simultaneously [29], we determined that the details of their relationships would be obvious when described in networks where individual interactions between PTMs and RSs may be explored in detail.
Networks: Our networks were built from relative frequency data, by applying Equations (1) and (2) using [30]. In the networks of each proteome, we determine the frequency magnitude by the size of the node: larger nodes describe more common occurrences. The left and right sides of the networks represent the PTM and RS populations (respectively), which were found in a proteome. The edges between the PTMs and RSs were calculated by the product of the PTM and RS frequencies. Because an interaction is not mutually exclusive, this calculation describes the interaction magnitude between the pair. Here, we note that the heavier edge weights describe more common interactions. The networks are read from the left-side PTMs, which interact with the RSs on the right side. We summarize the main results from the networks in [18].
Results and discussion
Heatmaps
We note that a single PTM observed in an isolated protein in the proteome may provide misleading information about its relevance to the proteome. We, therefore, apply a threshold to the frequency value to be able to distinguish the higher frequencies from the lower ones. In the Mt set, the range of PTM frequencies was 0.0003–0.21, and in non-Mt, the range was 0.0004–0.5, and we therefore defined the threshold to be 0.18, or the average of the midpoints of both ranges.
In Figure 4, we display the Mt and non-Mt heatmaps. Here, the counts of PTMs were eight and three for Mt and non-Mt, respectively. It is interesting to note that in non-Mt, the three PTMs, glycosylation, phosphorylation and methylation, are some of the more adaptive PTMs that are able to modify many different types of proteins [31] and have been observed to commonly interact together. We note from these heatmaps that related organisms generally appeared to have similar types and frequencies of PTMs. For example, the mammals of our data, Rattus norvegicus (rat), Mus musculus (mouse), Oryctolagus cuniculus (rabbit) and Homo sapiens (human) are closely clustered according to their PTM frequencies. In non-Mt, all mammals, including Canis familiaris (dog), were clustered together with the inclusion of Saccharomyces cerevisiae (yeast). Although Mt are highly conserved across organisms, we find that there is enough difference between PTM populations in the data to suggest that sequence similarity may not play much of a role. Extending this idea to the non-Mt protein data, we suggest that the clustering of mammal data in Figure 4 could be because of environmental conditions.
Mt is highly conserved across organisms, and so we may expect to see less diversity in PTMs in this set; however, we found that of the eight PTMs (shown in Figure 4), only glycosylation and phosphorylation were also common to the non-Mt set. The other PTMs may be involved in Mt-specific activities such as lipoyl for metabolism [32] and acetylation, which has been known to target large macromolecular complexes involved in diverse cellular processes for regulation [33].
In Figure 5, we note the associated amino acids that play roles as RSs in Mt and non-Mt. No frequency threshold was necessary because nearly all amino acids had strong frequencies (note the lighter colors). We observed that there was much more variety in selection for RSs in the non-Mt set than the Mt set. This signifies that there is more promiscuity in terms of PTMs interacting with diverse RSs in non-Mt and suggests that amino acids may have few restrictions in terms of their roles with PTMs. The PTMs of Mt appeared to interact with a specific type of RS. We will return to this observation in the networks, where we will see that PTMs generally interact with multiple types of RS in the non-Mt set.
In Figure 6, we note the compositions of all amino acids across the organismal proteomes. We note that the prominent RSs of Figure 5 are not necessarily the prominent amino acids of the same organisms. For instance, by the heatmap in the H. sapiens Mt proteome, the glycine (G), alanine (A), serine (S) and lysine (K) were prominent amino acids as RSs; however, they are not so as amino acids. The highest frequency magnitude in this organism was leucine (L), which was not found to be an RS. There are other similar observations to make from these heatmaps.
Domains and PTMs
At the heart of protein function are domains: the conserved parts of protein functional structures, which can evolve and exist independently of the rest of the protein chain. The functions of domain structures are thought to be context dependent and directed by PTM activity (i.e. phosphorylation) [34]. The difference between the prominences of the RS and amino-acid frequencies supports evidence that the location of an RS (perhaps, found near protein domains) may be more important than its basic biophysical properties. For example, in human, phosphorylation of p53 occurs at 13 serine and 5 threonine amino acids, which are distributed in the protein’s (functional) domain regions [35].
Protein conformations by PTMs create changes in behavior. For example, the DNA-binding domain of p53 is heavily influenced by changes in conformation from ubiquitination [35]. In [36] and [37], phosphorylation has been observed to disrupt FoxOs interaction with 14‐3‐3 proteins (likely at ww-domains [38]) to allow nuclear translocation of FoxO [39] and initiate programmed cell death (apoptosis).
PTMs that influence domains have been studied by the context of heart failure and arrhythmia as a result of functional defects in cardiac type 2 ryanodine receptors on the internal sarcoplasmic reticulum (SR). Specifically, the disease of this contractile protein (muscular) machinery has been attributed to regulation failure of the Calcium (Ca2+) release channels in the SR. Shao et al. discovered that carbonylation (a PTM) may be responsible for the (Ca2+) dysfunction, which was observed to disable two main lysine amino-acid sites (at positions 2190 and 2887), flanking the RyR2 (ryanodine receptor: a Ca2+ release channel) sub-domain site [40]. By disabling these lysine sites, the N-terminal and central protein domains of RyR2 (near two sub-domains at positions 2000–2500 and 2234–2750) were observed to be destabilized and unable to properly regulate Ca2+ for normal muscle function.
In a related study, [41], the activity of SERCA2a (a protein that undergoes a series of timed conformational changes to hydrolyze adenosine triphosphate and transport Ca2+ [42]) was studied in heart tissue. Here, the authors found that Ca2+ transport (regulated by the sarcoplasmic reticulum Ca2+ ATPase gene, SERCA2a) may be reduced or disabled when amino-acid sites are neutralized. For instance, four sites were studied, which reside in the protein’s domains; A-domain: {R164}, N- domain: {K476, K481} and P-domain: {R636}. The study found that Ca2+ transport was reduced or prevented by the paired modification (carbonylation/charge neutralization by conversion to glycines) of {R164, K481}, {K476, R636} and {R164, R636} (carbonylation/charge neutralized by conversion to tyrosines) to suggest that these amino acids behave as functional switches. In addition, the carbonylation/charge neutralization of {R164, K476, K481, R636} by conversion to tryptophan (which also increased the hydrophobic bulk of the sites) reduced the ability of SERCA2a to transport Ca2+. The above two findings support the notion that amino acids, making-up RSs residing near or inside protein domains, may be modified by PTMs to regulate the functions of domains. In our next section, we use networks to help us visualize some of these kinds of interactions.
Networks
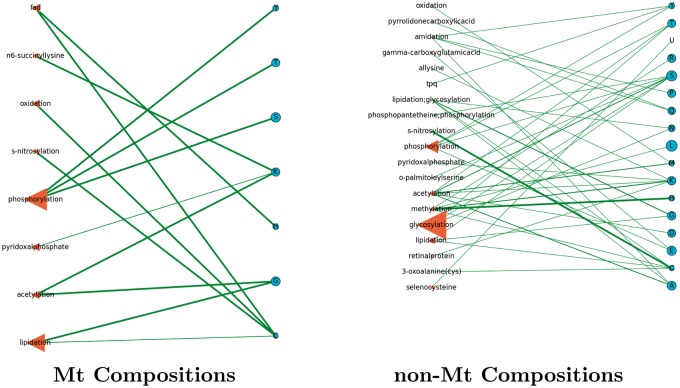
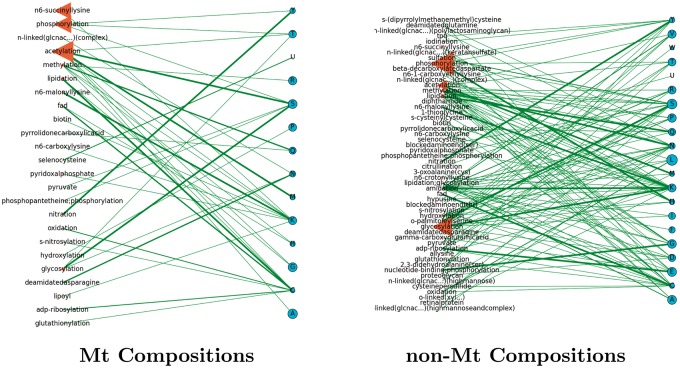
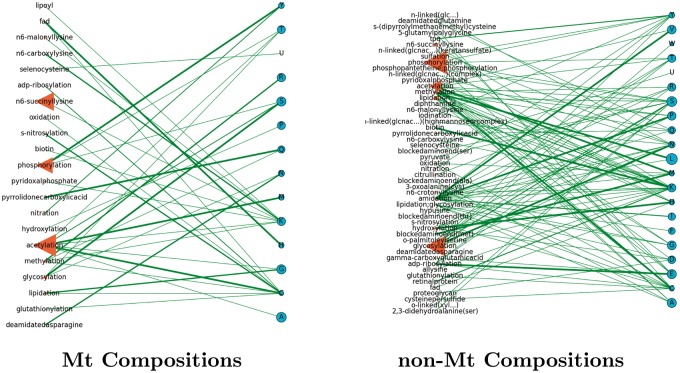
Network models help analyze the complex relationships between the interacting entities of our study. The models of Figures 7–17 (inclusive) describe the frequencies of PTM and RS occurrences, and the frequencies of their interactions. In Figure 18, we summarize the results by showing the number of PTMs and RSs per organism in an effort to describe their increase by organismal complexity. We show two networks for each organismal proteome, each of which has two types of nodes: PTMs (left) that interact with the RSs on the right. In these networks, we may determine the number of different interactions that a particular element may have by studying the degree (or number of connections) stemming from its node. In addition, the weight of the edge describes the relative frequency of the interaction between two elements.
Figure 7.
Arabidopsis thaliana: a network of PTM frequencies in Mt (left) and non-Mt (right) protein. We display the prominent PTMs across all organisms of our study having a frequency of at least 0.1.
Figure 8.
Aspergillus nidulans: a network of PTM frequencies in Mt (left) and non-Mt (right) protein. We display the prominent PTMs across all organisms of our study having a frequency of at least 0.1.
Figure 9.
Caenorhabditis elegans: a network of PTM frequencies in Mt (left) and non-Mt (right) protein. We display the prominent PTMs across all organisms of our study having a frequency of at least 0.1.
Figure 10.
Canis familiaris: a network of PTM frequencies in Mt (left) and non-Mt (right) protein. We display the prominent PTMs across all organisms of our study having a frequency of at least 0.1.
Figure 11.
Danio rerio: a network of PTM frequencies in Mt (left) and non-Mt (right) protein. We display the prominent PTMs across all organisms of our study having a frequency of at least 0.1.
Figure 12.
Homo sapiens: a network of PTM frequencies in Mt (left) and non-Mt (right) protein. We display the prominent PTMs across all organisms of our study having a frequency of at least 0.1.
Figure 13.
Mus musculus: a network of PTM frequencies in Mt (left) and non-Mt (right) protein. We display the prominent PTMs across all organisms of our study having a frequency of at least 0.1.
Figure 14.
Oryctolagus cuniculus: a network of PTM frequencies in Mt (left) and non-Mt (right) protein. We display the prominent PTMs across all organisms of our study having a frequency of at least 0.1.
Figure 15.
Rattus norvegicus: a network of PTM frequencies in Mt (left) and non-Mt (right) protein. We display the prominent PTMs across all organisms of our study having a frequency of at least 0.1.
Figure 16.
Saccharomyces cerevisiae: a network of PTM frequencies in Mt (left) and non-Mt (right) protein. We display the prominent PTMs across all organisms of our study having a frequency of at least 0.1.
Figure 17.
Xenopus laevis: a network of PTM frequencies in Mt (left) and non-Mt (right) protein. We display the prominent PTMs across all organisms of our study having a frequency of at least 0.1.
Figure 18.
We summarize the number of PTMs and RSs in the organismal networks. Here, we note that the higher organisms appear to have more PTMs and RSs. We note that the increasing numbers of PTMs and RSs are similar trends across the Mt and non-Mt data.
Mt and non-Mt networks
Although there were often cases of common PTMs observed between Mt proteomes (i.e. phosphorylation and acetylation), the Mt networks themselves presented a decided difference between kinds and types of PTM usages. In terms of PTM and RS connections from organism to organism, we found low to high levels of interactions between these nodes. For instance, in Figure 8 (A. nidulans) (fungi), the Mt proteome had only two Mt PTMs (methylation and pyridoxalphosphate), which interacted at the same lysine site. This was a sharp contrast to the Mt proteome of H. sapiens (Figure 12), where over 20 Mt PTMs were observed interacting with a host of different types of RSs and 7 (of this set) were observed to interact only with lysine. In Figure 9, C. elegans (worm) has six Mt PTMs interacting with a variety of RSs, of which four interacted with lysine. In Figure 7, Arabidopsis thaliana (mustard plant) had 14 Mt PTMs, of which five interacted with lysine. The networks describe many other similar features in terms of PTMs and RSs to show that the use of PTMs in the above fungi, worm and human proteomes are different.
For each organism, the rule of always having fewer Mt PTMs when compared with non-Mt data had few exceptions. Such an exception may be noted between the comparison of M. musculus (mouse) and R. norvegicus (rat) of Figures 13 and 15, respectively, which showed that mouse had more Mt PTMs but less non-Mt PTMs than Rat.
From their enlarged node sizes throughout nearly all Mt networks, acetylation and phosphorylation were prominent PTMs (often including glycosylation) that tended to interact with a few specific RSs. In the non-Mt networks, glycosylation and phosphorylation were prominent PTMs that tended to interact with a diverse set of RSs. We refer the reader once again to Figures 4 and 5 (heatmaps of PTM and RS compositions, respectively) to note the prominence of the above PTMs. We direct the reader to Figure 18 to show that higher organisms tended to use more PTMs and associated RSs than the others.
In Table 3, the proteomes have been ranked according the number of observed PTM types. We note that the majority of the proteomes have a similar ranking (see the non-gray cells). We also note that the organisms appear to become more complex as more PTMs are observed in the proteomes. For example, in Figure 9, C. elegans has fewer PTMs than the mammals, such as C. familiaris and H. sapiens of Figures 10 and 12, respectively.
Table 3.
A ranking of proteomes in terms of number of PTMs observed (Mt and non-Mt)
| Mt Rank | Org | Non-Mt Rank | Org |
|---|---|---|---|
| 2 | A. nidulans | 15 | A. nidulans |
| 4 | X. laevis | 22 | D. rerio |
| 6 | C. elegans | 27 | C. elegans |
| 8 | C. familiaris | 34 | S. cerevisiae |
| 9 | D. rerio | 37 | X. laevis |
| 11 | O. cuniculus | 42 | C. familiaris |
| 15 | A. thaliana | 46 | A. thaliana |
| 16 | S. cerevisiae | 49 | O. cuniculus |
| 31 | R. norvegicus | 114 | R. norvegicus |
| 33 | M. musculus | 131 | M. musculus |
| 34 | H. sapiens | 157 | H. sapiens |
The gray fields indicate that the ranking is not the same for both the Mt and non-Mt sets. The majority of proteomes have the same ranking in both sets.
We remark here that inequality may be the result of the types of environmental stresses that each organism may encounter in its environment. Because PTMs enable proteins to adapt to stress, the number of PTMs in an organism’s proteome could be a measurement of the kinds of stresses to which the organism may respond. It may be that there are many habitats where humans and mammals may thrive but are lethal to A. nidulans, A. thaliana, C. elegans and Danio rerio, such as arid environments. The correlation between the number of individual PTMs and organismal complexity may help to explain why the environments of lower organisms appear to be specialized in terms of warmth, moisture, humidity and others, in addition to the availability of food sources. Furthermore, in Figure 7, we note that A. thaliana has more PTMs than C. elegans (Figure 16). This could be explained by the concept that plants and fungi cannot easily remove themselves from their hostile environments and must, therefore, be able to survive their stresses by applying their arsenal of PTMs.
Bias analysis using gene ontologies
Environmental stresses may force proteins to alter their conformation by PTMs during stress responses. Because each protein has a specific function, they are likely to react in diverse ways to stresses. Some proteins, such as those which are found in Mt, may also have evolved adaptations to easily cope with types of oxidative stress [2]. To determine the types of PTMs that are involved with particular stresses, we studied the H. sapiens Mt and non-Mt proteins for their functions, as a function of their PTM interactions. We chose acetylation and phosphorylation because they were commonly encountered PTMs and were likely to be found in many different proteins. We made a list of all Mt and non-Mt proteins that were found to interact with acetylation (at least once) and another list of Mt and non-Mt proteins interacting only with phosphorylation (at least once). Using the method by [43], we extracted a list of functions from each protein on the acetylation and phosphorylation lists and applied them to Venn diagrams, found in the Supplementary Data: Supplementary Figures S1 and S2, and in Supplementary Tables S1–S16.
We determined that PTMs interacted with proteins that had some function in stress response. According to the diagrams, in Mt proteins, both acetylation and phosphorylation appeared to interact with proteins handling oxidative stress. Acetylation alone was specific to oxidative stress responses, whereas, phosphorylation alone was specific to general cellular responses to oxidative stresses. This is logical to have these PTMs in Mt proteins because these organelles produce cellular energy by oxidative processes.
In the non-Mt proteins, we noted that acetylation was involved uniquely with proteins that regulated cellular responses to stress, signaled apoptosis as a response to oxidative stress and were involved with the cellular response to heat. On the other hand, phosphorylation was typically involved with proteins that function to regulate cellular processes and control kinase signaling pathways, in addition to some general cellular responses to stresses. In the Supplementary Data, for our protein data, we provide information on their general functions.
Poisson approximation by the Chen–Stein method
Here, we discuss the increasing complexity of the networks of each organism of Figures 7–17. The complexity of a particular network increases according to the number of connections that exist between the PTMs and the RSs. Following a statistical approach similar to [44, 45], we provide P-values to support the notion that higher organisms tended to have more complex networks (i.e. having more interactions between their PTMs and RS nodes).
We now describe the test. Because more complex networks are characterized by having more PTMs, RSs and the connecting edges between them (PTM–RS pairs), we used the unique counts of these elements to compare the complexities of networks across the organisms by an, ‘all-against-all’ test. The data we collected are the following: counts of RSs (Mt and non-Mt), counts of PTMs (Mt and non-Mt) and counts of PTM–RS pairs (Mt and non-Mt). All these raw data are available in the Supplementary Data.
We discuss the test for the connecting edges between the PTM–RS pairs. For each of the sets of data, and that of Acanthamoeba castellanii for comparison, one count total for each of the 12 organisms was given. Let denote these 12 counts. We assume each xi is the sum of independent Bernoulli (binary) random variables. The numbers of such random variables equals the number of the (PTM, RS) combinations, which we will denote N.
We calculate each xi by the following. Where, , and , we used the following equation.
| (4) |
It is well known that if N is large, we may approximate the distribution of xi by either the Poisson distribution or a normal distribution. We also assume that independence of . For each pair of organisms (I and J), and provided that not both xi and xj equal zero, we compute an absolute Z-value where,
| (5) |
Here, we used the consistent approximation that the variance of xi is (or is close to) xi. In fact, xi is an estimated upper bound on the variance of xi because this variance is less than the mean of this variable. We therefore note that the value is a ‘conservative’ test statistic.
We have also used a Bonferroni inequality adjustment for simultaneous comparison of all 66 pairs of organisms, which makes these statistical tests even more conservative. The Poisson approximation element of the test can be used even in some cases of dependence. The conservative nature of the tests should allow for more than just slight dependence. Organisms I and J were considered different in terms of their complexity if the two-sided P-value was < , where α is the level of significance. For our purposes, α = 0.05 was sufficient, and the P-value for a single test for a particular pair I, J is given by the following:
| (6) |
We noted that a majority of the P-values were < , and we therefore concluded that most pairs of organisms differed in complexity. In Table 4, for the (PTM, RS) connections of the Mt organismal proteins, nearly all the P-values were significant to describe major differences in network complexities. Here, these tables are read starting from an organism in the left column, which is compared with those in its row. A significant value supports the notion that there are more edges in the network of the former organism than the latter. Only three tests were not significant (i.e A. nidulans by O. cuniculus, C. elegans by Xenopus laevis and H. sapiens by M. musculus). In these three tests, it was found that the latter of the pair of organisms presented a more complex network than the former according to the (PTM, RS) edges. In Table 5, we note the results for the non-Mt data. Only two tests (A. nidulans by S. cerevisiae and D. rerio by O. cuniculus) were not significant to support a departure of complexities between networks. The full results for the general PTM and RS complexity comparisons using this same statistical test are offered in the Supplementary Data.
Table 4.
The P-values from our Poisson approximation by the Chen–Stein method over (PTM, RS) pairs in Mt networks. Along the top and sides are the abbreviated names of the organisms. Significant P-values (i.e. values < ) are denoted by stars (*) to suggest that these pairs of organisms differ in complexity according to their networks.
| Ac | An | At | Ce | Cf | Dr | Hs | Mm | Oc | Rn | Sc | Xl | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ac | * | none | * | * | * | * | * | * | * | * | * | |
| An | * | * | * | * | * | * | 0.147 | * | * | * | ||
| At | * | * | * | * | * | * | * | * | * | |||
| Ce | * | * | * | * | * | * | * | 0.155 | ||||
| Cf | * | * | * | * | * | * | * | |||||
| Dr | * | * | * | * | * | * | ||||||
| Hs | 0.057 | * | * | * | * | |||||||
| Mm | * | * | * | * | ||||||||
| Oc | * | * | * | |||||||||
| Rn | * | * | ||||||||||
| Sc | * | |||||||||||
| Xl |
Table 5.
The P-values from our Poisson approximation by the Chen–Stein method over (PTM, RS) pairs in non-Mt networks. Along the top and sides are the abbreviated names of the organisms. Significant P-values (i.e. values < ) are denoted by stars (*) to suggest that these pairs of organisms differ in complexity according to their networks.
| Ac | An | At | Ce | Cf | Dr | Hs | Mm | Oc | Rn | Sc | Xl | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ac | * | * | * | * | * | * | * | * | * | * | * | |
| An | * | * | * | * | * | * | * | * | 0.40 | * | ||
| At | * | * | * | * | * | * | * | * | * | |||
| Ce | * | * | * | * | * | * | * | * | ||||
| Cf | * | * | * | * | * | * | * | |||||
| Dr | * | * | 0.002 | * | * | * | ||||||
| Hs | * | * | * | * | * | |||||||
| Mm | * | * | * | * | ||||||||
| Oc | * | * | * | |||||||||
| Rn | * | * | ||||||||||
| Sc | * | |||||||||||
| Xl |
Protein isoforms in organsims
Because much evolutionary time separates the complex organisms from the lower ones, conserved yet divergent isoform proteins are likely to exist. These isoforms may have originated from paralogous and alternatively spliced messenger RNA to create alternative gene products and functions from single coding sequences. It is known that alternative splicing is likely to encourage transcriptome diversity [46]. For instance, in [47], it was discussed that alternative splicing may have led to the larger divergence noted between the higher and lower organisms. In our own study, we also noted a wider divergence in PTM and RS usages between the higher and lower organisms.
During this evolutionary time, there is more opportunity for the generation of isoforms, which may use PTMs in diverse ways. For instance, in Figure 19, we note that the mammals, notably H. sapiens, M. musculus, R. norvegicus, have more isoform proteins than the other organisms according to UniProt. To gather these results, we searched for all proteins corresponding to each organism, and then we counted the number of isoforms. In addition to their abilities to respond to stresses, increasing PTM populations by organismal complexity may also be explained by their involvements with specific isoform-specific functionalities, such as RAS protein isoforms (monomeric GTPases acting as binary molecular switches for cellular regulation) [48] and 14‐3‐3 protein isoforms [49]. Also in Figure 19, we note that A. thaliana had a large number of isoforms (compared with the others), which may help to explain its complicated networks shown in Figure 7.
Figure 19.
The number of isoforms of the organisms in our study. These counts were prepared by querying all organismal proteins in UniProt and then determining how many isoform proteins were present. The increasing number of isoforms may help to explain the increasing number of PTMs in higher organisms. Note that A. nidulans has been omitted because of the lack of isoform information.
Notable PTMs
The most frequently occurring PTM in our network models was phosphoserine, among both the Mt and the non-Mt proteins. This particular PTM represents the phosphorylation of serine base in a protein’s amino-acid sequence and is one of the most common modifications to proteins that can alter functionality. Among other sites, such as threonine, tyrosine and histidine residues, serine is the most common type of phosphorylation. Serine phosphorylation, like other phosphorylations, can cause structural changes in proteins to activate or deactivate them.
Glycosylation is the result of a carbohydrate molecule that is added to a hydroxyl group, or another functional group of another molecule, (a glycosyl acceptor) in the protein. The majority of the proteins synthesized in the rough endoplasmic reticulum undergo glycosylation. Although we found traces of glycosylation in the Mt proteomes of our data, we generally found more glycosylation in the non-Mt proteomes. In plants, however, Mt have the function of photo respiration requiring glycosylation. Although there may be other more pertinent reasons, the rich glycosylation observed in the Mt proteome of A. thaliana (of Figure 7) may be because of performing aerobic respiration. Interestingly, no glycosylation was observed in the Mt proteome of C. elegans of Figure 9.
Acetyllysine is another important PTM that adds an acetyl group to a lysine residue in proteins. The acetylation of lysine (K) residue is considered as a regulating mechanism for various epigenetic factors [50, 51]. We observed higher amount of N6-acetyllsine in Mt proteins and was conserved across H. sapiens, M. musculus, O. cuniculus and R. norvegicus.
Conclusions
A bias is a preferential treatment of some element. Despite the present limitation of available PTM data across different species, our goal was to describe biases of PTM usage inherent to organisms using the most current available data. In this article, we used basic frequency information to produce evidence of the bias in the usage of PTMs, RSs and amino acids. We applied these frequency data (PTMs and RSs) to create heatmaps and networks, which gave clear details about the differences between organismal proteomes. From the heatmaps and the networks, we noted that PTMs and RSs have different compositions between proteomes. We observed that the non-Mt networks were generally more dense and more populated by PTMs and RSs than the Mt network of the same organism.
We noted that an organism’s PTM and RS bias is not likely explained by its amino-acid composition (from Figure 6) because the compositions were too similar between all organisms. Instead, this bias must come from another source, which we suggested was related to environmental stress response. Because PTMs enable stress response in protein, our study supports the notion that the environmental stresses of its habitat may likely play a role in an organism’s PTM and RS bias. This finding is strengthened by the discussion of the high number of PTMs and RSs that were observed in the networks of A. thaliana and A. nidulans (Figures 7 and 8, respectively). We note that the survival of these, and other plant organisms, may be based on their ability to tolerate their environmental stresses. Furthermore, we noted that the organismal complexity increased in tandem with the number of observed PTMs in both the Mt and non-Mt proteomes. This, we speculated, may be due in part to the ability of the more complicated organisms to inhabit regions that host a wider variety of environmental stresses than those habitats of the less complicated organisms, with the exception of plants.
Our study showed that PTMs such as acetylation and phosphorylation were common to the Mt proteomes, and glycosylation and phosphorylation were prominent across the non-Mt proteomes. Although many of the PTMs were common throughout our organismal data shown in Table 1, we noted that the individual organisms tended to interact with RSs in different ways. For instance, in all networks (Mt and non-Mt), the PTMs themselves did not interact consistently with the same RSs, across the organisms. In non-Mt proteomes, we observed that PTMs were more promiscuous in their interactions with RSs, and often a particular PTM would interact with several different RSs simultaneously. This was generally not the case in the Mt networks. Here, these PTMs tended to interact with the same RSs across the organisms. This finding may be partially explained by the conserved nature of Mt but says nothing about the environmental stresses which the organelles may have to tolerate from the organism’s habitat. Importantly, the differences in the frequencies of PTM and RS usage across the data may suggest a unique organismal mechanism, which supports our contention that environmental stress is likely the motivator of bias.
In future work, we intend to investigate the influence of stress on PTMs. In particular, in protein stress response systems, we intend to study the relationships between stresses and the PTMs of proteins that are related to specific functional groups across these and other organismal data.
Key Points
We develop a statistical tool to extract relative frequencies of PTM and RS compositions from the protein (Mt and non-MT) of diverse organisms.
Despite the highly related protein material between organisms, we use interaction networks and heatmaps to show that a clear bias exists between their PTMs and RSs.
We illustrate that the Mt and non-Mt protein networks become more complicated (more PTMs and RSs connections) as the organisms become more complex.
We discuss that environmental stress may have contributed to the PTM and RS bias.
Supplementary Material
Acknowledgment
We would like to thank the support staff in the UNO—Bioinformatics Core Facility. We would also like to thank Janyl Jumadinova for her help in proofing this manuscript.
Biographies
Oliver Bonham-Carter is a PhD candidate in bioinformatics at the College of Information Science and Technology at the University of Nebraska at Omaha.
Ishwor Thapa is a software applications developer in bioinformatics at the College of Information Science and Technology at the University of Nebraska at Omaha.
Steven From is a professor of statistics in the Department of Mathematics at the University of Nebraska at Omaha.
Dhundy Bastola is an associate professor in bioinformatics at the College of Information Science and Technology at the University of Nebraska at Omaha.
Oliver Bonham-Carter is a PhD candidate in bioinformatics at the College of Information Science and Technology at the University of Nebraska at Omaha.
Ishwor Thapa is a software applications developer in bioinformatics at the College of Information Science and Technology at the University of Nebraska at Omaha.
Steven From is a professor of statistics in the Department of Mathematics at the University of Nebraska at Omaha.
Dhundy Bastola is an associate professor in bioinformatics at the College of Information Science and Technology at the University of Nebraska at Omaha.
Funding
This work was funded by the grants from the NASA Nebraska Space Grant (2014–2015), the National Center for Research Resources (5P20RR016469) and the National Institute for General Medical Science (NIGMS), 8P20GM103427.
Supplementary data
Supplementary data are available online at http://bib.oxfordjournals.org/.
References
- 1.Khoury GA, Baliban RC, Floudas CA. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep 2011;1:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonham-Carter O, Pedersen J, Najjar L, et al. Modeling the effects of microgravity on oxidation in mitochondria: a protein damage assessment across a diverse set of life forms. Comput Biol Med 2014;53:179–89. [DOI] [PubMed] [Google Scholar]
- 3.Bonham-Carter O, Pedersen J, Najjar L, et al. Modeling the effects of microgravity on oxidation in mitochondria: a protein damage assessment across a diverse set of life forms. In: Data Mining Workshops (ICDMW), 2013 IEEE 13th International Conference on IEEE, 2013, pp. 250–7. [Google Scholar]
- 4.Peng L, Yuan Z, Li Y, et al. Ubiquitinated sirtuin 1 (sirt1) function is modulated during dna damage-induced cell death and survival. J Biol Chem 2015;290(14):8904–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 2015;16(1):18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levav-Cohen Y, Goldberg Z, Tan KH, et al. The p53-mdm2 loop: a critical juncture of stress response. In: Mutant p53 and MDM2 in Cancer. Volume 85 of the series Subcellular Biochemistry. Springer, Netherlands, 2014, pp. 161–86. [DOI] [PubMed] [Google Scholar]
- 7.Peuget S, Bonacci T, Soubeyran P, et al. Oxidative stress-induced p53 activity is enhanced by a redox-sensitive tp53inp1 sumoylation. Cell Death Differ 2014;21(7):1107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sommers JA, Suhasini AN, Brosh RM. Protein degradation pathways regulate the functions of helicases in the DNA damage response and maintenance of genomic stability. Biomolecules 2015;5(2):590–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sőti C, Csermely P. Protein stress and stress proteins: implications in aging and disease. J Biosci 2007;32(3):511–5. [DOI] [PubMed] [Google Scholar]
- 10.Belavy´ DL, Miokovic T, Armbrecht G, et al. Differential atrophy of the lower-limb musculature during prolonged bed-rest. Eur J Appl Physiol 2009;107(4):489–99. [DOI] [PubMed] [Google Scholar]
- 11.Guerra D, Crosatti C, Khoshro H, et al. Post-transcriptional and post-translational regulations of drought and heat response in plants: a spider’s web of mechanisms. Front Plant Sci 2015;6:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penfield S. Temperature perception and signal transduction in plants. New Phytol 2008;179(3):615–28. [DOI] [PubMed] [Google Scholar]
- 13.Späth GF, Drini S, Rachidi N. A touch of zen: post-translational regulation of the leishmania stress response. Cell Microbiol 2015;17(5):632–8. [DOI] [PubMed] [Google Scholar]
- 14.Shah SP, Lonial S, Boise LH. When cancer fights back: multiple myeloma, proteasome inhibition, and the heat shock response. Mol Cancer Res 2015;13(8):1163–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol 2015;25(6):354–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beltrao P, Bork P, Krogan NJ, et al. Evolution and functional cross-talk of protein post-translational modifications. Mol Syst Biol 2013;9(1):714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apweiler R, Bairoch A, Wu CH, et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res 2004;32(suppl 1): D115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonham-Carter O, Thapa I, Bastola D. Evidence of post translational modification bias extracted from the trna and corresponding amino acid interplay across a set of diverse organisms. In Proceedings of the 5th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics, Newport Beach, CA, USA. ACM, 2014, pp. 774–81. [Google Scholar]
- 19.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res 1999;27(8):1767–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavrov DV. Key transitions in animal evolution: a mitochondrial dna perspective. Integr Comp Biol 2007;47(5):734–43. [DOI] [PubMed] [Google Scholar]
- 21.Bonham-Carter O, Bastola DR. A text mining application for linking functionally stressed-proteins to their post-translational modifications. In: Bioinformatics and Biomedicine (BIBM), Washington DC, USA, 2015 IEEE International Conference on. IEEE, 2015, pp. 611–4. [Google Scholar]
- 22.Camara M, Bonham-Carter O, Jumadinova J. A multi-agent system with reinforcement learning agents for biomedical text mining. In: Proceedings of the International Conference on Bioinformatics, Computational Biology and Biomedical Informatics ACM, 2015. [Google Scholar]
- 23.Wheeler DL, Barrett T, Benson DA, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res 2007;35(suppl 1):D5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonham-Carter O, Ali H, Bastola D. A meta-genome sequencing and assembly preprocessing algorithm inspired by restriction site base composition. In: Bioinformatics and Biomedicine Workshops (BIBMW) Philadelphia, USA, 2012 IEEE International Conference on. IEEE, 2012, pp. 696–703. [Google Scholar]
- 25.Bonham-Carter O, Ali H, Bastola D. A base composition analysis of natural patterns for the preprocessing of metagenome sequences. BMC Bioinformatics 2013;14(Suppl 11):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonham-Carter O, Steele J, Bastola D. Alignment-free genetic sequence comparisons: a review of recent approaches by word analysis. Brief Bioinform 2013;15:890–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vinga S, Almeida J. Alignment-free sequence comparison—a review. Bioinformatics 2003;19(4):513–23. [DOI] [PubMed] [Google Scholar]
- 28.Kolde R. pheatmap: pretty heatmaps. r package version 0.7. 7. 2012. [Google Scholar]
- 29.Zand R, Li MX, Jin X, et al. Determination of the sites of posttranslational modifications in the charge isomers of bovine myelin basic protein by capillary electrophoresis-mass spectroscopy. Biochemistry 1998;37(8):2441–9. [DOI] [PubMed] [Google Scholar]
- 30.Schult DA, Swart P. Exploring network structure, dynamics,and function using networkx. In: Proceedings of the 7th Python in Science Conferences, SciPy 2008, Caltech, Pasadena, CA, Vol 2008, 2008, pp. 11–6. [Google Scholar]
- 31.Nothaft H, Szymanski CM. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol 2010;8(11):765–78. [DOI] [PubMed] [Google Scholar]
- 32.Sankaranarayanan R, Dock-Bregeon A-C, Romby P, et al. The structure of threonyl-trna synthetase-trna thr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell 1999;97(3):371–81. [DOI] [PubMed] [Google Scholar]
- 33.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009;325(5942):834–40. [DOI] [PubMed] [Google Scholar]
- 34.Lu CT, Huang KY, Su MG, et al. Dbptm 3.0: an informative resource for investigating substrate site specificity and functional association of protein post-translational modifications. Nucleic Acids Res 2012;41:D295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu B, Zhu WG. Surf the post-translational modification network of p53 regulation. Int J Biol Sci 2012;8(5):672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brent MM, Anand R, Marmorstein R. Structural basis for dna recognition by foxo1 and its regulation by posttranslational modification. Structure 2008;16(9):1407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehtinen MK, Yuan Z, Boag PR, et al. A conserved mst-foxo signaling pathway mediates oxidative-stress responses and extends life span. Cell 2006;125(5):987–1001. [DOI] [PubMed] [Google Scholar]
- 38.Schumacher B, Skwarczynska M, Rose R, Ottmann C. Structure of a 14‐3‐3σ–yap phosphopeptide complex at 1.15 å resolution. Acta Crystallogr F Struct Biol Cryst Commun 2010; 66(9):978–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aitken A. Post-translational modification of 14-3-3 isoforms and regulation of cellular function. In: Seminars in Cell & Developmental Biology, Vol. 22 Issue 7 Elsevier, 2011, pp. 673–80. [DOI] [PubMed] [Google Scholar]
- 40.Shao CH, Tian C, Ouyang S, et al. Carbonylation induces heterogeneity in cardiac ryanodine receptor function in diabetes mellitus. Mol Pharmacol 2012;82(3):383–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao CH, Capek HL, Patel KP, et al. Carbonylation contributes to serca2a activity loss and diastolic dysfunction in a rat model of type 1 diabetes. Diabetes 2011;60(3):947–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature 2002;418(6898):605–11. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Zhou X, Zhu J, et al. Go-function: deriving biologically relevant functions from statistically significant functions. Brief Bioinform 2012;13(2):216–27. [DOI] [PubMed] [Google Scholar]
- 44.Arratia R, Goldstein L, Gordon L. Poisson approximation and the chen-stein method. Stat Sci 1990;5:403–24. [Google Scholar]
- 45.Presman E. Approximation in variation of the distribution of a sum of independent bernoulli variables with a poisson law. Theory Probab Appl 1986;30(2):417–22. [Google Scholar]
- 46.Boue S, Letunic I, Bork P. Alternative splicing and evolution. Bioessays. Wiley Periodicals, Inc: 2003;25(11):1031–4. [DOI] [PubMed] [Google Scholar]
- 47.Brett D, Pospisil H, Valcárcel J, et al. Alternative splicing and genome complexity. Nat Genet 2002;30(1):29–30. [DOI] [PubMed] [Google Scholar]
- 48.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of ras. Nat Rev Mol Cell Biol 2012;13(1):39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenquist M, Sehnke P, Ferl RJ, et al. Evolution of the 14‐3‐3 protein family: does the large number of isoforms in multicellular organisms reflect functional specificity?. J Mol Evol 2000;51(5):446–58. [DOI] [PubMed] [Google Scholar]
- 50.Boyes J, Byfield P, Nakatani Y, et al. Regulation of activity of the transcription factor gata-1 by acetylation. Nature 1998;396(6711):594–8. [DOI] [PubMed] [Google Scholar]
- 51.Hernandez-Hernandez A, Ray P, Litos G, et al. Acetylation and mapk phosphorylation cooperate to regulate the degradation of active gata-1. EMBO J 2006;25(14):3264–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.