Abstract
Objective
Levels of Toll-like receptor 7 (TLR-7) are elevated in rheumatoid arthritis (RA), but the impact on RA is unknown because the endogenous ligand for TLR-7 has not been identified. The aim of this study was to identify a TLR-7 endogenous ligand and to determine its role in the pathogenesis of RA.
Methods
The presence of an endogenous TLR-7 ligand, microRNA let-7b (miR-let-7b), was examined by real-time polymerase chain reaction (PCR) analysis. Using RA knockdown cells, TLR-7–knockout mice, or antagonist, the specificity of miR-let-7b as a potential ligand for TLR-7 was tested. The mechanism by which ligation of miR-let-7b to TLR-7 promotes disease was investigated in RA myeloid cells by real-time PCR, enzyme-linked immunosorbent assay, and fluorescence-activated cell sorting. We also established the effect of ectopic miR-let-7b expression on arthritic joint inflammation.
Results
We found that a TLR-7 endogenous ligand resides mainly in RA synovial fluid macrophages. The GU-rich domain in miR-let-7b was found to be essential for TLR-7 ligation, since miR-147, the positive control for GU, was able to stimulate TLR-7+ myeloid cells, whereas miR-124, the negative, non-GU, control, was not. We demonstrated that miR-let-7b or exosomes containing miR-let-7b could transform the RA and/or mouse naive or antiinflammatory macrophages into inflammatory M1 macrophages via TLR-7 ligation. Consistently, we showed that miR-let-7b provokes arthritis by remodeling naive myeloid cells into M1 macrophages via TLR-7 ligation, since joint swelling and M1 macrophages are absent in TLR-7−deficient mice.
Conclusion
The results of this study underscore the importance of miR-let-7b ligation to TLR-7 in the joint during the effector phase of RA.
Toll-like receptors (TLRs) are pattern-recognition receptors that can distinguish self from non-self and are classified into 2 groups based on their distribution and ligand selection (1–3). TLRs 1, 2, 4, 5, and 6 are expressed on the cell surface and recognize microbi-al components, whereas the endosomal TLRs 3, 7, 8, and 9 detect nucleic acids (4–6). Studies performed in patients with rheumatoid arthritis (RA) (7–10) and in mice with experimental arthritis (11–13) strongly support the contribution of TLR-2/4 in the disease pathogenesis; however, nothing is known about the endogenous ligands that bind to the endosomal TLRs and their impact on RA pathology.
Fewer resources have been devoted to studying the role of endosomal TLRs in RA pathogenesis, as their endogenous ligands remain unknown. However, because TLR-7 is greatly elevated in synovial tissue (ST) lining and sublining macrophages from RA patients as compared to normal subjects and because its expression closely correlates with the Disease Activity Score in 28 joints (DAS28) and with the tumor necrosis factor (TNF) concentration in myeloid cells (14), there is an unmet need to investigate the significance of TLR-7 and its endogenous ligand in the pathogenesis of RA.
We have shown that human TLRs 7 and 8 respond to RNA extracted from RA synovial fluid (SF) and the synthetic nucleoside analog imidazoquinoline (R848) (14). Interestingly, single-stranded RNA (ssRNA) containing GU-rich or poly(U) sequences can trigger both TLR-7 and TLR-8; however, TLR-7 preferentially recognizes GU-rich RNA sequences, while TLR-8 has greater binding affinity for AU-rich oligos (15,16). In contrast to the TLR-7 data, TLR-8 expression in RA myeloid cells did not correlate with the DAS28, the TNF level, or the TLR-7 level (14). We therefore sought to identify a specific TLR-7 binding microRNA (miRNA) in RA SF.
MicroRNAs are short noncoding RNAs that are 19–24 nucleotides in length. To date, as many as 17 miRNAs (miRNAs 16, 23b, 26a, 34, 124a, 132, 142-3p/5p, 146a, 150, 155, 203, 221/222, 223, 323-3p, 363, and 498) have been identified in RA SF, ST, serum, and peripheral blood mononuclear cells, of which miRNA-146a (miR-146a) and miR-155 are the most abundant species and have therefore been the focus of many studies (17–19). It has been shown that miR-146a and miR-155 are modulated by TLRs, TNF, and interleukin-1β (IL-1β) in a variety of cell types, including myeloid cells, B cells, T cells, and RA fibroblasts (17–19). Previous studies showed that miR-146a has an antiinflammatory effect and is responsible for lipopolysaccharide (LPS) tolerance (20), and it also strongly abrogates bone destruction in collagen-induced arthritis (CIA) (17,18, 21). In contrast, miR-155−/− mice in a K/BxN arthritis model were shown to have impaired bone erosion due to defects in RANKL-mediated osteoclastogenesis (22).
We found that SF ssRNA, which can bind to RA TLR-7+ myeloid cells, is a GUUGUGU-rich miR-let-7b that is predominantly packaged in exosomes of RA SF macrophages. Interestingly, induction of cell death in RA SF macrophages can further accentuate the discharge of the exosomal miR-let-7b into the SF. We found that local expression of miR-let-7b contributes to arthritic joint inflammation through a mechanism that is dependent on the transformation of naive myeloid cells into M1 macrophages. Consistent with this notion, we found that miR-let-7b–incorporated exosomes are fully functional and that fusion of exosomal miR-let-7b into M2 myeloid cells promotes M1 macrophage differentiation. Our results thus suggest that TLR-7 and its endogenous ligand miR-let-7b may be a promising target for RA therapy.
MATERIALS AND METHODS
Extraction of GU-rich miRNA from RA specimens
Studies were approved by the Institutional Review Board of the University of Illinois at Chicago, and all donors gave informed written consent. RA patients were diagnosed according to the American College of Rheumatology 1987 revised criteria (23).
RNA was extracted from 1 ml of RA SF, osteoarthritis (OA) SF, RA plasma, and normal plasma. The RNA (10 ng) was reverse transcribed, and levels of miRNA were determined by real-time reverse transcription–polymerase chain reaction (RT-PCR) using a microRNA assay for hsa-let-7b according to the manufacturer’s instructions (Life Technologies). The copy number of exosomal miR-let-7b was calculated via a standard curve using the synthetic miR-let-7b (Gen-Script) and plotting the Ct values against the copy number, as described previously (24).
Mononuclear cells were isolated by Histopaque gradient centrifugation. Monocytes and T cells were isolated from normal and RA peripheral blood (PB) and/or SF using a negative selection kit (StemCell Technologies) (25,26), and neutrophils were isolated from normal PB (27). The copy number of miR-let-7b and miR-29a (Life Technologies) in exosomes was determined in PB T cells, in vitro differentiated macrophages, and neutrophils, as well as in RA SF macrophages and RA ST fibroblasts.
Isolation of exosomes from RA SF and RA ST
RA SF macrophages (4 × 106) or RA ST fibroblasts (0.33 gm) were cultured for 24 hours in 1 ml of RPMI 1640. Samples were then left untreated or were treated for 3 hours with 1 µM staurosporine to induce apoptosis, and then subjected to 3 freeze–thaw cycles to provoke necrosis. The medium was then collected, and exosomes were extracted using a total exosome isolation kit (Life Technologies).
RA myeloid cell treatment
To examine whether TLR-7 knockdown could impair miR-let-7b activation, differentiated macrophages from RA PB were transfected with 100 nM control or TLR-7 small interfering RNA (siRNA; Santa Cruz Biotechnology) for 72 hours using Lipofectamine RNAi-MAX (Life Technologies). Cells were then left untreated or were treated with R837 (InvivoGen; 1 µg/ml) as well as with 5 µg/ml of phosphorothioated miR-124, miR-147, and miR-let-7b (GenScript) for 6 hours, and the levels of TNF, IL-6, and IL-8 were quantified by real-time RT-PCR. To document whether chloroquine (InvivoGen), an inhibitor of endosomal acidification, or A151 (InvivoGen), a TLR-7 antagonist, could similarly block miR-let-7b activation, RA PB monocytes or RA PB differentiated macrophages were left untreated or were treated with 5 µg/ml of miR-let-7b alone, miR-let-7b plus chloroquine (50 µM), or miR-let-7b plus A151 (1 µM) for 6 hours, and the levels of TNF and IL-6 were quantified by real-time RT-PCR. To compare the potency of R837 and miR-let-7b, RA PB macrophages were left untreated or were treated with R837 (0.1 and 1 µg/ml) or miR-let-7b (0.25 and 5 µg/ml) for 18 hours, and the levels of TNF and IL-6 were determined by enzyme-linked immunosorbent assay (ELISA; R&D Systems).
To demonstrate the specificity of TLR-7 binding to miR-let-7b, THP-1 cells were transfected with 100 nM control, TLR-7, or TLR-8 siRNA (Santa Cruz Biotechnology) for 72 hours using Lipofectamine RNAiMAX according to the manufacturer’s instructions. Cells were then left untreated or were stimulated with miR-let-7b (5 µg/ml) for 6 hours, and the stimulatory effect of miR-let-7b was tested.
Western blot analysis
TLR-7 and TLR-8 knockdown studies were performed as described above, and the reduction of TLR-7 and/or TLR-8 levels was examined by Western blot analysis. Blots were probed with antibodies against TLR-7, TLR-8 (1:1,000 dilution), or actin (1:3,000 dilution) using Western blot blocking buffer (5% milk in Tris buffered saline–Tween). Activation of inducible nitric oxide synthase (iNOS) was examined in RA PB macrophages that had been left untreated or treated with R837 (1 µg/ml) or miR-let-7b (5 µg/ml) for 18 hours. Cell lysates were then probed with iNOS (1:1,000 dilution; Santa Cruz Biotechnology).
Flow cytometry
To document that miR-let-7b transforms naive cells into M1 macrophages, PB macrophages were left untreated or were treated with interferon-γ (IFNγ)/LPS (100 ng/ml each), R837 (1 µg/ml), or miR-let-7b (5 µg/ml) for 24 hours, and the frequency of CD127 cells was determined by fluorescence-activated cell sorter (FACS) analysis. To show that miR-let-7b ligation remodels M2 into M1 macrophages, mouse bone marrow cells were cultured for 7 days in macrophage–colony-stimulating factor (M-CSF) (20 ng/ml; M2). Cells were then left untreated or were treated with IFNγ (100 ng/ml), R837 (1 µg/ml), or 5 µg/ml of miR-124, miR-147, or miR-let-7b for 24 hours, and the frequency of CD80 was quantified by FACS analysis.
To establish that miR-let-7b provokes arthritis by recruiting F4/80+ cells into the joints, ankles from mice injected intraarticularly with adenovirus Ad-let-7b or Ad-control were harvested on day 7. Cells were then isolated from 6 ankle joints from 3 different mice (3 in the first run and 3 in the second) that had been crushed and vigorously vortexed in phosphate buffered saline (PBS) and filtered through a strainer. The total cell number was counted, and the percentage of F4/80 cells was determined by FACS analysis after staining with phycoerythrin-labeled F4/80 (eBioscience). Results are reported as the number of F4/80+ cells per ankle.
Treatment of mouse bone marrow cells
To substantiate the M0, M1, and M2 macrophage phenotype, mouse bone marrow cells were cultured in M-CSF plus granulocyte-macrophage colony-stimulating factor (GM-CSF) (10 ng/ml of each; M0), GM-CSF alone (20 ng/ml; M1), or M-CSF alone (20 ng/ml; M2) for 7 days. Subsequently, cells were stimulated with LPS (100 ng/ml) for 6 hours, and levels of TNF, IL-6, and IL-10 protein were determined by ELISA. To determine the pathogenic effect of miR-let-7b, bone marrow cells were cultured with M-CSF plus GM-CSF (10 ng/ml each; M0) or M-CSF alone (20 ng/ml; M2) for 7 days and were then treated with PBS, IFNγ (100 ng/ml), R837 (1 µg/ml), or with 5 µg/ml of miR-124, miR-147, or miR-let-7b for 6 hours, and the levels of TNF, IL-6, CCL2, and CCL5 were quantified by real-time RT-PCR. Alternatively, treatment was continued for 24 hours, and the levels of TNF and IL-6 were quantified by ELISA.
To demonstrate that miR-let-7b functions through TLR-7, bone marrow cells were cultured with M-CSF (20 ng/ml) for 7 days and were then left untreated or were treated with R837 (1 µg/ml) or miR-124/miR-147/miR-let-7b (5 µg/ml), in the presence or absence of A151 (1 µM), for 6 hours, and the TNF and IL-6 transcription levels were quantified.
The binding specificity of miR-let-7b was validated with bone marrow cells obtained from wild-type (WT) mice and TLR-7−/− mice (The Jackson Laboratory). Cells had been cultured in M-CSF (20 ng/ml) for 7 days prior to stimulation with PBS, IFNγ (100 ng/ml), R837 (1 µg/ml), or with 5 µg/ml of miR-124, miR-147, or miR-let-7b for 6 hours, and the TNF and IL-6 transcription levels were quantified.
Generation of exosomal miR-let-7b and determination of its functionality
To demonstrate that exosomes containing miR-let-7b are fully functional, bone marrow cells were differentiated with M-CSF (20 ng/ml) for 7 days. The cells were then left uninfected (mock) or were infected with 100 multiplicities of infection of Ad-control or Ad-let-7b (Vector BioLabs) for 4 days. Using a total exosome precipitation reagent (Life Technologies), the exosomes were extracted from the media in which the cells had been exposed to 3 freeze–thaw cycles. Bone marrow cells that had differentiated to M2 macrophages were stimulated with the exosomes obtained from an earlier experiment which contained no adenovirus (mock), or they were incorporated with Ad-control or Ad-let-7b for 6 hours. TNF and IL-6 levels were then determined by real-time RT-PCR.
Study protocol for miR-let-7b–induced arthritis
All animal studies were approved by the Animal Care and Use Committee of the University of Illinois at Chicago. WT C57BL/6 mice or TLR-7−/− mice (6–8 weeks old) were injected intraarticularly with 107 plaque-forming units of Ad-control or Ad-let-7b on day 0, and the joint circumference was measured from day 0 to 7, as described elsewhere (28–30). These experiments were independently repeated at least twice with the TLR-7−/− mice and 4 times with the WT mice.
Ankles to be used for histologic assessments were harvested on day 7 and were decalcified, formalin fixed, paraffin embedded, and sectioned. Slides were then deparaffinized in xylene, and antigens were unmasked by incubating the slides in proteinase K digestion buffer (Dako). Ankles were stained with F4/80 (1:100 dilution; Serotec), iNOS (1:200 dilution), or control IgG antibodies. Hematoxylin and eosin–stained ankle sections were scored by 2 observers (SK and MVV) who were blinded with regard to the experimental group. Each of the following features was scored on a 0–5 scale (where 0 = normal): inflammation, synovial lining thickness, bone erosion, myeloid cell recruitment to the joint, and their differentiation into M1 macrophages.
Messenger RNA (mRNA) or protein was extracted from WT and TLR-7−/− ankle homogenates, and levels of TNF, IL-1β, IL-6, CCL2, CCL5, iNOS, and transforming growth factor β (TGFβ) mRNA were quantified by real-time RT-PCR. Levels of TNF, IL-1β, IL-6, CCL2, CCL5, IFNγ, CXCL2, and IL-10 protein were determined by ELISA (R&D Systems).
Statistical analysis
One-way analysis of variance was used to compare multiple groups and was followed post hoc by Student’s 2-tailed t-test. The data were also analyzed by Student’s 2-tailed t-test for paired or unpaired comparisons between 2 groups. P values less than 0.05 were considered significant.
RESULTS
Expression of GU-rich oligos in RA SF
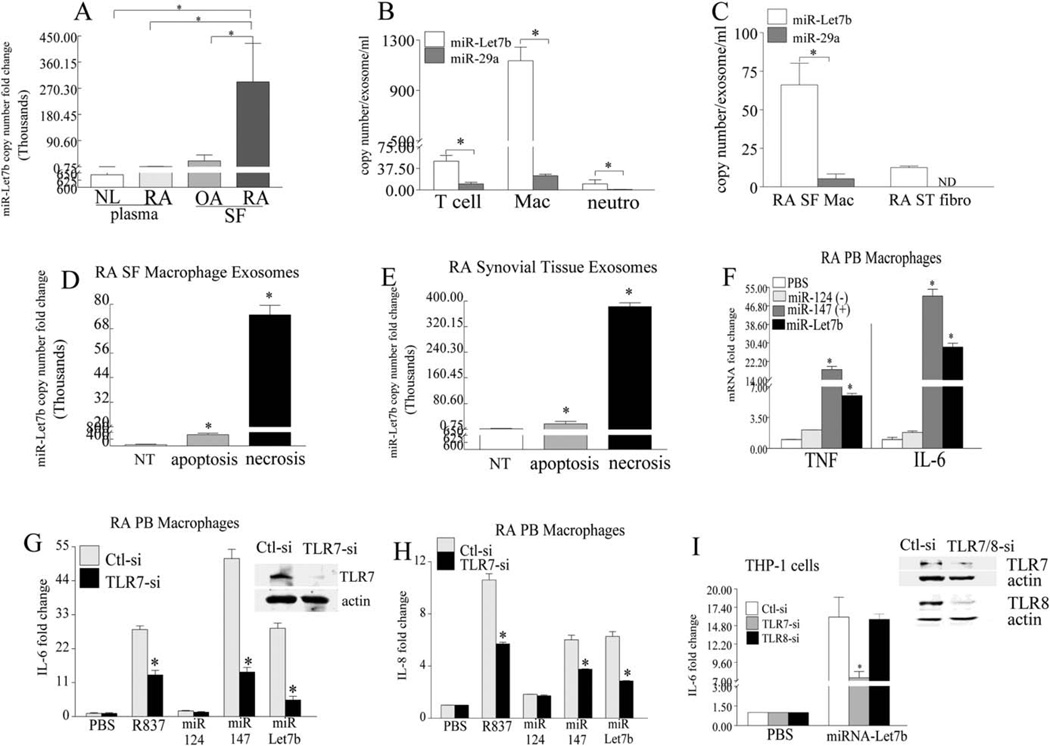
Consistent with the distribution of RA SF ssRNA (14), we found that miR-let-7b was markedly elevated in RA SF as compared to OA SF (14-fold), RA plasma (260-fold), and normal plasma (450-fold) (Figure 1A). We found that in blood, exosome levels of miR-let-7b were highest in monocyte-derived macrophages as compared to T cells and neutrophils (23-fold and 113-fold higher, respectively). Interestingly, a different GU-rich miRNA (miR-29a) was detected in exosomes released from all 3 cell types; however, the concentrations of exosomal miR-29a were significantly lower than those of miR-let-7b in exosomes discharged from T cells, in vitro-differentiated macrophages, and neutrophils (5-fold, 47-fold, and 12.5-fold, respectively) (Figure 1B). Consistent with the expression pattern of exosomal miR-let-7b in blood myeloid cells, miR-let-7b was predominantly expressed in exosomes released from RA SF macrophages as compared to RA ST fibroblasts (Figure 1C). While miR-29a was undetectable in the exosomes discharged from RA ST fibroblasts, its levels were greatly reduced compared to the exosomal miR-let-7b secreted from RA SF macrophages (13-fold) (Figure 1C).
Figure 1.
Detection of accentuated microRNA let-7b (miR-let-7b) expression in rheumatoid arthritis (RA) synovial fluid (SF) macrophages. A, Copy numbers of miR-let-7b in normal (NL) plasma, RA plasma, osteoarthritis (OA) SF, and RA SF (n = 10–13 samples per group), as determined by real-time reverse transcription-polymerase chain reaction (RT-PCR). B and C, Exosomal secretion of miR-let-7b and miR-29a in peripheral blood (PB) T cells, in vitro-differentiated macrophages (Mac), and neutrophils (neutro) (n = 3–5 samples per group) (B) as well as in RA SF macrophages and RA synovial tissue (ST) fibroblasts (fibro) (n = 3 samples per group) (C). D and E, Copy numbers of miR-let-7b in RA SF macrophages (D) or RA ST (E). Samples (n = 3 per group) were left untreated (NT) or were treated for 6 hours with 1 µM staurosporine to induce apoptosis or were subjected to repetitive freeze-thawing to promote necrosis. Exosomes released into the medium were extracted, and miR-let-7b copies were quantified. F, Levels of mRNA for tumor necrosis factor (TNF) and interleukin-6 (IL-6), as determined by real-time RT-PCR (n = 3 samples per group). RA PB in vitro-differentiated macrophages were left untreated (phosphate buffered saline [PBS] alone) or were treated for 6 hours with 5 µg/ml of miR-124, miR-147, or miR-let-7b. G and H, Western blot analysis of Toll-like receptor 7 (TLR-7) expression in control or knockdown cells and quantification of IL-6 (G) and IL-8 (H) transcription levels in in vitro-differentiated RA PB macrophages (n = 3 samples per group) after knockdown with small interfering RNA (siRNA). Cells were transfected for 72 hours with 100 nM control (Ctl-si) or TLR-7 (TLR7-si) siRNA and were left untreated or were stimulated for 6 hours with 1 µg/ml of R837 or with 5 µg/ml of miR-124, miR-147, or miR-let-7b. I, Western blot analysis and quantification of THP-1 cells transfected for 72 hours with 100 nM control, TLR-7, or TLR-8 siRNA Cells were left untreated or were stimulated for 6 hours with miR-let-7b, and IL-6 was quantified by real-time RT-PCR (n = 3 samples per group). The expression of TLRs 7 and 8 was markedly reduced after knockdown as compared to controls. Actin was used in Western blots as loading control. Fold change represents the increase over no treatment. Values are the mean ± SEM. * = P < 0.05. ND = not done.
We further found that exosomes released from RA SF macrophages were an important source of miR-let-7b storage (78 copies) and that cell death mediated by apoptosis (660 copies, 8-fold) or necrosis (74,900 copies, 950-fold) could further potentiate the discharge of exosomal miR-let-7b into RA SF (Figure 1D). We found that similar to RA SF, exosomes residing in RA ST contained miR-let-7b (2,330 copies) and that induced cell death could also increase the release of the miR-let-7b–incorporated exosomes from 7-fold to 163-fold (Figure 1E). These results show that the TLR-7 endogenous ligand, miR-let-7b, is primarily released from exosomes residing in RA SF macrophages and that its expression is markedly greater than that of the GU-rich miR-29a.
Selective binding of GU-rich miR-let-7b to TLR-7
Since in RA samples, levels of miR-let-7b were markedly higher than levels of miR-29a, our studies then focused on determining the impact of miR-let-7b on the pathogenesis of RA. We found that having a GU-rich motif could facilitate miRNA binding and activation of TLR-7. This finding was confirmed by the use of miR-124, a non–GU-rich oligo present in RA fibroblasts (17,31,32), as a negative control and miR-147 (GUGUG), a GU-rich oligo that is undetectable in RA samples, as a positive control. Results obtained from RA PB–differentiated macrophages demonstrated that while the GU-rich miR-let-7b (6–28-fold, respectively) and miR-147 (18–50-fold, respectively) could activate TNF and IL-6 transcription, miR-124 could not (Figure 1F). Consistent with this, we found that miR-let-7b– and miR-147–driven IL-6 or IL-8 transcription was impaired by TLR-7 knockdown as compared to the control counterparts (Figures 1G and H).
Since ssRNA can bind to both TLR-7 and TLR-8, we next sought to determine whether miR-let-7b could preferentially bind to TLR-7. We demonstrated that miR-let-7b explicitly ligated TLR-7, as knockdown of TLR-7, but not TLR-8, could modulate the miR-let-7b–mediated IL-6 transcription in THP-1 cells (Figure 1I). This suggests that TLR-7 is not randomly activated by nonspecific miRNA and that the highly expressed GU-rich miR-let-7b can bind to and activate TLR-7.
Transformation of naive RA myeloid cells into M1 macrophages following ligation of miR-let-7b
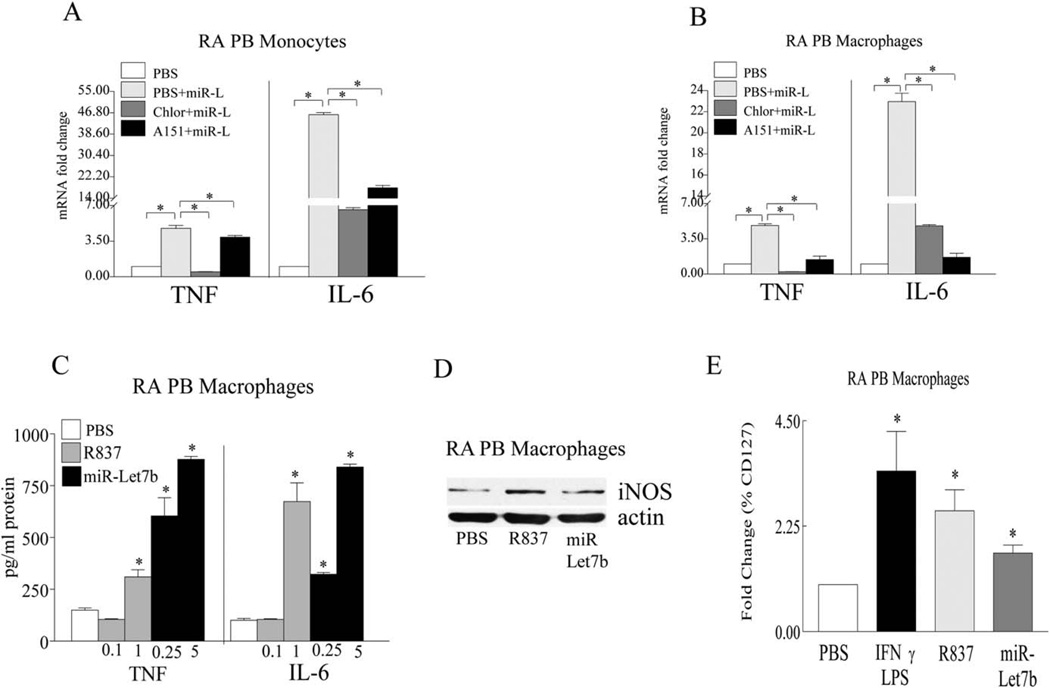
To confirm the results of the TLR-7 knockdown studies, we examined whether chloroquine and A151 treatment in RA PB monocytes and differentiated macrophages could impair endosomal function and TLR-7 activation. We found that both chloroquine and A151 therapy could effectively impair miR-let-7b–induced TNF and IL-6 production in RA monocytes and macrophages (Figures 2A and B). Next, different concentrations of miR-let-7b and the TLR-7 agonist R837 were used to assess miR-let-7b potency. We found that in RA macrophages, miR-let-7b could induce TNF production at a concentration of 0.25 µg/ml (0.034 mM), which is markedly lower than the levels required for R837 stimulation (1 µg/ml; 4.16 mM) (Figure 2C).
Figure 2.
Impaired miR-let-7b–induced M1 macrophage differentiation following treatment with a TLR-7 antagonist (A151) or an inhibitor of endosomal acidification (chloroquine). A and B, RA PB monocytes (A) or RA PB in vitro-differentiated macrophages (B) were left untreated (PBS) or were treated for 6 hours with 5 µg/ml of miR-let-7b (miR-L) alone, miR-let-7b plus chloroquine (50 µM; Chlor), or miR-let-7b plus A151 (1 µM), and the levels of mRNA for TNF and IL-6 were determined by real-time RT-PCR (n = 3 samples per group). C, RA PB in vitro-differentiated macrophages were left untreated (PBS) or were treated for 18 hours with R837 (0.1 or 1 µg/ml) and miR-let-7b (0.25 or 5 µg/ml), and the levels of TNF and IL-6 protein were determined by enzyme-linked immunosorbent assay (n = 5 samples per group). D, RA PB in vitro-differentiated macrophages were left untreated (PBS) or were treated for 18 hours with R837 (1 µg/ml) or miR-let-7b (5 µg/ml), and cell lysates were probed for inducible nitric oxide synthase (iNOS). A representative Western blot (n = 3 samples per group) is shown. Actin was used as a loading control. E, RA PB in vitro-differentiated macrophages were left untreated or were treated for 24 hours with interferon-γ (IFNγ) plus lipopolysaccharide (LPS) (100 ng/ml each), R837 (1 µg/ml), or miR-let-7b (5 µg/ml), and the frequency of CD127 was determined by fluorescence-activated cell sorter analysis (n = 3 samples per group). Values are the mean ± SEM. * = P < 0.05. See Figure 1 for other definitions.
Based on the potent effects of miR-let-7b on TNF and IL-6 transcription and secretion, we sought to determine whether miR-let-7b is capable of remodeling naive RA myeloid cells into proinflammatory M1 macrophages. We found that, similar to the M1 positive controls, IFNγ and LPS, miR-let-7b could transform naive myeloid cells from RA patients into M1 macrophages that expressed elevated levels of iNOS and the M1 cell surface marker CD127 (Figures 2D and E). Taken together, our results document that miR-let-7b can facilitate RA pathology by specifically binding to naive TLR-7+ myeloid cells and reconstructing them into proinflammatory M1 macrophages.
MicroRNA-let-7b conversion of mouse bone marrow M0 and M2 cells into M1 macrophages through TLR-7 ligation
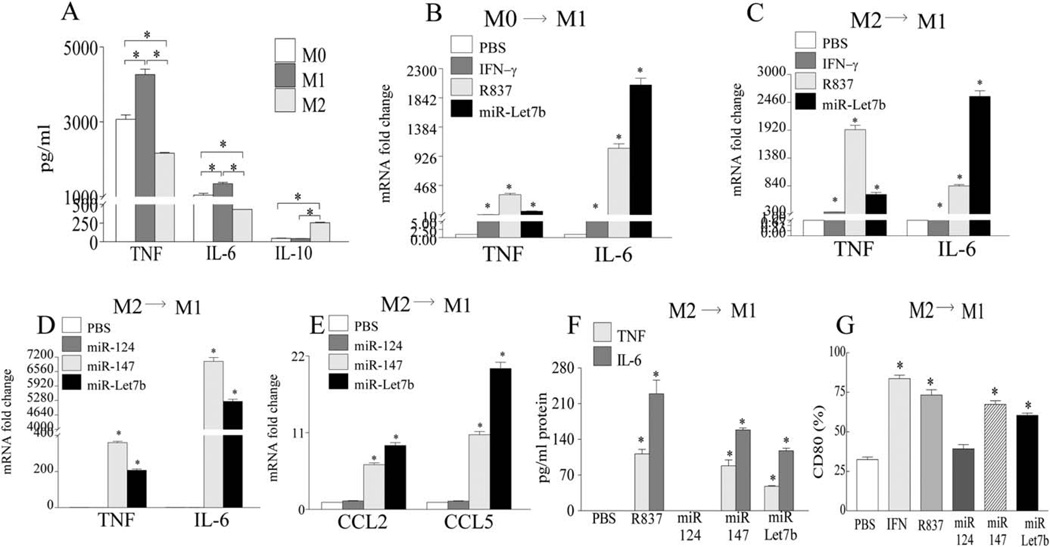
We found that GM-CSF-derived bone marrow M1 cells had the higher inflammatory (TNF and IL-6 production) and the lower antiinflammatory (IL-10 production) cytokine profile compared to M-CSF–derived bone marrow M2 macrophages, whereas GM-CSF–derived and M-CSF–derived M0 cells had a baseline profile that was intermediate to the phenotype detected in M1 and M2 myeloid cells (Figure 3A). When the effects of R837 and miR-let-7b on the mouse macrophage phenotype were examined, we found that, like the M1 positive control IFNγ, both TLR-7 ligands could potently transform M0 and M2 myeloid cells into M1 macrophages, as assessed by TNF and IL-6 transcription (Figures 3B and C). As in RA cells, only miRNAs with a GU-rich motif, such as miR-let-7b and miR-147, could strongly convert the mouse M2 macrophages into M1 cells (Figures 3C and D). This was documented by the transcription and/or production of TNF (50-fold protein for miR-let-7b and 90-fold protein for miR-147), IL-6 (100-fold protein for miR-let-7b and 150-fold protein for miR-147), CCL2, and CCL5, as well as the expression of the mouse M1 marker CD80 (2–2.5-fold), whereas the non–GU-rich miR-124 lacked this ability (Figures 3C–G).
Figure 3.
Transformation of mouse bone marrow naive (MO) and antiinflammatory (M2) macrophages into proinflammatory M1 myeloid cells following ligation of GU-rich miR-let-7b. A, Wild-type (WT) mouse bone marrow cells were cultured for 7 days in macrophage-colony-stimulating factor (M-CSF) plus granulocyte-macrophage CSF (GM-CSF) (10 ng/ml of each; MO), GM-CSF alone (20 ng/ml; Ml), or M-CSF alone (20 ng/ml; M2). Cells were then stimulated for 6 hours with lipopolysaccharide (LPS), and the levels of TNF, IL-6, and IL-10 protein were determined by enzyme-linked immunosorbent assay (ELISA). B and C, WT mouse bone marrow cells were cultured for 7 days with M-CSF plus GM-CSF (10 ng/ml each; MO) (B) or M-CSF (20 ng/ml; M2) (C), treated for 6 hours with PBS, interferon-γ (IFNγ; 100 ng/ml), R837 (1 µg/ml), or miR-let-7b (5 µg/ml), and the levels of mRNA for TNF and IL-6 were determined by real-time RT-PCR. D and E, Bone marrow cells were cultured for 7 days with M-CSF (20 ng/ml; M2), treated for 6 hours with PBS or with 5 µg/ml of miR-124, miR-147, or miR-let-7b, and the levels of mRNA for TNF and IL-6 (D) as well as for CCL2 and CCL5 (E) were determined by real-time RT-PCR. F and G, WT mouse bone marrow cells were cultured for 7 days in M-CSF (20 ng/ml; M2) and were then left untreated (PBS) or were stimulated for 24 hours with IFNγ (for protein assessment; 100 ng/ml) as well as with R837 (1 µg/ml) or with 5 µg/ml of miR-124, miR-147, or miR-let-7b, and the levels of TNF and IL-6 protein were determined by ELISA (F) and the frequency of CD80 was determined by fluorescence-activated cell sorting (G). Values are the mean ± SEM of 3 samples per group. * = P < 0.05. See Figure 1 for other definitions.
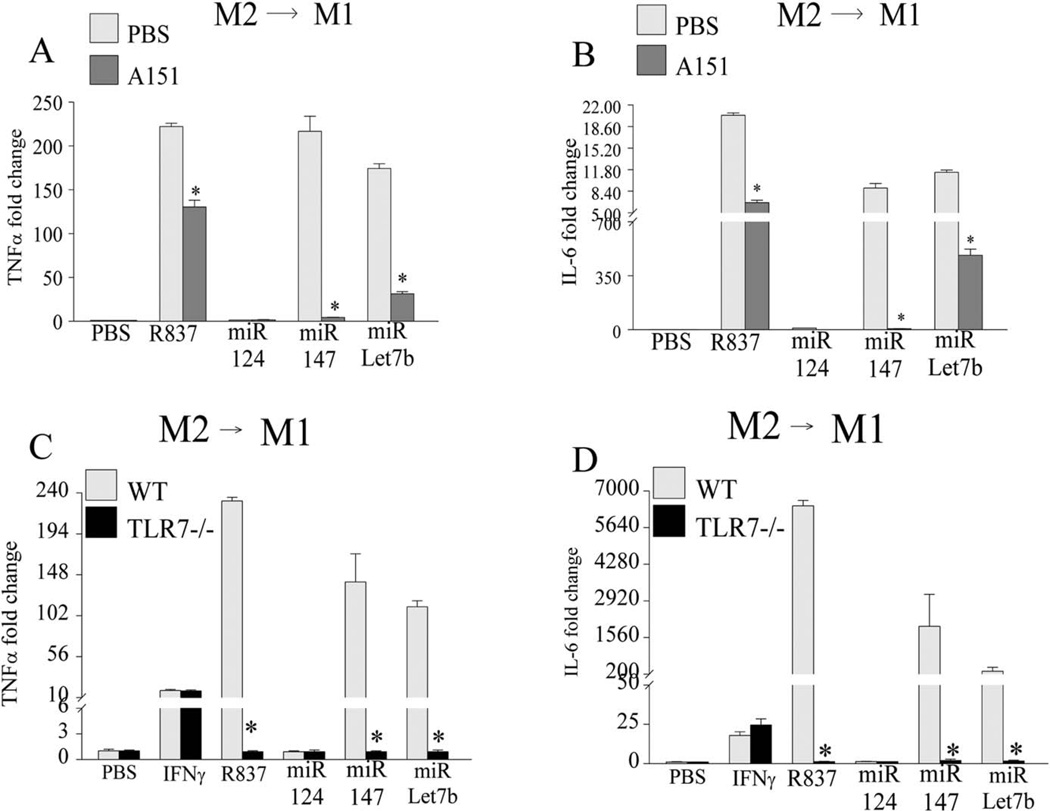
The M1 maturation process driven by miR-let-7b and miR-147 was found to be mediated through TLR-7 ligation, given that the use of a TLR-7 antagonist (A151) or TLR-7−/− mice was shown to impair this function (Figures 4A–D). However, TLR-7 deficiency had no effect on the remodeling of murine M2→M1 macrophages driven by IFNγ (Figures 4C and D). Thus, our results demonstrate that miR-let-7b is capable of polarizing WT M0 and M2 cells into M1 macrophages, and since this effect is lost in TLR-7−/− murine cells, it would indicate that ligation of miR-let-7b to TLR-7 is indispensable for shifting the balance toward M1 macrophage differentiation.
Figure 4.
Dysregulation of mouse bone marrow M2→M1 remodeling provoked by miR-let-7b via the use of TLR-7 antagonist or knockout mice. A and B, Bone marrow cells were cultured for 7 days with macrophage-colony-stimulating factor (M-CSF) (20 ng/ml; M2) and were left untreated (PBS) or were treated for 6 hours with 1 µg/ml of R837 in the presence or absence of A151 (1 µM), as well as with 5 µg/ml of miR-124, miR-147, or miR-let-7b in the presence or absence of A151, and the levels of TNF (A) and IL-6 (B) transcription were determined. C and D, Bone marrow cells obtained from C57BL/6 wild-type (WT) mice and TLR-7−/− mice were cultured for 7 days in M-CSF (20 ng/ml; M2), treated for 6 hours with PBS, interferon-γ (IFNγ; 100 ng/ml), R837 (1 µg/ml), and with 5 µg/ml of miR-124, miR-147, or miR-let-7b, and the transcription levels of TNF (C) and IL-6 (D) were determined by real-time RT-PCR. Values are the mean ± SEM of 3 samples per group. * =P < 0.05. See Figure 1 for other definitions.
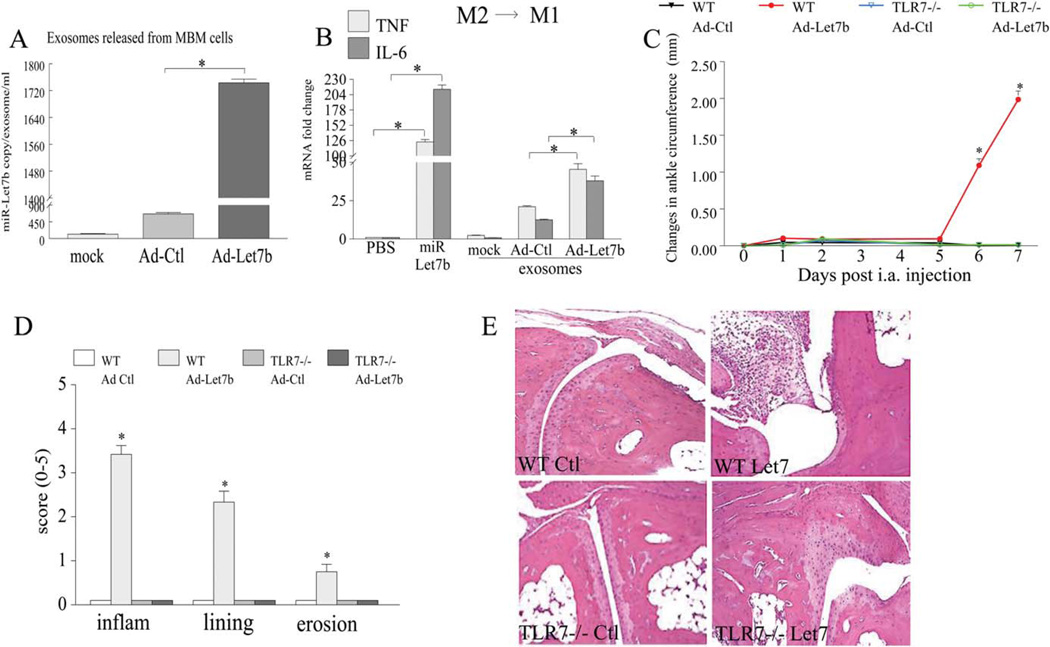
Full functionality of miR-let-7b–incorporated exosomes and strong promotion of arthritis by local ligation of miR-let-7b to TLR-7
To demonstrate that exosomes containing miR-let-7b are fully functional, exosomes were extracted from mouse bone marrow progenitor cells 4 days after infection with Ad-control or Ad-let-7b (Figure 5A). M2-derived bone marrow macrophages were then stimulated with exosomes obtained from these 3 different treatment groups (Figure 5B). We found that, similar to the synthetic miR-let-7b, while exosomes extracted from uninfected or control-infected cells were unable to alter the M2 macrophage phenotype, exosomes incorporated with miR-let-7b were capable of reconstructing M2→M1 macrophages and accentuating the TNF and IL-6 levels by 2–3-fold (Figure 5B). These results suggest that exosomes can protect, transport, and deliver miR-let-7b to its final destination and that miR-let-7b–incorporated exosomes are fully functional. We then found that ligation of TLR-7 by local injection of miR-let-7b resulted in markedly elevated joint inflammation, lining thickness, and bone lesions in WT mice as compared to control mice (Figures 5C–E). However, joint swelling was not affected in TLR-7−/− mice injected with Ad-let-7b or Ad-control, indicating that ligation of miR-let-7b to TLR-7 is responsible for the induction of arthritis (Figures 5C–E).
Figure 5.
Full functionality of exosomes containing miR-let-7b, their potent differentiation of M2 myeloid cells into M1 macrophages, and the fostering of arthritic joint inflammation through the local expression of miR-let-7b. A, Mouse bone marrow (MBM) cells were differentiated for 7 days with macrophage-colony-stimulating factor (M-CSF; 20 ng/ml), cells were left uninfected (mock) or were infected for 4 days with 100 multiplicities of infection of adenovirus control (Ad-Ctl) or Ad-miR-let-7b (Ad-let-7b), exosomes were extracted, and miR-let-7b copy numbers were determined (n = 3 samples per group). B, Bone marrow cells differentiated to M2 macrophages were left untreated (PBS) or were stimulated for 6 hours with miR-let-7b or with the exosomes isolated from the media in A, which contained no adenovirus (mock) or had incorporated Ad-Ctl or Ad-let-7b, and the levels of mRNA for TNF and IL-6 were determined by real-time RT-PCR (n = 3 samples per group). C, Changes in ankle circumference were assessed in wild-type (WT) mice or TLR-7−/− mice that had been injected intraarticularly (IA) with 107 PFU of Ad-Ctl or Ad-let-7b. Ankles were harvested on day 7 (n = 8 mice/16 ankles per group). D, Hematoxylin and eosin (HAE)-stained ankles from all 4 experimental groups were scored for inflammation (inflam), synovial lining thickness, and bone erosion (n = 6 ankles per group). Values are the mean ± SEM. * = P < 0.05. E, Representative H&E-stained sections of ankle joints from the 4 experimental groups (n = 6 ankles per group) are shown. Original magnification × 200. See Figure 1 for other definitions. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/journal/doi/10.1002/art.39544/abstract.
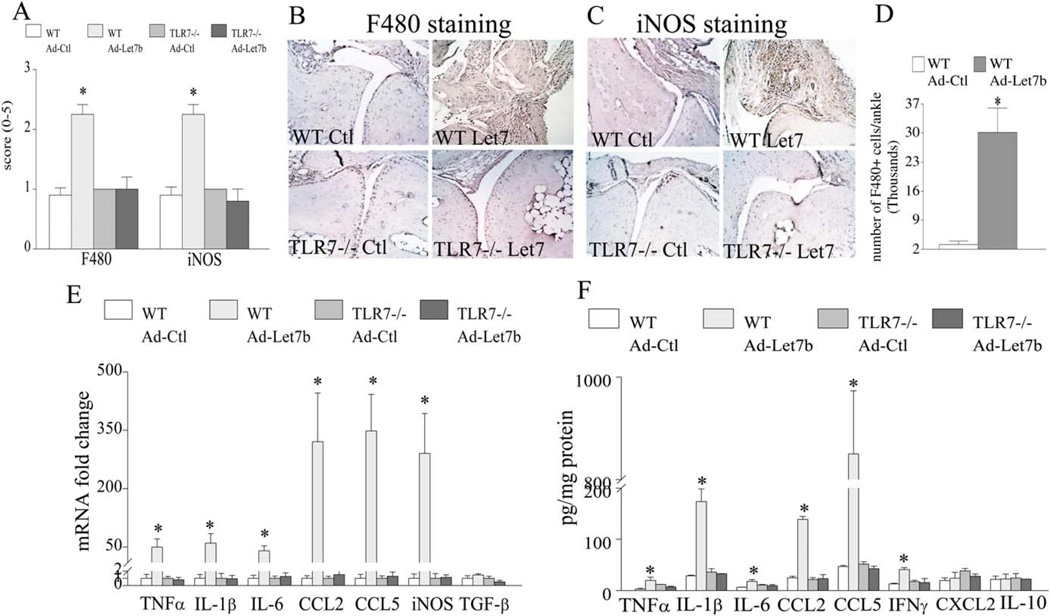
Histologic and FACS analyses demonstrate that ligation of miR-let-7b in WT mice resulted in 2.5–10-fold higher levels of infiltrated F4/80+ myeloid cells into the ankle joint as compared to the joints of Ad-control mice (Figures 6A, B, and D). However, in contrast to the local expression of miR-let-7b in WT mice, ectopic injection of Ad-let-7b and Ad-control in TLR-7−/− mice did not yield a marked increase in joint monocyte recruitment or myeloid cell conversion to iNOS+ M1 macrophages (Figures 6A–C). Confirming the clinical and histologic data, levels of mRNA and protein for the M1 macrophage markers, such as TNF (50-fold and 7-fold, respectively), IL-1 (60-fold and 6-fold), IL-6 (40-fold and 3-fold), CCL2 (320-fold and 5.5-fold), CCL5 (350-fold and 18-fold), and iNOS (290-fold mRNA for the NOS2 gene), were highly elevated in WT mice that received local injection of miR-let-7b as compared to those that received Ad-control injection (Figures 6E and F). In contrast, the levels of M2 macrophage markers, such as TGFβ and IL-10, remained unchanged in all 4 treatment groups (Figures 6E and F). Collectively, our results demonstrate that miR-let-7b can provoke arthritis by transforming the recruited naive myeloid cells into proinflammatory M1 macrophages and that this function is nullified in TLR-7−/− mice.
Figure 6.
Provocation of joint inflammation via miR-let-7b attraction of myeloid cells into the joints and their reconstruction into proinflammatory M1 macrophages. A, Wild-type (WT) or TLR-7−/− mice were ectopically injected with adenovirus control (Ad-Ctl) or Ad-miR-let-7b (Ad-let-7b), joints were harvested on day 7 and stained with anti-F4/80 and anti-inducible nitric oxide synthase (anti-iNOS) antibodies, and the staining was scored. B and C, Representative sections of ankle joints from the 4 experimental groups (n = 6 ankles per group) stained with F4/80 (B) and iNOS (C) are shown. Original magnification × 200. D, Ankles from WT mice injected intraarticularly with Ad-Ctl or Ad-let-7b were harvested on day 7. Six ankle joints from 3 different mice (3 in the first run and 3 in the second) were crushed, vigorously vortexed in PBS, and filtered through a strainer. The total cell number was determined by fluorescence-activated cell sorting, and cells were then isolated and stained with phycoerythrin-labeled F4/80. Results are shown as the number of F4/80+ cells per ankle (n = 6 samples per group). E and F, Ankle homogenates from the 4 experimental groups were prepared, and the levels of mRNA for TNF, IL-lβ, IL-6, CCL2, CCL5, iNOS, and transforming growth factor β (TGFβ) were determined by real-time RT-PCR (E), and the levels of TNF, IL-lβ, IL-6, CCL2, CCL5, interferon-γ (IFNγ), CXCL2, and IL-10 protein were determined by enzyme-linked immunosorbent assay (F) (n = 5 ankles per group). Values are the mean ± SEM. * = P < 0.05. See Figure 1 for other definitions.
DISCUSSION
Our research group pioneered the field of RA by documenting that ssRNA extracted from RA SF is a potential TLR-7 ligand that can selectively stimulate TLR-7+ cells (14). We found that TLR-7, but not TLR-8, expressing cells could bind to miR-let-7b that resided primarily in RA SF macrophages and that cell death could further enhance the release of the exosomal miR-let-7b into the SF. We showed that local ligation of miR-let-7b to TLR-7 provoked arthritis by transforming naive joint myeloid cells from WT, but not TLR-7−/−, mice into proinflammatory M1 macrophages, thus supporting the critical importance of miR-let-7b and TLR-7 function in the pathology of RA.
A recent study showed that treatment with hairpin TLR-7 RNA given before arthritis onset ameliorates rat CIA due to a reduction in joint vascularity and vascular endothelial growth factor production from fibroblasts (33). A second study showed that resolution of CIA in the TLR-7−/− mouse was due to a decrease in the IL-17 response and an elevation of Treg cell numbers (34). Similar to our findings in RA, the 2 studies of experimental arthritis highlight the critical role of TLR-7 in disease pathology (33,34); however, these studies define the TLR-7 mechanism of function in RA fibroblasts and T cells and not in myeloid cells. Since neither group of investigators was able to identify the TLR-7 endogenous ligand (33,34), it is not clear whether TLR-7 blockade has direct effects on joint fibroblasts and Th-17 cells or whether the effect is mediated indirectly through other factors. Hence, studies were designed to reveal the TLR-7 endogenous ligand in RA samples and to investigate the mechanism by which ligation of this endogenous ligand regulates arthritis.
Earlier studies identified 17 miRNAs in RA that are expressed in PB mononuclear cells, ST, and SF (32). When the previously reported miRNAs in RA PB, ST, and more specifically, SF (133a, 142-3p, 146a, 155, 203, 221/222, 223, and 323-3p) were screened for GU-rich oligos, none of them fulfilled the TLR-7 binding criteria. Interestingly, we found novel GU-rich miRNA packaged in exosomes secreted from RA SF macrophages and RA ST fibroblasts. The most abundant GU-rich miRNA detected in the exosomes shed from RA SF macrophages was miR-let-7b, which was 13-fold higher than miR-29a. While exosomal miR-let-7b levels were 5-fold lower in RA ST fibroblasts than in RA SF macrophages, miR-29a was undetectable in RA ST fibroblasts. Similarly, when exosomal secretion of miR-let-7b and miR-29a was assessed in blood, miR-let-7b was found to be expressed mainly in monocyte-derived macrophages as compared to T cells and neutrophils, and its levels were 5–47-fold higher than the level of miR-29a in all 3 cell types. In contrast to our finding of miR-let-7b expression mainly in RA SF macrophages, other investigators have reported that miR-16, miR-132, miR-146a, and miR-223 are markedly elevated in RA plasma as compared to SF and can therefore be used as disease biomarkers (35). However, consistent with the miR-let-7b distribution, miR-16, miR-146a, miR-155, and miR-223 expression levels are significantly higher in RA SF than in OA SF (35).
We found that cell death induced through apoptosis or necrosis of RA SF macrophages or RA ST can accentuate the release of miR-let-7b–incorporated exosomes. Consistent with our findings, other investigators have shown that miR-let-7b is discharged from injured dying neurons; however, while ligation of miR-let-7b to TLR-7 potentiates RA inflammation through M1 differentiation without affecting myeloid cell death, miR-let-7b secreted during neurodegeneration in Alzheimer’s disease was shown to accelerate neural cell death via TLR-7 (36).
Confirming the notion that macrophages are involved in exosomal miR-let-7b release into RA ST and SF, other investigators have demonstrated that macrophages discharge exosomes that can “shuttle” the oncogenic miRNA into the recipient cancerous cells (37). The mechanism involved in the secretion of exosomes is essentially cell type-specific, since intracellular Ca2+ increases (38) and HIF-1α–mediated hypoxia (39) can trigger exosome shedding in cancer cells, whereas in HeLa cells, the Rab GTPase family is responsible for this process (40). Previous studies have shown that miRNA are selectively packaged into exosomes, since the miRNA profiles in the exosomes are different from those in the parental cells (41), suggesting that exosomes transport and deliver a distinct set of miRNAs depending on what needs to be communicated to the receipt cells.
We found that in myeloid cells, miR-let-7b preferentially binds to TLR-7 and not TLR-8 due to its GU-rich oligo content, rather than AU-rich oligos. Our results showed that having a GU-rich domain is essential for TLR-7 ligation, as M1 polarization induced by miR-let-7b and miR-147 was impaired by TLR-7 knockdown, knockout, and antagonists, suggesting that the GU-rich miR-let-7b present in RA can bind and activate TLR-7.
It is known that miRNA can inhibit the translation of their target genes through mRNA degradation (17–19,31,32); however, it is unclear how miRNA interacts with proteins. MicroRNA let-7b can directly bind to TLR-7, or it may require a carrier protein for its binding, and hence, adaptor proteins need to be available in the miR-let-7b–packaged exosome. Our data demonstrated that miR-let-7b–incorporated exosomes were fully functional and contained the necessary components, since fusion of exosomal miR-let-7b into M2 myeloid cells could differentiate these cells into M1 macrophages. An elegant study by Valadi et al (41) revealed that exosomal RNA discharged from mouse mast cells could be transferred into human mast cells, since new mouse proteins were found in the human recipient cells. The findings by those investigators substantiate our findings by showing that exosomal miRNA delivered to the recipient cell are fully functional and that the “exosomal shuttle miRNA” is a novel mechanism of communication between the cells (41).
This study is the first to show that the local expression of miR-let-7b in the WT mice provokes joint swelling, as detected on day 6 postinjection. We speculate that the delay in disease onset is partly due to the time required for the discharge and fusion of exosomal miR-let-7b into target cells prior to the manifestation of the clinical signs. The histologic studies showed that miR-let-7b ectopically expressed in WT mice promotes joint inflammation, synovial lining thickness, and minimal bone lesions and that this effect was blunted in TLR-7−/− mice, while no pathologic features were detected in WT or TLR-7−/− mice that received a local control injection. In contrast, previous studies have shown that administration of miR-let-7 markedly diminishes the size of the tumors that developed in a murine model of lung cancer by reducing the expression of Ras oncogene and cyclin-dependent kinase (42,43). We found that local injection of miR-let-7b provoked joint inflammation by recruiting myeloid cells from the circulation into the joint and further remodeling them into M1 macrophages. The miR-let-7b–driven joint M1 macrophages produced high levels of proinflammatory factors, such as TNF, IL-1, IL-6, CCL2, and CCL5, and expressed the M1 marker iNOS. Our histologic findings also indicated that almost all of the infiltrated joint myeloid cells driven by miR-let-7b were of the M1 phenotype, and this mechanism of action could be further amplified by enhancing the production of IFNγ in the joint.
We found that one of the integral RA SF ssRNAs that bind to TLR-7+ myeloid cells is a GU-rich miR-let-7b that can potently promote arthritic joint inflammation through M1 macrophage differentiation. Our results indicate that interruption of miR-let-7b binding to joint TLR-7 can be used as a target for RA therapy.
Acknowledgments
Supported in part by the Department of Veterans Affairs (MERIT award 1I01BX002286), the NIH (grants AR-056099 and AR-065778), the Department of Defense (award PR093477), and the Arthritis Foundation (Innovative Research grant).
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Shahrara had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Kim, Shahrara.
Acquisition of data. Kim, Chen, Essani, Elshabrawy, Volin, Shahrara.
Analysis and interpretation of data. Kim, Chen, Essani, Elshabrawy, Volin, Volkov, Swedler, Arami, Sweiss, Shahrara.
REFERENCES
- 1.Brentano F, Kyburz D, Schorr O, Gay R, Gay S. The role of Toll-like receptor signalling in the pathogenesis of arthritis. Cell Immunol. 2005;233:90–96. doi: 10.1016/j.cellimm.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 2.Drexler SK, Sacre SM, Foxwell BM. Toll-like receptors: a new target in rheumatoid arthritis? Expert Rev Clin Immunol. 2006;2:585–599. doi: 10.1586/1744666X.2.4.585. [DOI] [PubMed] [Google Scholar]
- 3.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 5.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 6.Hoshino K, Sugiyama T, Matsumoto M, Tanaka T, Saito M, Hemmi H, et al. IκB kinase α is critical for interferon-α production induced by Toll-like receptors 7 and 9. Nature. 2006;440:949–953. doi: 10.1038/nature04641. [DOI] [PubMed] [Google Scholar]
- 7.Iwahashi M, Yamamura M, Aita T, Okamoto A, Ueno A, Ogawa N, et al. Expression of Toll-like receptor 2 on CD16+ blood monocytes and synovial tissue macrophages in rheumatoid arthritis. Arthritis Rheum. 2004;50:1457–1467. doi: 10.1002/art.20219. [DOI] [PubMed] [Google Scholar]
- 8.Radstake TR, Roelofs MF, Jenniskens YM, Oppers-Walgreen B, van Riel PL, Barrera P, et al. Expression of Toll-like receptors 2 and 4 in rheumatoid synovial tissue and regulation by proinflammatory cytokines interleukin-12 and interleukin-18 via interferon-γ. Arthritis Rheum. 2004;50:3856–3865. doi: 10.1002/art.20678. [DOI] [PubMed] [Google Scholar]
- 9.Sorensen LK, Havemose-Poulsen A, Sonder SU, Bendtzen K, Holmstrup P. Blood cell gene expression profiling in subjects with aggressive periodontitis and chronic arthritis. J Periodontol. 2008;79:47785. doi: 10.1902/jop.2008.070309. [DOI] [PubMed] [Google Scholar]
- 10.Huang QQ, Pope RM. The role of Toll-like receptors in rheumatoid arthritis. Curr Rheumatol Rep. 2009;11:357–364. doi: 10.1007/s11926-009-0051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdollahi-Roodsaz S, Joosten LA, Roelofs MF, Radstake TR, Matera G, Popa C, et al. Inhibition of Toll-like receptor 4 breaks the inflammatory loop in autoimmune destructive arthritis. Arthritis Rheum. 2007;56:2957–2967. doi: 10.1002/art.22848. [DOI] [PubMed] [Google Scholar]
- 12.Abdollahi-Roodsaz S, Joosten LA, Helsen MM, Walgreen B, van Lent PL, van den Bersselaar LA, et al. Shift from Toll-like receptor 2 (TLR-2) toward TLR-4 dependency in the erosive stage of chronic streptococcal cell wall arthritis coincident with TLR-4—mediated interleukin-17 production. Arthritis Rheum. 2008;58:3753–3764. doi: 10.1002/art.24127. [DOI] [PubMed] [Google Scholar]
- 13.Abdollahi-Roodsaz S, Joosten LA, Koenders MI, van den Brand BT, van de Loo FA, van den Berg WB. Local interleukin-1-driven joint pathology is dependent on Toll-like receptor 4 activation. Am J Pathol. 2009;175:2004–2013. doi: 10.2353/ajpath.2009.090262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chamberlain ND, Kim SJ, Vila OM, Volin MV, Volkov S, Pope RM, et al. Ligation of TLR7 by rheumatoid arthritis synovial fluid single strand RNA induces transcription of TNFα in monocytes. Ann Rheum Dis. 2013;72:418–426. doi: 10.1136/annrheumdis-2011-201203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demaria O, Pagni PP, Traub S, de Gassart A, Branzk N, Murphy AJ, et al. TLR8 deficiency leads to autoimmunity in mice. J Clin Invest. 2010;120:3651–3662. doi: 10.1172/JCI42081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imbertson LM, Beaurline JM, Couture AM, Gibson SJ, Smith RM, Miller RL, et al. Cytokine induction in hairless mouse and rat skin after topical application of the immune response modifiers imiquimod and S-28463. J Invest Dermatol. 1998;110:734–739. doi: 10.1046/j.1523-1747.1998.00174.x. [DOI] [PubMed] [Google Scholar]
- 17.Duroux-Richard I, Jorgensen C, Apparailly F. miRNAs and rheumatoid arthritis: promising novel biomarkers. Swiss Med Wkly. 2011;141:w13175. doi: 10.4414/smw.2011.13175. [DOI] [PubMed] [Google Scholar]
- 18.Miao CG, Yang YY, He X, Xu T, Huang C, Huang Y, et al. New advances of microRNAs in the pathogenesis of rheumatoid arthritis, with a focus on the crosstalk between DNA methylation and the microRNA machinery. Cell Signal. 2013;25:1118–1125. doi: 10.1016/j.cellsig.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 19.Duroux-Richard I, Jorgensen C, Apparailly F. What do microRNAs mean for rheumatoid arthritis? [review] Arthritis Rheum. 2012;64:11–20. doi: 10.1002/art.30651. [DOI] [PubMed] [Google Scholar]
- 20.Nahid MA, Pauley KM, Satoh M, Chan EK. miR-146a is critical for endotoxin-induced tolerance: implication in innate immunity. J Biol Chem. 2009;284:34590–34599. doi: 10.1074/jbc.M109.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakasa T, Shibuya H, Nagata Y, Niimoto T, Ochi M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. 2011;63:1582–1590. doi: 10.1002/art.30321. [DOI] [PubMed] [Google Scholar]
- 22.Bluml S, Bonelli M, Niederreiter B, Puchner A, Mayr G, Hayer S, et al. Essential role of microRNA-155 in the pathogenesis of autoimmune arthritis in mice. Arthritis Rheum. 2011;63:1281–1288. doi: 10.1002/art.30281. [DOI] [PubMed] [Google Scholar]
- 23.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 24.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickens SR, Chamberlain ND, Volin MV, Pope RM, Mandelin AM, II, Shahrara S. Characterization of CCL19 and CCL21 in rheumatoid arthritis. Arthritis Rheum. 2011;63:914–922. doi: 10.1002/art.30232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pickens SR, Chamberlain ND, Volin MV, Pope RM, Talarico NE, Mandelin AM, II, et al. Characterization of interleukin-7 and interleukin-7 receptor in the pathogenesis of rheumatoid arthritis. Arthritis Rheum. 2011;63:2884–2893. doi: 10.1002/art.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyum A. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand J Clin Lab Invest Suppl. 1968;97:77–89. [PubMed] [Google Scholar]
- 28.Chen Z, Kim SJ, Chamberlain ND, Pickens SR, Volin MV, Volkov S, et al. The novel role of IL-7 ligation to IL-7 receptor in myeloid cells of rheumatoid arthritis and collagen-induced arthritis. J Immunol. 2013;190:5256–5266. doi: 10.4049/jimmunol.1201675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickens SR, Chamberlain ND, Volin MV, Gonzalez M, Pope RM, Mandelin AM, II, et al. Anti-CXCL5 therapy ameliorates IL-17-induced arthritis by decreasing joint vascularization. Angiogenesis. 2011;14:443–455. doi: 10.1007/s10456-011-9227-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SJ, Chen Z, Chamberlain ND, Essani AB, Volin MV, Amin MA, et al. Ligation of TLR5 promotes myeloid cell infiltration and differentiation into mature osteoclasts in rheumatoid arthritis and experimental arthritis. J Immunol. 2014;193:3902–3913. doi: 10.4049/jimmunol.1302998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ceribelli A, Nahid MA, Satoh M, Chan EK. MicroRNAs in rheumatoid arthritis. FEBS Lett. 2011;585:3667–3674. doi: 10.1016/j.febslet.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ammari M, Jorgensen C, Apparailly F. Impact of microRNAs on the understanding and treatment of rheumatoid arthritis. Curr Opin Rheumatol. 2013;25:225–233. doi: 10.1097/BOR.0b013e32835d8385. [DOI] [PubMed] [Google Scholar]
- 33.Chen SY, Shiau AL, Li YT, Lin YS, Lee CH, Wu CL, et al. Suppression of collagen-induced arthritis by intra-articular lentiviral vector-mediated delivery of Toll-like receptor 7 short hairpin RNA gene. Gene Ther. 2012;19:752–760. doi: 10.1038/gt.2011.173. [DOI] [PubMed] [Google Scholar]
- 34.Alzabin S, Kong P, Medghalchi M, Palfreeman A, Williams R, Sacre S. Investigation of the role of endosomal Toll-like receptors in murine collagen-induced arthritis reveals a potential role for TLR7 in disease maintenance. Arthritis Res Ther. 2012;14:R142. doi: 10.1186/ar3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murata K, Yoshitomi H, Tanida S, Ishikawa M, Nishitani K, Ito H, et al. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2010;12:R86. doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehmann SM, Kruger C, Park B, Derkow K, Rosenberger K, Baumgart J, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 37.Yang M, Chen J, Su F, Yu B, Lin L, Liu Y, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Savina A, Furlan M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278:20083–20090. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 39.King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 41.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 42.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]