Abstract
Understanding how multiple stressors interact is needed to predict the dynamical outcomes of diverse biological systems, ranging from drug-resistant pathogens that are combated and treated with combination drug therapies to ecosystems impacted by environmental toxicants or disturbances. Nevertheless, extensive studies of higher-order (more than two component) interactions have been lacking. Here, we conduct experiments using 20 three-drug combinations and their effects on the bacterial growth of Escherichia coli. We report our measurements of growth rates in single, pairwise and triple-drug combinations. To uncover emergent interactions, we derive a simple framework to calculate expectations for three-way interactions based on the measured impact of each individual stressor and of each pairwise interaction. Using our framework, we find that (i) emergent antagonisms are more common than emergent synergies and (ii) emergent antagonisms are more common and emergent synergies are more rare than would be inferred from measures of net effects that do not disentangle pairwise interactions from three-way interactions.
Keywords: multiple stressors, higher-order interactions, synergy, antagonism, drug resistance, antibiotics
1. Introduction
Drugs are now a pervasive part of our everyday environment and have both helpful and harmful effects on biological systems from the molecular up to the individual, population and whole ecosystem level [1]. In the clinic, drugs are used to combat pathogens but resistance to drugs such as antibiotics is becoming more common, largely because these drugs are used so pervasively throughout our environment, from hand soaps to agriculture. One strategy for countering antibiotic resistance is to use drugs in combination to more effectively kill pathogens, to combat drug-resistant strains and to help slow the evolution of resistance [2–5]. Ideally, this strategy would be used in tandem with the development of new drugs, but pharmaceutical companies are not investing heavily in antibiotics [6]. Thus, there is a compelling case for the critical importance of devising effective antibiotic combinations for use in the clinic. Past work has largely focused on two-drug interactions [2,3,7,8], but multi-drug therapies in the clinic are increasingly moving in the direction of higher-order combinations (those involving three or more drugs). Indeed, some of the best-known drug treatments, including HIV drug cocktails [9] and treatments for Mycobacterium tuberculosis infections [10], involve three-drug combinations.
To find effective higher-order drug combinations, one of the key challenges is to correctly identify the type and magnitude of drug interactions because combining non-interacting drugs does not leverage the benefits of certain interaction types. A useful categorization when two drugs are combined is: (i) synergistic—the interaction of the two drugs enhances the effect expected based on each drug alone with no interactions; (ii) antagonistic—the interaction reduces the expected effects; or (iii) additive—there is no interaction and the combined effect matches the expected effect. Synergistic drug combinations are useful because they kill bacteria more effectively and are advantageous for individual patients, while antagonistic drug combinations are useful because they may be able to help slow the evolution of antibiotic resistance [4,11,12]. In this paper, we extend ideas about these categorizations to higher-order interactions, including both net (arising from either pairwise or three-way combinations) and emergent (not arising solely from pairwise combinations) interactions.
Empirical studies of ecotoxicology and ecological disturbances have examined the nature of interactions in the laboratory [13–15] and in the wild [16,17]. Both synergistic [18–25] and antagonistic [23,26–29] interactions have been uncovered in a wide range of organisms, environments and systems that range from gene epistasis [30–32] to predator–prey interactions [33]. Nevertheless, higher-order interactions that involve more than two stressors are still poorly understood. Moreover, because three-drug combinations have received substantially less attention than two-drug combinations, there are many basic questions about three-way and higher-order interactions that remain, such as: Are there emergent properties that arise in three-drug combinations that cannot be predicted from the pairwise parts? As we increase the number of drugs, do we increase the proportion of synergistic or antagonistic interactions?
This lack of understanding and answers for three-way and higher-order interactions is partly due to the difficulty of obtaining measurements for the effects of all single, pairwise and higher-order combinations and partly due to the lack of a rigorous quantitative and conceptual framework that distinguishes between net and emergent interactions. Here, we help address both issues by using a tractable, empirical laboratory system in which Escherichia coli is exposed to environments of antibiotic combinations that allows measurements of effects of all subsets of drug combinations, and by developing an explicit and rigorous theoretical framework that encompasses both net and emergent higher-order interactions.
Empirically, as a model system for studying and addressing outstanding questions about interactions among stressors, antibiotics and bacteria offer several advantages: (i) the control of levels of the drugs in the environment and of fluctuations in concentration across time and space; (ii) the use of a specific ordered set or randomized set of antibiotics; and (iii) knowledge about the mechanisms of action for the antibiotics used in this study, allowing the selection of specific pathways in the bacteria that we want to disrupt. Moreover, the use of well-designed experiments in highly simplified microbial systems can make even complex problems tractable [34].
We also develop a novel theoretical framework to understand and to quantify which higher-order combinations of stressors produce emergent interactions, meaning that the interaction does not arise from single and pairwise interactions alone. For example, in the system of drugs as stressors, a three-drug synergy or antagonism may not actually be the result of all three drugs in combination. Instead, such interactions may come from an interaction of just two of the drugs. On the other hand, it may be possible that synergy can arise in a three-drug combination in which no two-drug interactions show synergy, i.e. an emergent synergy. This is an important distinction because true synergies—those that only emerge with all three drugs—could provide novel treatments, whereas a three-way synergy that merely arises from a synergistic pairwise interaction would not be particularly novel. Such superficial three-drug synergies may be detrimental to the patient because additional drugs might be added that are in actuality not needed for increased pathogen killing efficiency. As a result, it is challenging to determine which specific drug combinations are most clinically relevant [3,5,35].
Key advances were made in the few previous studies that focused on three (or more) drug interactions. In particular, Wood et al. [36] applied maximum entropy methods to six combinations of three antibiotics that varied across a range of concentrations. In this way, they searched for three-way interactions that could not be predicted from pairwise interactions. As part of this study, they discovered a simple and highly informative algebraic metric related to the one for emergent interactions we derive below. More recently, Zimmer et al. [37] used a framework that incorporates interaction coefficients as part of a model based on Hill functions to show how to increase predictive power for three-way interactions based on limited information about pairwise interactions across a range of drug concentrations.
Our work shares some core goals and similarities with these previous approaches. Although our study is currently more limited with regard to understanding ranges of drug concentrations as studied by Wood et al. [36] and Zimmer et al. [37], there are a few central contributions represented by our approach. First, our study provides a clear conceptual derivation for the simple algebraic measures presented below, and in so doing, further reveals a way to differentiate between the emergent interaction (an interaction that only exists when all three drugs are present) and the net interaction (the overall interaction in comparison to single-drug effects). Second, in contrast to other higher-order interaction studies, we also rescale the raw magnitude of our metrics in order to compare information and categories that correspond to baselines for synergistic and antagonistic interactions, as previously done by Segre and colleagues for pairwise interactions [38]. Third, we consider a much larger set of three-antibiotic combinations (20 as opposed to six [36]), though we only take measurements at fixed concentrations for these combinations. We also identify higher levels of net and emergent three-way (E3) interactions, including both synergy and antagonism, than previous studies on higher-order interactions, as explained in the Discussion section. As we will demonstrate below, the distinction between net versus emergent interaction provides compelling results, especially in the case of antagonistic interactions for combinations of antibiotics.
More generally, for any system involving more than two stressors, it is not obvious a priori whether the higher-order combination will interact in ways that are easily predictable from their single or pairwise effects. Although the theory and experiments in this paper have been developed using antibiotics and bacteria as an example, we suggest the terms and concepts developed here could be usefully translated, at least as a starting point, to think about other stressors and systems. For instance, studies of how multiple predators affect prey population dynamics may provide a strong correspondence because the survival rates of prey species are analogous to the growth rates of bacteria in multiple drug environments (e.g. [39]) and appear in the multiple predator effect (MPE) metrics in the same exact form as in our metrics for net interactions (i.e. the deviation from additivity (DA) measure) below. Notably, MPE methods do not differentiate between net and truly emergent interactions or rescale the magnitude of their metrics to assess the information about interactions, which is why we think our framework might also be generalized to resolve questions about MPEs. In the Discussion, we provide more details on how the framework with drugs as stressors can be applied to MPEs and other systems that cover similar concepts to those described in this paper.
In the remainder of the paper, we explain our approach for quantifying and understanding higher-order interactions. In doing so, we create a framework to classify higher-order combinations of stressors by examining net three-drug interactions and comparing them with all three pairwise interactions for each three-drug combination. Next, we use this framework to uncover interactions among a set of six antibiotics by examining systematically all 20 of the possible three-drug combinations. In experiments with wild-type E. coli, we measure growth rates of bacteria in single, pairwise and three-drug combinations, and we identify emergent three-drug combinations that require all three antibiotics to yield synergistic or antagonistic interactions.
2. Material and methods
2.1. Experimental data
2.1.1. Escherichia coli strain and growth conditions
The strain of E. coli used in these experiments is BW25113, the wild-type strain (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) [40] derived from the strain W1485 background [41]. All experiments were conducted using LB media (10 g l−1 tryptone, 5 g l−1 yeast extract and 10 g l−1 NaCl). Frozen glycerol cultures, stored at −80°C, were made by inoculating from a culture made from a single colony. Experiments were started by inoculating from a resuspension of this glycerol culture in MC buffer stored at 4°C and grown for 5 h in a 37°C incubator before being used to seed overnight cultures. Overnight cultures were seeded by inoculating 975 μl cultures with 25 μl of a 10−4 dilution of the over-day culture. After 18 h of incubation at 37°C in a shaker at 215 r.p.m., the optical density at 600 nm (OD600) was measured.
2.1.2. Antibiotics
Antibiotics included in this survey were chosen to cover a broad range of biochemical classifications [42]. Drugs included are clindamycin hydrochloride (Sigma C-5269), ciprofloxacin hydrochloride (MP Biomedicals 199020), tobramycin sulfate (Sigma T-1783), streptomycin sulfate (Sigma Aldrich S-6501), cefoxitin sodium salt (Fluka C4786) and erythromycin (Sigma Aldrich E-6376). Mechanisms of action, abbreviations and dosages are listed in table 1.
Table 1.
List of all antibiotics used in the study, abbreviation, dose used and mechanism of action.
| drug (abbreviation) | dose range (µg ml−1) | main mechanism(s) of action |
|---|---|---|
| clindamycin (CLI) | 31.5 | protein synthesis, 50S |
| ciprofloxacin (CPR) | 0.010–0.013 | DNA gyrase |
| tobramycin (TOB) | 1.3 | aminoglycoside, protein synthesis, 30S |
| streptomycin (STR) | 4.5–5.0 | aminoglycoside, protein synthesis, 30S |
| cefoxitin (FOX) | 1.16 | cell wall |
| erythromycin (ERY) | 12.0–14.0 | protein synthesis, 50S |
2.1.3. Growth measurements of no drug and single, double and triple antibiotic combinations
A range of concentrations was first tested for each individual drug to determine the appropriate non-lethal concentration required to reduce growth by 15–35% compared with the no drug control (LB). We choose this amount of reduction in growth rate because larger reductions would yield lethality in two-drug combinations, making three-drug effects irrelevant, and also because smaller reductions may make it difficult to tease apart additivity and antagonism. The range from 15 to 35% was necessitated by the variability in antibiotic sensitivity of single-drug treatments in our empirical system, representing a limit to our ability to choose the exact same reduction in growth rate across all drugs and experiments. We use an additive design, meaning that we test bacterial response to a set concentration of X (denoted [X]), a set concentration of Y (denoted [Y]), a set concentration of Z (denoted [Z]) and all pairwise combinations, [X] + [Y], [Y] + [Z] and [X] + [Z], and the triple combination [X] + [Y] + [Z]. This additive design is standard in the field of drug interactions [3], and we use the additive design to standardize with other drug interaction studies.
To measure the effect of the triple combination of drugs versus the pairwise and individual effects, each experiment was performed using a no-drug control, a control for each individual drug at the previously determined concentration, pairwise combinations of the three drugs and a triple-drug combination (electronic supplementary material, table S1). We also examine dose dependence for ten different drugs, including the six drugs used in the triple combination experiments. For each drug, we measure effects of concentrations [X], [2X] and [3X] (electronic supplementary material, table S2) according to the metrics defined here (equations (2.2) and (2.3)). These data show that our methods usually classify drugs as additive with themselves, meaning that the growth rates predicted by wXwX or wXwXwX based on concentration [X] match with the measured effects from drug concentrations of [2X] and [3X], respectively.
In all cases, drugs were added to appropriate concentrations in 1 ml LB (inoculated as described above), and 100 μl was aliquoted into four to six wells of a 96-well plate. Cell densities were then determined after 18 h of incubation (as described above) using optical density analysis at 600 nm. Optical density readings were used to calculate growth percentages as compared to the no-drug control. Each three-drug combination experiment was repeated at least three times, but due to minor changes in drug concentrations across different experiments, the number of replicates reported for each specific concentration of drugs may be two in some cases. Data are represented as median, minimum and maximum for repeated three-drug combination experiments. Graphs of triple antibiotic interactions were produced using Matlab v. R2013a.
2.2. Theoretical framework
2.2.1. Pairwise interaction measure
In previous work on pairwise interactions of drugs [35] and metabolic genes [38], drug interactions have often been classified using the definition of Bliss Independence (BI) [3]. BI defines drugs to be independent (or additive) when the per cent change in bacterial fitness in the presence of one drug (X) does not depend upon the presence or the absence of the other drug (Y) (see equation (2.1)). The per cent change in fitness can be measured by the relative fitness, defined as the ratio of growth of the bacteria in the presence of a drug relative to the growth when no drug is present. For instance, in the presence of drug X, the bacteria has relative fitness, 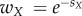 , where SX is the selection coefficient that measures how a specific drug concentration of X affects the bacterial fitness. We follow the same notation for other drugs Y or Z or any combination of drugs. According to BI, in the presence of two non-interacting drugs, the fitness of the bacteria is
, where SX is the selection coefficient that measures how a specific drug concentration of X affects the bacterial fitness. We follow the same notation for other drugs Y or Z or any combination of drugs. According to BI, in the presence of two non-interacting drugs, the fitness of the bacteria is 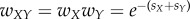 , where the XY subscript denotes a combination of drugs X and Y (denoted by X + Y in our figures) and is consistent with previous notation.
, where the XY subscript denotes a combination of drugs X and Y (denoted by X + Y in our figures) and is consistent with previous notation.
This non-interacting case is called additive because the selection coefficients add together, SX + SY. Consequently,
| 2.1 |
measures the DA. By definition, this measure is zero when X and Y are additive. When this measure is positive and sufficiently large in magnitude (see below), the interaction is called antagonistic because the drugs are working against each other such that the bacterial growth is higher than would be expected based on the two single-drug effects and no interactions between the drugs (i.e. additivity). When this measure is negative and sufficiently large in magnitude, the interaction is synergistic because the drugs are working together such that the bacterial growth rate is lower than would be expected based on the two single-drug effects and no drug interactions. The additive range defined below in terms of the rescaled measure is based on conservative values described by Yeh et al. [35]. When considering pairwise combinations of X and Y with a third drug, Z, there are two more pairwise DA measures that describe X interacting with Z, ɛX,Z = wXZ − wXwZ, and Y interacting with Z, ɛY,Z = wYZ − wYwZ.
2.2.2. Three-way interaction measures
We now develop two interaction measures for three-drug combinations. The first is the direct extension of the DA measure for two drugs in equation (2.1) [36,43,44]. When the drugs do not interact, the fitness of the bacteria exposed to the three-drug combination should be equal to the product of the fitnesses of bacteria exposed to each single drug alone, i.e. wXYZ = wXwYwZ. Therefore, DA, which measures the net interaction, is given by
| 2.2 |
To characterize emergent interactions that arise when all three drugs are combined, we need to assess how much of the three-drug interaction does not originate from pairwise interactions. To measure how much of the interaction arises from a single pairwise interaction, we recognize that a single pairwise interaction creates the entire three-drug interaction when the third drug is additive with the other two drugs. That is, the interaction between drugs X, Y and Z is due solely to the interaction between drugs X and Y when drug Z is additive with their combination (e.g. wXYZ = wXYwZ). For this special case, ɛX,Y,Z = wXYwZ − wXwYwZ = wZ(wXY − wXwY) = wZɛX,Y based on equations (2.1) and (2.2). By identical reasoning, the contribution coming solely from the interaction of drugs Y and Z is wXɛY,Z and from the interaction of drugs X and Z is wYɛX,Z. For three drugs, these three terms represent all three of the possible pairwise interactions among single drugs. Therefore, all three of these pairwise contributions need to be subtracted from the overall three-drug interaction (electronic supplementary material, figure S1) in order to introduce a new measure that we term the E3 interaction
| 2.3 |
When all of the pairwise interactions are additive (i.e. wXY = wXwY etc.), no part of the three-way interaction could possibly originate from pairwise interactions, and the E3 measure reduces to the DA measure 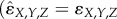 ), as it must. Substituting equations (2.1) and (2.2) into equation (2.3) allows the E3 measure to be expressed purely in terms of relative fitnesses
), as it must. Substituting equations (2.1) and (2.2) into equation (2.3) allows the E3 measure to be expressed purely in terms of relative fitnesses
| 2.4 |
In summary, by construction our new E3 measure provides a simple calculation for capturing the part of the three-drug interaction that is emergent and not due to pairwise interactions, while the DA measure captures whether there is an interaction at all.
2.2.3. Re-scaled three-way interaction measures
Following Segre et al. [38], we rescaled both of our interaction measures, DA (ɛX,Y,Z in equation (2.2)) and E3 (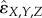 in equation (2.4)), by dividing them by the absolute value of the same functional form as the unscaled metrics, but with wXYZ replaced by 0 when the unscaled metric is negative (synergistic) and by the minimum value of the single-drug fitnesses, min (wx, wY, wZ), when the unscaled metric is positive (antagonistic) [43,44]. Effectively, this rescaling allows us to characterize the degree of synergy relative to the extreme lethal synergy case—when the combination of drugs completely kills the bacteria so that fitness (wXYZ) is 0, even though no single drug completely killed the bacteria. All positive interactions are rescaled by the case of buffering antagonism—when drugs combine to have the same effect as the single drug with the strongest effect.
in equation (2.4)), by dividing them by the absolute value of the same functional form as the unscaled metrics, but with wXYZ replaced by 0 when the unscaled metric is negative (synergistic) and by the minimum value of the single-drug fitnesses, min (wx, wY, wZ), when the unscaled metric is positive (antagonistic) [43,44]. Effectively, this rescaling allows us to characterize the degree of synergy relative to the extreme lethal synergy case—when the combination of drugs completely kills the bacteria so that fitness (wXYZ) is 0, even though no single drug completely killed the bacteria. All positive interactions are rescaled by the case of buffering antagonism—when drugs combine to have the same effect as the single drug with the strongest effect.
Our re-scaled E3 measure is similar to a term introduced by Darroch [45] and Kroonenberg & Andersen [46], who considered regression models and interaction terms. Importantly, our measure differs from these regression models because we assume nonlinear exponential fitness functions rather than linear approximations implied by linear dependencies for single-drug effects in these previous regression models. This linear approximation will be especially problematic for drug interactions that push growth rates more towards lethality (often synergy) or wild-type (often antagonism) and correspond to the exact regions where this linear approximation must break down. Moreover, these previous studies always re-scale according to our synergistic case, meaning they are using the wrong baseline or scale bar for antagonistic interactions. Finally, they either assume or hypothesize that their measure is always zero, meaning they assume there is no interaction before even comparing with data.
2.2.4. Cut-off values for determining three-way interaction types
Because of the variability between different interaction measures, and between fractional inhibitory concentration index data [47], cut-off values for determining three-way interaction types must be chosen cautiously. Our method for determining the type of three-way interaction follows previous work [35,38]. To calculate our rescaled three-way interaction measures, we use the median of replicate measurements for each experiment at a single antibiotic concentration. We then use the conservative cut-off values of rescaled ɛX,Y,Z (or rescaled 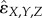 ) > 0.5 for antagonism and rescaled ɛX,Y,Z (or rescaled
) > 0.5 for antagonism and rescaled ɛX,Y,Z (or rescaled 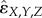 ) <−0.5 for synergy. Note that rescaled interaction measures tend to range from values of −1 (synergistic lethality) to 1 (complete antagonistic buffering). Hence, any value between −0.5 and 0.5 represents an additive interaction. These cut-off values are also based on natural breaks in the histogram distribution for the rescaled epsilon value and are consistent with the conservative values described for pairwise interactions by Yeh et al. [35].
) <−0.5 for synergy. Note that rescaled interaction measures tend to range from values of −1 (synergistic lethality) to 1 (complete antagonistic buffering). Hence, any value between −0.5 and 0.5 represents an additive interaction. These cut-off values are also based on natural breaks in the histogram distribution for the rescaled epsilon value and are consistent with the conservative values described for pairwise interactions by Yeh et al. [35].
3. Results
3.1. Identification of triple-drug interactions
Comparing the two distinct interaction measures, as depicted using schematics with idealized data (figures 1 and 2) and explained above we found that three-drug combinations can in principle have several distinct effects—additive according to either or both measures, synergistic according to either or both measures, and antagonistic according to either or both measures (electronic supplementary material, figure S2 and table S1). Experimental data are shown in triple-drug combination figures for examples of both synergistic (figure 3a) and antagonistic (figure 3b) interactions according to our E3 interaction measure, which captures three-drug interactions that do not arise from pairwise interactions.
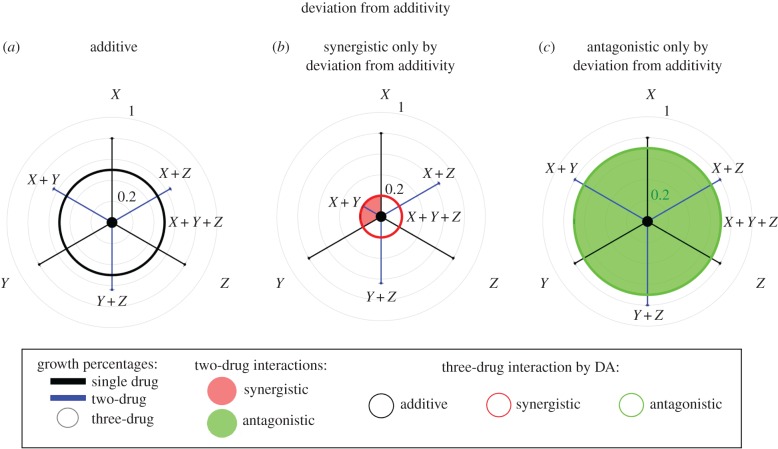
Figure 1.
Schematic of triple-drug combination effects according to DA. Relative growth rates for bacteria populations in one-, two- and three-drug combinations are shown as increasing from 0% growth in the centre to 100% growth (no-drug control) at the largest outer circle of the polar graph. Growth rate is shown for single-drug (black bars), two-drug (blue bars with corresponding single-drug treatments adjacent) and triple-drug combinations (radius of inner circle). Shading of the inner circle indicates nature of pairwise interactions (red, synergistic; green, antagonistic; no shading, additive). Outline colouring of inner circle indicates measure of DA (black, additive; red, synergistic; green, antagonistic). Three possible triple-drug combination effects are depicted schematically for theoretical drugs X, Y and Z according to measures of DA. (a) Strictly additive interactions of all pairwise combinations and three-drug combination. (b) Synergy of three drugs according to DA. (c) Antagonistic interaction of three drugs according to DA.
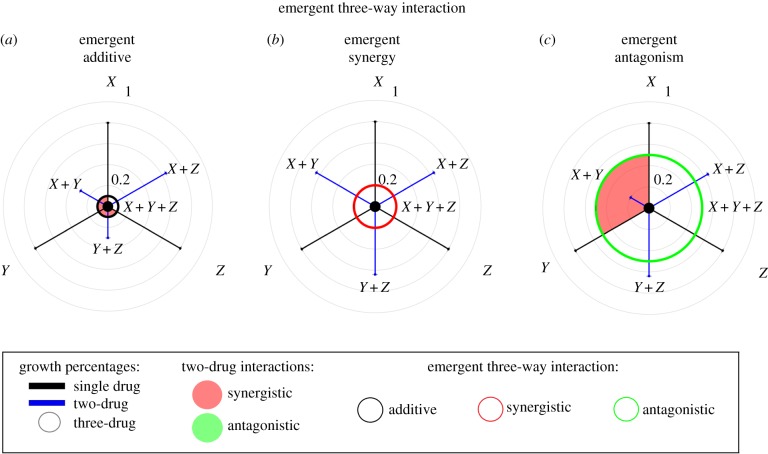
Figure 2.
Schematic of triple-drug combination effects according to E3 interactions. Relative growth rates for bacteria populations in one-, two- and three-drug combinations are shown as increasing from 0% growth in the centre to 100% growth (no-drug control) at the largest outer circle of the polar graph. For the E3 interaction measure, the single, two- and triple-drug combination growth rates, as well as pairwise interactions, are shown with the same colour coding as used for the DA measure (figure 1), but with outline colouring of the inner circle indicating measure of E3 (black, additive; red, synergistic; green, antagonistic). Three possible triple-drug combination effects are depicted schematically for theoretical drugs X, Y and Z according to measures of E3. (a) Interactions combine additively according to E3, even though several pairwise interactions are synergistic. (b) Emergent synergy of three drugs according to E3. (c) Emergent antagonism of three drugs according to E3.
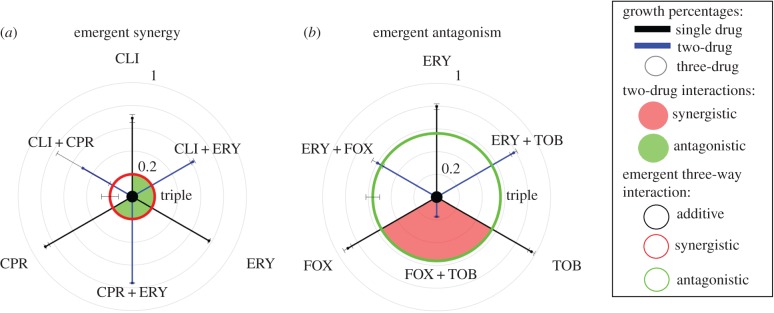
Figure 3.
Emergent synergistic and antagonistic interactions in triple antibiotic combinations. Data for triple antibiotic figures are shown according to E3 interaction measures. Figures are presented as described in figures 1 and 2. Data are represented as median ± minimum/maximum. (a) The combination of ciprofloxacin 0.013 µg ml−1 (CPR), clindamycin 31.5 µg ml−1 (CLI) and erythromycin 14 µg ml−1 (ERY) interacts synergistically in three-drug combinations according to both DA and emergent (E3) measures. (b) The combination of ERY 14 µg ml−1, cefoxitin 1.16 µg ml−1 (FOX) and tobramycin 1.3 µg ml−1 (TOB) interacts antagonistically in three-drug combinations according to the E3 measure and additively according to the DA measure. Only data for one concentration of each antibiotic are shown, although each three-drug combination was tested in a minimum of three independent experiments (see Material and methods).
Overall, examination of all 20 of our three-drug combinations revealed that 35% exhibit emergent, higher-order interactions, including two emergent synergistic interactions (figure 3a and electronic supplementary material, figure S2) and five emergent antagonistic interactions (figure 3b and electronic supplementary material, figure S2).
3.2. Deviation from additivity versus emergent three-way interactions
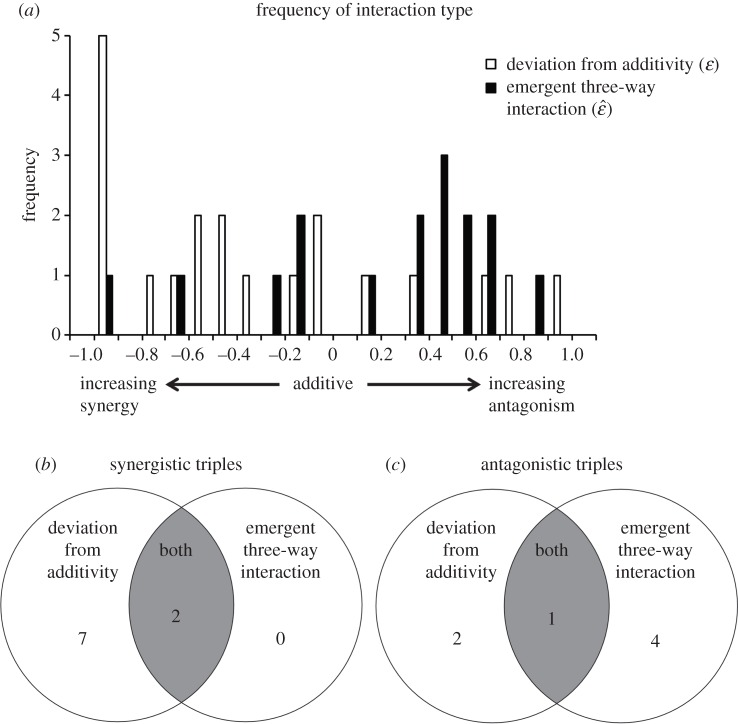
Our data show that when we categorized interaction types according to the DA measure, the distribution of interactions was skewed towards synergistic effects (figure 4a, skewness = 0.71). Conversely, when we used the E3 measure, the distribution skewed towards antagonistic effects (figure 4a, skewness = −0.98). We also compared the number of synergistic and antagonistic interactions according to each method (figure 4b,c). We found nine of 20 cases to be synergistic according to the DA measure. That is, the growth of the triple combination was often lower than the expected growth based on single-drug effects. However, only two of these cases were synergistic according to the E3 measure. Because the E3 measure leads to fewer synergistic classifications than the DA measure, the synergistic effects measured by DA are often the result of synergistic pairwise interactions, rather than an emergent interaction of the three drugs. Conversely, only three of 20 combinations were antagonistic according to the DA measure whereas five of 20 cases were found to be antagonistic according to the E3 measure, meaning that the growth of bacteria under triple-drug combinations was considerably higher than the expected growth based on pairwise interactions. Frequencies of specific antibiotics involved in emergent synergistic and antagonistic three-drug interactions are given in the electronic supplementary material, figure S3.
Figure 4.
Comparing interaction measures for triple-drug combinations. Comparison of measures of DA and E3 interactions are based on 20 triple-drug combination experiments. Of the 20 combinations, E3 is non-applicable in four cases because of both two-drug and three-drug lethality. Thus, it is impossible to see the effect of the third drug. DA is applicable in all cases because this measure ignores two-drug effects. (a) Overlapping histograms for measures of DA (white bars) and E3 interactions (black bars) with bin size of 0.1. (b) Venn diagram comparing overlap between synergistic three-drug interactions according to measures of DA and E3 interactions. (c) Venn diagram comparing overlap between antagonistic three-drug interactions according to measures of DA and E3 interactions. In both (b) and (c), the shaded area indicates the combinations which are fully synergistic or antagonistic, that is, synergistic in both DA and E3 measurements, or antagonistic in both measurements.
In the case of synergies, every emergent synergy was also considered synergistic as measured by DA (figure 4b). This was not true for antagonism, where most emergent antagonistic combinations (four out of five as measured by E3) were not also antagonistic by the DA measure (figure 4c). In addition, there were only two triple-drug combinations that were antagonistic solely from the DA measure, compared to seven triple-drug combinations that were synergistic solely from the DA measure.
4. Discussion
In this paper, we have conducted comprehensive experiments to measure all single, pairwise and three-way interactions for a set of six antibiotics. We also developed a metric to quantify whether there are interactions that arise only when all three drugs are present and are not simply a result of pairwise combinations, and we refer to this type of interaction as an E3 interaction. Using these experiments and theory, we systematically investigated three-drug interactions for our set of six antibiotics and showed that our new E3 measure is conceptually distinct and yields considerably different results from the default method for analysing interactions according to the DA measure. As we explained, this DA measure quantifies net three-way interactions that could be arising from interactions among either drug pairs or all three drugs. Thus, the E3 measure can be used to identify three-component interactions that have emergent properties—whether synergistic or antagonistic.
Notably, we find higher levels of emergent drug interactions than previous work, including an impressive study by Wood et al. that involved six three-way antibiotic combinations at a range of concentrations (in contrast to our 20 three-way combinations at fixed concentrations). Interactions in these data were searched for by Wood et al. [36] using maximum entropy methods and more recently by Zimmer et al. [37] using a model based on Hill functions and interaction coefficients. Those studies concluded that the vast majority of three-way interactions are additive or can be predicted from pairwise interactions. This important difference in results and conclusions is likely because neither of these previous studies performed a rescaling based on maximum entropy or other methods to set a baseline expectation for synergy or antagonism as done here or in previous pairwise studies [35,38]. Indeed, some of us recently re-analysed Wood et al.'s data using a few rescaling methods and did find the existence of several higher-order emergent interactions [48].
In addition, these previous studies did not explicitly distinguish between net interactions and emergent interactions as done here, and thus did not look for patterns that we find, such as emergent (E3) higher-order interactions tending to be more antagonistic than net (DA) higher-order interactions. Indeed, our results suggest emergent synergies in three-drug combinations are infrequent but do exist. Before further investigation of these emergent synergies, however, it is important to note that there are cases of emergent synergies that may not be clinically advantageous. From a clinical standpoint, designing the most effective drug treatments requires using three-drug combinations that have a net interaction—producing more effect than expected based on single drugs—and that have an emergent interaction—all three drugs produce more effect than expected based on pairwise interactions. Otherwise, there is little to be gained by using the three-drug combination because more drugs would be used than may really be necessary. Consequently, the optimal three-drug combinations are likely those that show the same type of interaction according to both the DA and E3 measures, corresponding to the shaded intersection region in the Venn diagrams in figure 4b,c. We see that there are two such fully synergistic interactions (and one such fully antagonistic interaction) among the 20 combinations we studied.
This relative paucity of synergistic interactions can be better understood by taking a wider perspective and by looking at the entire histogram of the E3 measure that reveals the overall distribution of interaction types. Our study shows that when taking into account pairwise (as opposed to single-drug) effects, the amount of antagonism increases, as seen by the rightward shift of the distribution in figure 4a. According to the DA measure, it may appear that the addition of more drugs leads to greater synergism. However, when we look for emergent properties of three-component interactions according to the E3 measure, we find that emergent interactions are in fact more often antagonistic. Natural populations, such as soil environments, often have many different species, including multi-drug-resistant strains that produce many different antibiotics [49,50]. It therefore seems likely that in the wild the outcome of many interacting antibiotics would be antagonistic to attenuate the effects suffered by natural bacteria. Given that synergistic and antagonistic interactions are roughly equally represented among pairwise interactions [35], there is no obvious explanation for this potential paradox.
Interestingly, within the drug literature, synergistic interactions appear to garner more attention and research than antagonistic ones. In the clinic, the goal is to use the synergy as a positive to eradicate the entire population of harmful bacteria due to the greater killing efficiency of synergistic drugs. However, from a basic rather than applied perspective, there is no reason to place primacy on synergism because the percentage of antagonistic interactions, in both drug–drug and ecological-driver effects, is roughly equal to the percentage of synergistic interactions. (In drug studies, 26% were found to be synergistic and 37% antagonistic [35]; in terrestrial systems, 35% synergistic and 42% antagonistic [39]; and in marine systems, 36% synergistic and 38% antagonistic [51]). Moreover, our study suggests that antagonism becomes more frequent than synergism when searching for emergent higher-order interactions. Furthermore, antagonism itself is relevant and interesting when examining drug combinations, given that antagonistic drug combinations may be better at slowing down the evolution of resistance [12] and decreasing the likelihood of resistance evolving [11].
A natural extension of the methods and results in this paper would be to allow stressors to be adjusted across a large gradient by changing drug concentrations, thereby covering cases like those studied by others such as Wood et al. [36] and Zimmer et al. [37], to determine whether our findings about the prevalence of synergistic and antagonistic interactions still hold. Dose-dependent interactions with three or more drugs [37,52–54] have been studied by extending the Loewe additivity measure that classifies two-drug interactions based on lines of equivalent growth rates (isobolograms) across a range of concentrations of the combined drugs. In this regard, a study by Jonker et al. [55] uses Loewe additivity and provides a model that can test whether the interaction type is independent of the absolute concentrations of the combined drugs or dependent only upon the dose ratios of the drugs. Further work is required to extend our framework in order to identify emergent interactions when there is a gradient of drug concentrations.
Another possibility to consider for combinations of drugs is that stressors can occur in sequence, rather than simultaneously, and with different timings [16]. If drug interactions depend on the sequence and timing with which the drugs are administered, these factors could be optimized for pathogen treatments for patients. Therefore, sequential multi-stressor interactions are a topic that could be explored in the future by using and extending our new framework on emergent interactions.
This framework could also be generalized to other systems with three or more component interactions that require quantitative analysis. As an example of our framework applied to more than three components, we derive the emergent four-way interaction measure in the electronic supplementary material text S1. Although our experiments were carried out in a bacteria–antibiotics system, the questions addressed here about interactions are also relevant to larger scale systems in terms of ecological drivers and survival of populations, species and biodiversity. Most of the core questions in ecology revolve around how species interact with each other and the environment. Although several meta-analyses of ecological two-stressor interactions have been conducted, three-stressor interactions have been examined much less frequently (but see [51,56]). Crain and colleagues conducted a comprehensive review of how a third stressor affects two-stressor interactions in marine systems. In their paper, they quantified the pairwise interactions in the presence and absence of the third stressor, and they found that the addition of a third stressor often caused the interaction between the first two stressors to become more synergistic [51]. Similarly, Chen and colleagues used three-way ANOVA to analyse an empirical study with three stressors on zooplankton and amphibians. They found that the three-stressor combination had synergistic effects that harmed survival and reproduction of the two study species [56]. However, in these studies, it is difficult to determine what an emergent interaction is, because the effects of the three stressors combined together are not explicitly compared with all single and all pairwise stressor interactions. More discussion on the comparison of three-way ANOVA and our E3 interaction measure can be found in the electronic supplementary material, text S2.
Within the field of ecology, analogous to the idea of emergent properties from three-drug combinations is the idea of emergent effects from multiple predators, known as MPEs. There are very few empirical studies that examine higher-order interactions that result from three or more predators, and even fewer that examine the entire factorial of combinations (single predator effects, all pairwise effects and a three-predator effect). Indeed, we could find only three such studies, and these studies involved a single prey species [57–59]. In MPE studies that look for interactions among three predators, multiplicative models are typically used and are equivalent to the DA measure, where the fitness parameters are survival rates [57–60]. However, as discussed throughout this paper, DA is not capable of distinguishing truly emergent interactions that require all three predators and do not arise from pairwise interactions. Moreover, these studies have no equivalent method to our E3 measure for quantifying and identifying emergent interactions. In this sense, the generalized emergent measure (E3) we propose here should be highly informative in deciding whether complex predator–prey relationships are actually a result of subsets of simpler interactions (with less components). Indeed, recent work by Cheng et al. [61] shows the importance of subsets of three-way interactions for understanding emergence in a five species predator–prey model and suggests similar approaches may be fruitful.
Applying these ideas to ecological systems, however, will also require additional considerations. For example, Barrios-O'Neill et al. [62] examined how multiple predators affect prey when the predators are of different trophic levels and are allowed to evolve. They found that when there is a secondary predator one trophic level up, MPEs behave differently when involving native versus invasive predators, demonstrating how predator–prey systems and food web dynamics are often more complex than the interactions presented here using drugs and bacteria. We recognize this as a limitation of our system being applied to ecological questions that involve multiple species and trophic levels because our study involves only one species and one trophic level of ‘predator’ (antibiotics). Furthermore, antibiotics do not evolve in the ways predators do, thus simplifying our system but also limiting its generality. Nevertheless, we suggest that the measure derived here could aid in the quantification of interactions among large numbers of species, but careful reasoning needs to be applied to interpret the results for macro-organism studies on predator–prey interactions that contain a larger variety of behaviours and responses.
Although applying this framework across other systems will require further work, we can already identify intriguing connections between the E3 measure developed here and formulae from other fields. It is analogous to theoretical physics concepts such as the 3-point connected correlation or Ursell function in quantum field theory, and the third joint cumulant in statistical physics and statistics [56,57]. However, our measure differs from these because it is not defined with respect to expectation values, and the combination of two variables is through addition and not multiplication. In the previously mentioned work by Wood et al. [36], they insightfully noted that their maximum entropy calculations for determining interaction type were well described by a simple algebraic formula that they identified as the Isserlis theorem. In the case of three-way interactions, this formula can be re-expressed to show its equivalence to our emergent interaction measure E3. However, these two approaches yield substantially different interaction metrics for characterizing four-way (or higher) interactions since our method naturally includes all possible lower-order effects in the metric, whereas Wood and colleagues considered only pairwise and single-drug effects (see the electronic supplementary material, text S1). Importantly, Wood et al. [36] found this formula phenomenologically by using a computational maximum entropy method but did not provide a general derivation as above. Lastly, based on our starting definition of no interaction (i.e. BI), standard three-way ANOVA (even when log transformed) is not equivalent to our correct measure of emergent interactions (see the electronic supplementary material, text S2 and table S3).
In summary, we provide a tool for defining emergent interactions of multiple stressors. In the past, statistical methods, such as ANOVA and maximum entropy [36,57,59,63,64], have been used to search for interactions. The advantage of our approach to understanding higher-order emergent interactions by deriving a simple algebraic formula is that it: (i) explicitly distinguishes between net and emergent interactions, (ii) decomposes the interaction into its natural component pieces with exactly specified and empirically measurable coefficients (e.g. single-drug fitnesses) instead of an increasing number of free parameters that require fitting, (iii) greatly simplifies calculations, helping to avoid technical mistakes and rounding errors while also taking less time computationally, (iv) leads to re-scaling that allows consistency with previous pairwise analyses [38] and clearer identification of interactions, and (v) naturally generalizes to higher-order interactions in a way that unambiguously incorporates the effects of all combinations of subsets of components. In conclusion, using our framework to analyse higher-order interactions may be important for revealing emergent properties in medical, environmental and ecological systems.
Supplementary Material
Acknowledgements
We thank Tina Manzhu Kang and Borna Shirvani for assistance in the laboratory, and Alex Hall, Daniel Barrios-O'Neill and three anonymous reviewers through Axios Review for comments on the manuscript.
Data accessibility
The datasets for this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
P.J.Y. conceived of the project. C.B., V.M.S. and P.J.Y. designed experiments. C.B., Z.M., C.W., C.M. and E.V. performed experiments. C.B., E.T., Z.M., C.W., V.M.S. and P.J.Y. analysed the data. E.T., V.M.S., J.H.M. and P.J.Y. contributed reagents/materials/analysis tools. C.B., E.T., V.M.S. and P.J.Y. wrote the paper.
Competing interests
We declare we have no competing interests.
Funding
For funding we thank NIH Initiative to Maximize Student Development (C.B.), James S. McDonnell Foundation Complex Systems Scholar Award (V.M.S.), NSF DBI Career Award (V.M.S.) and UCLA Faculty Career Development Award (P.J.Y.).
References
- 1.Carson R. 1962. Silent spring. Boston, MA: Houghton Mifflin. [Google Scholar]
- 2.Loewe S. 1953. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung 3, 285–290. [PubMed] [Google Scholar]
- 3.Bliss CI. 1939. The toxicity of poisons applied jointly. Ann. Appl. Biol. 26, 585–615. ( 10.1111/J.1744-7348.1939.Tb06990.X) [DOI] [Google Scholar]
- 4.Yeh PJ, Hegreness MJ, Aiden AP, Kishony R. 2009. Drug interactions and the evolution of antibiotic resistance. Nat. Rev. Microbiol. 7, 460–466. ( 10.1038/nrmicro2133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalan L, Wright G. 2011. Antibiotic adjuvants: multicomponent anti-infective strategies. Expert Rev. Mol. Med. 13, e5 ( 10.1017/S1462399410001766) [DOI] [PubMed] [Google Scholar]
- 6.Cooper MA, Shlaes D. 2011. Fix the antibiotics pipeline. Nature 472, 32 ( 10.1038/472032a) [DOI] [PubMed] [Google Scholar]
- 7.Greco W, Bravo G, Parsons JC. 1995. The search for synergy: a critical review from a response surface perspective. Pharmoacol. Rev. 47, 331–385. [PubMed] [Google Scholar]
- 8.Chou TC. 2006. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 58, 621–681. ( 10.1124/pr.58.3.10) [DOI] [PubMed] [Google Scholar]
- 9.Hammer S, et al. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. New Engl. J. Med. 337, 725–733. ( 10.1056/NEJM199709113371101) [DOI] [PubMed] [Google Scholar]
- 10.Diacon AH, et al. 2012. 14-day bactericidal activity of PA-824, bedaquiline, pyrazinamide, and moxifloxacin combinations: a randomised trial. Lancet 380, 986–993. ( 10.1016/S0140-6736(12)61080-0) [DOI] [PubMed] [Google Scholar]
- 11.Michel J, Yeh P, Chait R, Moellering R, Kishony R. 2008. Drug interactions modulate the potential for evolution of resistance. Proc. Natl Acad. Sci. USA 105, 14 918–14 923. ( 10.1073/pnas.0800944105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hegreness M, Shoresh N, Damian D, Hartl D, Kishony R. 2008. Accelerated evolution of resistance in multidrug environments. Proc. Natl Acad. Sci. USA 105, 13 977–13 981. ( 10.1073/pnas.0805965105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mora C, Metzger R, Rollo A, Myers RA. 2007. Experimental simulations about the effects of overexploitation and habitat fragmentation on populations facing environmental warming. Proc. R. Soc. B 274, 1023–1028. ( 10.1098/rspb.2006.0338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cramp RL, Reid S, Seebacher F, Franklin CE. 2014. Synergistic interaction between UVB radiation and temperature increases susceptibility to parasitic infection in a fish. Biol. Lett. 10, 20140449 ( 10.1098/rsbl.2014.0449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folt CL, Chen CY, Moore MV, Burnaford J. 1999. Synergism and antagonism among multiple stressors. Limnol. Oceanogr. 44, 864–877. ( 10.4319/lo.1999.44.3_part_2.0864) [DOI] [Google Scholar]
- 16.Ross KA, Taylor JE, Fox MD, Fox BJ. 2004. Interaction of multiple disturbances: importance of disturbance interval in the effects of fire on rehabilitating mined areas. Austral Ecol. 29, 508–529. ( 10.1111/j.1442-9993.2004.01405.x) [DOI] [Google Scholar]
- 17.Christensen MR, Graham MD, Vinebrooke RD, Findlay DL, Paterson MJ, Turner MA. 2006. Multiple anthropogenic stressors cause ecological surprises in boreal lakes. Glob. Change Biol. 12, 2316–2322. ( 10.1111/j.1365-2486.2006.01257.x) [DOI] [Google Scholar]
- 18.Relyea RA. 2003. Predator cues and pesticides: a double dose of danger for amphibians. Ecol. Appl. 13, 1515–1521. ( 10.1890/02-5298) [DOI] [Google Scholar]
- 19.Boone MD, Semlitsch RD, Little EE, Doyle MC. 2007. Multiple stressors in amphibian communities: effects of chemical contamination, bullfrogs, and fish. Ecol. Appl. 17, 291–301. ( 10.1890/1051-0761(2007)017%5B0291:MSIACE%5D2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 20.Dávalos A, Nuzzo V, Blossey B. 2014. Demographic responses of rare forest plants to multiple stressors: the role of deer, invasive species and nutrients. J. Ecol. 102, 1222–1233. ( 10.1111/1365-2745.12279) [DOI] [Google Scholar]
- 21.Metz MR, Varner JM, Frangioso KM, Meentemeyer RK, Rizzo DM. 2013. Unexpected redwood mortality from synergies between wildfire and an emerging infectious disease. Ecology 94, 2152–2159. ( 10.1890/13-0915.1) [DOI] [PubMed] [Google Scholar]
- 22.Pringle RM, Kimuyu DM, Sensenig RL, Palmer TM, Riginos C, Veblen KE, Young TP. 2015. Synergistic effects of fire and elephants on arboreal animals in an African savanna. J. Anim. Ecol. 84, 1637–1645. ( 10.1111/1365-2656.12404) [DOI] [PubMed] [Google Scholar]
- 23.Lakeman-Fraser P, Ewers RM. 2014. Untangling interactions: do temperature and habitat fragmentation gradients simultaneously impact biotic relationships? Proc. R. Soc. B 281, 20140687 ( 10.1098/rspb.2014.0687) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shears NT, Ross PM. 2010. Toxic cascades: multiple anthropogenic stressors have complex and unanticipated interactive effects on temperate reefs. Ecol. Lett. 13, 1149–1159. ( 10.1111/j.1461-0248.2010.01512.x) [DOI] [PubMed] [Google Scholar]
- 25.Gooding RA, Harley CD, Tang E. 2009. Elevated water temperature and carbon dioxide concentration increase the growth of a keystone echinoderm. Proc. Natl Acad. Sci. USA 106, 9316–9321. ( 10.1073/pnas.0811143106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darling ES, McClanahan TR, Côté IM. 2010. Combined effects of two stressors on Kenyan coral reefs are additive or antagonistic, not synergistic. Conserv. Lett. 3, 122–130. ( 10.1111/j.1755-263X.2009.00089.x) [DOI] [Google Scholar]
- 27.Treasure AM, Chown SL. 2014. Antagonistic effects of biological invasion and temperature change on body size of island ectotherms. Divers. Distrib. 20, 202–213. ( 10.1111/ddi.12153) [DOI] [Google Scholar]
- 28.Annala M, Mykrä H, Tolkkinen M, Kauppila T, Muotka T. 2014. Are biological communities in naturally unproductive streams resistant to additional anthropogenic stressors? Ecol. Appl. 24, 1887–1897. ( 10.1890/13-2267.1) [DOI] [PubMed] [Google Scholar]
- 29.O'Regan SM, Palen WJ, Anderson SC. 2014. Climate warming mediates negative impacts of rapid pond drying for three amphibian species. Ecology 95, 845–855. ( 10.1890/13-0916.1) [DOI] [PubMed] [Google Scholar]
- 30.Vowels JJ, Thomas JH. 1992. Genetic analysis of chemosensory control of Dauer formation in Caenorhabditis elegans. Genetics 130, 105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottlieb S, Ruvkun G. 1994. daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics 137, 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gagiano M, van Dyk D, Bauer FF, Lambrechts MG, Pretorius IS. 1999. Msn1p/Mss10p, Mss11p and Muc1p/Flo11p are part of a signal transduction pathway downstream of Mep2p regulating invasive growth and pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Microbiol. 31, 103–116. ( 10.1046/j.1365-2958.1999.01151.x) [DOI] [PubMed] [Google Scholar]
- 33.Billick I, Case TJ. 1994. Higher order interactions in ecological communities: what are they and how can they be detected? Ecology 75, 1529–1543. ( 10.2307/1939614) [DOI] [Google Scholar]
- 34.Dai L, Korolev KS, Gore J. 2013. Slower recovery in space before collapse of connected populations. Nature 496, 355–358. ( 10.1038/nature12071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeh P, Tschumi A, Kishony R. 2006. Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 38, 489–494. ( 10.1038/ng1755) [DOI] [PubMed] [Google Scholar]
- 36.Wood K, Nishida S, Sontag ED, Cluzel P. 2012. Mechanism-independent method for predicting response to multidrug combinations in bacteria. Proc. Natl Acad. Sci. USA 109, 12 254–12 259. ( 10.1073/pnas.1201281109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmer A, Katzir I, Dekel E, Mayo AE, Alon U. 2016. Prediction of multidimensional drug dose responses based on measurements of drug pairs. Proc. Natl Acad. Sci. USA 113, 10 442–10 447. ( 10.1073/pnas.1606301113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segre D, DeLuna A, Church G, Kishony R. 2005. Modular epistasis in yeast metabolism. Nat. Genet. 37, 77–83. ( 10.1038/ng1489) [DOI] [PubMed] [Google Scholar]
- 39.Darling ES, Côté IM. 2008. Quantifying the evidence for ecological synergies. Ecol. Lett. 11, 1278–1286. ( 10.1111/j.1461-0248.2008.01243.x) [DOI] [PubMed] [Google Scholar]
- 40.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645. ( 10.1073/pnas.120163297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 20060008. ( 10.1038/msb4100050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott G, Kyi M. 2001. Handbook of essential antibiotics. Amsterdam, The Netherlands: Harwood Academic. [Google Scholar]
- 43.Sanjuán R, Elena SF. 2006. Epistasis correlates to genomic complexity. Proc. Natl Acad. Sci. USA 103, 14 402–14 405. ( 10.1073/pnas.0604543103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanjuán R, Nebot MR. 2008. A network model for the correlation between epistasis and genomic complexity. PLoS ONE 3, e2663 ( 10.1371/journal.pone.0002663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darroch JN. 1974. Multiplicative and additive interaction in contigency tables. Biometrika 61, 207–214. ( 10.1093/biomet/61.1.207) [DOI] [Google Scholar]
- 46.Kroonenberg PM, Anderson CJ. 2006. Additive and multiplicative models for three-way contingency tables: Darroch (1974) revisited. In Multiple correspondence analysis and related methods (ed. Blasius MGJ.), pp. 455–486. New York, NY: Chapman & Hall/CRC. [Google Scholar]
- 47.Odds FC. 2003. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 52, 1 ( 10.1093/jac/dkg301) [DOI] [PubMed] [Google Scholar]
- 48.Tekin E, Beppler C, White C, Mao Z, Savage VM, Yeh PJ. 2016. Enhanced identification of synergistic and antagonistic emergent interactions among three or more drugs. J. R. Soc. Interface 13, 20160332. ( 10.1098/rsif.2016.0332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D'Costa VM, McGrann KM, Hughes DW, Wright GD. 2006. Sampling the antibiotic resistome. Science 311, 374–377. ( 10.1126/science.1120800) [DOI] [PubMed] [Google Scholar]
- 50.Walsh F, Duffy B. 2013. The culturable soil antibiotic resistome: a community of multi-drug resistant bacteria. PLoS ONE 8, e65567 ( 10.1371/journal.pone.0065567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crain CM, Kroeker K, Halpern BS. 2008. Interactive and cumulative effects of multiple human stressors in marine systems. Ecol. Lett. 11, 1304–1315. ( 10.1111/j.1461-0248.2008.01253.x) [DOI] [PubMed] [Google Scholar]
- 52.Berenbaum M. 1978. Method for testing for synergy with any number of agents. J. Infect. Dis. 137, 122–130. ( 10.1093/infdis/137.2.122) [DOI] [PubMed] [Google Scholar]
- 53.Yu G, Baeder DY, Regoes RR, Rolff J. 2016. Combination effects of antimicrobial peptides. Antimicrob. Agents Chemother. 60, 1717–1724. ( 10.1128/AAC.02434-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jilek BL, Zarr M, Sampah ME, Rabi SA, Bullen CK, Lai J, Shen L, Siliciano RF. 2012. A quantitative basis for antiretroviral therapy for HIV-1 infection. Nat. Med. 18, 446–451. ( 10.1038/nm.2649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jonker MJ, Svendsen C, Bedaux JJ, Bongers M, Kammenga JE. 2005. Significance testing of synergistic/antagonistic, dose level-dependent, or dose ratio-dependent effects in mixture dose-response analysis. Environ. Toxicol. Chem. 24, 2701–2713. ( 10.1897/04-431R.1) [DOI] [PubMed] [Google Scholar]
- 56.Chen CY, Hathaway KM, Thompson DG, Folt CL. 2008. Multiple stressor effects of herbicide, pH, and food on wetland zooplankton and a larval amphibian. Ecotoxicol. Environ. Saf. 71, 209–218. ( 10.1016/j.ecoenv.2007.08.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sokol-Hessner L, Schmitz OJ. 2002. Aggregate effects of multiple predator species on a shared prey. Ecology 83, 2367–2372. ( 10.1890/0012-9658(2002)083%5B2367:AEOMPS%5D2.0.CO;2) [DOI] [Google Scholar]
- 58.Gunzburger MS, Travis J. 2005. Effects of multiple predator species on green treefrog (Hyla cinerea) tadpoles. Can. J. Zool. 83, 996–1002. ( 10.1139/z05-093) [DOI] [Google Scholar]
- 59.Van Son TC, Thiel M. 2006. Multiple predator effects in an intertidal food web. J. Anim. Ecol. 75, 25–32. ( 10.1111/j.1365-2656.2005.01019.x) [DOI] [PubMed] [Google Scholar]
- 60.McCoy MW, Stier AC, Osenberg CW. 2012. Emergent effects of multiple predators on prey survival: the importance of depletion and the functional response. Ecol. Lett. 15, 1449–1456. ( 10.1111/ele.12005). [DOI] [PubMed] [Google Scholar]
- 61.Cheng H, Yao N, Huang ZG, Park J, Do Y, Lai YC. 2014. Mesoscopic interactions and species coexistence in evolutionary game dynamics of cyclic competitions. Sci. Rep. 4, 7486 ( 10.1038/srep07486). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barrios-O'Neill D, Dick JTA, Emmerson MC, Ricciardi A, MacIsaac HJ, Alexander ME, Bovy HC. 2014. Fortune favours the bold: a higher predator reduces the impact of a native but not an invasive intermediate predator. J. Anim. Ecol. 83, 693–701. ( 10.1111/1365-2656.12155) [DOI] [PubMed] [Google Scholar]
- 63.Soluk DA, Collins NC. 1988. Synergistic interactions between fish and stoneflies: facilitation and interference among stream predators. Oikos 52, 94–100. ( 10.2307/3565987) [DOI] [Google Scholar]
- 64.Sih A, Englund G, Wooster D. 1998. Emergent impacts of multiple predators on prey. Trends Ecol. Evol. 13, 350–355. ( 10.1016/S0169-5347(98)01437-2) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets for this article have been uploaded as part of the electronic supplementary material.