Superresolution fluorescence microscopy of vertebrate kinetochore proteins reveals that outer kinetochore components assume diverse distribution patterns, including a ring-like configuration encircling the CENP-A–marked centromeric chromatin on the metaphase spindle in Xenopus egg extracts.
Abstract
The kinetochore is often depicted as having a disk-like architecture in which the outer layer of proteins, which engage microtubules and control checkpoint signaling, are built on a static inner layer directly linked to CENP-A chromatin. Here, applying three-dimensional (3D) structural illumination microscopy (SIM) and stochastic optical reconstruction microscopy (STORM) to Xenopus egg extracts and tissue culture cells, we report various distribution patterns of inner and outer kinetochore proteins. In egg extracts, a configuration in which outer kinetochore proteins surround the periphery of CENP-A chromatin is common, forming an ∼200-nm ring-like organization that may engage a bundle of microtubule ends. Similar rings are observed in Xenopus tissue culture cells at a lower frequency but are enriched in conditions in which the spindle is disorganized. Although rings are rare in human cells, the distribution of both inner and outer kinetochore proteins elongates in the absence of microtubule attachment in a manner dependent on Aurora B. We propose a model in which the 3D organization of both the outer and inner kinetochore regions respond to the progression from lateral to end-on microtubule attachments by coalescing into a tight disk from less uniform distributions early in prometaphase.
INTRODUCTION
Kinetochores attach chromosomes to microtubules and act as signaling hubs to control mitotic progression (Foley and Kapoor, 2013; Sacristan and Kops, 2015). The site of kinetochore assembly is epigenetically defined by the histone H3 variant CENP-A, which recruits the constitutive centromere–associated network (CCAN) proteins (Perpelescu and Fukagawa, 2011; Westhorpe and Straight, 2013). During mitosis, the CCAN further recruits a network composed of the KNL1 complex (KNL1C), the Mis12 complex (Mis12C), and the Ndc80 complex (Ndc80C; the KMN complex). Ndc80C is responsible for microtubule end-on attachment, whereas the KNL1 protein acts as a major scaffold for the transient spindle assembly checkpoint (SAC) proteins (including Bub1, BubR1, Bub3, Mad1, Mad2, the Rod/ZW10/Zwilch [RZZ] complex, and Spindly; Varma and Salmon, 2012; Silio et al., 2015) and the motor proteins dynein and CENP-E, which capture the lateral sides of microtubules (Rieder and Alexander, 1990; Wood et al., 1997; Kapoor et al., 2006). These motors and checkpoint proteins are localized to a “fibrous corona” region that can extend >100 nm into the cytoplasm from the outer kinetochore plate (McEwen et al., 1998; Hoffman et al., 2001). It remains to be established how components are organized in the large vertebrate kinetochore, which binds 15–30 microtubules, to execute diverse context- and cell cycle–dependent functions.
In the classic repeat subunit model, kinetochores are composed of arrays of microtubule-binding units, each built on one CENP-A nucleosome and associating CCAN, with the capacity to bind one microtubule fiber (Zinkowski et al., 1991; Santaguida and Musacchio, 2009). However, recent simulation analysis predicts that each microtubule fiber is not necessarily constrained to one binding unit but that instead microtubule-binding proteins form a dynamic lawn (Zaytsev et al., 2014). Superresolution analysis and low-salt unfolding of chicken centromeric chromatin led to a “boustrophedon” model of centromere chromatin folding (Ribeiro et al., 2010). The absence of repeating units in electron microscope (EM) tomographic reconstructions of vertebrate kinetochores led to a model of the outer kinetochore as a flexible network of intertwining fibers (Dong et al., 2007). EM tomography has visualized 25- to 75-nm fibers connecting centromeric chromatin directly to individual microtubule protofilaments as the predominant microtubule-interacting structure across species (McIntosh et al., 2008, 2013). Multiple microtubule-binding sites appear to cluster in each animal kinetochore, but the molecular identity of these fibers and how the various microtubule-interacting complexes are organized in three dimensions remains unknown.
The shape and composition of each kinetochore change, depending on its microtubule attachment status. A subset of CCAN proteins and the centromere itself stretch in response to microtubule-dependent forces (Wan et al., 2009; Suzuki et al., 2011, 2014), and stretching of CREST and Ndc80 signals has been reported in kinetochores with merotelic attachments (Cimini et al., 2001; Knowlton et al., 2006). In the absence of microtubule attachment, the corona expands beyond CENP-A chromatin (Thrower et al., 1996; Hoffman et al., 2001). Recent work using correlative EM and light microscopy along with live analysis supports a functional role for this transient expansion in establishing bipolar attachments (Magidson et al., 2015), and structured illumination microscopy (SIM) imaging shows that the human Ndc80 homologue Hec1, along with CENP-T, adopts an extended morphology on chromosomes that lack microtubule attachments (Magidson et al., 2016). In Xenopus egg extracts, we showed that kinetochore proteins responsible for lateral attachment and SAC signaling, but not those generating end-on attachment (Ndc80), expand to form micrometer-scale filamentous structures and strengthen the SAC signal (Wynne and Funabiki, 2015). Unexpectedly, among CCAN proteins, CENP-C is an integral component of these expanded kinetochores. It remains to be tested whether CENP-C expansion is unique to Xenopus egg extracts.
RESULTS AND DISCUSSION
Superresolution microscopy shows heterogeneous outer kinetochore organization in Xenopus egg extracts, including ∼200-nm rings
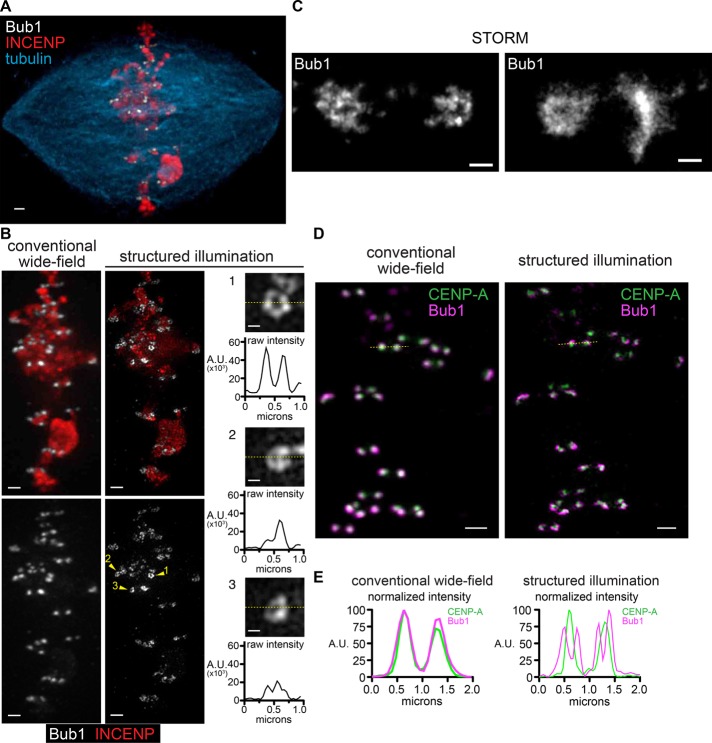
In an attempt to visualize kinetochores in Xenopus egg extracts using three-dimensional (3D) SIM, we observed that a subset of kinetochore proteins exhibit ring-like configurations on replicated sperm chromosomes in a metaphase spindle (Figure 1). Immunofluorescence recognizing Bub1, the outer kinetochore component, showed pairs of sister kinetochores that appeared as solid foci by conventional deconvolution microscopy due to the subdiffraction size of kinetochores (<200 nm; Figure 1, A and B). However, when these samples were imaged using 3D SIM, Bub1 signals showed a heterogeneous distribution, which often had regions of substantially reduced intensity in the center (Figure 1B), sometimes approaching background intensity levels (Figure 1B, inset 1). Similar ring-like distributions of Bub1 were observed using stochastic optical reconstruction microscopy (STORM; Figure 1C), suggesting that the rings observed in SIM were not an artifact of the image reconstruction. When samples were costained with antibodies against CENP-A and imaged with SIM, the CENP-A signals formed small foci adjacent to Bub1 and often in the center of the Bub1 rings (Figure 1, D and E). The dramatic difference in the configuration seen for Bub1 and CENP-A suggests that 3D SIM has adequate resolution in the xy-plane to distinguish kinetochore components.
FIGURE 1:
Superresolution imaging of kinetochore proteins in Xenopus egg extracts. (A) Bipolar spindle assembled in Xenopus egg extract, imaged using conventional wide-field deconvolution microscopy. Scale bar, 1 μm. (B) Comparison of images of the same spindle shown in A produced using conventional deconvolution (left) and SIM (middle). Scale bars, 1 μm. Higher-magnification images of single kinetochores shown with intensity scaling from 0 to the maximum value in each image; scale bars, 0.2 μm. Line scans corresponding to the dotted yellow line in each are shown below. (C) Kinetochores from samples as in A and B imaged using direct STORM. (D) Bub1 (magenta) and CENP-A (green) on a metaphase spindle. Images were produced using conventional deconvolution (left) and SIM (right). Scale bars, 1 μm. (E) Intensity plots for the yellow dotted lines in D in which values are normalized separately to 100 for the maximum intensity in each channel.
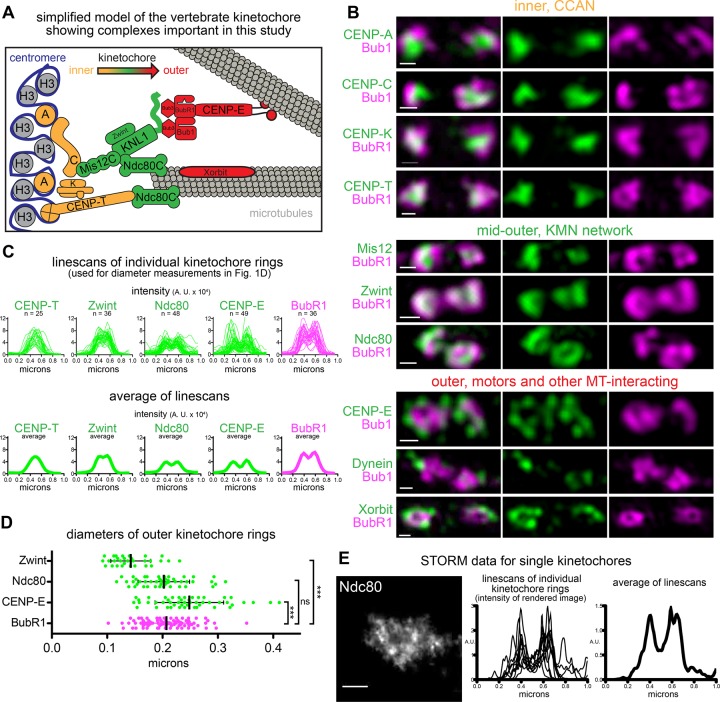
We next systematically tested the localization of components throughout the kinetochore by 3D SIM (Figure 2A). Whereas Mad2 did not show apparent kinetochore localization in the metaphase spindle, other checkpoint proteins—BubR1 and Mad1—exhibited ring-like configurations like that of Bub1 (Figure 2, B and C, and Supplemental Figure S1, A–D; Chen et al., 1998; Sharp-Baker and Chen, 2001; Chen, 2002). The CCAN members CENP-C, -K, and -T formed small foci that rarely showed reduced intensities in the center (Figure 2B, top). Like CENP-A, foci of these CCAN proteins often localized to the region of reduced signal in the center of the Bub1/BubR1 rings. The pattern of the KMN components differed among proteins. Whereas ring-shaped signals were also rare for Mis12, they were more apparent for Zwint, and Ndc80 signals formed robust rings with similar frequencies as BubR1 (Figures 2B, middle, and Supplemental Figure S1, A and B). The outermost microtubule-interacting proteins—CENP-E, dynein, and Xorbit/CLASP—localized on the external side of the rings either in foci (dynein) or discontinuous rings (CENP-E and Xorbit; Figure 2B, bottom).
FIGURE 2:
Systematic imaging of kinetochore proteins in Xenopus egg extracts. (A) Schematic of Xenopus kinetochore components emphasizing their location relative to the centromere (inner) and microtubule (outer). (B) SIM images of pairs of sister kinetochores, indicated as inner and outer kinetochore components (green), costained with Bub1 or BubR1 (magenta). Scale bars, 0.2 μm. (C) Line scans of kinetochores with ring-shaped BubR1 stained as in B. Top, overlays; bottom, average intensity. Average BubR1 line scan from Zwint-stained kinetochores. (D) Ring diameters calculated by interpolating individual line scans from C using cubic splines and finding the distances between the two intensity peaks. Each point represents an independent diameter measurement of one kinetochore; mean and SD are shown in black. Mann–Whitney test: ***p < 0.0001; ns, p = 0.4 (n = 30, 48, 48, and 90 for Zwint, Ndc80, CENP-E, and BubR1, respectively). (E) Kinetochores stained for Ndc80 and imaged with STORM (left; scale bar, 0.2 μm); individual (middle) and average (right) line scans generated for ring-shaped kinetochores.
We noticed that when signals showed ring-like configurations, the size of the ring correlated with the location of the protein along the inner–outer kinetochore axis. When line scans of multiple kinetochores with ring-like Bub1 or BubR1 distributions were averaged, the average showed two peaks for outer kinetochore signals, as expected for rings, whereas centromeric proteins such as CENP-T showed only a single peak (Figure 2C). Further, whereas average plots of BubR1, Ndc80, and CENP-E showed well-resolved peaks, there was only a small decrease in the average signal separating the Zwint peaks. This result is consistent with the limit of resolution of SIM, which is at best 115 nm for Alexa 488, because ring configurations with diameters close to this distance would not be well resolved using this system. Next we calculated the diameters of the ring-like signals by measuring the peak-to-peak distances of individual line scans and found that Ndc80, Bub1, and BubR1 produced rings with average diameters close to 200 nm (202, 184, and 207 nm, respectively), whereas the average diameters of Zwint (143 nm) and CENP-E (248 nm) rings were significantly smaller and larger (Figure 2D). Ndc80 visualized using STORM generated similar kinetochore line scans (Figure 2E and Supplemental Figure S2). Owing to the higher resolution of STORM, these line scans were noisier, but when multiple line scans of ring-like kinetochores were averaged, two strong peaks were seen separated by ∼200 nm, consistent with our measurements using SIM and with our STORM images of Bub1 (Figure 1C). The ring diameter measurements are roughly double the distances between each protein and CENP-A measured along the inner–outer kinetochore axis (Supplemental Figure S1E; Wan et al., 2009; Varma et al., 2013). A ring configuration can be explained by at least two models, which depend on the orientation of the kinetochore. Rings may result from en face views of kinetochores if the radial organization of the kinetochore correlates with the organization of complexes along the inner to outer kinetochore axis (Supplemental Figure S1F and model 2 in Supplemental Figure S4D). Alternatively, in side views of kinetochores, common when a bipolar spindle is orthogonal to the light path, rings suggest that outer kinetochore proteins surround the centromeric surface of the kinetochore (model 1 in Supplemental Figure S4D). If outer kinetochore material surrounds the inner centromere surface, this could be a minimal level of the kinetochore expansion that we recently characterized in extracts (Wynne and Funabiki, 2015).
Kinetochore rings may engage a bundle of microtubules
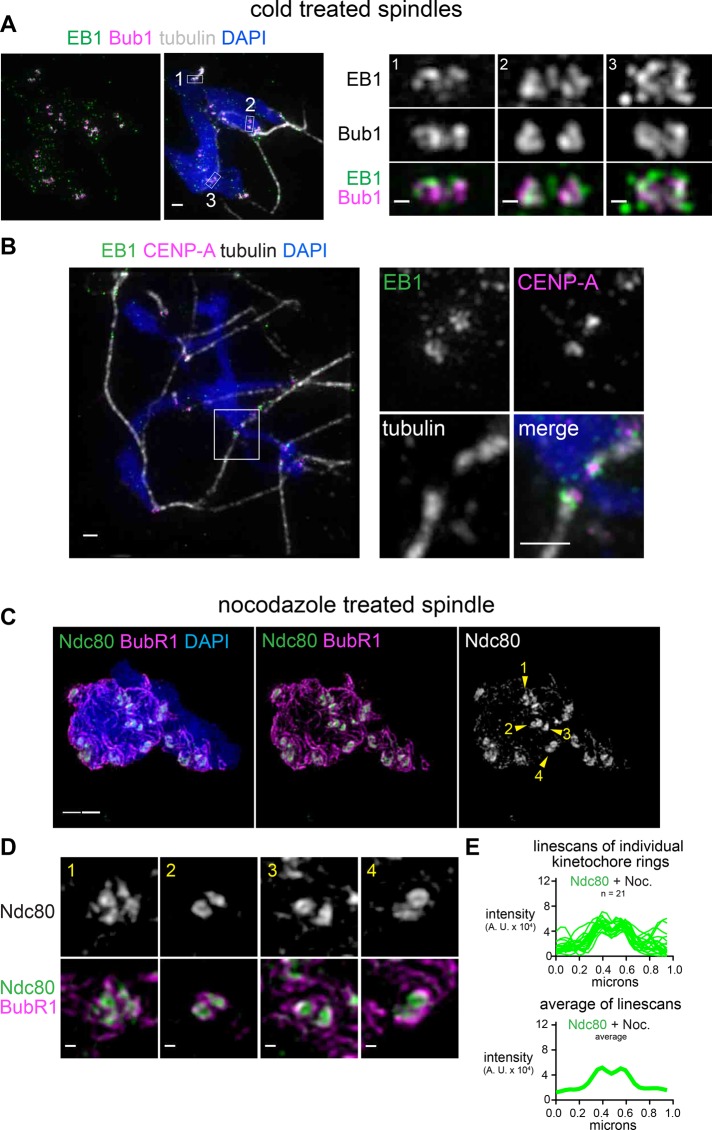
The critical question raised by the observation of rings is how these structures might engage microtubules. The apparent rings could be kinetochore proteins engaged in end-on attachments, or they could be outer kinetochore proteins that fail to contact microtubule ends and are excluded from the center of the rings, potentially by a dense bundle of microtubules. Direct visualization of kinetochore microtubules is difficult in extracts due to the large number of nonkinetochore microtubules. In an attempt to visualize how kinetochore rings interact with microtubule ends, we fluorescently conjugated recombinant EB1 protein and added it to extracts along with fluorescent tubulin. A high dose (33 μM) of nocodazole, which completely depolymerized microtubules, eliminated EB1 staining at the kinetochore (Supplemental Figure S3A), confirming that the EB1 localization at the kinetochore depends on microtubules, as previously reported (Tirnauer et al., 2002). When nonkinetochore microtubules were selectively depolymerized using cold treatment, which did not affect the ring distribution of Ndc80 and resulted in long filaments of dense tubulin staining leading away from kinetochores (Supplemental Figure S3, B and C), EB1 was colocalized with Bub1 in ring-like configurations (Figure 3A) or surrounding CENP-A foci at the ends of microtubule bundles (Figure 3B) but not within the “holes” in the rings. This suggests that the ends of microtubules may engage with the ring structures by forming hollow bundles at the ends. Further, the observation of rings of fluorescent EB1 suggests that the ring-like distribution is not an artifact of immunofluorescence.
FIGURE 3:
Kinetochore rings may engage a hollow bundle of microtubules but do not depend on microtubules. (A, B) Spindles assembled in the presence of Alexa 488–labeled EB1 and incubated on ice for 2 min (scale bar, 1 μm). Pairs of sister kinetochores are shown at higher magnification (scale bars, 0.2 μm), with locations indicated by white boxes. (C, D) Mitotic chromosomes assembled in extract treated with 33 mM nocodazole to depolymerize all microtubules (scale bar, 1 μm). Pairs of sister kinetochores are shown at higher magnification (scale bars, 0.2 μm), with locations indicated by arrowheads. All images are maximum-intensity projections of SIM data. (E) Lines scans of ring-shaped Ndc80 signals in nocodazole-treated extract. Top, overlays; bottom, average intensity.
If the rings are engaging microtubule ends, one possibility is that the microtubule bundle is inducing the ring organization. However, Ndc80 maintained a ring-like configuration even when microtubule assembly was completely inhibited by a high dose (33 μM) of nocodazole (Figure 3, C–E) and we did not detect a significant change in the diameter of the rings in this condition (188 nm, p = 0.24 by the Mann–Whitney test, n = 21). Together these data suggest that metaphase kinetochores in Xenopus egg extracts form ∼200-nm rings that engage bundles of microtubule ends and that this kinetochore structure can induce a ring configuration at the end of a microtubule bundle (Supplemental Figure S1F).
Xenopus somatic tissue culture cells show diverse kinetochore configurations, including rings in disorganized spindles
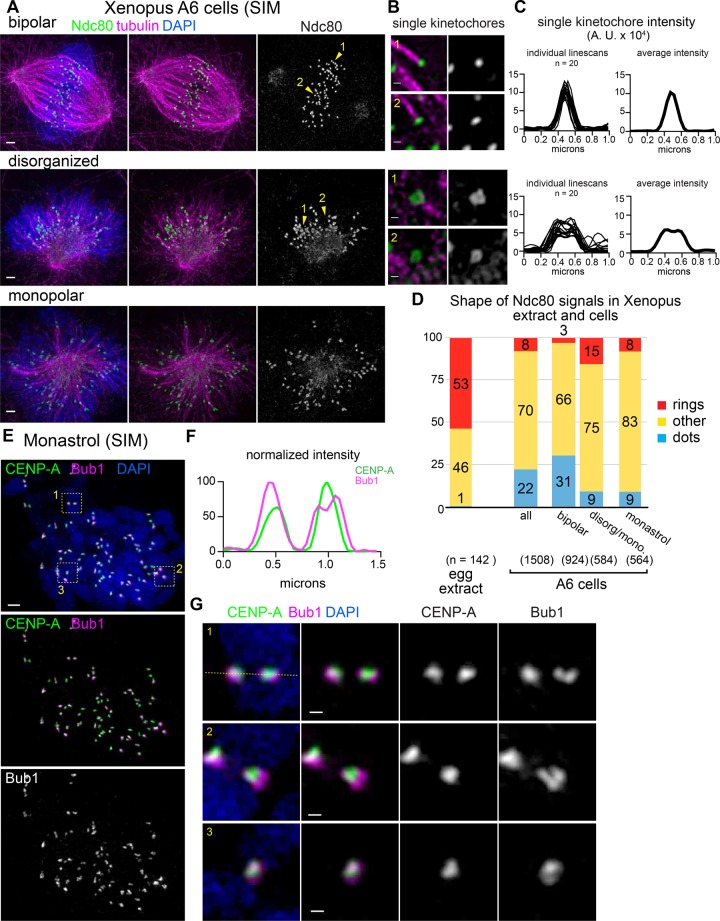
We next tested whether rings or other nondisk configurations could be seen in the A6 Xenopus laevis kidney cell line. Immunofluorescence using the same antibody used in extract showed smaller Ndc80 foci with a different range of configurations, which depended on the state of the mitotic spindle (Figure 4, A–E). In mitotic cells with well-organized bipolar spindles, unlike in Xenopus egg extracts, Ndc80 proteins are often seen as uniform punctate dots at the end of clear kinetochore fibers (Figure 4, A and B, top). In contrast, mitotic cells with a disorganized spindle harbored multiple kinetochores that had nonpunctate signals, including rings (Figure 4, A and B, middle). Kinetochores with ring-like organization in A6 cells were not associated with robust kinetochore fibers, consistent with them representing either en face views or transient structures that occur before end-on attachment. Line scans of kinetochores showed a clear difference, with wider signals in disorganized spindles and a small decrease in the average signal in the center (Figure 4C). Similar kinetochore configurations were also apparent in rare monopolar spindles (Figure 4A, bottom). To analyze further the kinetochore distribution in monopolar spindles, we treated cells with monastrol and found a reduced number of dot-like Ndc80 distributions, but we did not see the same increase in rings (Figure 4D and Supplemental Figure S3D). Bub1 signals again showed nonpunctate distributions, and small foci of CENP-A could sometimes be seen in the regions of lower Bub1 intensity (Figure 4, E–G).
FIGURE 4:
Diverse kinetochore configurations, including rings, are also seen in Xenopus cells. (A, B) Xenopus A6 kidney cells were synchronized using RO-3306, released into medium containing MG132, and processed for immunofluorescence and classified based on spindle morphology (scale bars, 1 μm). Single kinetochores and adjacent microtubules are shown at higher magnification (scale bars, 0.2 μm). Locations of single kinetochores are indicated by numbered yellow arrowheads. (C) The 1-μm line scans of individual kinetochores were overlaid (left) or averaged (right) to illustrate the difference in the ring-like kinetochore configurations seen in disorganized and monopolar spindles. (D) Quantification of kinetochore shapes in Xenopus extract and A6 cells. (E) Xenopus A6 cells were treated with monastrol and processed for immunofluorescence (scale bar, 1 μm). Locations of higher-magnification images of kinetochores are indicated by numbered yellow boxes. (F, G) Higher-magnification images of CENP-A and Bub1 staining in sister kinetochores (scale bars, 0.2 μm); line scan indicated by the dotted yellow line in G. All images are maximum-intensity projections of SIM data.
Together these data show that outer kinetochore components in cells can exhibit nondisk configurations. These configurations are not as common in cells as in extracts but can be found in disorganized spindles. Because these distributions are more common in disorganized bipolar spindles than in monopolar spindles, they may be promoted by early attachments in the context of a bipolar spindle, perhaps by merotelic attachments or lateral attachments.
The core kinetochore, as well as the outer kinetochore, alters its organization in response to microtubule attachments
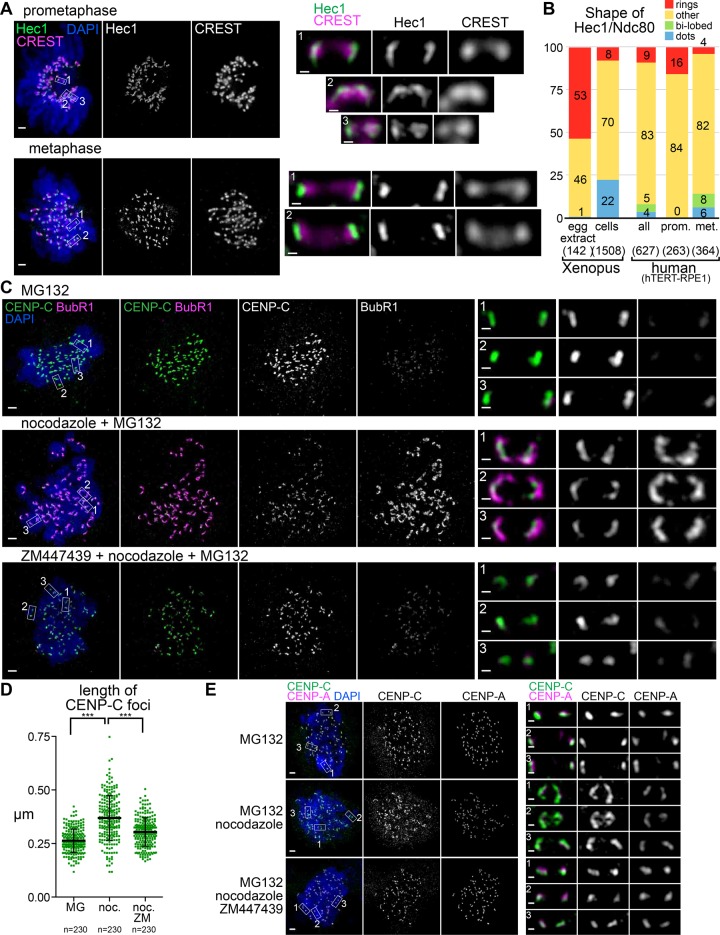
We next monitored kinetochore protein distribution in human retinal pigmented epithelial cells immortalized with telomerase (hTERT-RPE1). Consistent with recent work showing elongated Hec1 (human Ndc80) signals in unattached kinetochores in Taxol-treated cells by SIM (Magidson et al., 2016), we saw dramatic differences in Hec1 distribution between metaphase and prometaphase (Figure 5, A and B). In metaphase, Hec1 showed relatively uniform elongated signals perpendicular to the axes connecting the sister kinetochores. In contrast, in prometaphase cells, Hec1 signals were expanded into longer arcs or even into ring-like configurations. The total volume of Hec1 staining was greater in prometaphase, consistent with the increased amounts of Hec1 seen previously on cells that have not aligned on the metaphase plate (Magidson et al., 2015; Supplemental Figure S4A). We also noted that in metaphase cells, some Hec1 signals appeared to exhibit a bilobed distribution (Figure 5B and Supplemental Figure S4D), which may be related to a ring-like configuration seen in Xenopus.
FIGURE 5:
Centromere and core kinetochore configurations are both sensitive to microtubule attachment status. (A) hTERT-RPE1 cells in prometaphase and metaphase. Locations of higher- magnification images of sister kinetochore pairs (scale bars, 0.2 μm) indicated by boxes. (B) Quantification of kinetochore shapes in human cells and Xenopus (C–E) hTERT-RPE1 cells arrested in mitosis for 4 h in the presence or absence of nocodazole and ZM447439 (scale bars, 1 μm). Locations of higher-magnification images of sister kinetochore pairs (scale bars, 0.2 μm) indicated by boxes. Quantification of CENP-C foci lengths from nuclei in C is shown in D.
In the absence of microtubule attachment, it is well established that outer kinetochore proteins expand, and we previously observed that CENP-C, but not another CCAN member, CENP-T, also coexpands in Xenopus egg extracts. It is unknown whether CENP-C shows expansion in human cells. We arrested cells in mitosis using MG132 with or without treatment with 3.3 μM nocodazole, which is sufficient to depolymerize all microtubules in cells. As expected, this treatment caused a large increase in BubR1 localization to kinetochores, which formed “collars” (Figure 5C). The intensities of CENP-C signals were reduced, and they were organized into elongated crescents that underlay the BubR1 signals. This result is consistent with the centromere being deformed by the expansion of the outer kinetochore, or it may reflect a codependent alteration of the inner and outer kinetochore like the one we characterized in Xenopus, in which expansion of CENP-C required the outer kinetochore. Because kinetochore expansion in Xenopus egg extracts is sensitive to Aurora B activity, we treated cells with the Aurora B inhibitor ZM447439. On inhibition of Aurora B activity, we saw a dramatic reduction in BubR1 localization (Figure 5C, bottom). In this condition, the CENP-C signals became less elongated, with lengths similar to those of metaphase spindles (Figure 5D). Like CENP-C, CENP-A signals could also be seen to elongate after nocodazole treatment, and again this change in shape depended on Aurora B activity (Figure 5E). Thus, in contrast to Xenopus extracts, human centromeres show uniform reorganization on unattached kinetochores that appears positioned to support the elongated outer kinetochore. In addition, this change in centromere configuration is clearly distinct from the stretching caused by merotelic attachment characterized previously (Cimini et al., 2001) or the ring-like conformation of outer kinetochore components we describe here because it occurs in the absence of microtubules. However it extends the structural plasticity in response to microtubule attachment status known to exist in the kinetochore into the centromere.
A model of the dynamic 3D architecture of the kinetochore
Prevailing 3D architectural models of the kinetochore (Wan et al., 2009; Ribeiro et al., 2010; O’Connell et al., 2012; Zaytsev et al., 2014) predict a configuration in which a semihomogeneous layer of microtubule-binding and SAC proteins form on the top of the lawn of CENP-A–marked chromatin. A majority of schematics consistent with these models depict the kinetochore in a way that the surface size of centromeric chromatin, the inner kinetochore, and the outer kinetochore are the same and microtubule-binding units are homogeneously distributed within a kinetochore. Our analysis using superresolution microscopy suggests that such an architectural model is not necessarily universal and that the kinetochore has a capacity to exhibit more dynamic structural changes.
One of the most striking observations we report here is that the outer kinetochore components can form a ring-like configuration in the metaphase spindle assembled in Xenopus egg extracts. Prevalence of the ring configuration in Xenopus egg extracts may reflect characteristics of the spindle unique to eggs, in which spindle microtubules assemble by chromatin-dependent pathways, the majority of spindle microtubules are nonkinetochore microtubules, and kinetochore microtubule fibers (k-fibers) are much less robust than those in tissue culture cells. Whereas kinetochores oscillate in the metaphase plate in many systems, those in egg extracts are static despite the presence of fast poleward microtubule flux (2 μm/min; Maddox et al., 2003). Thus microtubule ends attached to these kinetochores are continuously in the polymerization state, consistent with EB1 localization. A ring-like configuration may enhance stable attachment on rapidly polymerizing microtubules by enriching microtubule binding units and increased avidity. Another aspect of the ring configuration is that ring size differs, depending on the protein. Thus the ring size of CENP-E, responsible for lateral attachment, is bigger than the ring size of Ndc80. This may reflect that CENP-E engages the side of microtubule bundles, whereas Ndc80 attaches the ends. It is tempting to speculate that such a ring stabilizes a microtubule bundle. A ring configuration has also been shown for the 16 Saccharomyces cerevisiae kinetochores, which encircle central spindle microtubules in metaphase, and so some features of this ring architecture may be widely conserved (Haase et al., 2013).
We did not detect many rings in human hTERT-RPE1 or Xenopus tissue culture cells, but the frequency of rings was consistently higher in disorganized or prometaphase cells. Thus we propose that the ring conformation is a transient structure in tissue culture cells and that in extracts, which are known to have persistent lateral attachments (Ohi et al., 2003), and cells with disorganized spindles, it is stabilized. This ring organization may reflect a specific class of prometaphase kinetochores in which end-on and lateral attachments both exist, which is then resolved into a more punctate disk structure as lateral attachments are lost or converted to end-on (Supplemental Figure S4). In Caenorhabditis elegans oocytes, there are well-characterized ring structures that form between chromosomes, which make lateral attachments to microtubules and generate a plus end–directed force that is essential to chromosome alignment and accurate segregation (Wignall and Villeneuve, 2009; Dumont et al., 2010; Muscat et al., 2015). Our study illustrates the morphological and functional diversity of vertebrate kinetochores, which may adapt to the specific requirements associated with distinct developmental programs.
MATERIALS AND METHODS
Kinetochore assembly and SAC assays in Xenopus egg extracts
To assemble kinetochores, 1600/μl demembranated sperm were added to cytostatic factor (CSF)-arrested meiotic metaphase II X. laevis egg extracts (Murray, 1991) and cycled through interphase using 0.3 mM CaCl2 for 90 min, followed by addition of 1 volume of fresh CSF-arrested extract for 45 min at 20°C. For nocodazole treatment, 33 μM nocodazole was added immediately after addition of fresh CSF extract.
Immunofluorescence and antibodies
For immunofluorescence, mitotic spindles or chromosomes in the absence of microtubules were fixed in 2% formaldehyde, spun down onto glass coverslips, treated with methanol for 3–5 min, rehydrated in TBS-TX (10 mM Tris, 150 mM NaCl, 0.1% Triton X-100), and blocked overnight in AbDil (TBS-TX, 2% bovine serum albumin) before antibody incubations (Desai et al., 1999; Funabiki and Murray, 2000). Human, histidine-tagged, recombinant EB1 and bovine tubulin were conjugated to Alexa 488, -594, or -647 fluorophores (Life Technologies, Carlsbad, CA) and added to extracts as in Tirnauer et al. (2002) at 0.33 μM (EB1) or 0.2 μM (tubulin) before cycling. Primary and secondary antibodies for immunofluorescence were diluted in AbDil. Samples stained with fluorescently labeled rabbit primary antibodies (done for both BubR1 and Bub1) after staining with a different rabbit primary antibody, which was detected using secondary anti-rabbit antibodies, were blocked with 100 μg/ml rabbit immunoglobulin G for 30 min before addition of the direct-labeled primary antibody without intervening wash steps, and control single-stained samples were prepared to ensure specificity of the fluorescence signal (unpublished data). For immunoblots, nitrocellulose membranes were blocked with autoclaved 4% nonfat milk in phosphate-buffered saline (PBS) at room temperature, and primary antibodies were diluted in Odyssey Blocking Buffer (927-40000; Li-Cor, Lincoln, NE) with 0.1% Tween 20 and detected with IRDye 800CW and 680LT secondary antibodies using the Odyssey Infrared Imaging System (Li-Cor). For antibodies against X. laevis proteins, Bub1 and BubR1 antibodies were generated according to Chen (2002) and Sharp-Baker and Chen (2001), and CENP-A antibodies were generated according to Maddox et al. (2003). Dasra antibodies were reported previously (Sampath et al., 2004). CENP-C, -E, -K, -T, and dynein antibodies were provided by A. F. Straight (Stanford University School of Medicine). Mis12, Ndc80, and Zwint antibodies were provided by P. T. Stukenberg (University of Virginia Medical Center). Mad1 antibodies were provided by R. H. Chen (Institute of Molecular Biology, Academia Sinica) and T. Lorca (Spanish National Cancer Research Centre), who also provided CENP-E antibodies. Mad2 and Mps1 antibodies were provided by Y. Mao (Columbia University Medical Center). Xorbit antibodies were provided by R. Heald (University of California, Berkeley). For antibodies against human proteins, rabbit CENP-C antibodies were provided by W. Earnshaw (University of Edinburgh) and P. Warburton (The Mount Sinai School of Medicine), guinea pig CENP-C antibodies were provided by K. Yoda (Nagoya University) and H. Kimura (Tokyo Institute of Technology) (Ando et al., 2002), and CENP-A antibodies were provided by G. Goshima (Nagoya University). BubR1 (MAB3612; Millipore, Billerica, MA) and ACA (15-235; Antibodies Inc., Davis, CA) antibodies were purchased.
Alexa 488-EB1 signals were reproducibly retained even in the presence of nocodazole when combined with a specific set of antibodies (e.g., Ndc80, CENP-T) but not with CENP-A or Bub1 antibodies. We do not know the reason for this discrepancy, but we report here only the condition in which Alexa488-EB1 depends on microtubules.
Cell culture and immunofluorescence
hTERT-RPE1 cells were grown in DMEM/F-12 (Life Technologies) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA) and penicillin/streptomycin (Life Technologies). For immunofluorescence, cells grown on coverslips were treated for 4 h with 10 nM MG132 and dimethyl sulfoxide or 3.3 μM nocodazole with or without 2 μM ZM447439, fixed in 2% paraformaldehyde in PBS for 10 min, and permeabilized with PBS containing 0.5% Triton X-100 for 10 min. Incubations with antibody and 4′,6-diamidino-2-phenylindole were done as for extract, as described earlier.
Xenopus A6 cells (a gift of D. Shechter, Albert Einstein College of Medicine, Yeshiva University) were grown at room temperature in 60% L-15 (Leibovitz) medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10% FBS (Atlanta Biologicals), penicillin/streptomycin, and sodium pyruvate (Life Technologies). Cells were synchronized at the G2-to-M boundary by 9 μM RO-3306 for 12 h and then released into M phase by changing to medium containing 10 μM MG132 or 100 μM monastrol (Tocris Bioscience, Minneapolis, MN). Immunofluorescence was done as described, but for improved microtubule preservation, Xenopus cells were fixed in cold methanol for 10 min and permeabilization was omitted.
Microscopy and image analysis
All 3D SIM microscopy data were acquired using a DeltaVision OMX V4/Blaze 3D SIM superresolution microscope (GE Healthcare, Chicago, IL) maintained by the Rockefeller University Bio-Imaging Resource Center (BIRC). This OMX system is fitted with a 100×/1.40 numerical aperture (NA) UPLSAPO oil objective (Olympus, Tokyo, Japan) and three Evolve electron-multiplying charge-coupled device (EMCCD) cameras (Photometrics, Tucson, AZ) that were used in EM gain mode fixed at 170 electrons/count. Immersion oils ranging in refractive index from 1.512 to 1.518 were used, depending on the ambient temperature and fluorochromes used. The 405-, 488-, and 568-nm laser lines were used for excitation, and the corresponding emission filter sets were 436/31, 528/48, and 609/37 nm, respectively. Two identical stacks of optical sections with 125-nm spacing were collected for each data set, first using conventional wide-field illumination of all channels and then using structured illumination of selected channels. The system produces an effective xy pixel size of 40 nm in the 3D SIM mode. After acquisition, all data sets were processed using softWoRx, version 6.1 software (GE Healthcare), using optical transfer functions (OTFs) generated from point spread functions acquired with 100-nm (green and red) or 170-nm (blue) FluoSpheres (Invitrogen/Life Technologies). Wide-field data sets were deconvolved using constrained iterative deconvolution, and structured illumination data sets were reconstructed as described (Schermelleh et al., 2008), using channel-specific k0 values, custom OTFs, and Wiener filters of 0.005, 0.002, and 0.002 for the blue, green, and red channels, respectively. Image registration was performed with parameters refined using 100-nm TetraSpeck beads (Invitrogen/Life Technologies). Image registration parameters and OTFs were frequently refined by the BIRC staff. Imaris software (Bitplane, Zurich, Switzerland) and ImageJ (National Institutes of Health, Bethesda, MD) were used for 3D visualization and generation of projection images for visualization. Image segmentation and quantification of intensity, volume, and extent were done using MATLAB (MathWorks, Natick, MA).
STORM data were acquired using a Nikon TiE inverted microscope with Perfect Focus mechanism using 647-nm, 120-mW laser illumination with the total internal reflection fluorescence angle adjusted to fully illuminate kinetochores closest to the coverslip, a 100×/1.49 NA APO-TIRF oil objective with Nikon Type NF immersion oil, and an Andor DU-897 EMCCD camera controlled with Elements software (Nikon, Tokyo, Japan). All STORM data sets were collected using ∼30,000 images of spontaneous blinking Alexa 647– or Cy5-labeled secondary antibodies under 80–100% laser power at the maximum frame rate of the camera (16 ms). Samples were prepared on 18-mm circular coverslips and mounted in a Chamlide CMB chamber (CM-B18-1; Quorum Technologies, East Sussex, United Kingdom) containing 1 ml of STORM buffer (5.6 mg/ml glucose oxidase [G2133; Sigma-Aldrich], 0.34 mg/ml catalase [106810; Roche], 1.43 M 2-mercaptoethanol, 50 mM Tris-HCl, pH 8.0, 10 mM NaCl, and 10% glucose). STORM localizations were generated (fit to an integrated Gaussian point spread function), drift corrected (by cross-correlation), filtered to remove high-uncertainty (>20 nm) localizations, and rendered (by normalized Gaussian fitting) using NSTORM Elements software (Nikon) and the ThunderSTORM ImageJ plug-in (Ovesny et al., 2014). Line scans of STORM images were generating from normalized Gaussian renderings of STORM localization data with magnification of 16× (final pixel size, 5 nm).
Supplementary Material
Acknowledgments
We thank R. H. Chen, W. Earnshaw, G. Goshima, R. Heald, H. Kimura, T. Lorca, Y. Mao, N. Gray, D. Shechter, A. Straight, P. T. Stukenberg, P. E. Warburton, and K. Yoda for reagents and A. North and K. Thomas of the Rockefeller University Bio-Imaging Resource Center for help with data acquisition and processing. D.W. was supported by a National Research Service Award Postdoctoral Fellowship (F32GM103147). This work was supported by grants from the National Institutes of Health to H.F. (R01GM075249) and to the Bio-Imaging Resource Center (S10RR031855) from the National Center for Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Abbreviations used:
- CCAN
constitutive centromere–associated network
- KMN
KNL1, Mis12, and Ndc80 complexes
- SAC
spindle assembly checkpoint
- SIM
structured illumination microscopy
- STORM
stochastic optical reconstruction microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-02-0130) on May 11, 2016.
REFERENCES
- Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R-H. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J Cell Biol. 2002;158:487–496. doi: 10.1083/jcb.200204048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R-H, Shevchenko A, Mann M, Murray AW. Metaphase arrest induced by an excess of the spindle checkpoint protein Xmad2 is independent of Xmad1. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Murray A, Mitchison TJ, Walczak CE. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J, Oegema K, Desai A. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat Cell Biol. 2010;12:894–901. doi: 10.1038/ncb2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Murray AW. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Haase J, Mishra PK, Stephens A, Haggerty R, Quammen C, Taylor IIRM, Yeh E, Basrai MA, Bloom KS. A 3D map of the yeast kinetochore reveals the presence of core and accessory centromere-specific histone. Curr Biol. 2013;23:1939–1944. doi: 10.1016/j.cub.2013.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- Maddox P, Straight A, Coughlin P, Mitchison TJ, Salmon ED. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J Cell Biol. 2003;162:377–382. doi: 10.1083/jcb.200301088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, He J, Ault JG, O’Connell CB, Yang N, Tikhonenko I, McEwen BF, Sui H, Khodjakov A. Unattached kinetochores rather than intrakinetochore tension arrest mitosis in taxol-treated cells. J Cell Biol. 2016;212:307–319. doi: 10.1083/jcb.201412139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, Paul R, Yang N, Ault JG, O’Connell CB, Tikhonenko I, McEwen BF, Mogilner A, Khodjakov A. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat Cell Biol. 2015;17:1134–1144. doi: 10.1038/ncb3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Hsieh CE, Mattheyses AL, Rieder CL. A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma. 1998;107:366–375. doi: 10.1007/s004120050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Grishchuk EL, Morphew MK, Efremov AK, Zhudenkov K, Volkov VA, Cheeseman IM, Desai A, Mastronarde DN, Ataullakhanov FI. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, O’Toole E, Zhudenkov K, Morphew M, Schwartz C, Ataullakhanov FI, Grishchuk EL. Conserved and divergent features of kinetochores and spindle microtubule ends from five species. J Cell Biol. 2013;200:459–474. doi: 10.1083/jcb.201209154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Muscat CC, Torre-Santiago KM, Tran MV, Powers JA, Wignall SM. Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. Elife. 2015;4:e06462. doi: 10.7554/eLife.06462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell CB, Khodjakov A, McEwen BF. Kinetochore flexibility: creating a dynamic chromosome–spindle interface. Curr Opin Cell Biol. 2012;24:40–47. doi: 10.1016/j.ceb.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R, Coughlin ML, Lane WS, Mitchison TJ. An inner centromere protein that stimulates the microtubule depolymerizing activity of a KinI kinesin. Dev Cell. 2003;5:309–321. doi: 10.1016/s1534-5807(03)00229-6. [DOI] [PubMed] [Google Scholar]
- Ovesny M, Krizek P, Borkovec J, Svindrych Z, Hagen GM. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci USA. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacristan C, Kops GJ. Joined at the hip: kinetochores, microtubules, and spindle assembly checkpoint signaling. Trends Cell Biol. 2015;25:21–28. doi: 10.1016/j.tcb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MGL, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp-Baker H, Chen RH. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J Cell Biol. 2001;153:1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silio V, McAinsh AD, Millar JB. KNL1-Bubs and RZZ provide two separable pathways for checkpoint activation at human kinetochores. Dev Cell. 2015;35:600–613. doi: 10.1016/j.devcel.2015.11.012. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Badger BL, Wan X, DeLuca JG, Salmon ED. The architecture of CCAN proteins creates a structural integrity to resist spindle forces and achieve proper Intrakinetochore stretch. Dev Cell. 2014;30:717–730. doi: 10.1016/j.devcel.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Hori T, Nishino T, Usukura J, Miyagi A, Morikawa K, Fukagawa T. Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. J Cell Biol. 2011;193:125–140. doi: 10.1083/jcb.201012050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower DA, Jordan MA, Wilson L. Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell Motil Cytoskeleton. 1996;35:121–133. doi: 10.1002/(SICI)1097-0169(1996)35:2<121::AID-CM5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Tirnauer JS, Grego S, Salmon ED, Mitchison TJ. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol Biol Cell. 2002;13:3614–3626. doi: 10.1091/mbc.E02-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, Salmon ED. The KMN protein network–chief conductors of the kinetochore orchestra. J Cell Sci. 2012;125:5927–5936. doi: 10.1242/jcs.093724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, Wan X, Cheerambathur D, Gassmann R, Suzuki A, Lawrimore J, Desai A, Salmon ED. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J Cell Biol. 2013;202:735–746. doi: 10.1083/jcb.201304197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu S-T, Yen TJ, McEwen BF, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe FG, Straight AF. Functions of the centromere and kinetochore in chromosome segregation. Curr Opin Cell Biol. 2013;25:334–340. doi: 10.1016/j.ceb.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wignall SM, Villeneuve AM. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat Cell Biol. 2009;11:839–844. doi: 10.1038/ncb1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood KW, Sakowicz R, Goldstein LS, Cleveland DW. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- Wynne DJ, Funabiki H. Kinetochore function is controlled by a phospho-dependent coexpansion of inner and outer components. J Cell Biol. 2015;210:899–916. doi: 10.1083/jcb.201506020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev AV, Sundin LJ, DeLuca KF, Grishchuk EL, DeLuca JG. Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J Cell Biol. 2014;206:45–59. doi: 10.1083/jcb.201312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.