A novel approach based on the convex-hull algorithm is used for parallel analysis of growth dynamics and relative spatiotemporal protein concentration along flexible filopodial protrusions. Testing of filopodia formation in silico, in vitro, and in vivo validates the robustness and sensitivity of the proposed approach.
Abstract
Filopodia are dynamic, actin-rich structures that transiently form on a variety of cell types. To understand the underlying control mechanisms requires precise monitoring of localization and concentration of individual regulatory and structural proteins as filopodia elongate and subsequently retract. Although several methods exist that analyze changes in filopodial shape, a software solution to reliably correlate growth dynamics with spatially resolved protein concentration along the filopodium independent of bending, lateral shift, or tilting is missing. Here we introduce a novel approach based on the convex-hull algorithm for parallel analysis of growth dynamics and relative spatiotemporal protein concentration along flexible filopodial protrusions. Detailed in silico tests using various geometries confirm that our technique accurately tracks growth dynamics and relative protein concentration along the filopodial length for a broad range of signal distributions. To validate our technique in living cells, we measure filopodial dynamics and quantify spatiotemporal localization of filopodia-associated proteins during the filopodial extension–retraction cycle in a variety of cell types in vitro and in vivo. Together these results show that the technique is suitable for simultaneous analysis of growth dynamics and spatiotemporal protein enrichment along filopodia. To allow readily application by other laboratories, we share source code and instructions for software handling.
INTRODUCTION
Filopodia formation, elongation, and subsequent retraction are orchestrated via elaborate spatiotemporal control of actin polymerization dynamics (Dunaevsky et al., 1999; Matus et al., 2000). This is achieved by local and transient enrichment or activation of actin-regulatory proteins during a particular phase of the extension–retraction cycle (Galic et al., 2014). Consistently, changes in activity and concentration of individual actin-regulatory proteins that control these regulatory modules have been shown to selectively alter initiation frequency, final length, or lifetime of filopodia (Luo et al., 1996; Cheadle and Biederer, 2012; Galic et al., 2014). The picture that has emerged from these experiments is that the life cycle of filopodia is composed of separate actin-polymerization modules with unique features that transit from one to the next in a stereotypic manner (Svitkina et al., 2003; Galic et al., 2014). However, although many proteins that control specific aspects of the resulting filopodial growth–retraction cycle have been described, our understanding of the process has remained incomplete due to the lack of automated image analysis software suitable to accurately measure growth dynamics and correlate with relative protein localization along protrusions that bend, laterally shift, or tilt (i.e., change in angle relative to cell).
Excellent software solutions have been published to determine cell shape (Blum, 1967; Styner et al., 2003; Xiong et al., 2010; Tsygankov et al., 2014) and/or analyze filopodial density, elongation speed, or form (Styner et al., 2003; Costantino et al., 2008; Fanti et al., 2011; Nilufar et al., 2013; Hendricusdottir and Bergmann, 2014; Tsygankov et al., 2014; Tarnok et al., 2015). Similarly, image analysis tools are available to measure spatiotemporal protein concentration in cellular structures such as the leading edge (Machacek et al., 2009; Barry et al., 2015). However, no software has been presented that unifies analysis of filopodial growth dynamics and information on spatiotemporal protein concentration along the full filopodial length. This is relevant because proteins that control the filopodial growth cycle show not only a temporal but also a spatially restricted localization along filopodia (Hotulainen et al., 2009; Galic et al., 2014). Here we developed automated image analysis software that integrates measurements of filopodial growth dynamics and relative protein concentrations along the filopodial length for up to three protein channels. Specifically, the software, which is based on a novel approach using the convex-hull algorithm, adapts in each frame to the changing filopodial shape to extract the accurate growth parameters and relative protein concentration along the complete filopodial length. We validate our software in silico, showing accurate growth measurements for various imaging conditions and filopodial geometries. To test our approach in living cells, we quantify filopodia formation in a series of in vitro and in vivo model systems. We reliably detect filopodial growth dynamics, spatiotemporal differences in the relative concentration of fluorescently labeled proteins involved in filopodia formation, and changes in filopodial length and lifetime upon overexpression of known regulators of filopodial dynamics. Together these experiments provide evidence that our software reliably analyzes growth dynamics and relative spatiotemporal protein localization along filopodia independent of bending, lateral shift, or tilting.
RESULTS
Working principle of the image analysis software
Quantification starts by preparing selected movies for image analysis. The software, which is operated via a graphical user interface (GUI), provides the user with the possibility to adjust a series of tracking parameters (e.g., range of bending angle, interframe movement) and optional manual corrections (e.g., cropping, rotation, deleting unwanted objects) to maximize flexibility and robustness for the particular experimental setting (Supplemental Figure S1 and Supplemental Movie S1). Considering that fluorescence intensities of individual channels are not altered by the software, standard preprocessing steps for ratiometric image analysis (e.g., background subtraction, bleach adjustment, and potential bleedthrough correction) should be performed before loading the movies using one of the many excellent software solutions available elsewhere (Abramoff et al., 2004; Courtney et al., 2015).
Once parameters are adjusted, the movie is binarized to increase computation speed. To maximize accuracy, individual channels of multicolor movies are merged before binarization. Next the outline of the binarized image is determined, and a convex hull is generated to encompass the outline of the binarized object. The hypothesis is that the protruding filopodial tip will become a vertex belonging to the convex hull (Figure 1, A and B). The script is then initiated by manually determining which vertex belongs to the filopodial tip, as well as the base of the filopodium (Figure 1A, left). After this initiation, the filopodial tip is being tracked frame by frame throughout the whole movie. This is achieved by finding in the subsequent frame the vertex that is nearest to the vertex of the filopodial tip. To avoid wrong annotations, the new vertex in the subsequent frame can be located only inside a specified radius from the initial position (Figure 1A, middle). In each frame, an imaginary line is then drawn between the tip and the base point (Figure 1A, middle). Using this tip–base line (i.e., axis), nodes where the line can bend are introduced (the number of nodes is one of the parameters that can be selected by the user). Next, at each nodal point, the intensity values are measured in transverse direction to the axis, and the position of the nodal points is updated to follow the position with maximal pixel values. The newly positioned nodal points are then connected to approximate the shape of the filopodia in each frame of the movie. At each pixel along the adjusted trace (i.e., filopodial backbone), an imaginary line perpendicular to the local direction of the trace is drawn (Figure 1A, right, and Supplemental Movie S2). The three highest intensity values are then averaged to estimate the protein concentration for all channels at each position along the filopodial backbone. For spatially comparing two fluorescence proteins in a symmetric manner, the ratio of the corresponding protein intensities is visualized using a log scale for every pixel along the filopodial backbone.
FIGURE 1:
Image analysis software to correlate protein recruitment kinetics and growth dynamics in filopodia. (A) Schematic representation of the working principle of the image analysis software. In step 1, the user selects the base and tip of the filopodia. In step 2, the software takes control and tracks the tip and shape of the filopodia. In step 3, ratiometric analysis is performed along the adapted filopodial backbones and data are exported. (B) Screenshot of convex-hull algorithm used by image analysis software to detect filopodial structures. (C) Schematic overview of filopodial extension–retraction cycle (top) and summary of the parameters that can be analyzed with the image analysis software (bottom).
Once analysis is completed, all information regarding filopodial tracking and spatially resolved fluorescence intensity is visualized in the GUI as ratiometric kymographs and also exported for documentation and further analysis as images (in .png and .fig format) depicting kymographs, as well as in an Excel sheet containing all data. Information that can be extracted with the image analysis software includes kinetic parameters (e.g., filopodial length, lifetime, and growth rates), as well as absolute and relative fluorescence intensities along the filopodia length throughout the full extension–retraction cycle (Figure 1C).
Testing image analysis software in silico
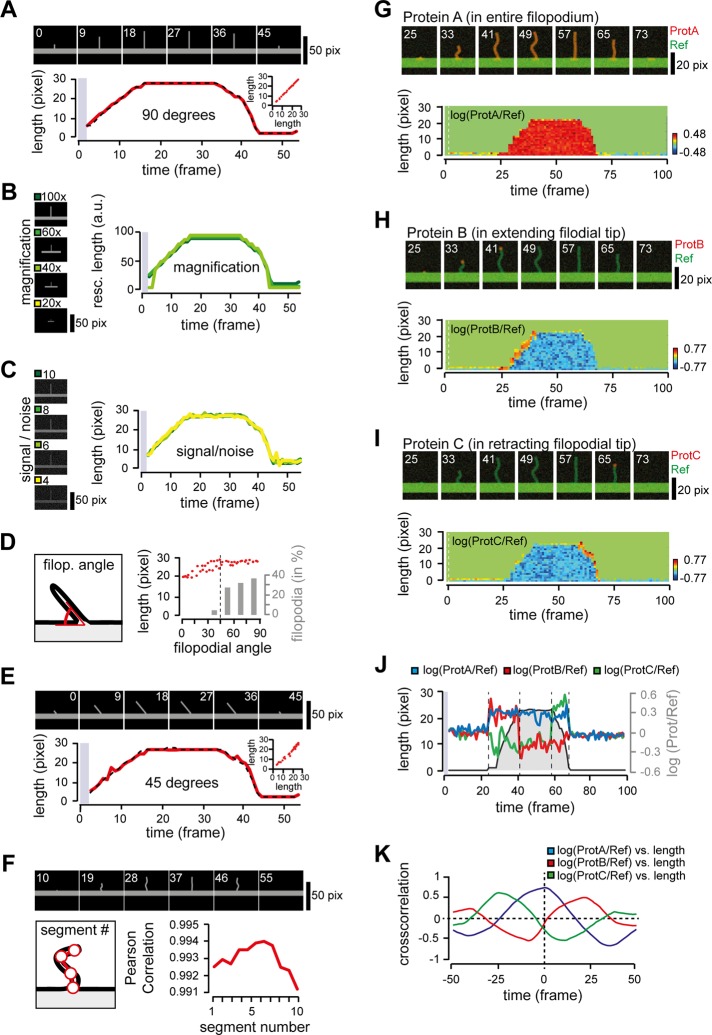
To validate the image analysis software in a controlled manner, we decided to start by generating a series of simulated filopodia emerging from a base, in which individual shape and signal parameters were systematically altered one after the other. In a first step, we tested the software using a filopodium that extends and retracts perpendicular (i.e., at 90°) to the base. We found a strong overlap for manually (Figure 2A, dotted line) and automatically (red line) measured filopodial length, arguing that the script reliably detects the extending structure. In this experiment, we considered formation of a 2-μm-long filopodia with a pixel resolution analogous to images acquired on a complementary metal-oxide semiconductor (CMOS) camera with a 100× objective (i.e., 64.5 nm/pixel). To test the sensitivity of the software for different magnifications, we next generated movies of filopodia with a constant maximal length, with pixel resolution corresponding to images acquired with 60×, 40×, and 20× objectives, respectively. We found the software capable of accurately tracking extension and retraction of 2-μm-long filopodia using 100×, 60×, and 40× objectives but not for a 20× objective, where tracking initiation failed (Figure 2B). To test the sensitivity of the software for different fluorescence intensity, we then systematically reduced the gray values of individual filopodia while keeping the background noise level constant (mean 10, variance 10) and found that the image analysis reliably detects protruding filopodia with signal-to-noise level >4 (Figure 2C).
FIGURE 2:
In silico analysis of filopodial dynamics. (A) Analysis of protrusion length for a filopodium growing orthogonally from the base. Manually (dotted line) and automatically (red line) measured filopodial lengths. The image analysis software uses the first two frames for tracking adjustments (gray box). Inset, scatterplot analysis of manual (x-axis) and automatic (y-axis) measurements of filopodial length, with a Pearson r = 0.9996. (B) Analysis of filopodial growth dynamics using different objective magnifications. Left, an ∼2-μm-long filopodium in a constant field of view (96 × 72 pixel size) acquired with a 100× (dark green), 60× (green), 40× (light green), or 20× (yellow) objective using a CMOS camera with pixel size of 64,5 nm. Right, trace length rescaled before plotting according to the used magnification. Note that acquisition with a 20× objective (yellow) did not provide sufficient pixel resolution for image analysis and is thus missing. (C) Systematic changes in signal intensity show robust response of image analysis software. Filopodia with constant background noise (average 10; variance 10) and mean gray values of 100 (dark green), 80 (green), 60 (light green), and 40 (yellow) were analyzed. (D) Systematic changes in the tilting angle of filopodium with a constant length (left) show accurate length measurements (in red) for angles of >45° from the base. Analysis of angles at which filopodia emerge from dendrites in cultured hippocampal neurons is shown in gray bars. Cells were transfected at 8 d in vitro with a cytosolic marker and imaged 24 h later. Note that >95% of all filopodia emerge at an angle >45° from the dendrite axis (dashed vertical line). (E) Analysis of protrusion length for filopodium extending and retracting at exactly 45° from the base. Manually (dotted line) and automatically (red line) measured filopodial lengths. Inset, scatterplot analysis of manual (x-axis) and automatic (y-axis) measurements of filopodial length shows a Pearson r = 0.9940. (F) Measurement of filopodial length with increasing number of segments. Graph depicts Pearson’s r of manually vs. automatically measured filopodial length as a function of segment number. Note that segment number should not exceed the total filopodial length, as this will result in reduced measurement accuracy. (G–I) Examples of simulated signal enrichment showing a reference channel (green) together with signal channels (red) for enrichment of protein A in the entire filopodium (G), protein B only in the extending tip (H), and protein C only in the retracting tip (I). Bottom, quantification of relative protein signal intensity during the extension–retraction cycle, showing relative enrichment of protein A in the entire filopodium (G), of protein B in the extending tip (H), and of protein C in the retracting tip (I). The first two frames, used for tracking adjustments, are separated by the dashed white line. (J) Scatterplot of filopodial length during the extension–retraction cycle (black) and the relative intensity for the three most distal pixels of the protrusions for proteins A (blue line), B (red line), and C (green line). (K) Cross-correlation analysis for filopodial length and average signal of the three most distal pixels of proteins A (blue line), B (red line), and C (green line). Scale bars, 50 pixels (A, E, F), 20 pixels (G–I).
This far, simulated filopodia were elongating and retracting perpendicular to the base. However, filopodia are dynamic structures that undergo extension–retraction cycles at different angles and also bend. Whereas filopodial deviations are rather modest between frames, these movements sum up throughout the full movie, precluding a simple line-scan approach that may seem suitable to analyze the previous scenarios. To test how the image analysis software responded to tilting, we systematically changed the angle between an extended filopodium (with constant length) and the base. We found that the image analysis software reliably detects filopodial length at all angles (Figure 2D). For filopodial angles of 45 to 90° relative to the base, we found a deviation of <5%, likely caused by errors introduced when preparing the tilt series as well as the pixilation of the images. Consistent with these results, the length of a filopodium emerging at an angle of exactly 45° from the dendritic shaft could be measured accurately (Figure 2E). Using cultured neurons as reference, we found that >95% of all filopodia emerging from dendritic arbors elongate at an angle of 45–90° relative to the dendritic shaft (Figure 2D, gray).
Next we sought to investigate whether nodal points (i.e., points connecting the tip of the filopodium with the base into equidistant segments) will further improve accuracy when measuring a bending filopodium. We found that increasing the number of segments is indeed suitable to adapt to shape changes in bending filopodia and reduce measurement errors that would arise from a simple tip-to-base measurement (Figure 2F).
To test the capability of the image analysis software to integrate growth dynamics and relative protein concentration along the filopodial length, we then generated a series of ratiometric movies. Using the bending filopodium described earlier, we created a reference channel in which signal intensity in the filopodium was half the intensity of the base (Figure 2, G–I, green) and merged this reference channel with signal channels where the protein was enriched throughout the whole filopodium (Figure 2G, red), present only in the extending tip (Figure 2H, red) or limited to the retracting tip (Figure 2I, red), respectively. Finally, constant noise was added to all frames in each channel, and the resulting dual-color movies were subjected to image analysis. We found that the software not only tracks protrusion length, but also measures the relative protein concentration spatially resolved along the filopodial length (Figure 2, G–I, graphs).
Using the average of the most distal 3 pixels of the filopodia, we then plotted growth dynamics against the relative protein intensity at the filopodial tip. As expected, the image analysis software detected protein A present throughout the whole extension–retraction cycle (Figure 2J, blue), whereas protein B was present only during extension (Figure 2J, red) and protein C only during the retraction (Figure 2J, green) of the protrusion. To further validate these changes, we then performed a spatiotemporal cross-correlation comparing protein concentration with protrusion length. For protein A, which is present during the whole extension–retraction cycle, we found the relative maximum centered at 0 (Figure 2K, blue). In contrast, the maximal peaks for protein B (Figure 2K, red, present only during extension) and protein C (Figure 2K, green, only during retraction) are shifted in opposite directions, arguing that spatiotemporal cross-correlation may be suitable to correlate relative protein recruitment dynamics with the extension–retraction cycle.
Testing image analysis software in vitro
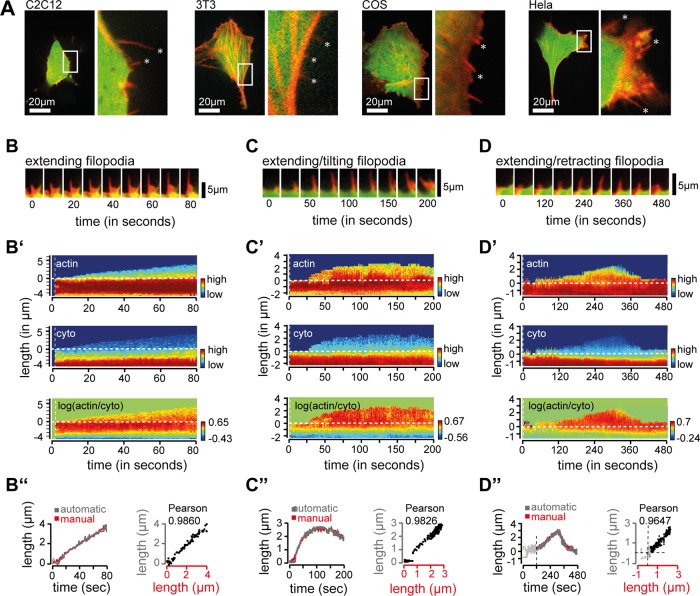
Next we sought to validate the image analysis software in living cells. In a first step, we tested for the presence of dynamic filopodia in a series of cultured cell lines (Figure 3A and Supplemental Figure S2). Based on the abundance of dynamic filopodia, we proceeded with our experiments using COS cells. For image analysis, individual cells were transfected with f-tractin, a marker for filamentous actin (Schell et al., 2001), and a cytosolic reference and imaged at a frequency of 1 Hz. We found slowly extending filopodia (Figure 3B) and filopodia that emerge and tilt (Figure 3C), as well as filopodia that emerge and subsequently retract (Figure 3D). Consistent with images in the time series (Figure 3, B–D, top), ratiometric analysis of individual structures showed enrichment of actin in filopodia and actin cortex but not in the cytosol (Figure 3, B–D, middle). To validate the accuracy of the measurements, we then compared manually and automatically measured changes in filopodial length. We found a high degree of correlation between manually and automatically measured filopodia, arguing that the image analysis software reliably tracks filopodia (Figure 3, B–D, bottom).
FIGURE 3:
In vitro analysis of filopodial dynamics in cultured cells. (A) Test for filopodia formation in cultured COS, C2C12, HeLa, and 3T3 cells. Individual cell lines were transfected with f-tractin (red), a marker for filamentous actin, and a cytosolic reference (green). (B) Filopodium forming on COS cells. Time series of COS cell transfected with f-tractin and a cytosolic reference. (B′) Analysis of filopodial growth dynamics and relative protein concentration of f-tractin (top) and cytosolic reference (middle) and ratio of f-tractin vs. cytosolic reference (bottom). The first two frames, used for tracking adjustments, are separated by the vertical dashed white line. (B′′) Analysis of alignment accuracy. Bottom left, overlay of automatically (gray) and manually (red) traced filopodia. Scatter plot analysis of manual (x-axis, red) and automatic (y-axis, gray) measurements of filopodial length show a Pearson r = 0.9860. (C) Filopodium forming and tilting; time-series analysis, as before. Protein concentration (C′) and comparison of manual and automatic filopodial length (C′′). (D) Filopodium forming and retracting, time-series analysis, as before. Protein concentration (D′) and comparison of manual and automatic filopodial length (D′′). Note that values from 0–120 s before filopodial initiation (D′′, gray) were not used for Pearson r. Scale bars, 20 μm (A), 5 μm (B–D).
Encouraged by these results, we then sought to test whether the image analysis software is suitable to detect changes in filopodial dynamics and relative protein concentration in cultured mouse hippocampal neurons (Figure 4A). In this primary cell system, dynamic exploratory filopodia were previously shown to form along the dendrite to sample the environment and initiate new transsynaptic contacts (Dunaevsky et al., 1999; Matus et al., 2000). In time-series experiments, we found formation of filopodial and mushroom-shaped protrusions along the dendrite (Figure 4B). Using kymographs of line scans to visualize the dynamics of these structures, we found filopodial structures to be transient, whereas spine-like structures remain stable during the acquisition time (Figure 4, B and C), which is consistent with previous work (Dailey and Smith, 1996; Ziv and Smith, 1996). Intriguingly, we further observed substantial amounts of structural rearrangements and lateral dynamics for both structures (Figure 4D), causing the protrusions to exit the axis of the line scan. This is relevant because it illustrates the limitations of a static, one-dimensional approach to analyzing such dynamic structures that bend and tilt.
FIGURE 4:
In vitro analysis of filopodial dynamics in cultured hippocampal neurons. (A) Representative image of primary hippocampal neuron. Neuron was transfected with fluorescent marker 14 d after plating and imaged 24 h later. (B) Dendritic protrusions can be divided into transient (filopodium, top) and static (spine, bottom) protrusions. (C) Vertical kymograph of filopodium (top) and spine (bottom) show differences in growth dynamics. (D) Horizontal kymograph of filopodium (left) and spine (right). Note that both protrusions show substantial lateral movement. (E) Time series of a filopodium forming on the dendrite shaft (top) and measurements using the image analysis software (bottom). (F) Differences in filopodial dynamics upon overexpression of constitutively active Rho GTPases in neurons. Time series of filopodia forming on a neuron transfected with control (black), constitutively active Cdc42 (blue), and constitutively active Rac1 (red). Note the differences in length and lifetime. (G) F-BAR domain of srGAP2 is enriched in the filopodium. Time series of neurons transfected at day in vitro (DIV) 8 with a construct encoding the F-BAR domain of srGAP2 (red) and a cytosolic reference (green) and imaged 24 h later. (G′) Analysis of growth dynamics and relative protein concentration of the F-BAR domain of srGAP2 (top), cytosolic reference (middle), and ratio of F-BAR domain of srGAP2 vs. cytosolic reference (bottom). The first two frames, used for tracking adjustments, are separated by the vertical dashed white line. (G′′) Analysis of srGAP2 recruitment (red) and filopodial elongation (blue) during the extension–retraction cycle. Note that enrichment of the F-BAR domain of srGAP2 and filopodia formation coincide. (H) Actin enrichment precedes filopodia elongation. Time series of cell transfected at DIV 8 with f-tractin (red) and a cytosolic reference (green) and imaged 24h later. Note the formation of actin-rich patches before filopodia elongation. (H′) Analysis of growth dynamics and relative protein concentration of actin (top), cytosolic reference (middle), and ratio of actin vs. cytosolic reference (bottom). (H′′) Analysis of actin recruitment (red) and filopodia elongation (blue) during the extension–retraction cycle. Note that actin-enrichment precedes filopodia formation. Scale bars, 50 μm (A), 2 μm (B, C, F), 500 nm (D), 1 μm (E), 5 μm (G, H).
In a first series of experiments, we transfected cultured hippocampal neurons 8 d after plating with a cytosolic fluorescent reporter and imaged 14–24 h later for 10 min using 10-s intervals. We can reliably monitor different phases of the extension–retraction cycle, maximal length, and total lifetime of an individual dendritic filopodium (Figure 4E). We then separately expressed constitutively active Cdc42 (i.e., Cdc42Q61L), as well as constitutively active Rac1 (i.e., Rac1Q61L; Miller and Johnson, 1994; Best et al., 1996). Compared to control cells (Figure 4F, black), we found that Cdc42Q61L expression increased lifetime and maximal filopodial length (Figure 4F, blue), whereas expression of Rac1Q61L increased membrane ruffling and reduced the lifetime of protrusions emerging from these dendritic ruffles (Figure 4F, red), arguing that the software can reliably analyze changes in filopodial dynamics in neurons.
We then sought to validate filopodial dynamics in relation to relative protein concentration in neurons. As previously published (Coutinho-Budd et al., 2012), we found the fluorescently labeled F-BAR domain of srGAP2 (amino acids 1–501) enriched over a cytosolic reference in elongating dendritic filopodia (Figure 4G). Consistently, the image analysis software showed that the F-BAR domain of srGAP2 was enriched in the filopodium throughout the full extension–retraction cycle (Figure 4G′). To test the correlation between the F-BAR domain of srGAP2 and filopodial growth dynamics, we combined ratiometric and dynamic analysis and found protein enrichment to coincide with filopodial elongation (Figure 4G′′). In a separate set of experiments, we then expressed f-tractin together with a cytosolic reference. As previously described (Ketschek and Gallo, 2010; Galic et al., 2014), we found the formation of actin-rich patches to precede filopodial elongation (Figure 4H). Consistent with these findings, we observed relative enrichment of actin before filopodial expansion in the ratiometric images (Figure 4H′), as well as in the combined ratiometric and dynamic analysis (Figure 4H′′), thus arguing that the software is suitable for correlative analysis of protein enrichment and filopodial growth dynamics in neurons.
Testing image analysis software in vivo
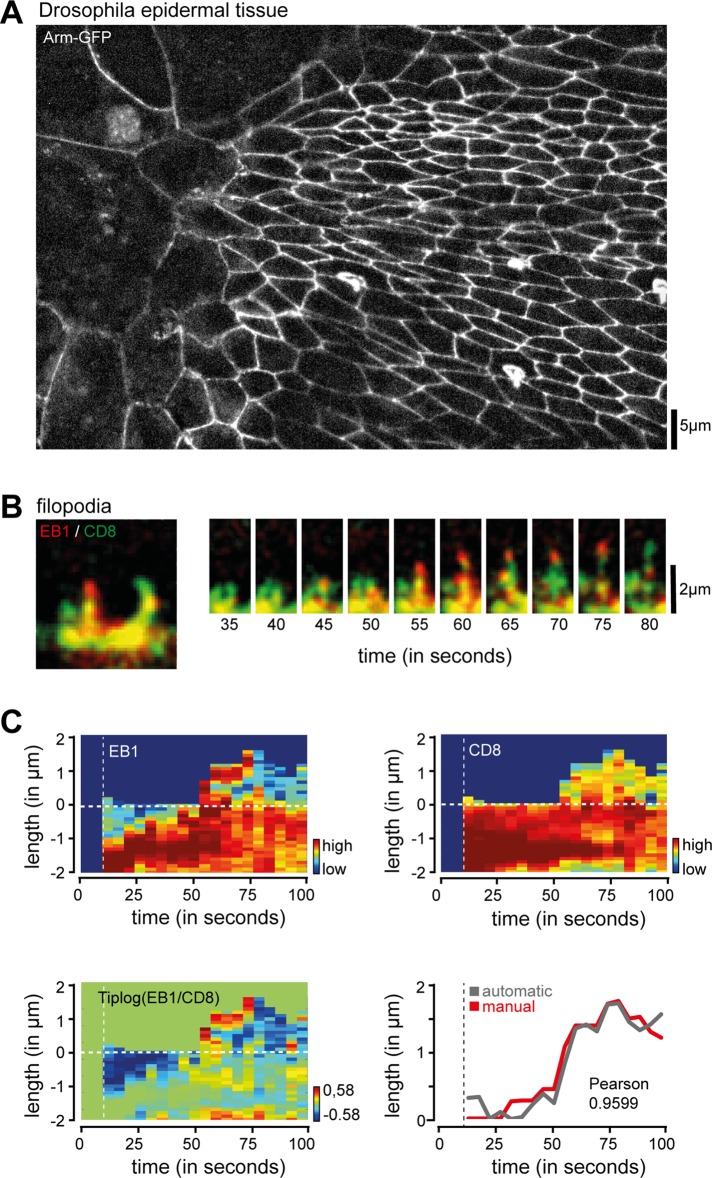
Finally, we sought to test whether the image analysis software can also be applied to analyze filopodial dynamics in cellular ensembles. We took advantage of abdominal epidermis formation in Drosophila, a model system for collective cell migration in vivo (Figure 5A). In this model system, individual cells within the epithelial sheet create small filopodial protrusions at the leading edge during dorsal collective cell migration. Using red fluorescent protein (RFP)–tagged CD8 as membrane reference together with EB1–green fluorescent protein (GFP), a microtubule plus end–binding protein (Tirnauer and Bierer, 2000; Matis et al., 2014), we found invasion of microtubule into elongating filopodia (Figure 5, B and C), providing evidence that the image analysis software is suitable for image analysis of not only single cells in vitro but also subcellular structures that form in this developing tissue.
FIGURE 5:
In vivo analysis of filopodial dynamics in epithelial sheets. (A) Migrating cells in epithelial sheet of Drosophila abdomen. Individual cell boundaries were visualized with Arm-GFP. (B) Time series of cells expressing EB1 (red) and membrane marker CD8 (green) during filopodial elongation. Note the enrichment of the microtubule plus-end marker EB1 in elongating structures. (C) Analysis of growth dynamics and relative protein concentration for EB1 (top left), CD8 (top right), and ratio of EB1 vs. CD8 (bottom left). Bottom right, overlay of automatically (gray) and manually (red) traced filopodial length. The first two frames, used for tracking adjustments, are separated by the vertical dashed line. Scale bars, 5 μm (A), 2 μm (B).
Limitations of the software
Although the experiments so far indicate that the software can track filopodia in various cell types, they do not address its limitations. Probably the main concern when tracking filopodia is that these dynamic structures may leave the plane of acquisition. To test how the software responds to this scenario, we took advantage of HeLa cells, which carry long filopodial structures that rapidly move along the cell surface (Planchon et al., 2011). Consistent with previous work, we found HeLa cells covered with long and rapidly tilting filopodial protrusions (Figure 6, A and B). Using the image analysis software, we then analyzed these structures. We found long-lasting filopodia, likely reflective of protrusions attached to the substrate (Figure 6C), as well as dynamic filopodia that either partially (Figure 6D) or completely (Figure 6E) left the plane during image acquisition. Of note, when comparing static and dynamic filopodia, we found substantial differences in relative fluorescence intensity of actin along filopodia over time, which can be used as a quality control to exclude filopodia bending out of the plane of acquisition (Figure 6, C and D). Furthermore, we found striking differences in growth/retraction rates in filopodia leaving the acquisition window, which cannot be achieved under physiological conditions (Figure 6E). As a safety measure, we thus included a feature in the software in which abrupt changes in protrusion length (i.e., due to tilting out of the acquisition plane) will trigger an error message. To allow analysis of protrusions that leave but then rapidly reenter the acquisition plane, we also introduced a manual option that makes it possible to continue tracking. However, analysis of temporally unresolved filopodia should be used only when one is absolutely certain that the structure entering the plane is the same as the one that left.
FIGURE 6:
Limitations of image analysis software. (A) Scanning electron microscope images of HeLa cell. Note the large amount of filopodial structures covering the surface. (B) HeLa cell transfected with filamentous actin. Confocal section of cell (top), kymograph of line scan through filopodia (middle), and color-coded overlay of time series (bottom). Note the substantial amount of filopodial movement. (C) Image analysis of long-lived filopodia. Time series of cells transfected with f-tractin (red) and cytosolic marker (green), as well as plot depicting analysis of filopodial growth dynamics and relative concentration of f-tractin along the filopodium. The first two frames, used for tracking adjustments, are separated by the vertical dashed white line. No change in length was visible during the acquisition interval. (D) Filopodia buckling during image acquisition. Time series and analysis as before. Note the strong differences in fluorescence intensity due to helical buckling of the filopodium. (E) Filopodia leaving the confocal plane. Time series and analysis. Scale bars, 10 μm (A, left), 4 μm (A, right), 10 μm (B, top), 2 μm (B, middle and bottom), 5 μm (C–E).
What other scenarios are conceivable in which image analysis will not work? Tracking stops when fluorescence intensity is too low to generate a binary mask for the convex-hull algorithm. In an experiment, this may occur when filopodia leave the acquisition plane or if photobleaching reduces the fluorescence signal beyond the detection level. However, whereas all other cases will lead to a completion of the tracking script, not all filopodia are suitable for analysis. This includes protrusions that loop back to the cell body (here, the most distal vertex is not the tip of the filopodium), as well as filopodia that cross paths with other objects. Analysis of such protrusions should thus be excluded.
Other concerns that should be taken into account when preparing images for analysis are real and artificial movements of the filopodial base. Artificial movements—for instance, induced by lateral drift of the sample during image acquisition—can be corrected using available software (e.g., imagej.net/Linear_Stack_Alignment_with_SIFT). In contrast, real movements of the base, such as seen with filopodia growing on lamellipodial structures, are more challenging, as relative movements of both dynamic structures need to be put into relation to each other. As shown in Supplemental Figure S2B, filopodia can emerge from extending lamellipodia. Here, using the lamellipodial edge as baseline may look like a suitable solution to determine the relative filopodial growth. However, as shown in Supplemental Figure S2A, filopodia-like structures can also be created as lamellipodia retract. Here a baseline created from the lamellipodial edge would register these rather static structures wrongfully as rapidly growing filopodia. To avoid wrong annotations, we thus recommend, in cases in which the base and the filopodia are both in motion, generating two parallel measurements—one of the filopodium and one of the adjacent baseline. To generate a line scan of the baseline, one simply selects a tip position, sets the detection region to 0 pixels, and increases the bending threshold above the maximal length of the adjacent filopodium.
Finally, it is worth mentioning that our software analyzes one filopodium at a time. Although proper statistical analysis is feasible for sample size of ∼100–200 filopodia, manual input required for image analysis may pose challenges for larger data sets (>1000 filopodia).
DISCUSSION
Here we introduced novel image analysis software that unifies analysis of growth dynamics and spatiotemporal protein concentration in filopodia. The software, which is operated using a GUI, provides the user with the possibility to customize image analysis for up to three channels for the particular biological setting (for details, see Supplemental User Manual and Supplemental Movie S2). The inclusion of tracking parameters and additional features for manual fine-tuning maximize robustness while allowing flexible adjustments to the particular experimental setting. As output, the software provides kymographs containing single channels and ratiometric information in .png and .fig formats, as well as all analyzed parameters (e.g., filopodial length, relative protein intensities along the filopodial length, protocol of used tracking parameters) on separate Excel working sheets for documentation and further analysis.
Systematic validation of the image analysis software in silico provides evidence that it can be used independent of tilting and bending of filopodia, as well as over a wide range of fluorescence signal intensities and magnifications (Figure 2). Using a series of model systems, we further show that the image analysis software can successfully be used to investigate filopodial dynamics in vitro (Figures 3 and 4) and in vivo (Figure 5). Comparing various cell types, we found substantial differences in filopodial dynamics. For instance, filopodia in C2C12 cells often appeared as lamellipodia retreated, whereas filopodia in COS cells actively extended from the cell body (Supplemental Figure S2, A and B). Furthermore, we also found helical buckling inside filopodia (Figure 6D), which was recently described as being used by cells to generate traction (Leijnse et al., 2015). Together these results suggest that various subforms of filopodial protrusions may exist.
Using primary neurons as model system, we then tested the image analysis software in a more detailed manner. We clearly could distinguish different phases of the extension–retraction cycle, as well as maximal length and lifetime of individual exploratory filopodia (Figure 4, C and E). Measured filopodial size and growth dynamics are in agreement with published experimental and theoretical work (Portera-Cailliau et al., 2003; Mogilner and Rubinstein, 2005; Zhuravlev and Papoian, 2009). Considering that filopodia were described as growing at an average velocity of 0.05–0.1 μm/s (Argiro et al., 1985; Mogilner and Rubinstein, 2005), acquisition rates of ∼1 Hz are recommended for precise tracking and measurements of growth rates, as well as for intermittent tip fluctuations during elongation and retraction. Expressing a cytosolic control versus small Rho GTPases, we further found that total filopodial length and lifetime were substantially expanded upon overexpression of constitutive active Cdc42 (Figure 4F, blue), whereas expression of constitutive active Rac1 caused formation of ruffle-like structures from which protrusions with reduced lifetime emerged (Figure 4F, red lines). These results, which are consistent with published results in neurons and other model systems (Dumontier et al., 2000; Nakayama et al., 2000; Chen et al., 2006), show the feasibility of the image analysis software to quantify changes in filopodial dynamics in vitro. In line with previous work (Schell et al., 2001; Coutinho-Budd et al., 2012), our dual-color experiments showed filopodial enrichment for the F-BAR domain of srGAP2, as well as for f-tractin, over a cytosolic reference. Of greater importance, however, we found that recruitment of the F-BAR domain of srGAP2 and filopodia formation coincide (Figure 4G), whereas actin-patch formation preceded filopodia formation (Figure 4H), suggesting that this platform is suitable for spatiotemporal cross-correlation analysis. Finally, we also tested the image analysis software in vivo. Using Drosophila epithelial sheet formation as a model system, we found spatially restricted recruitment of EB1 during filopodia formation (Figure 5), showing that our software can also be used for combined analysis of growth dynamics and protein concentration along filopodia in tissue.
Comparing Pearson’s r of manually and automatically measured filopodial length showed an average value of >0.9 both in silico and in vitro, arguing that the software is accurately estimating filopodial length (Figures 2 and 3). Considering that filopodia are not the only finger-like extensions formed by cells, it is plausible to envision that the software would also be suitable for analysis of other forms of cellular protrusions. However, because such extending structures may differ in geometry—for instance, neurites in PC12 cells create a growth cone (Supplemental Figure S3, A–C)—other approaches may yield better results.
Taken together, these experiments show that the image analysis software is well suited to reliably analyze and correlate growth dynamics and spatiotemporal protein concentration during the extension–retraction cycle independent of bending or relative growth angle of filopodia. Unlike previously published work, which is capable of analyzing either growth parameters such as elongation speed and filopodial shape (Styner et al., 2003; Costantino et al., 2008; Fanti et al., 2011; Nilufar et al., 2013; Hendricusdottir and Bergmann, 2014; Tsygankov et al., 2014; Tarnok et al., 2015) or relative spatiotemporal protein concentration (Machacek et al., 2009; Barry et al., 2015), this approach allows us to track filopodial dynamics and combine these shape measurements with spatiotemporal information on relative protein concentration along filopodia. However, although the experiments provide evidence for the versatility and robustness of the software, we stress that the image analysis is only as good as the quality of the images. For instance, increasing the signal-to-noise ratio not only will improve tracking accuracy, but it may also reduce the variance in the ratiometric image analysis. Further, even if the signal is strong, filopodia may still tilt out of the imaging plane (Figure 6). Although amendments to the software allow exclusion of some of these unwanted events, it remains the responsibility of the user to generate data suitable for filopodia analysis.
MATERIALS AND METHODS
Downloading and handling image analysis software
The latest version of the source code can be found at https://campus.uni-muenster.de/index.php?id=13794&L=1. On download, install all .m, .fig, and .xls files from the package in the work folder. To run the software, we recommend MATLAB version 2015b (or above). A detailed description on handling the image analysis software (Supplemental Figure S1 and Supplemental Movie S1) is given in the Supplemental User Manual.
Culturing and imaging 3T3, COS, HeLa, and C2C12 cells
Individual cell lines were plated on glass coverslips using DMEM containing 4.5 g/l d-glucose, GlutaMax-I, and pyruvate (31966-021; Life Technologies, Carlsbad, CA), 10% fetal bovine serum (L11-044; Biochrom AG, Berlin, Germany) and 1% penicillin/streptomycin (10.00 U/ml) (12212; Biochrom AG) and incubated at 37 degree and 5% CO2. Transfection was accomplished with Lipofectamine 2000 (11668-027; Life Technologies) following the manufacturer’s protocol and imaged 16–24 h later.
Culturing and imaging primary hippocampal neurons
Sterilized glass coverslips were functionalized with poly-d-lysine in hydrobromide solution (70 μg/ml; P6407; Sigma-Aldrich, St. Louis, MO) for 1 h and rinsed off with double-distilled H2O. Mouse hippocampal neurons were prepared as previously described (Wienisch and Klingauf, 2006). In brief, primary neurons prepared from embryonic day 18 mice were plated onto coverslips using Neurobasal Medium, 21103-049; Life Technologies), supplemented with B27 (17504-044; Life Technologies), penicillin/streptomycin (12212; Biochrom AG) and 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (15630; Life Technologies). Transfection was accomplished with Lipofectamine 2000 (11668-027; Life Technologies) following the manufacturer’s protocol. After 14–24 h of expression, cells were imaged each 10 s for 10 min at 37°C. All animal protocols were approved by the Veterinär- und Lebensmittelüberwachungsamt Münster.
Culturing and imaging PC12 cells
Glass coverslips were coated with 50 μl of poly-d-lysine in hydrobromide solution (P6407; Sigma-Aldrich) and incubated at 37°C for 30 min. After incubation, coverslips were washed twice with sterile water and again incubated with 100 μl of collagen solution (in 20 mM acetic acid) at 37°C for 45 min. Next coverslips were washed twice with phosphate-buffered saline (PBS; 10010-023; Life Technologies). PC12 cells were cultured on coated coverslips using DMEM containing 4.5 g/l d-glucose (31053-028; Life Technologies), 10% fetal bovine serum (FBS: L11-044; Biochrom AG), 5% horse serum (B15-023; PAA, GE Lifesciences, Fairfield, CT), 1% Glutamax (35050-061; Life Technologies) and 1% penicillin/streptomycin (10.00 U/ml; 12212; Biochrom AG) and incubated at 37°C and 5% CO2. Transfection was accomplished 1 d after plating with Lipofectamine 2000 (11668-027; Life Technologies) following manufacturer’s protocol. At 24 h after transfection, culture medium was exchanged with differentiation medium containing DMEM with 4.5 g/l d-glucose (31053-028Life Technologies), 0.67% FBS (Biochrom AG, L11-044), 0.33% horse serum (B15-023; PAA), 1% GlutaMax, 1% penicillin/streptomycin (10,00 U/ml; 12212; Biochrom AG), and 1 μl of rat nerve growth factor-β (N2513; Sigma-Aldrich). The differentiating PC12 cells were imaged 5 d after plating at 10-s intervals.
Culturing and imaging Drosophila epithelial cells
The following fly lines, obtained from the Bloomington Drosophila Stock Center (Bloomington, IN), were used: ci-GAL4, Arm::GFP, UAS-CD8::RFP, and UAS-EB1::GFP. For live imaging, pupae were removed from incubation at 25°C at 10 min before the desired time after puparium formation (APF). Pupae were dissected to open a small window in their pupal cases to provide visual access to the live dorsal pupal abdomen.
Plasmid constructs for mammalian cells
Constructs for Cdc42(Q61L) (Miller and Johnson, 1994), Rac1(Q61L) (Best et al., 1996), f-tractin (Schell et al., 2001), and the F-BAR domain of srGAP2 (Coutinho-Budd et al., 2012) were previously published.
Fluorescence microscopy
Images for Figure 4 were captured using a CMOS camera (Orca Flash 4.0, C11440-22C; Hamamatsu Photonics, Hamamatsu, Japan) mounted on the side port of an inverted microscope (Eclipse Ti; Nikon, Tokyo, Japan) using a 100× objective and a binning of 1 × 1. Images for Figure 5 were captured using an inverted Zeiss Observer.Z1 microscope equipped with a CSU-X1 spinning-disk scanning unit (Yokogawa Denki, Musashino, Japan), an AxioCam MRm3 charge-coupled device (CCD) camera using a 63× objective, and a binning of 1 × 1. All other images were captured using an electron-multiplying CCD camera (IXON Ultra, DU897-U-CSO-BV; Andor, Oxford Instruments, Belfast, UK), mounted on the side port of an inverted microscope (Eclipse Ti) equipped with a Yokogawa CSU-X1 spinning-disk scanning unit using a 60× or 100× objective and a binning of 1 × 1.
Electron microscopy
Cultured HeLa cells were washed three times with PBS (10010-023; Life Technologies) containing 4% sucrose (S7903; Sigma-Aldrich) and then incubated for fixation in 2.5% glutaraldehyde (R1011; Agar Scientific, Essex, UK) on the basis of 4% sucrose in 1× PBS (10010-023; Life Technologies) for 2 h. With removal of all glutaraldehyde, the sample was washed four times with PBS containing 4% sucrose. Then the sample was incubated in PBS with 1% osmium tetraoxide (7436.1; Roth, Dautphetal, Germany) for 1 h. To reduce the amount of water in the sample, it was transferred from 30% of ethanol to 50, 70, 90, and twice to 99.9% of ethanol, each incubating for 20 min. After successful critical point drying, the sample was mounted on an aluminum slide using LeitC (G3300; Plano, Wetzlar, Germany), gassed out, and coated with 3.5-nm platinum-carbon while being rotated at an angle of 65° in an evaporation chamber (BAF 300; Balzers Union). The scanning electron microscope images were taken with a CamScan (CS2; Cambridge Scanning Company) at a tilting angle of 47° and a working distance of 42 mm.
Supplementary Material
Acknowledgments
We thank the Polleux lab for the srGAP2 construct and members of the Galic, Matis, and Klingauf labs for helpful comments and suggestions. This work was supported by funds from the Deutsche Forschungsgemeinschaft to M.M. (EXC-1003, SPP-1782) and M.G. (EXC-1003).
Abbreviations used:
- APF
after puparium formation
- CCD
charge-coupled device
- CMOS
complementary metal-oxide-semiconductor
- DIV
days in vitro
- EMCCD
electron-multiplying CCD
- GFP
green fluorescent protein
- GUI
graphical user interface
- RFP
red fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-06-0406) on August 17, 2016.
REFERENCES
- Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Argiro V, Bunge MB, Johnson MI. A quantitative study of growth cone filopodial extension. J Neurosci Res. 1985;13:149–162. doi: 10.1002/jnr.490130111. [DOI] [PubMed] [Google Scholar]
- Barry DJ, Durkin CH, Abella JV, Way M. Open source software for quantification of cell migration, protrusions, and fluorescence intensities. J Cell Biol. 2015;209:163-180. doi: 10.1083/jcb.201501081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best A, Ahmed S, Kozma R, Lim L. The Ras-related GTPase Rac1 binds tubulin. J Biol Chem. 1996;271:3756–3762. doi: 10.1074/jbc.271.7.3756. [DOI] [PubMed] [Google Scholar]
- Blum H. A transformation for extracting new descriptors of shape. In: Wathen-Dunn W, editor. Models for the Perception of Speech and Visual Form: Proceedings of a Symposium; Cambridge, MA. MIT Press; 1967. pp. 362–380. [Google Scholar]
- Cheadle L, Biederer T. The novel synaptogenic protein Farp1 links postsynaptic cytoskeletal dynamics and transsynaptic organization. J Cell Biol. 2012;199:985–1001. doi: 10.1083/jcb.201205041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TJ, Gehler S, Shaw AE, Bamburg JR, Letourneau PC. Cdc42 participates in the regulation of ADF/cofilin and retinal growth cone filopodia by brain derived neurotrophic factor. J Neurobiol. 2006;66:103–114. doi: 10.1002/neu.20204. [DOI] [PubMed] [Google Scholar]
- Costantino S, Kent CB, Godin AG, Kennedy TE, Wiseman PW, Fournier AE. Semi-automated quantification of filopodial dynamics. J Neurosci Methods. 2008;171:165–173. doi: 10.1016/j.jneumeth.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Courtney J, Woods E, Scholz D, Hall WW, Gautier VW. MATtrack: a MATLAB-based quantitative image analysis platform for investigating real-time photo-converted fluorescent signals in live cells. PLoS One. 2015;10:e0140209. doi: 10.1371/journal.pone.0140209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho-Budd J, Ghukasyan V, Zylka MJ, Polleux F. The F-BAR domains from srGAP1, srGAP2 and srGAP3 regulate membrane deformation differently. J Cell Sci. 2012;125:3390–3401. doi: 10.1242/jcs.098962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey ME, Smith SJ. The dynamics of dendritic structure in developing hippocampal slices. J Neurosci. 1996;16:2983–2994. doi: 10.1523/JNEUROSCI.16-09-02983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumontier M, Hocht P, Mintert U, Faix J. Rac1 GTPases control filopodia formation, cell motility, endocytosis, cytokinesis and development in Dictyostelium. J Cell Sci. 2000;113:2253–2265. doi: 10.1242/jcs.113.12.2253. [DOI] [PubMed] [Google Scholar]
- Dunaevsky A, Tashiro A, Majewska A, Mason C, Yuste R. Developmental regulation of spine motility in the mammalian central nervous system. Proc Natl Acad Sci USA. 1999;96:13438–13443. doi: 10.1073/pnas.96.23.13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti Z, Martinez-Perez ME, De-Miguel FF. NeuronGrowth, a software for automatic quantification of neurite and filopodial dynamics from time-lapse sequences of digital images. Dev Neurobiol. 2011;71:870–881. doi: 10.1002/dneu.20866. [DOI] [PubMed] [Google Scholar]
- Galic M, Tsai FC, Collins SR, Matis M, Bandara S, Meyer T. Dynamic recruitment of the curvature-sensitive protein ArhGAP44 to nanoscale membrane deformations limits exploratory filopodia initiation in neurons. Elife. 2014;3:e03116. doi: 10.7554/eLife.03116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricusdottir R, Bergmann JH. F-dynamics: automated quantification of dendrite filopodia dynamics in living neurons. J Neurosci Methods. 2014;236:148–156. doi: 10.1016/j.jneumeth.2014.08.016. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Llano O, Smirnov S, Tanhuanpaa K, Faix J, Rivera C, Lappalainen P. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol. 2009;185:323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketschek A, Gallo G. Nerve growth factor induces axonal filopodia through localized microdomains of phosphoinositide 3-kinase activity that drive the formation of cytoskeletal precursors to filopodia. J Neurosci. 2010;30:12185–12197. doi: 10.1523/JNEUROSCI.1740-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leijnse N, Oddershede LB, Bendix PM. Helical buckling of actin inside filopodia generates traction. Proc Natl Acad Sci USA. 2015;112:136–141. doi: 10.1073/pnas.1411761112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN. Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature. 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matis M, Russler-Germain DA, Hu Q, Tomlin CJ, Axelrod JD. Microtubules provide directional information for core PCP function. Elife. 2014;3:e02893. doi: 10.7554/eLife.02893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A, Brinkhaus H, Wagner U. Actin dynamics in dendritic spines: a form of regulated plasticity at excitatory synapses. Hippocampus. 2000;10:555–560. doi: 10.1002/1098-1063(2000)10:5<555::AID-HIPO5>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Miller PJ, Johnson DI. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A, Rubinstein B. The physics of filopodial protrusion. Biophys J. 2005;89:782–795. doi: 10.1529/biophysj.104.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. 2000;20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilufar S, Morrow AA, Lee JM, Perkins TJ. FiloDetect: automatic detection of filopodia from fluorescence microscopy images. BMC Syst Biol. 2013;7:66. doi: 10.1186/1752-0509-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portera-Cailliau C, Pan DT, Yuste R. Activity-regulated dynamic behavior of early dendritic protrusions: evidence for different types of dendritic filopodia. J Neurosci. 2003;23:7129–7142. doi: 10.1523/JNEUROSCI.23-18-07129.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MJ, Erneux C, Irvine RF. Inositol 1,4,5-trisphosphate 3-kinase A associates with F-actin and dendritic spines via its N terminus. J Biol Chem. 2001;276:37537–37546. doi: 10.1074/jbc.M104101200. [DOI] [PubMed] [Google Scholar]
- Styner M, Gerig G, Lieberman J, Jones D, Weinberger D. Statistical shape analysis of neuroanatomical structures based on medial models. Med Image Anal. 2003;7:207–220. doi: 10.1016/s1361-8415(02)00110-x. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnok K, Gulyas M, Bencsik N, Ferenc K, Pfizenmaier K, Hausser A, Schlett K. A new tool for the quantitative analysis of dendritic filopodial motility. Cytometry A. 2015;87:89–96. doi: 10.1002/cyto.a.22569. [DOI] [PubMed] [Google Scholar]
- Tirnauer JS, Bierer BE. EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J Cell Biol. 2000;149:761–766. doi: 10.1083/jcb.149.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsygankov D, Bilancia CG, Vitriol EA, Hahn KM, Peifer M, Elston TC. CellGeo: a computational platform for the analysis of shape changes in cells with complex geometries. J Cell Biol. 2014;204:443–460. doi: 10.1083/jcb.201306067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienisch M, Klingauf J. Vesicular proteins exocytosed and subsequently retrieved by compensatory endocytosis are nonidentical. Nat Neurosci. 2006;9:1019–1027. doi: 10.1038/nn1739. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Kabacoff C, Franca-Koh J, Devreotes PN, Robinson DN, Iglesias PA. Automated characterization of cell shape changes during amoeboid motility by skeletonization. BMC Syst Biol. 2010;4:33. doi: 10.1186/1752-0509-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuravlev PI, Papoian GA. Molecular noise of capping protein binding induces macroscopic instability in filopodial dynamics. Proc Natl Acad Sci USA. 2009;106:11570–11575. doi: 10.1073/pnas.0812746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv NE, Smith SJ. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron. 1996;17:91–102. doi: 10.1016/s0896-6273(00)80283-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.