Abstract
Recently, single-cell molecular analysis has been leveraged to achieve unprecedented levels of biological investigation. However, a lack of simple, high-throughput single-cell methods has hindered in-depth population-wide studies with single-cell resolution. We report a microwell-based cytometric method for simultaneous measurements of gene and protein expression dynamics in thousands of single cells. We quantified the regulatory effects of transcriptional and translational inhibitors on cMET mRNA and cMET protein in cell populations. We studied the dynamic responses of individual cells to drug treatments, by measuring cMET overexpression levels in individual non-small cell lung cancer (NSCLC) cells with induced drug resistance. Across NSCLC cell lines with a given protein expression, distinct patterns of transcript-protein correlation emerged. We believe this platform is applicable for interrogating the dynamics of gene expression, protein expression, and translational kinetics at the single-cell level - a paradigm shift in life science and medicine toward discovering vital cell regulatory mechanisms.
Molecular analysis at the single-cell level may reveal unprecedented insight into sub-cellular-level activities to deepen the understanding of molecular and cellular biology. Many single-cell platforms have been introduced1,2, but they are typically complex systems and often limited by the number of processed cells, resulting in low throughput and difficulty of population-wide studies at the single-cell level. Recent studies have highlighted the significant heterogeneity in the genome3, transcriptome4, and proteome5 across cancer cell populations. Characterizing this heterogeneity is critical to understanding tumor progression6. For example, patients with non-small cell lung cancer (NSCLC) undergoing treatment with epidermal growth factor receptor (EGFR)-targeted tyrosine kinase inhibitors (TKIs, e.g. erlotinib) typically develop drug resistance within a year, through either a secondary mutation to EGFR (e.g. T790M) or overexpression of cMET7–9. It has been hypothesized that clinical progression occurs as treatments drive tumor evolution, leading initially rare resistant cells to dominate the tumor9–11. Unfortunately, the vast majority of molecular analysis approaches, ranging from complementary DNA (cDNA) microarrays to mass-spectrometry-based proteomics, are “bulk assays,” which acquire a single averaged measurement from hundreds to millions of cells. As a result, masked signals are observed from rare cell populations. Single-cell analyses12–14, made possible by recent technological advances in sensitivity and throughput, have enabled characterization of the heterogeneity of complex biological systems15,16. We hypothesize that concurrent analysis of mRNA and protein at the single-cell level will enable effective discovery and characterization of cancer sub-populations, such as rare cells employing post-transcriptional mechanisms to increase cMET abundance.
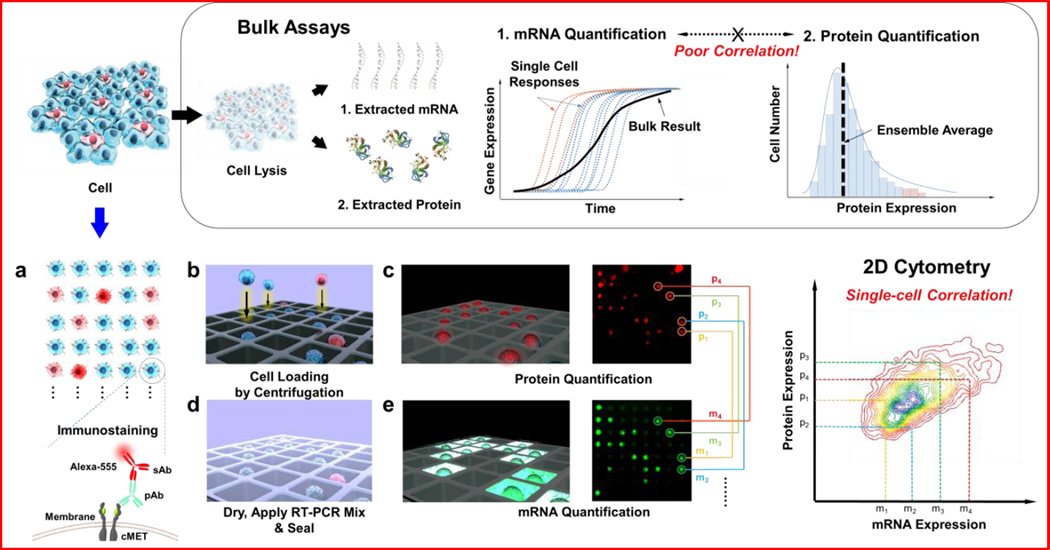
Concurrent transcript and protein quantification is conceptually attractive because of its potential to determine properties of biological systems that are not accurately represented by either mRNA or protein analysis alone17,18. For example, mRNA and protein levels may not be correlated because of post-transcriptional control of the protein translation rate, the variation in the half-lives of specific proteins or mRNAs, or intracellular or extracellular control of degradation of either the mRNA or the protein product19. Currently, there does not exist a high-throughput, single-cell approach for measuring both transcript and protein levels from single cells, although many novel technologies have been developed for either transcript20,21 or proteomic studies22–25 at the single-cell level. Existing single-cell transcript profiling methods using cDNA microarrays and mRNA sequencing (mRNA-Seq) have not been compatible with protein abundance detection26. Likewise, approaches for single-cell protein detection, such as fluorescence-activated cell sorting (FACS) and affinity arrays27, are not compatible with mRNA measurement. Recent developments in single-cell analysis13,28 using a microwell device allow isolation of cells into physically quarantined confinements29,30, and dynamic single-cell analysis platforms generate a vast amount of quantitative experimental biology data12,13. In these single-cell analysis platforms, the isolation of cells allows for in situ permeabilization and detection of an expressed gene of interest by amplification in individual cells. Here, we achieved further advancement of the protein quantification process by incorporating immunostaining, which allows simultaneous measurements of mRNA and protein. This single-cell analysis device represents a platform to study patterns of gene expression, protein expression, and translation kinetics at the single-cell level (Fig. 1).
Figure 1.
Schematic diagrams of simultaneous quantification of mRNA and protein for single-cell level analysis. Conventional analytical methods for individual mRNA and protein quantification rely on bulk assays, with which either population-based analysis or correlation between mRNA and protein cannot be studied at the single-cell level. Our strategy for single-cell-level analysis and correlation studies utilizes a microwell device for single-cell loading and analysis. Individual cells stained with immunofluorescence antibody for the target protein were loaded into single wells on the microwell device, followed by imaging of the protein fluorescence intensities from the immunostained cells. The cells on the device were then directly lysed, and their target transcripts were amplified and tagged with a fluorescent probe via RT-PCR. Each window of the microwell device was imaged, and mRNA fluorescence intensities were extracted. The extracted fluorescence signals from protein and mRNA were analyzed and displayed for correlation using MATLAB software.
We first applied the device to quantify protein and transcript levels of cMET and its associated protein product, hepatocyte growth factor receptor (HGFR, or cMET), in HCC827 NSCLC cells. cMET was selected as an initial target because of its role in mediating resistance to EGFR-targeted therapies in NSCLC. As shown in Fig. 1, cells were first immunostained with anti-CMET antibody and a fluorescent secondary antibody, and then settled into the massive microwell array (25,600 wells). The size of the individual microwells (~20 µm) and initial loading cell density (~110 cell/µL) were previously optimized for single-cell loading. Fluorescence images of the immunostained cells were collected, and the intensities were extracted with a MATLAB program. Following measurements of protein abundance, cells were lysed within their wells, and transcript expression was quantified by measuring the fluorescence intensity via on-chip reverse transcription polymerase chain reaction (RT-PCR), as previously described29. The amplification procedure was initially optimized off-chip using conventional reverse transcription quantitative PCR (RT-qPCR), which indicated that 40 rounds of amplification were optimal to achieve maximum dynamic range (Fig. S1). The use of 40 cycles allowed pseudo-linear amplification of the target gene and accurate quantification of the original amount of the target gene. Simultaneous quantification of both transcript and proteome expression was possible because of landmarks designed into the microwell device that allow arrays to be registered and individual cells to be located (Fig. S2).
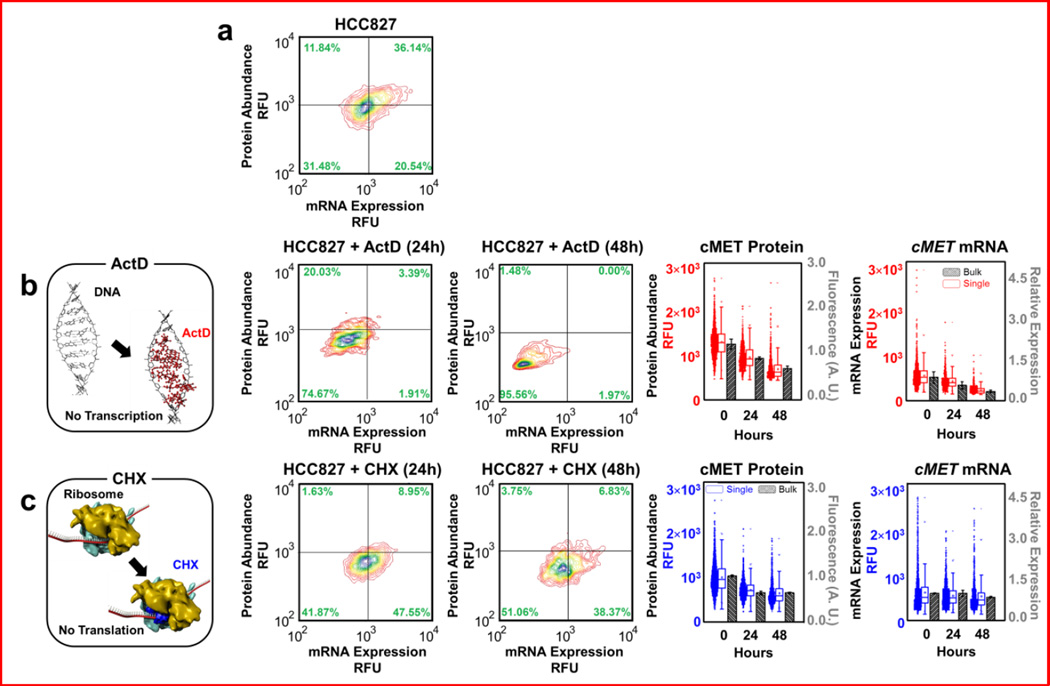
We next validated that the microwell approach could reliably investigate the relationships between mRNA and protein abundances, by treating HCC827 cells with either Actinomycin D (ActD) to inhibit transcription (Fig. 2a,b) or cycloheximide (CHX) to inhibit translation (Fig. 2a,c). IC50 calculations were performed to determine appropriate doses (Fig. S3). We first treated HCC827 cells with 20-nM ActD and then measured surviving cells at 24 h and 48 h. Since ActD inhibits mRNA synthesis and is thus somewhat mimetic of transcription-level regulation, we hypothesized that treated cells would have alterations to both mRNA and protein31 (Fig. 2b). As expected, levels of cMET mRNA decreased by 27% after 24 h and by 46% after 48 h. The level of CMET protein was reduced by 46% after 24 h and by 82% after 48 h, compared to untreated cells at 24 and 48 h. We note that the effect of ActD treatment was initially more apparent as a change in cMET mRNA expression, while alterations to protein abundance were delayed. This distinction between the time scales of change in mRNA and protein abundances likely arose from differences in the half-lives of cMET mRNA relative to cMET protein. We next demonstrated that the platform could detect scenarios in which changes in mRNA and protein levels were discordant, as in cases where regulation is post-transcriptional. Since CHX inhibits protein biosynthesis32 post-transcriptionally, we hypothesized that treatment with CHX should decrease cells’ protein levels, but not their transcript levels. As hypothesized, treatment with CHX had little effect on cMET mRNA levels, but significantly reduced protein abundance (Fig. 2c). The level of protein was reduced by 29% in surviving cells after 24 h and by 36% after 48 h, compared to untreated cells at 24 and 48 h. In contrast, mRNA levels decreased only 12% after 24 h and by 14% after 48 h. These estimates matched bulk RT-qPCR and enzyme-linked immunosorbent assay (ELISA) measurements. Importantly, our approach enabled the measurement of how the distribution of transcript and protein levels changed in single cells. Notably, there are significant cell-to-cell variations in transcript and protein that are not possible to capture in bulk cell qPCR and ELISA assays. Cell-to-cell variation has been regarded as “noise” in conventional bulk assays and defined as the standard deviation divided by the mean. This “noise” interpretation in our single-cell assays exceeded 20 percent, which may represent important cellular heterogeneity information.
Figure 2.
Experimental results of simultaneous mRNA and protein quantification from HCC827 and its derivatives. Validation with conventional (bulk) methods is also shown. A total of 6,300 single cells was analyzed, with an average of ~1,000 cells displayed per plot.
(a) Parental HCC827 single-cell level measurements of cMET mRNA and its corresponding protein. Concurrent analysis of cMET mRNA and protein at the single-cell level was employed to study dynamic responses of individual cells to various drug treatments. Levels of mRNA versus levels of protein are plotted.
(b) Schematic diagram of the transcriptional inhibitory mechanism ActD. Next, single-cell-level measurements of cMET mRNA and cMET protein after treating HCC827 cells with ActD for 24 h and 48 h, respectively. ActD treatment led to significant decreases in mRNA expression level and consequently protein abundance. Corresponding bulk cMET protein measurement of HCC827 treated with ActD for 24 h and 48 h using ELISA, and bulk cMET mRNA measurement of HCC827 treated with ActD for 24 h and 48 h using RT-PCR.
(c) Schematic diagram of the translational inhibitory mechanism CHX. Next, single-cell-level measurements of cMET mRNA and cMET protein after treating HCC827 cells with CHX for 24 h and 48 h, respectively. cMET protein levels decreased on average, while mRNA levels did not change significantly. Corresponding bulk cMET protein measurement of HCC827 treated with CHX for 24 h and 48 h using ELISA, and bulk cMET mRNA measurement of HCC827 treated with CHX for 24 h and 48 h using RT-PCR.
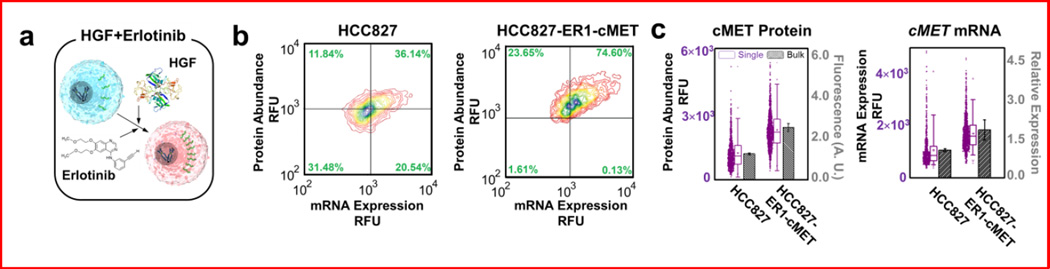
As noted, acquisition of resistance to EGFR-targeted therapeutics in patients with NSCLC8,33–35 often occurs through overexpression of the cMET oncogene. In a previous study of acquired drug resistance5, transient exposure to hepatocyte growth factor (HGF) in combination with erlotinib treatment led to stable ligand-independent erlotinib resistance in HCC827. Using this approach, we derived an erlotinib-resistant cell line (Fig. S4). Resistant cells (termed HCC827−ER1) were much more resistant to erlotinib (IC50 > 5 µM) than the parental cell line (IC50 < 0.01 µM) (Fig. S4b). Single-cell analysis indicated changes in cMET mRNA and protein expression relative to the parental cells, with the HCC827−ER1 cells showing higher levels of cMET protein and cMET mRNA expression than the parental line (Fig. 3). However, our ability to observe both transcript and protein levels in single cells reveals interesting facets of resistance that are not readily observable in bulk assays. Notably, within the parental population, there existed cells whose mRNA transcript levels were nearly as high as those in the derived HCC827−ER1 cells. However, the average cMET protein level of the HCC827−ER1 cells was higher than even the most extreme cells in the parental population, suggesting that in addition to an increase in transcription of cMET, there may also have been additional alterations that allowed for enhanced translation or decreased protein degradation. The ability to investigate these sorts of subtle questions about the relationships between transcript and protein is only possible with analytic platforms that concurrently measure both mRNA and protein levels, as introduced here. Protein distributions were verified by flow cytometry (Fig. S5).
Figure 3.
(a) Schematic diagram of the emergence of drug resistance by treating HGF and erlotinib to parental HCC827 cells. (b) Next, single-cell-level measurements of cMET mRNA and cMET protein for HCC827−ER1, an HCC827 derivative grown in the presence of HGF and erlotinib are compared with its parental cell line – HCC827. The cells grown under selective pressure exhibited higher levels of mRNA and cMET protein expression than did the parental cells. (c) Corresponding bulk cMET protein measurement of parental and overexpressed HCC827 using ELISA, and bulk cMET mRNA measurement of parental and overexpressed HCC827 using RT-PCR. Notably, average protein and mRNA levels from single-cell analyses match that of bulk measurements, while conserving wide cell-to-cell variations.
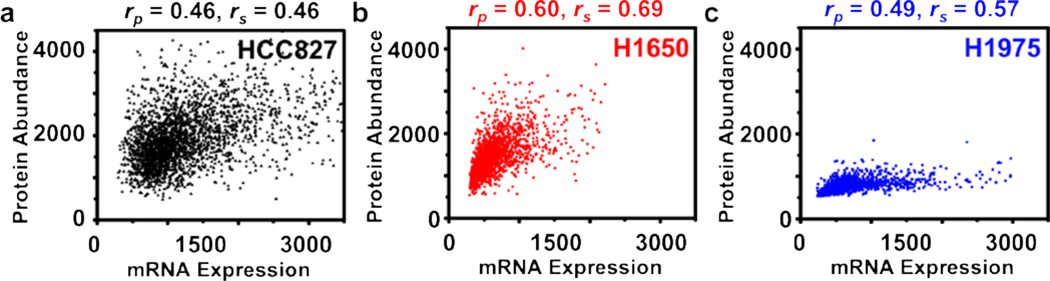
Likewise, three NSCLC cell lines (HCC827, H1650, and H1975) were chosen to interrogate their cMET transcript-protein correlations. H1975 has been previously shown to have relatively high erlotinib resistance (~10 µM), while H1650 and HCC827 have medium and low erlotinib resistance (~1 µM and ~10 nM, respectively). As shown in Fig. 4, we asserted that the three NSCLC cell lines exhibit the distinct transcript-protein correlation pattern, namely that H1975 and H1650 display tighter correlations than that of HCC827. The Spearman rank coefficient weighed this hypothesis by quantifying their correlation values: 0.69 and 0.57 for H1650 and H1975, respectively, while HCC827 has lower correlation with a value of 0.46. This result implies that in a given protein expression, the transcript-protein correlation pattern can be a characteristic signature of a given cell line. Currently there is no consensus on cMET transcript/protein correlation. One study by Watermann et al.36 revealed no explicit connection was found between the expression on protein level and mRNA level of NSCLC, whereas Ozasa et al.37 indicated strong correlation between RT-PCR results and western blot results of drug-resistant small-cell lung cancer cells (SCLC). Other than lung cancer, human gastric carcinoma showed some levels of correlation between cMET gene and protein expression based on individual measurements on cMET mRNA and protein by Huang et al.38 In light of the above-mentioned research, we concluded that further investigation on the correlation study is imperative.
Figure 4.
Simultaneous transcript and protein correlation from NSCLC cell lines (HCC827, H1650, and H1975) with different EGFR-TKI drug resistance responses. (a) Across HCC827, a scattered correlation pattern was observed with a Pearson correlation coefficient (denoted as rp) of 0.46 and a Spearman rank coefficient (denoted as rs) of 0.46. The total number of cells analyzed in the plot was 1,922. (b, c) In comparison, much tighter cMET transcript-protein correlation patterns were observed in H1650 and H1975 cells, which exhibited higher EGFR-TKI drug resistance response than that of HCC827. Pearson correlation coefficients and Spearman rank coefficients were calculated and confirmed their tighter correlation (rp = 0.60 and 0.69, rs = 0.69 and 0.57, respectively). These results indicate that a correlation pattern is a characteristic signature of a given cell line. The total number of cells analyzed was 1,922 for H1650 and 1,344 for H1975.
We have demonstrated a reliable two-dimensional microwell-based method to quantify both protein and mRNA levels directly in single cells, using immunostaining and one-step RT-qPCR, respectively. Measuring cell populations treated with translation inhibitor, CHX, showed a clear shift in transcript-to-protein abundance, while treatment with transcription inhibitor, ActD, showed a uniform decrease in both transcript and protein abundance. In addition, analysis of cMET in individual NSCLC cells that were either sensitive to, or resistant to, EGFR-targeted therapy revealed significant variation, both across cell lines and across cells within each cell line. We also note that validation with well-known technologies such as immunoFISH (fluorescent in situ hybridization) may further enhance the proposed platform’s reliability.
Keeping this in mind, we envision that cytometric platforms like the one introduced here may ultimately allow researchers to investigate the evolution of cancerous tissues and/or circulating tumor cells, enabling detailed monitoring of tumor progression and their point-of-care applications39.
EXPERIMENTAL SECTION
Antibody Staining
We quantified protein abundance by staining the cells with an immunoassay. Prior to immunostaining, cells were fixed in 2% (cold and freshly prepared) paraformaldehyde in DPBS (Dulbecco’s phosphate buffered saline, Invitrogen, Carlsbad, California, USA) and permeablized with TritonX-100 (Sigma-Aldrich, St. Louis, MO, USA) at concentration of 0.2% for 5 minutes at room temperature. The prepared cells were washed twice with 10 mL of 1× DPBS, then resuspended and blocked with 1% bovine serum albumin (BSA, Life Technologies, Carlsbad, California, USA) in DPBS for 10 min at 4°C. After centrifugation at 1,500 rpm for 3 min, the supernatant DPBS was removed, and the cells were resuspended in primary antibody solution diluted in 1:100 DPBS on ice for 10 min. Each 1 µL of primary antibody (rabbit polyclonal IgG provided at 200 µg/mL for detection of Met of human origin, Met Antibody [C-12], sc-10, Santa Cruz Biotechnology, Dallas, TX, USA) was maintained against 106 cells. After incubation, the cells were washed three times with 1 mL of 1% BSA in DPBS to prevent non-specific binding. The cells then were incubated on ice for 10 min with secondary antibody (Anti-rabbit IgG (H+L), F(ab')2 Fragment with Alexa Fluoro® 555 conjugate, A-21430, Thermo Fisher Scientific, Waltham, MA, USA) solution diluted in 1:500 DPBS. According to the provider’s recommendation, the solution composition used was 0.4 µL of the original secondary antibody solution per million cells. Finally, two washing steps with 1 % BSA DPBS and two subsequent washing steps with DPBS were performed to wash away non-specifically bound secondary antibody. The primary antibody (cMET rabbit polyclonal antibody) labels cMET, and the secondary antibody labels the primary antibody with a fluorescent tag (Alexa Fluor 555). Prepared cells were loaded into the fabricated microwells by centrifugation at 3,000 rpm for 10 min. Cells are deposited from a suspension onto the microwells to a density of ~1 cell per well, according to previous characterization29. Then, the fluorescence intensity was measured for each well to analyze cMET abundance.
On-chip and off-chip Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Off-chip reactions were performed with the same reaction mix, but loaded into conventional 96-well plates (Applied Biosystems, Warrington, UK) with a reaction volume of 50 µL and cycled in a bench-top real-time PCR machine (CFX 96 Thermocycler, Bio-Rad, Hercules, CA, USA). After fluorescence images were acquired for proteome quantification, the whole chips were completely dried at 40°C for 20 min and an RT-PCR master mix (CellsDirect™ One-Step qRT-PCR, Thermo Fisher Scientific, Waltham, MA, USA), consisting of 2× reaction mix and polymerases (SuperScript® III RT/Platinum® Taq Mix, Life Technologies) in addition to Taqman Probe® (Life Technologies) for targeting cMET and DEPC-treated water, was applied and centrifuged for microwell delivery at 3,000 rpm for 10 min. Then, the microwells were capped with a small piece of adhesive PCR sealant film (Bio-Rad, Hercules, CA, USA) and spun down at 3,000 rpm for 10 min to prevent water evaporation during PCR thermocycling. Mineral oil (White Mineral Oil Heavy, Dodge Oil, Maywood, CA, USA) was also applied around the device for additional evaporation prevention. Lysis of cells and subsequent reverse transcription for cDNA synthesis were performed in the microwell at 50°C for 45 min. Then, the microwell device went through 40 rounds of thermocycling to amplify the transcribed cDNA (Fig. S1). The first ten cycles consisted of denaturation (at 95°C) and annealing and extension (at 65°C) for specific priming, and the remaining cycles were maintained at 90 °C and 60°C for general amplification.
Transcriptional/Translational Inhibition
We employed two chemicals – Actinomycin D (ActD) and cycloheximide (CHX), which are known to inhibit transcription and translation in eukaryotic cells, respectively. ActD, also known as Dactinomycin, is a type of polypeptide antibiotic that interferes with DNA transcription. ActD acts as a competitive inhibitor by intercalating into GC rich sequences in DNA and stabilizing cleavable topoisomerase-I, II complexes with DNA, thereby preventing RNA polymerase from binding and initiating RNA polymerization. In this experiment, we treated HCC827 cells with ActD to halt transcription and evaluate the time course effect of depletion of cMET mRNA on the level of its corresponding protein, cMET. CHX is a widely used reagent that inhibits protein synthesis in eukaryotic cells only. CHX has been shown to block the elongation phase of eukaryotic translation by binding to the ribosome and inhibiting eEF2-mediated translocation. However, the details of its mechanism, such as the exact binding region of CHX and how it interacts with eEF2, still remain unclear. In this experiment, we treated HCC827 cells with CHX to halt protein synthesis and thus evaluate the effect of post-transcriptional regulation on the level of mRNA and its corresponding protein. Therefore, these inhibitory effects allow us to manipulate the level of mRNA and protein, so that we can evaluate the reliability of the microwell approach in investigating the relationship between mRNA and protein abundances can be properly evaluated. The parental NSCLC cell line, HCC827 cells, were grown in a 25-cm2 cell culture flask. After reaching 80% confluency, the culture media were changed with new media consisting of 1) RPMI media without growth factors for cell cycle arrest, and 2) either 325 nM CHX (Cycloheximide Ready-made solution, Sigma-Aldrich, St. Louis, MO, USA) or 20 nM ActD (ActinomycinD from Streptomyces, Sigma-Aldrich, St. Louis, MO, USA) for translational and transcriptional inhibition, respectively. After 24 h and 48 h of normal incubation, the cells were harvested and underwent immunostaining and RT-PCR to determine the expression levels of both protein and mRNA.
cMET Overexpressed Cell-line
Erlotinib was purchased from LC Laboratories (Woburn, MA, USA). As shown in Fig. S4a, HCC827 parental cells were expanded with 50 ng/mL of hepatocyte growth factor (HGF). 1-µM erlotinib was simultaneously added to these cells and replenished every 3 days. 15 days after addition of HGF and erlotinib, HGF was removed from the media; however, the cells were continuously maintained in erlotinib - these cells are called HCC827−ER1. A cell proliferation assay (MTS) was performed to determine cell viability against erlotinib, as in Fig. S4b. HCC827 parental cells were very susceptible to the presence of erlotinib, while HCC827−ER1 cells were found to be resistant even at high concentrations of erlotinib (> 5 µM). In Fig. S4c, lysates of HCC827 parental and HCC827−ER1 were immunoblotted to detect indicated proteins including cMET, phosphorylated cMET, AKT, phosphorylated AKT, ERK2, phosphorylated ERK ½, ERBB3, phosphorylated ERBB3, and Actin. HCC827−ER1 expressed more cMET than the parental HCC827 cell line. In particular, protein data were directly compared to flow cytometry data, as shown in Fig. S5. All data shown herein were acquired in triplicate to demonstrate robustness, as shown in Fig. S6.
Poisson Distribution of Cell Seeding in the Microwell
By seeding n cells among m wells in a microwell array, the cell distribution across wells would be expected to follow a Poisson random variable X. Then given a particular microwell, the probability that it contains at most one cell (i.e., one cell or no cells) is described by:
In our analysis, most experiments were performed under cell concentration conditions such that at least 99.93% of wells contained at most 1 cell.
Supplementary Material
Acknowledgments
This project was supported by the U.S. National Institutes of Health (NIH) Award U54CA151459 (Center for Cancer Nanotechnology Excellence and Translation (CCNE-T)) and by PSOC-MCSTART U54CA143907.
Footnotes
Note: Supplemental information is available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
S.-m.P., J.Y.L., S.H.., I.K.D.., P.M., and L.P.L. conceived and designed the experiments; S.-m.P., J.Y.L., and S.H., H.L., and Q.P. performed the experiments and analyzed experimental results; S.-m.P., J.Y.L., S.H., S.H.L., I.K.D., P.M., and L.P.L. contributed to data analysis and interpretation; S.S., S.H.L., H.L., performed gene expression analysis and contributed to experimental design; K.L., A.M.W., and S.M.M. prepared and provided biological reagents and cell lines; and S.-m.P., J.Y.L., P.M., and L.P.L. wrote the paper with contributions from all authors.
REFERENCES
- 1.Lecault V, White AK, Singhal A, Hansen CL. Microfluidic single cell analysis: from promise to practice. Curr. Opin. Chem. Biol. 2012;16:381–390. doi: 10.1016/j.cbpa.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 2.Yin H, Marshall D. Microfluidics for single cell analysis. Curr. Opin. Biotechnol. 2012;23:110–119. doi: 10.1016/j.copbio.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramskold D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turke AB, et al. Preexistence and Clonal Selection of MET Amplification in EGFR Mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bean J, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc. Natl. Acad. Sci. USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 9.Mumenthaler SM, et al. Evolutionary modeling of combination treatment strategies to overcome resistance to tyrosine kinase inhibitors in non-small cell lung cancer. Mol Pharm. 2011;8:2069–2079. doi: 10.1021/mp200270v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyo-eun CB, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med. 2015;21:440–448. doi: 10.1038/nm.3841. [DOI] [PubMed] [Google Scholar]
- 11.Mumenthaler SM, et al. The Impact of Microenvironmental Heterogeneity on the Evolution of Drug Resistance in Cancer Cells. Cancer Inform. 2015;14:19. doi: 10.4137/CIN.S19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Carlo D, Wu LY, Lee LP. Dynamic single cell culture array. Lab Chip. 2006;6:1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 13.Di Carlo D, Wu LY, Lee LP. Dynamic single cell culture array. Lab Chip. 2006;6:1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 14.Di Carlo D, Aghdam N, Lee LP. Single-cell enzyme concentrations, kinetics, and inhibition analysis using high-density hydrodynamic cell isolation arrays. Anal. Chem. 2006;78:4925–4930. doi: 10.1021/ac060541s. [DOI] [PubMed] [Google Scholar]
- 15.Bhang HE, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat Med. 2015;21:440–448. doi: 10.1038/nm.3841. [DOI] [PubMed] [Google Scholar]
- 16.Hudak CS, et al. Pref-1 marks very early mesenchymal precursors required for adipose tissue development and expansion. Cell Rep. 2014;8:678–687. doi: 10.1016/j.celrep.2014.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Y, et al. How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim. Biophys. Sin. 2008;40:426–436. doi: 10.1111/j.1745-7270.2008.00418.x. [DOI] [PubMed] [Google Scholar]
- 18.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 19.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 20.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu S, et al. Capillary sieving electrophoresis/micellar electrokinetic capillary chromatography for two-dimensional protein fingerprinting of single mammalian cells. Anal Chem. 2004;76:4044–4049. doi: 10.1021/ac0498314. [DOI] [PubMed] [Google Scholar]
- 23.Irish JM, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 24.Xu F, et al. Single-cell chemical proteomics with an activity-based probe: identification of low-copy membrane proteins on primary neurons. Angew Chem Int Ed Engl. 2014;53:6730–6733. doi: 10.1002/anie.201402363. [DOI] [PubMed] [Google Scholar]
- 25.Hughes AJ, et al. Single-cell western blotting. Nat Methods. 2014;11:749–755. doi: 10.1038/nmeth.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu M, Singh AK. Single-cell protein analysis. Curr. Opin. Biotechnol. 2012;23:83–88. doi: 10.1016/j.copbio.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang JB, Fan HC, Behr B, Quake SR. Genome-wide Single-Cell Analysis of Recombination Activity and De Novo Mutation Rates in Human Sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimov IK, et al. Discriminating cellular heterogeneity using microwell-based RNA cytometry. Nat. Commun. 2014;5 doi: 10.1038/ncomms4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong Y, Ogunniyi AO, Love JC. Massively parallel detection of gene expression in single cells using subnanolitre wells. Lab Chip. 2010;10:2334–2337. doi: 10.1039/c004847j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sobell HM. Actinomycin and DNA transcription. Proc Natl Acad Sci U S A. 1985;82:5328–5331. doi: 10.1073/pnas.82.16.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider-Poetsch T, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balak MN, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor - Mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin. Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 34.Kosaka T, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin. Cancer Res. 2006;12:5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi S, et al. EGFR mutation and resistance of non–small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 36.Watermann I, et al. Improved diagnostics targeting c-MET in non-small cell lung cancer: expression, amplification and activation? Diagnostic Pathology. 2015;10:1–12. doi: 10.1186/s13000-015-0362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozasa H, et al. Significance of c-MET overexpression in cytotoxic anticancer drug-resistant small-cell lung cancer cells. Cancer Sci. 2014;105:1032–1039. doi: 10.1111/cas.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang TJ, Wang JY, Lin SR, Lian ST, Hsieh JS. Overexpression of the c-met protooncogene in human gastric carcinoma--correlation to clinical features. Acta Oncol. 2001;40:638–643. doi: 10.1080/028418601750444204. [DOI] [PubMed] [Google Scholar]
- 39.Park SM, Sabour AF, Son JH, Lee SH, Lee LP. Toward integrated molecular diagnostic system (i MDx): principles and applications. IEEE Trans Biomed Eng. 2014;61:1506–1521. doi: 10.1109/TBME.2014.2309119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.