A DEAD-box helicase regulator has a novel role in centrosome and cilia function, ensuring proper pericentrin recruitment, microtubule organization, and ciliary beating. Independent of mRNA export function, Gle1 forms a toroid structure around the mother centriole. This affects human development and disease mechanisms.
Abstract
Control of organellar assembly and function is critical to eukaryotic homeostasis and survival. Gle1 is a highly conserved regulator of RNA-dependent DEAD-box ATPase proteins, with critical roles in both mRNA export and translation. In addition to its well-defined interaction with nuclear pore complexes, here we find that Gle1 is enriched at the centrosome and basal body. Gle1 assembles into the toroid-shaped pericentriolar material around the mother centriole. Reduced Gle1 levels are correlated with decreased pericentrin localization at the centrosome and microtubule organization defects. Of importance, these alterations in centrosome integrity do not result from loss of mRNA export. Examination of the Kupffer’s vesicle in Gle1-depleted zebrafish revealed compromised ciliary beating and developmental defects. We propose that Gle1 assembly into the pericentriolar material positions the DEAD-box protein regulator to function in localized mRNA metabolism required for proper centrosome function.
INTRODUCTION
The centrosome and its related organelle cilium coordinate critical signals that regulate cell function and influence development. As the major microtubule-organizing center (MTOC) in animal cells, the centrosome influences cell shape, polarity, and motility by regulating microtubule (MT) organization (Doxsey, 2001; Bornens, 2002; Nigg, 2002). The centrosome consists of a pair of centrioles and the associated protein-dense pericentriolar material (PCM). Unlike the centriole, which shows a rigid, ninefold-symmetric cylindrical structure of MTs, the PCM displays as an amorphous structure under the electron microscope (Rieder and Borisy, 1982). Recent advances in superresolution microscopy reveal that individual PCM components adopt a higher-order organization around centrioles (Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012; Sonnen et al., 2012). Understanding how the structure and function of the PCM are controlled is critical to dissecting the roles of centrosomes in different biological and pathological contexts.
The centrosome is not a static organelle. As cells enter mitosis, centrosomes drastically increase in size through the expansion of the PCM (known as centrosome maturation). The PCM plays a critical role in mitosis, as it nucleates MTs (Boveri, 1900; Gould and Borisy, 1977) and enables centrosomes to become robust MTOCs for organization of the spindle apparatus. The PCM is also involved in centriole duplication (Dammermann et al., 2004; Loncarek et al., 2008) and basal body formation (Martinez-Campos et al., 2004; Moser et al., 2010). In differentiated cells, the “mother” centriole (the older of the pair) can transform into a basal body, from which two types of MT structures—motile and primary (nonmotile) cilia—nucleate and protrude from the cell surface. Nonmotile primary cilia are present in nearly all cells and are believed to act as a sensory “antenna” for the cell. Motile cilia exhibit a rhythmic beating motion and are restricted to certain populations of cells, where they serve a number of functions ranging from the simple movement of extracellular debris to the complex process that establishes proper left–right asymmetry in vertebrate body planes.
In our prior studies, we found that inositol 1,3,4,5,6-pentakisphosphate 2-kinase (Ipk1) is enriched at the centrosome and basal body in zebrafish embryos, allowing localized production of inositol hexakisphosphate (IP6) required for proper ciliary beating and length maintenance (Sarmah et al., 2007). Depletion of Ipk1 randomizes both left–right patterning and the asymmetry of normal heart tube placement, with Ca2+ fluxes also altered at the Kupffer’s vesicle, an organ implicated in axial specification during early development (Sarmah et al., 2005). Ipk1 also distinctly marks the nuclear envelope (NE) in budding yeast and plants (York et al., 1999; Lee et al., 2015) in close proximity to Gle1, a well-characterized conserved IP6-binding protein (Alcazar-Roman et al., 2006, 2010; Weirich et al., 2006; Lee et al., 2015).
Gle1 is a multifunctional essential regulator of DEAD-box proteins (Dbps) throughout multiple phases of the mRNA life cycle, including mRNA export, translation, and stress granule formation. Gle1 function has been most carefully dissected at the site of nucleocytoplasmic transport: nuclear pore complexes (NPCs) embedded in the NE. At the cytoplasmic face of NPCs, IP6-bound Gle1 stimulates the RNA-dependent ATPase activity of Dbp5 (Alcazar-Roman et al., 2006; Weirich et al., 2006) to trigger the “remodeling” of mRNA-ribonucleoprotein particles (mRNPs) and confer directionality on mRNA export through NPCs (Alcazar-Roman et al., 2006; Weirich et al., 2006; Tran et al., 2007; Montpetit et al., 2011). Gle1 also regulates efficient translation initiation and termination through the DEAD-box proteins Ded1 (the Saccharomyces cerevisiae orthologue of human DDX3) and Dbp5, respectively (Bolger et al., 2008; Bolger and Wente, 2011). Under stress conditions, human Gle1 is localized to stress granules, where it interacts with DDX3 and modulates the distribution of mRNAs between the states of active and repressed translation (Aditi et al., 2015). The recruitment and specificity of Gle1 function appear to be dictated by localized interaction partners: at NPCs, by interaction with specific NPC proteins (nucleoporins [Nups]; Rayala et al., 2004; Kendirgi et al., 2005), and in translation initiation by associating with eIF3 (Bolger et al., 2008).
There are also established connections between developmental diseases and misregulation of Gle1 function (Nousiainen et al., 2008; Kaneb et al., 2015). Mutations in GLE1 have been causally linked to a human congenital disorder, lethal congenital contracture syndrome 1 (LCCS1) (Nousiainen et al., 2008). In a zebrafish model of LCCS1, we showed that apoptosis of organ precursors, including neural precursors, contributes to the pathogenesis of LCCS1 (Jao et al., 2012). Different GLE1 mutations have also been linked to a familial form of amyotrophic lateral sclerosis, with a disease mechanism apparently distinct from that of LCCS1, in which haploinsufficiency potentially affects the disease pathology in these patients (Kaneb et al., 2015).
Here we report that Gle1 is localized to centrosomes and basal bodies and is a novel PCM component that forms toroid-like organization around the outer layer of the PCM. Down-regulation of Gle1 activity leads to defects in both centrosome integrity and ciliary motility. These are reflected in the loss of a major PCM component, pericentrin (PCNT), from the centrosome and compromises in centrosomal MT nucleation, as well as a defect in the asymmetry of heart looping. Of importance, the centrosomal perturbations caused by Gle1 deficiency are independent of defects in mRNA export. We propose that Gle1-regulated mRNA metabolism at the centrosome mediates proper assembly and function of this essential organelle and that impaired Gle1 function at the centrosome/basal body potentially contributes to the pathogenesis of human disorders linked to GLE1 mutations.
RESULTS
Gle1 localizes to the mother centriole and basal body
Our prior studies show that human Gle1 shuttles between the nucleus and cytoplasm with steady-state localization at NPCs (Kendirgi et al., 2003) and is also at cytoplasmic stress granules under stress conditions (Aditi et al., 2015). On the basis of the requirement for IP6 at basal bodies of cilia in zebrafish (Sarmah et al., 2005, 2007), we hypothesized that Gle1 also plays a role at this essential organelle. We therefore used confocal fluorescence microscopy to examine the localizations of Gle1 and centrosomal components in zebrafish (Figure 1A) and human hTERT-immortalized retinal pigment epithelial cells (RPE-1; Figure 1B). Enhanced green fluorescent protein (eGFP)–hGle1 fluorescence was significantly coenriched with Cetn4–Tag red fluorescent protein (RFP) at centrosomes in zebrafish (Figure 1A, arrowhead) (Of note, with mosaic labeling, not all of the cilia are labeled with both GFP and RFP.) Moreover, endogenous Gle1 was also concentrated at basal bodies and centrosomes in RPE-1 cells (Figure 1B), as detected by respective localization at the base of the acetylated tubulin-positive cilium and overlapping with PCNT in all centrosomes examined.
FIGURE 1:
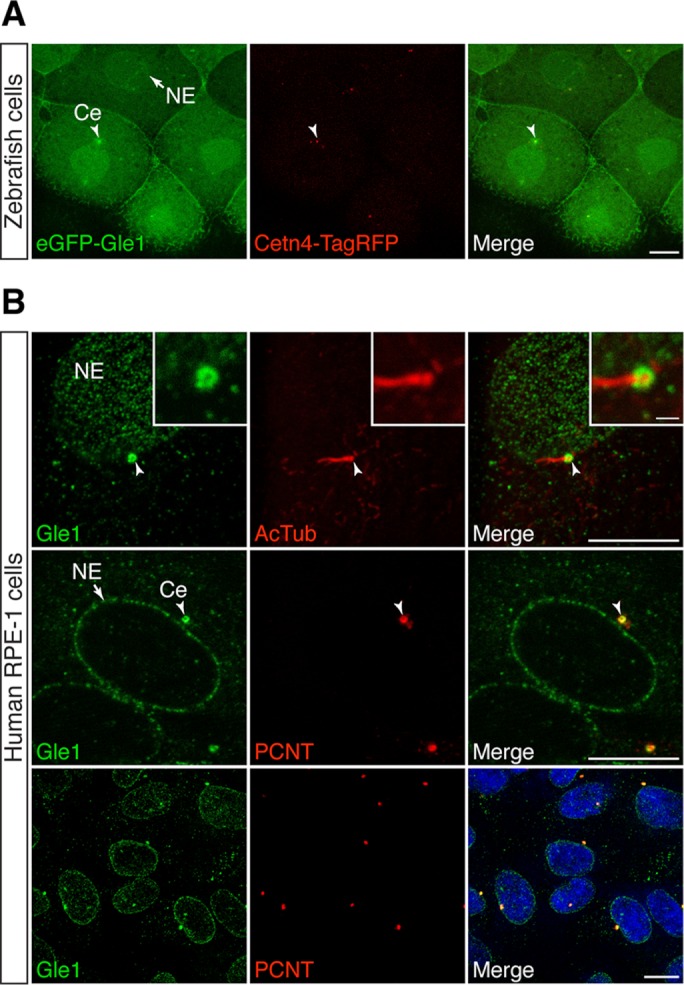
Gle1 is localized to the centrosome and basal body. (A) eGFP-tagged human Gle1 and Cetn4-TagRFP were transiently expressed from the injected RNA in the cells of dome-stage zebrafish embryos. eGFP-Gle1 localized to the NE (arrow) and the centrosome (Ce; arrowhead). (B) Confocal images of serum-starved human RPE-1 cells stained with antibodies against Gle1 and acetylated tubulin (AcTub; top) or Gle1 and PCNT (middle and bottom). Endogenous Gle1 localized to the ciliary base (top; arrowhead) and the centrosome (middle and bottom; arrowhead) as well as to NE (arrow). Scale bars: 10 μm (main images), 1 μm (insets).
To further elucidate the centrosomal association of Gle1 at different cell cycle stages, we examined Gle1 localization in CETN-GFP RPE-1 cells, in which the intensity and number of GFP-tagged centriolar distal lumen protein CETN foci are a reflection of cell cycle stages (Loncarek et al., 2008). In G1 cells, immunofluorescence microscopy showed that Gle1 was predominantly present in one of the centrioles (Figure 2A)—the one with a brighter CETN signal, which in general represents the mother centriole (Loncarek et al., 2008). In late G1/S phase, when the daughter centriole became the new “mother” centriole and two centrioles started to duplicate, the Gle1 signals started to encompass the dimmer, new mother centriole (Figure 2A). In G2/M cells, where there were two pairs of centrioles present, each pair with one mother (brighter) and one newly formed daughter centriole, Gle1 was detected predominantly around the mother centriole (Figure 2A). Similar centrosomal localizations were also observed with a different anti-Gle1 antibody (Supplemental Figure S1A). This toroid-like distribution pattern of Gle1 around the mother centriole was reminiscent of that for PCM, which is predominantly at the mother centriole (Wang et al., 2011). Indeed, we observed a similar mother centriole–dominant, ring-like distribution pattern for one of the major PCM components, PCNT (Figure 2B). Of interest, Gle1 and PCNT localizations at the mother centriole extensively overlapped (see later discussion).
FIGURE 2:
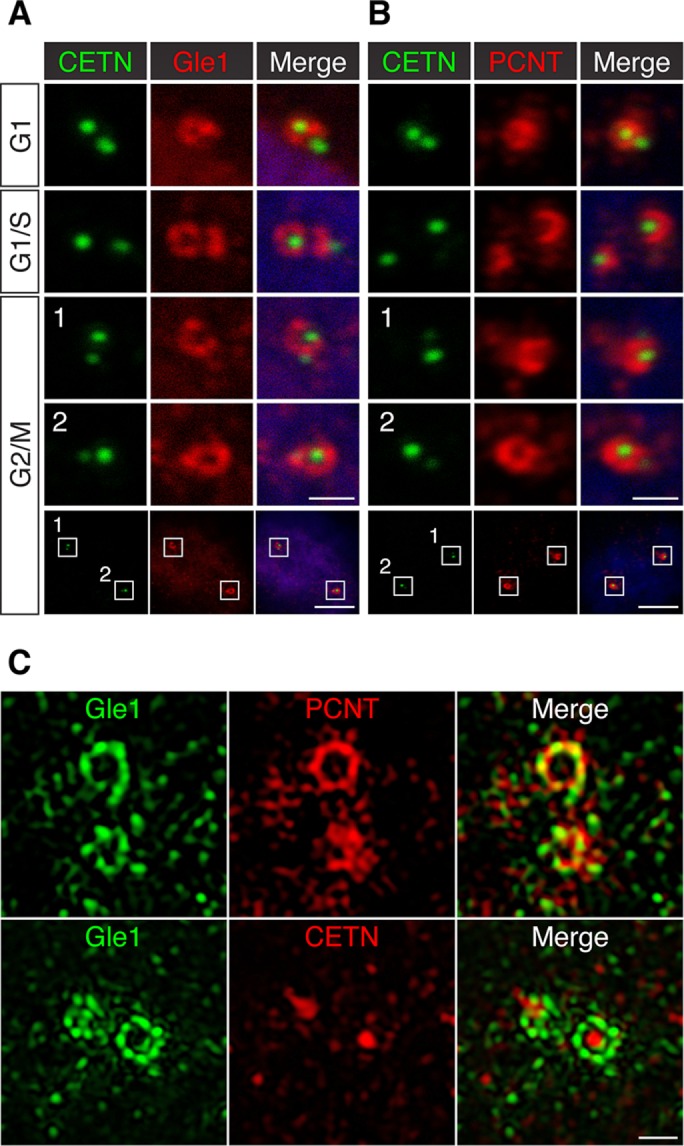
Gle1 is enriched around the mother centriole and intercalated with PCNT. (A, B) Confocal images of CETN-GFP RPE-1 cells stained with antibodies against Gle1 (A) or PCNT (B) show that Gle1 and PCNT are predominantly localized around the mother centriole at different cell cycle stages. (C) 3D-SIM images of interphase human RPE-1 cells stained with antibodies to Gle1 and PCNT (top) or Gle1 and CETN (bottom). The PCNT antibody recognizes the N-terminal portion of PCNT. Scale bars, 1 μm (A, B, all but bottom), 5 μm (A,B, bottom), 0.5 μm (C).
Gle1 enriches in the outermost layers of PCM intercalated with PCNT
The PCM-like distribution pattern of Gle1 around the mother centriole raised the possibility that Gle1 is a PCM component. Recent advances in superresolution microscopy reveal that the PCM adopts a layered organization around centrioles (Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012; Sonnen et al., 2012). In interphase, PCM proteins form concentric rings or fibril structures around the centriolar core in human centrosomes (Lawo et al., 2012; Sonnen et al., 2012). To examine the centrosomal Gle1 localization at subdiffraction resolutions, we conducted immunofluorescence with three-dimensional structured illumination microscopy (3D-SIM). As shown in Figure 2C, in RPE-1 cells, Gle1 formed a toroid-like, beads-on-string structure around the mother centriole, reminiscent of several PCM components (Lawo et al., 2012; Sonnen et al., 2012). Of interest, Gle1 toroids adopted distinct density masses that resemble the ninefold symmetry of the centriole. The Gle1 toroids did not completely overlap but instead intercalate with the PCNT signals (Figure 2C) and appeared to be also located at a similar distance from the centriolar core as did the PCNT toroid. This suggested that both Gle1 and PCNT occupied similar domains at the outer layer of the PCM. Similar topological structures of Gle1 were also observed in U-2 OS cells (Supplemental Figure S1B).
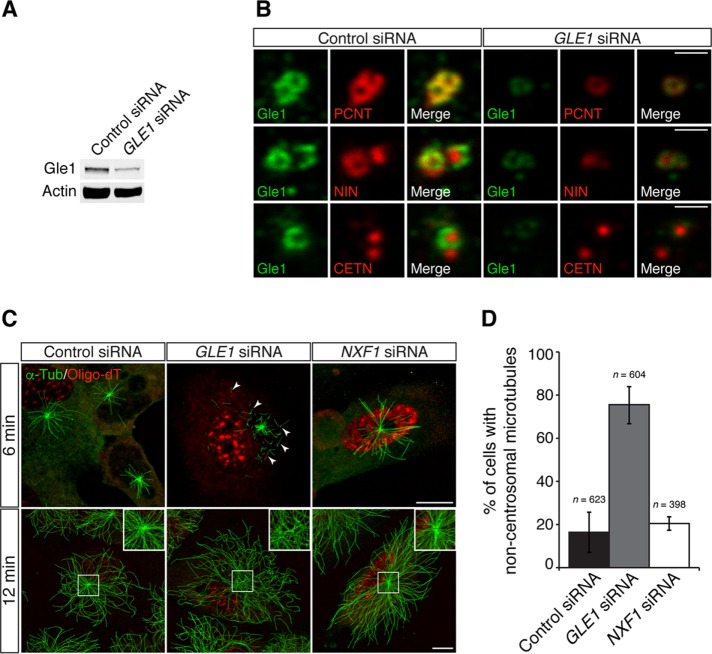
Gle1 is required for the centrosomal localization of PCNT and the formation of subdistal appendages
Because Gle1 localized to a pericentriolar region in close proximity with PCNT, we investigated centrosome organization by examining the localization of PCNT and CETN in control and GLE1 small interfering RNA (siRNA)–treated RPE-1 cells. Immunoblot analysis showed that the Gle1 level was reduced to ∼ 50% in a population of GLE1 siRNA–treated cells (Figure 3A). By immunofluorescence microscopy in GLE1-knockdown cells, PCNT signals at the centrosome were reduced, whereas the CETN levels in the centriolar lumen were not affected (Figure 3B). Thus the centriolar core was not perturbed when Gle1 levels decreased. We also observed that the level of ninein (NIN), a component of the subdistal appendages of the mother centriole, was reduced upon the loss of centrosomal Gle1 (Figure 3B). Together these results indicated that Gle1 is required for proper PCNT positioning in the PCM, as well as for the formation of centriolar structures unique to the mature/mother centriole, such as the subdistal appendage.
FIGURE 3:
Gle1 is required for the PCNT, NIN, and MT organization at the centrosome. (A) Total cell lysates of scrambled control siRNA– or GLE1 siRNA no. 7 (Hs_GLE1L_7 FlexiTube siRNA)–transfected cells were analyzed by immunoblotting for Gle1. Actin served as a loading control. A 30-μg amount of total protein was loaded per lane. (B) Human RPE-1 cells transfected with scrambled control siRNA or GLE1 siRNA no. 7 were processed for indirect immunofluorescence microscopy with antibodies against Gle1, as well as against PCNT (top), NIN (middle), or CETN (bottom). (C) Human RPE-1 cells transfected with scrambled control siRNA, GLE1 siRNA no. 7, or NXF1 siRNA were subjected to a MT regrowth assay, fixed at the indicated time points, and stained with antibodies to α-tubulin (α-Tub), followed by in situ hybridization using Cy3-labeled oligo-dT probes to label poly(A)-containing RNA. In GLE1-knockdown cells, increased ectopic cytoplasmic MT nucleation (6 min, GLE1 siRNA, arrowheads), and few detectable MTs anchored at the centrosome (12 min, GLE1 siRNA) were observed. (D) Quantification of the MT nucleation events in the MT regrowth assay between RPE-1 cells transfected with scrambled control siRNA, GLE1 siRNA no. 7, or NXF1 siRNA. Values are mean ± SEM, and n is the number of cells analyzed in each condition. Scale bar, 1 μm (B), 10 μm (C). Using a second GLE1 siRNA (i.e., GLE1 siRNA no. 4, Hs_GLE1L_4 FlexiTube siRNA) recapitulated the phenotypes (see Supplemental Figure S2).
Down-regulation of GLE1 results in an aberrant MT organization independent of mRNA export defects
PCNT is one of the major PCM components required for anchoring the γ-tubulin ring complex, which templates MT nucleation at the centrosome (Dictenberg et al., 1998; Moritz et al., 2000; Takahashi et al., 2002; Zimmerman et al., 2004). To determine the functional consequence of diminished PCNT levels at the centrosome upon Gle1 depletion, we tested whether MT nucleation was affected upon GLE1 knockdown by a MT regrowth assay. RPE-1 cells were chilled on ice for 50 min to completely depolymerize MTs. The cells were then rewarmed to 30°C to induce MT reassembly. At 6 min after rewarming, strong MT asters formed from the centrosome in the control cells. However, in GLE1 siRNA–treated cells, the MT asters were small (Figure 3C, top). In addition, a majority of the GLE1 knockdown cells also showed numerous MTs nucleated in the cytoplasm away from the centrosome (Figure 3, C, top, and D). After a longer recovery period (12 min), extensive MT regrowth was observed in both control cells and GLE1-knockdown cells. The control cells displayed radially arranged MTs originating mainly at the centrosome. However, in the GLE1-knockdown cells, few MTs were focused at the centrosome (Figure 3C, bottom). Similar reduction in centrosomal levels of PCNT and NIN, but not CETN, and defects in MT organization were observed in knockdown cells using two different GLE1 siRNAs (Supplemental Figure S2 and Figure 3), indicating that these defects are not due to off-target effects of siRNA knockdown.
To test whether defects in MT nucleation/anchoring were an indirect consequence of a general inhibition of mRNA export, we analyzed cells with the knockdown of another essential mRNA export factor, NXF1 (Kang and Cullen, 1999). By in situ hybridization with oligo-dT, nuclear accumulation of poly(A)+ RNA was clearly detected in both GLE1- and NXF1-knockdown cells, reflecting inhibited nuclear mRNA export (Figure 3C). However, down-regulation of NXF1 did not phenocopy the MT defects in GLE1-knockdown cells (Figure 3C). The percentage of cells with noncentrosomal microtubules was similar in control and NXF1 siRNA–treated cells (Figure 3D). In contrast, >70% of the GLE1 siRNA–treated cells showed noncentrosomal microtubules. Thus, independent of its roles in mRNA export, we concluded that Gle1 is uniquely required for organizing PCNT (and likely other PCM components) at the centrosome for proper MT nucleation and anchoring.
Gle1 knockdown alters ciliary beating and establishment of normal left–right heart looping
Based on the established roles for IP6 in cilia function, the shared localization of Ipk1 and Gle1 to basal bodies, and the observation that Gle1 directly binds IP6 (Alcazar-Roman et al., 2010; Montpetit et al., 2011), we speculated that Gle1 might play a role in cilia function. We used zebrafish to investigate this possibility in a whole-animal model, testing whether ciliary beating in the Kupffer’s vesicle (KV) of early embryos was affected upon loss of Gle1 activity using antisense morpholinos (gle1MOs). Of importance, these gle1MOs were well characterized in prior studies and did not have off-target affects (Jao et al., 2012). To assess the ciliary beating in vivo, we transiently expressed Arl13b-eGFP and Centrin4 (Cetn4)-TagRFP to label the ciliary axonemes and basal bodies, respectively. The beating motion of the GFP-positive motile cilia in the KV rendered a distinct fan-like structure under a fluorescence microscope (Figure 4A, top). To confirm that this imaging strategy was capable of assessing ciliary beating defects, we reduced Ipk1 activity with a well-characterized antisense morpholino, ipk1MO1, from our previous study (Sarmah et al., 2005). Consistent with our previous kymography analysis (Sarmah et al., 2005), the stalled ciliary beating defect was readily observed as a distinct loss of the fan-like morphology (Figure 4A, middle). Of interest, the Gle1 knockdown resulted in a ciliary beating defect similar to that observed in ipk1 morphants (Figure 4A, bottom).
FIGURE 4:
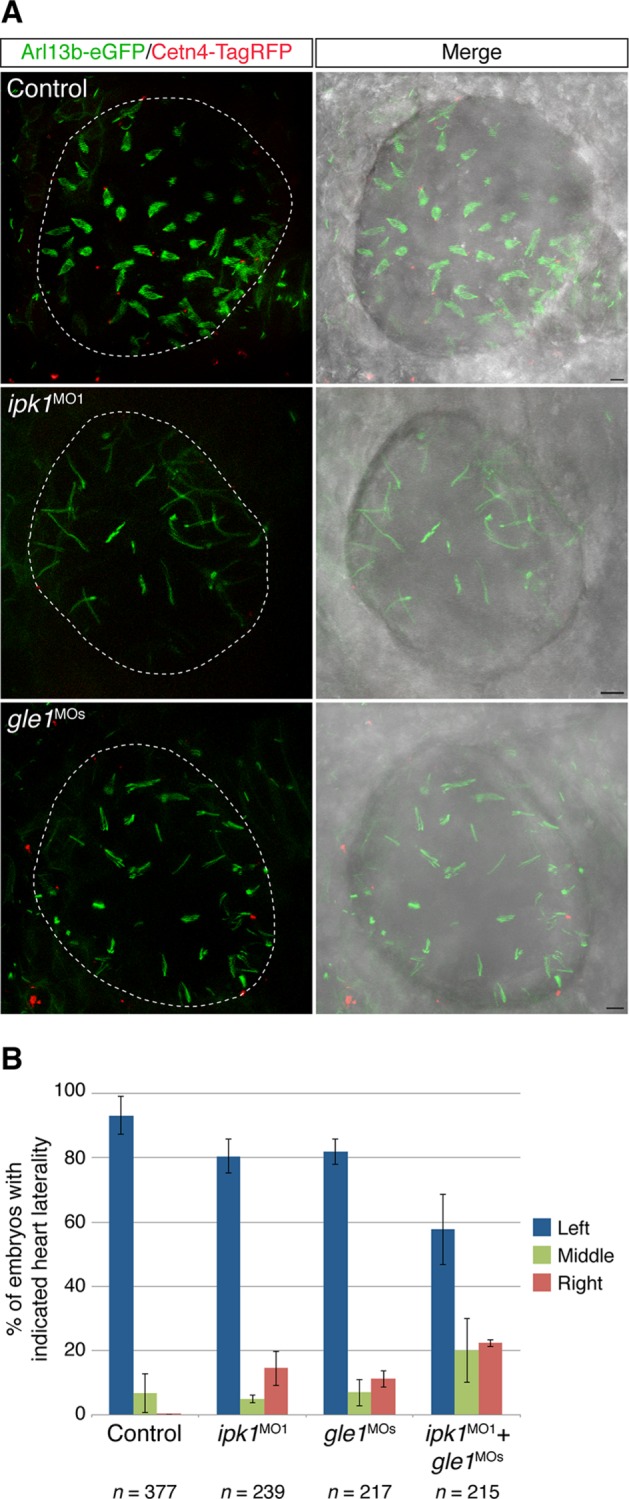
Depletion of Gle1 reduces ciliary beating and alters left–right asymmetry in zebrafish. (A) The axoneme and the basal body of the cilia were mosaically labeled by Arl13b-eGFP and Cetn4-TagRFP, respectively, in uninjected, ipk1MO1-injected, or gle1MOs-injected embryos. Motile cilia in the Kupffer’s vesicle were imaged at six- to nine-somite stages from the dorsal side, anterior to the right. Normal ciliary beating is detected by a fan-like morphology in the control. Scale bar, 5 μm. (B) Tg(-5.1mnx1:TagRFP) embryos were injected with suboptimal doses of translation-blocking ipk1 morpholino (ipk1MO1, 4 ng), a mix of two nonoverlapping translation-blocking gle1 morpholinos (gle1MOs, 1.5 ng each), or a combination of both ipk1MO1 and gle1MOs at the one-cell stage. The direction of the heart looping (TagRFP+) was assessed at 60–72 h post fertilization. Three independent injections were performed for each condition. Values are mean ± SEM; n is the number of embryos analyzed in each condition.
Impaired KV ciliary beating disrupts directional fluid flow within the KV and consequently perturbs left–right (LR) organ patterning, including heart laterality (Essner et al., 2005; Sarmah et al., 2005). We reported heart laterality defects due to impaired KV ciliary beating upon Ipk1 knockdown (Sarmah et al., 2005, 2007). It is not surprising that gle1 morphants also exhibited heart laterality defects (i.e., the heart looped to the midline or to the right instead of to the left; Figure 4B). Of importance, the laterality defects were exacerbated when Ipk1 and Gle1 activities were both knocked down with combined suboptimal doses of both morpholinos (Figure 4B). Thus both Ipk1 and Gle1 activities were involved in normal ciliary beating and heart loop left–right asymmetry determination.
DISCUSSION
Here we report that Gle1, a conserved key regulator of several DEAD-box RNA helicases, is localized to the centrosome, forms a toroid-like structure around the mother centriole, and is required for proper PCM composition. We discover an unexpected role of Gle1 in recruiting a major PCM component, PCNT, and the subdistal appendage protein, NIN, to the mother centriole. We also observe Gle1 at the basal body and that lack of Gle1 results in altered ciliary beating and vertebrate left–right body asymmetry.
Analyzing the effects of reduced Gle1 levels on centrosomal structure and function reveals several specific perturbations. It is striking that cytoplasmic, noncentrosomal MT nucleation increased dramatically upon the loss of Gle1. This phenotype resembles the MT regrowth defect of cells with ablated centrosomes (Khodjakov et al., 2000). Both phenotypes are likely a direct consequence of dispersing the γ-tubulin complex, the scaffold for initiating MT nucleation, into the cytoplasm, with the former caused by the disruption of the centrosome itself (Khodjakov et al., 2000) and the latter by the failure of PCNT and PCM assembly at the mother centriole (Figure 3).
In regard to ciliary beating and randomization of heart looping, Gle1 is also necessary for the assembly of subdistal appendage protein NIN at the centrosome (Figure 3B). Of importance, the subdistal appendage at the mother centriole and the basal foot at the ciliary base form at similar positions along the proximal-distal length of the centriole and share some common components. Thus they have been suggested to have similar etiologies (Anderson, 1972; Kodani et al., 2013). Of importance, basal feet are required for maintaining coordinated ciliary beating (Nakagawa et al., 2001; Kunimoto et al., 2012). Thus, with reduced Gle1, the integrity of the subdistal appendage and basal foot is likely compromised, leading to the impaired ciliary motility. Future experiments employing transmission electron microscopy will be able to test this model. Considering our prior discovery of a role for IP6 production in ciliary beating and length maintenance and regulation of left–right asymmetry (Sarmah et al., 2005, 2007), we further conclude that Gle1 is at least one of the IP6 binding targets responsible for the ipk1 cellular and developmental phenotypes.
How does Gle1 influence PCNT assembly into the centrosome? There are at least two possible mechanisms. These alternatives are not mutually exclusive. First, Gle1 might act as a structural scaffold to mediate the interaction of PCNT and other PCM components at the mother centriole. Like many centrosomal proteins, Gle1 also contains a coiled-coil domain, which is required for its self-association and mRNA export function at the NPC (Folkmann et al., 2013).
Second, all known functions of Gle1 are intimately linked to the modulation of DEAD-box RNA helicases in mRNA export or translation (Alcazar-Roman et al., 2006; Weirich et al., 2006; Bolger et al., 2008; Bolger and Wente, 2011). Given that perturbations on centrosomes are independent of defects in nuclear mRNA export (Figure 3), Gle1 might regulate unknown DEAD-box protein(s) and a subset of mRNAs at the centrosome. Of interest, a global analysis of mRNA localization shows that the transcript of Drosophila pericentrin-like gene CP309/PLP is localized to the centrosome (Lecuyer et al., 2007). Thus it is tempting to speculate that Gle1 indeed regulates the localized translation of PCNT mRNA and facilitates the assembly of PCNT into the centrosome. Alternatively, Gle1 might facilitate the trafficking of PCNT mRNA to the centrosome. We propose that Gle1 serves as a unique link between the modulation of centrosome function and the regulation of RNA metabolism at different subcellular compartments. As such, cellular coordination between efficient gene expression and continued cell division is potentially facilitated.
Our 3D-SIM data show that Gle1 adapts a toroid structure at the outer layer of the PCM around the mother centriole (Figure 2C and Supplemental Figure S1B). However, it is unclear how Gle1 is localized to the mother centriole. Others reported the presence of some Nups at the cilia (Kee et al., 2012), and we documented human Gle1 interactions with at least two Nups, including Nup155 and hCG1 (Rayala et al., 2004; Kendirgi et al., 2005). Thus Gle1 might be localized to centrosomes and basal bodies via interactions with centrosome/basal body–localized Nups. Nup155 and hCG1 localization at centrosomes or basal bodies has not been reported and will be the focus of future studies. However, Nup155 interacts in a specific subcomplex with Nup93 and Nup188 (Grandi et al., 1997; Hawryluk-Gara et al., 2008). Of interest, a recent study demonstrated that both Nup93 and Nup188 are localized at centrosomes and basal bodies and are required for ciliary function (Del Viso et al., 2016). We propose that Nup155 is part of the Nup93/Nup188 centriole/basal body complex and provides a specific binding site for Gle1.
Given the role of Gle1 in cilia and centrosomes, it is noteworthy that LCCS1, the congenital disorder caused by mutations in human Gle1, does not have clinical features similar to ciliopathies (Herva et al., 1985, 1988; Vuopala and Herva, 1994; Nousiainen et al., 2008). The lack of ciliopathy phenotypes could be due to the fact that Gle1 plays essential roles outside of centrosomes/cilia (e.g., in mRNA export). Indeed, in our prior studies, we linked the LCCS1 mutant allele (Gle1-FinMajor) to altered Gle1 function at the nuclear pore complex during mRNA export (Folkmann et al., 2013). Thus the pleiotropic LCCS1 effects upon the loss or alteration of Gle1 might mask the ciliopathy phenotypes, which would have been observed otherwise if the defects were more restricted to only centrosomes and cilia.
Regarding the disease mechanisms underlying LCCS1 in neural precursor death in the zebrafish model of LCCS1 (Jao et al., 2012), the results in this study highlight the importance of considering the effect of Gle1 deficiency on centrosome/ciliary function and whether some of the pathological features are the result of centrosome/ciliary dysfunction during neurogenesis. Concordant with this notion, during neurogenesis in many different biological systems, ciliary structures of neural precursors undergo remarkable remodeling events, such as asymmetric inheritance of ciliary remnants (Paridaen et al., 2013), differential positioning of cilia (Wilsch-Brauninger et al., 2012), and abrupt abscission of ciliary membranes (Das and Storey, 2014). Determining whether Gle1 dysfunction affects any of these events involving plasticity of ciliary structures during neurogenesis may shed new light on the disease pathology caused by Gle1 dysfunction.
In sum, Gle1 is another unique “moonlighting” factor (e.g., Madarampalli et al., 2015) that plays important roles in centrosome/basal body function independent from its roles at the NPC in mRNA export. This work suggests that local regulation of RNA metabolism at the centrosome and basal body is a potential new layer of control in centrosome function and ciliary signaling. With the intimate connection between Gle1 and components of the inositol signaling Ipk1/IP6 and the importance of inositol pathways in cell and developmental signaling (Bielas et al., 2009; Sarmah and Wente, 2010; Xu et al., 2016), an exciting future challenge is to further elucidate the interplay between RNA metabolism, Gle1, and inositol signaling at the centrosome and basal body.
MATERIALS AND METHODS
Zebrafish husbandry
Wild-type AB and Tg(-5.1mnx1:TagRFP) fish were bred and maintained using standard procedures (Westerfield, 2000). Embryos were obtained by natural spawning and staged as described (Kimmel et al., 1995). All animal research was approved by the Institutional Animal Care and Use Committee, Office of Animal Welfare Assurance, Vanderbilt University.
Plasmids and RNA synthesis
The coding sequences of zebrafish arl13b and centrin4 (cetn4) were amplified from total RNA by reverse transcriptase PCR as described (Jao et al., 2012) and cloned into a Gateway Entry vector (Kwan et al., 2007). The plasmids with arl13b-eGFP, cetn4-TagRFP, and eGFP-GLE1, under the control of the CMV/SP6 promoter, were generated by MultiSite Gateway cloning (Kwan et al., 2007; Jao et al., 2012). For arl13b-eGFP and cetn4-TagRFP RNA, the template DNA was linearized by NotI digestion and purified using a QIAprep column (Qiagen, Germantown, MD). Capped RNA was synthesized using an mMESSAGE mMACHINE SP6 kit (Invitrogen, Grand Island, NY) and purified using an RNeasy Mini kit (Qiagen) following manufacturers’ instructions.
Microinjections
Antisense morpholino oligonucleotides (MOs) and RNA were microinjected as described (Jao et al., 2012). Briefly, one-cell-stage embryos from wild-type or Tg(-5.1mnx1:TagRFP) fish were injected with ∼ 1 nl of MOs or RNA. To label the cilia and centrosomes in early zebrafish embryos, ∼30 pg of arl13b-eGFP and cetn4-TagRFP RNA were injected into one-cell-stage embryos. To knock down ipk1 and gle1, translation-blocking ipk1 morpholino (ipk1MO1; Sarmah et al., 2005) and a mix of drgle1ATG1 MO and drgle1UTR1 MO (gle1MOs; Jao et al., 2012) were injected, respectively, at suboptimal doses (4 ng/embryo for ipk1MO1 and 1.5 ng each/embryo for gle1MOs). The direction of heart looping was assessed at 60–72 h postfertilization as described (Sarmah et al., 2005) in the Tg(-5.1mnx1:TagRFP) embryos, where their hearts were fluorescently labeled with TagRFP.
Cell culture, siRNA, and transfection
The hTERT-immortalized retinal pigment epithelial cell line (RPE-1; a gift from Irina Kaverina, Vanderbilt University, Nashville, TN) and CETN-GFP RPE-1 cells (a gift from Alexey Khodjakov, Wadsworth Center, New York State Department of Health, Rensselaer Polytechnic Institute, Albany, NY; Uetake et al., 2007) were maintained in DMEM/F12 and U-2 OS cells in DMEM. All cells were supplemented with 10% fetal bovine serum (FBS) and maintained in a humidified incubator at 37°C in 5% CO2. To induce ciliogenesis in Figure 1B, cells were grown in DMEM/F12 with 0.5% FBS for 48 h. For siRNA transfection, cells were transfected with 40 nM scrambled control siRNA, GLE1 siRNA (Predesigned siRNA from Qiagen; Hs_GLE1L_4 FlexiTube siRNA, and Hs_GLE1L_7 FlexiTube siRNA), or 20 nM NXF1 siRNA (GE Dharmacon, Lafayette, CO), using DharmaFECT 1 transfection reagent (GE Dharmacon) and following the manufacturer’s instructions. siRNA-transfected cells were analyzed 72 h posttransfection.
Antibodies
Guinea pig anti-human Gle1 antibody (1:300, ASW48) was previously reported (Aditi et al., 2015). Rabbit anti-CETN antibody (1:1500; Gayek and Ohi, 2014) was a gift from Ryoma Ohi (Vanderbilt University, Nashville, TN). The following antibodies were purchased commercially: rabbit anti-Gle1 (1:300, ab96007; Abcam, Cambridge, MA; target region, amino acids 414–656), rabbit anti-PCNT (1:500; ab4448; Abcam), rabbit anti-NIN (1:500, ab4447; Abcam), mouse anti–γ-tubulin (1:300; GTU-88; Sigma-Aldrich), rat anti–α-tubulin (1:500; YOL1/34; Accurate Chemical, Westbury, NY), mouse anti–β-actin (1:10,000; A5441; Sigma-Aldrich, St. Louis, MO), and rabbit anti–glyceraldehyde-3-phosphate dehydrogenase (1:1000, 2118L; Cell Signaling Technology, Danvers, MA). Secondary antibodies were Alexa Fluor 488, 568, or 647 (1:400; Invitrogen).
Immunofluorescence
Cells grown on coverslips were fixed in ice-cold methanol at −20°C for 5 min or 4% paraformaldehyde (for rabbit anti-Gle1 antibody), rinsed in 1× phosphate-buffered saline (PBS) briefly, and incubated in blocking solution (2% goat serum, 0.1% Triton X-100, and 10 mg/ml of bovine serum albumin in 1× PBS) at room temperature for 1 h. Primary antibodies were diluted in blocking solution and incubated at room temperature for 1 h. After washes with 1× PBS, the cells were incubated with secondary antibodies for 1 h. Nuclei were counterstained with 1 μM TO-PRO-3 dye (Invitrogen). Coverslips were mounted using Vectashield HardSet mounting medium (Vector Labs, Burlingame, CA).
Microscopy
Confocal microscopy was performed using a Leica TCS SP5 laser-scanning confocal microscope equipped with a 100×/numerical aperture (NA) 1.47 HCX PL APO oil-immersion objective. Images were processed using LAS AF (Leica, Wetzlar, Germany) and ImageJ (National Institutes of Health, Bethesda, MD) software. Superresolution microscopy was performed using a 3D-SIM system (OMX v.4; Applied Precision/GE Healthcare, Marlborough, MA) equipped with a 60×/NA 1.45 Plan-Apochromat oil-immersion objective (Olympus, Tokyo, Japan). The 3D image stacks (0.125 μm apart) were captured with sequential excitation of fluorophores and reconstructed using the softWoRx 5.0 software package (Applied Precision) with the following parameters: Wiener filter constant, 0.001; background intensity offset, 50.00; and custom k0 angles. For final display of cropped centrosomal regions, 3D-SIM micrographs were further resampled (6× Preserve Detail algorithm, Photoshop CC; Adobe, San Jose, CA). Brightness and contrast were adjusted using Photoshop CC in accordance with journal policy. Figures were composed with Adobe Illustrator CC.
Oligo-dT in situ hybridization
Cells were incubated with 1 ng/μl Cy3-labeled 50mer oligo-dT DNA in hybridization buffer containing 125 μg/ml tRNA, 1 mg/ml single-stranded DNA, 1% bovine serum albumin, 10% dextran sulfate, 50% formamide, and 5× saline sodium citrate (SSC; 0.75 M sodium chloride and 75 mM sodium citrate, pH 7.0) at 37°C for 1.5 h, followed by washes with 2× SCC (5 min ×2 at room temperature) and 1× PBS (10 min ×2 at room temperature) before mounting. For cells subjected to both antibody staining and in situ hybridization, immunofluorescence was performed first.
Microtubule regrowth assays
Cells were chilled on ice for 50 min and then recovered at 30°C. At various time points after recovery, cells were fixed and permeabilized in 4% paraformaldehyde, 0.1% glutaraldehyde, and 0.2% Triton X-100 in 1× PBS for 15 min before the immunofluorescence procedure.
Supplementary Material
Acknowledgments
We thank the Wente laboratory for discussions, Irina Kaverina for the RPE-1 cells, Alexey Khodjakov for the CETN-GFP RPE-1 stable cell line, and Ryoma Ohi for anti-CETN antibody. Experiments were performed in part through the use of the VUMC Cell Imaging Shared Resource (supported by National Institutes of Health Grants CA68485, DK20593, DK58404, DK59637, and EY08126). This work was supported by the March of Dimes Foundation (FY-10-360 to S.R.W.) and the National Institutes of Health (5R37GM051219 to S.R.W.).
Abbreviations used:
- MT
microtubule
- PCM
pericentriolar material
- PCNT
pericentrin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-09-0675) on November 9, 2016.
REFERENCES
- Aditi, Folkmann AW, Wente SR. Cytoplasmic hGle1A regulates stress granules by modulation of translation. Mol Biol Cell. 2015;26:1476–1490. doi: 10.1091/mbc.E14-11-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar-Roman AR, Bolger TA, Wente SR. Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1. J Biol Chem. 2010;285:16683–16692. doi: 10.1074/jbc.M109.082370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- Anderson RG. The three-dimensional structure of the basal body from the rhesus monkey oviduct. J Cell Biol. 1972;54:246–265. doi: 10.1083/jcb.54.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas SL, Silhavy JL, Brancati F, Kisseleva MV, Al-Gazali L, Sztriha L, Bayoumi RA, Zaki MS, Abdel-Aleem A, Rosti RO, et al. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat Genet. 2009;41:1032–1036. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger TA, Folkmann AW, Tran EJ, Wente SR. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008;134:624–633. doi: 10.1016/j.cell.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger TA, Wente SR. Gle1 is a multifunctional DEAD-box protein regulator that modulates Ded1 in translation initiation. J Biol Chem. 2011;286:39750–39759. doi: 10.1074/jbc.M111.299321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Boveri T. Über die Natur der Centrosomen. Zellen-Studien 4, Jena, Germany: G. Fischer. 1900.
- Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Das RM, Storey KG. Apical abscission alters cell polarity and dismantles the primary cilium during neurogenesis. Science. 2014;343:200–204. doi: 10.1126/science.1247521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Viso F, Huang F, Myers J, Chalfant M, Zhang Y, Reza N, Bewersdorf J, Lusk CP, Khokha MK. Congenital heart disease genetics uncovers context-dependent organization and function of nucleoporins at cilia. Dev Cell. 2016;38:478–492. doi: 10.1016/j.devcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S. Re-evaluating centrosome function. Nat Rev Mol Cell Biol. 2001;2:688–698. doi: 10.1038/35089575. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- Folkmann AW, Collier SE, Zhan X, Aditi, Ohi MD, Wente SR. Gle1 functions during mRNA export in an oligomeric complex that is altered in human disease. Cell. 2013;155:582–593. doi: 10.1016/j.cell.2013.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Glover DM. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012;2:120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayek AS, Ohi R. Kinetochore-microtubule stability governs the metaphase requirement for Eg5. Mol Biol Cell. 2014;25:2051–2060. doi: 10.1091/mbc.E14-03-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould RR, Borisy GG. The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J Cell Biol. 1977;73:601–615. doi: 10.1083/jcb.73.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Dang T, Pane N, Shevchenko A, Mann M, Forbes D, Hurt E. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell. 1997;8:2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk-Gara LA, Platani M, Santarella R, Wozniak RW, Mattaj IW. Nup53 is required for nuclear envelope and nuclear pore complex assembly. Mol Biol Cell. 2008;19:1753–1762. doi: 10.1091/mbc.E07-08-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herva R, Conradi NG, Kalimo H, Leisti J, Sourander P. A syndrome of multiple congenital contractures: neuropathological analysis on five fetal cases. Am J Med Genet. 1988;29:67–76. doi: 10.1002/ajmg.1320290109. [DOI] [PubMed] [Google Scholar]
- Herva R, Leisti J, Kirkinen P, Seppanen U. A lethal autosomal recessive syndrome of multiple congenital contractures. Am J Med Genet. 1985;20:431–439. doi: 10.1002/ajmg.1320200303. [DOI] [PubMed] [Google Scholar]
- Jao LE, Appel B, Wente SR. A zebrafish model of lethal congenital contracture syndrome 1 reveals Gle1 function in spinal neural precursor survival and motor axon arborization. Development. 2012;139:1316–1326. doi: 10.1242/dev.074344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneb HM, Folkmann AW, Belzil VV, Jao LE, Leblond CS, Girard SL, Daoud H, Noreau A, Rochefort D, Hince P, et al. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Hum Mol Genet. 2015;24:1363–1373. doi: 10.1093/hmg/ddu545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Cullen BR. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendirgi F, Barry DM, Griffis ER, Powers MA, Wente SR. An essential role for hGle1 nucleocytoplasmic shuttling in mRNA export. J Cell Biol. 2003;160:1029–1040. doi: 10.1083/jcb.200211081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendirgi F, Rexer DJ, Alcazar-Roman AR, Onishko HM, Wente SR. Interaction between the shuttling mRNA export factor Gle1 and the nucleoporin hCG1: a conserved mechanism in the export of Hsp70 mRNA. Mol Biol Cell. 2005;16:4304–4315. doi: 10.1091/mbc.E04-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kodani A, Salome Sirerol-Piquer M, Seol A, Garcia-Verdugo JM, Reiter JF. Kif3a interacts with Dynactin subunit p150 Glued to organize centriole subdistal appendages. EMBO J. 2013;32:597–607. doi: 10.1038/emboj.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunimoto K, Yamazaki Y, Nishida T, Shinohara K, Ishikawa H, Hasegawa T, Okanoue T, Hamada H, Noda T, Tamura A, et al. Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell. 2012;148:189–200. doi: 10.1016/j.cell.2011.10.052. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol. 2012;14:1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lee HS, Lee DH, Cho HK, Kim SH, Auh JH, Pai HS. InsP6-sensitive variants of the Gle1 mRNA export factor rescue growth and fertility defects of the ipk1 low-phytic-acid mutation in Arabidopsis. Plant Cell. 2015;27:417–431. doi: 10.1105/tpc.114.132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madarampalli B, Yuan Y, Liu D, Lengel K, Xu Y, Li G, Yang J, Liu X, Lu Z, Liu DX. ATF5 connects the pericentriolar materials to the proximal end of the mother centriole. Cell. 2015;162:580–592. doi: 10.1016/j.cell.2015.06.055. [DOI] [PubMed] [Google Scholar]
- Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J Cell Biol. 2004;165:673–683. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, Agard DA. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol. 2012;14:1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- Moser JJ, Fritzler MJ, Ou Y, Rattner JB. The PCM-basal body/primary cilium coalition. Semin Cell Dev Biol. 2010;21:148–155. doi: 10.1016/j.semcdb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Yamane Y, Okanoue T, Tsukita S, Tsukita S. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol Biol Cell. 2001;12:1687–1697. doi: 10.1091/mbc.12.6.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Centrosome aberrations: cause or consequence of cancer progression. Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Nousiainen HO, Kestila M, Pakkasjarvi N, Honkala H, Kuure S, Tallila J, Vuopala K, Ignatius J, Herva R, Peltonen L. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat Genet. 2008;40:155–157. doi: 10.1038/ng.2007.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paridaen JT, Wilsch-Brauninger M, Huttner WB. Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell. 2013;155:333–344. doi: 10.1016/j.cell.2013.08.060. [DOI] [PubMed] [Google Scholar]
- Rayala HJ, Kendirgi F, Barry DM, Majerus PW, Wente SR. The mRNA export factor human Gle1 interacts with the nuclear pore complex protein Nup155. Mol Cell Proteomics. 2004;3:145–155. doi: 10.1074/mcp.M300106-MCP200. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Borisy GG. The centrosome cycle in PtK2 cells: asymmetric distribution and structural changes in the pericentriolar material. Biol Cell. 1982;44:117–132. [Google Scholar]
- Sarmah B, Latimer AJ, Appel B, Wente SR. Inositol polyphosphates regulate zebrafish left-right asymmetry. Dev Cell. 2005;9:133–145. doi: 10.1016/j.devcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Sarmah B, Wente SR. Zebrafish inositol polyphosphate kinases: new effectors of cilia and developmental signaling. Adv Enzyme Regul. 2010;50:309–323. doi: 10.1016/j.advenzreg.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah B, Winfrey VP, Olson GE, Appel B, Wente SR. A role for the inositol kinase Ipk1 in ciliary beating and length maintenance. Proc Natl Acad Sci USA. 2007;104:19843–19848. doi: 10.1073/pnas.0706934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Yamagiwa A, Nishimura T, Mukai H, Ono Y. Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol Biol Cell. 2002;13:3235–3245. doi: 10.1091/mbc.E02-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, Khodjakov A, Sluder G. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J Cell Biol. 2007;176:173–182. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuopala K, Herva R. Lethal congenital contracture syndrome: further delineation and genetic aspects. J Med Genet. 1994;31:521–527. doi: 10.1136/jmg.31.7.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WJ, Soni RK, Uryu K, Tsou MF. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J Cell Biol. 2011;193:727–739. doi: 10.1083/jcb.201101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) Eugene: University of Oregon Press; 2000. [Google Scholar]
- Wilsch-Brauninger M, Peters J, Paridaen JT, Huttner WB. Basolateral rather than apical primary cilia on neuroepithelial cells committed to delamination. Development. 2012;139:95–105. doi: 10.1242/dev.069294. [DOI] [PubMed] [Google Scholar]
- Xu Q, Zhang Y, Wei Q, Huang Y, Hu J, Ling K. Phosphatidylinositol phosphate kinase PIPKIgamma and phosphatase INPP5E coordinate initiation of ciliogenesis. Nat Commun. 2016;7:10777. doi: 10.1038/ncomms10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol Biol Cell. 2004;15:3642–3657. doi: 10.1091/mbc.E03-11-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.