Human chromatin assembly factor-1 contains a domain (p150N) dispensable for histone deposition but required for normal levels of Ki-67 accumulation on the mitotic perichromosomal layer. This activity requires the sumoylation-interacting motif within p150N. Thus p150N coordinates both interphase and mitotic nuclear structures via Ki67.
Abstract
Chromatin assembly factor 1 (CAF-1) deposits histones during DNA synthesis. The p150 subunit of human CAF-1 contains an N-terminal domain (p150N) that is dispensable for histone deposition but promotes the localization of specific loci (nucleolar-associated domains [NADs]) and proteins to the nucleolus during interphase. One of the p150N-regulated proteins is proliferation antigen Ki-67, whose depletion also decreases the nucleolar association of NADs. Ki-67 is also a fundamental component of the perichromosomal layer (PCL), a sheath of proteins surrounding condensed chromosomes during mitosis. We show here that a subset of p150 localizes to the PCL during mitosis and that p150N is required for normal levels of Ki-67 accumulation on the PCL. This activity requires the sumoylation-interacting motif within p150N, which is also required for the nucleolar localization of NADs and Ki-67 during interphase. In this manner, p150N coordinates both interphase and mitotic nuclear structures via Ki67.
INTRODUCTION
Eukaryotic chromosomes must condense for segregation during mitosis and then decondense and readopt interphase configurations in the next cell cycle. However, mechanisms that link higher-order mitotic and interphase genome organization remain poorly understood. Of note, cellular heterochromatin is highly enriched at specific sites in interphase nuclei, at the nuclear lamina, in pericentric foci, and in perinucleolar regions (reviewed in Politz et al., 2016). Therefore major questions relate to how these loci are relocalized after each mitosis and what mitotic molecules might aid in this.
Nucleolar-associated domains (NADs) are genomic loci that interact frequently with the nucleolar periphery (Németh et al., 2010; van Koningsbruggen et al., 2010; Padeken and Heun, 2014; Matheson and Kaufman, 2015; Pontvianne et al., 2016). NADs are highly enriched for repetitive DNA satellites and histone modifications associated with heterochromatic silencing, such as H3K27me3, H3K9me3, and H4K20me3 (Németh et al. 2010; Politz et al. 2016). One protein shown to regulate NAD localization is the p150 subunit of chromatin assembly factor 1 (CAF-1; Smith et al., 2014). CAF-1 is a highly conserved three-subunit protein complex that deposits histone (H3/H4)2 tetramers onto replicating DNA during S phase of the cell cycle (Smith and Stillman, 1989; Kaufman et al., 1995; Krude, 1995) and is particularly important for DNA replication and maintenance of histone marks at heterochromatic loci (Reese et al., 2003; Quivy et al., 2004, 2008; Sarraf and Stancheva, 2004; Houlard et al., 2006; Dohke et al., 2008; Huang et al., 2010; Baldeyron et al., 2011). In addition to these functions, the N-terminal domain of p150 (p150N) regulates the association of NADs to the perinucleolar region and also regulates the nucleolar localization of several proteins, including the proliferation antigen Ki-67 (Smith et al., 2014).
Ki-67 also controls heterochromatin organization (Sobecki et al., 2016) and affects chromosome condensation (Takagi et al., 1999; Kametaka et al., 2002). Ki-67 is highly expressed in proliferating cells and minimally expressed in quiescent cells (Gerdes et al., 1983, 1984), and meta-analyses of numerous clinical studies have validated Ki-67 as a prognostic marker in grading tumors (Luo et al., 2015; Pezzilli et al., 2016; Pyo et al., 2016; Richards-Taylor et al., 2016). Similar to Ki-67, p150 is also highly expressed in proliferating cells and minimally expressed in quiescent cells and has been proposed as an alternative cellular proliferation marker in clinical studies (Polo et al., 2004).
Recent studies demonstrate that Ki-67 is required for formation of the perichromosomal layer (PCL; Booth et al., 2014; Sobecki et al., 2016), a layer of proteins that surrounds all condensed chromosomes during mitosis (reviewed in Van Hooser et al., 2005). At the PCL, Ki-67’s status as a large, charged protein that possesses “surfactant”-like properties keeps individual mitotic chromosomes dispersed rather than aggregated upon nuclear envelope disassembly, thereby ensuring normal kinetics of anaphase progression (Cuylen et al., 2016). Similarly, acute depletion experiments show that Ki-67 is important for maintaining mitotic chromosome structure and function (Takagi et al., 2016). Because p150N regulates interphase Ki-67 localization, we investigated whether p150N also affects Ki-67 localization during mitosis.
RESULTS AND DISCUSSION
Ki-67 regulates α-satellite localization to the perinucleolar region
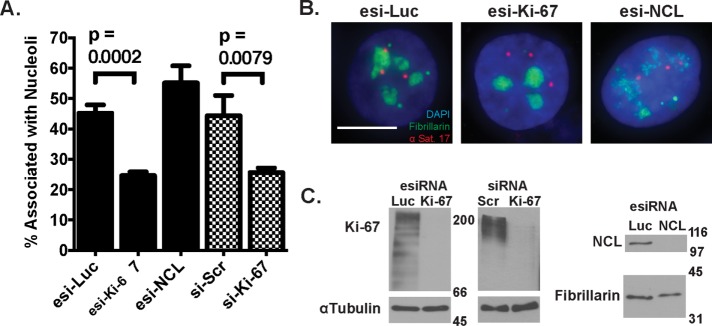
Recent studies suggest that Ki-67 regulates repetitive DNA localization to the nucleolus. Ki-67 depletion decreased the nucleolar association of a LacO array proximal to the rDNA repeats on chromosome 13 (Booth et al., 2014) and the centromeric histone variant CENP-A (Sobecki et al., 2016). To determine whether Ki-67 also regulates satellite repeat association with the nucleolus, we transfected small interfering RNAs (siRNAs) into HeLa cells and performed immuno–fluorescent in situ hybridization (FISH) to visualize the position of α-satellite DNA from chromosome 17 (αSat 17) relative to the nucleolar protein fibrillarin. HeLa cells transfected with siRNAs directed against luciferase or a scramble control displayed ∼45% of αSat 17 alleles associated with nucleoli, which represents a high-frequency interaction characteristic of a NAD locus (Németh et al., 2010; van Koningsbruggen et al., 2010; Smith et al., 2014). In contrast, cells transfected with two different siRNA reagents directed against distinct regions of the Ki-67 mRNA displayed αSat 17 association frequencies to ∼25% (Figure 1, A and B).
FIGURE 1:
α-Satellite associations with nucleoli are reduced upon depletion of Ki-67 but not nucleolin. (A) Immuno-FISH analyses of the association between α-satellite DNA from chromosome 17 (αSat 17, red) and fibrillarin (green) was performed in HeLa cells transfected for 72 h with in vitro–diced esiRNAs targeting luciferase, Ki-67, or NCL (black bars) or with synthetic duplex siRNAs (checked bars) targeting Ki-67 or a scrambled sequence control (si-Scr). The percentage of αSat 17 alleles colocalized with fibrillarin is indicated, with means and SD error bars from three replicate experiments. p values comparing association frequencies in cells treated with esi-Luc (669 alleles) vs. esi-Ki-67 (573 alleles) and si-SCR (540 alleles) with si-Ki-67 (567 alleles) are indicated. (B) Representative FISH images of cells analyzed in A. Scale bar, 10 μm. (C) Immunoblot analyses of cells from A. Blots were probed with antibodies recognizing Ki-67, α-tubulin (loading control), NCL, or fibrillarin (loading control), as indicated.
We also depleted HeLa cells of nucleolin (NCL), a nucleolar protein that primarily localizes to the perinucleolar region (Bugler et al., 1982) and regulates transcription, folding, and assembly of rRNA (Ginisty et al., 1998, 1999; Roger et al., 2002; Rickards et al., 2007). Depletion of NCL altered nucleolar morphology (Figure 1B), as previously reported (Ma et al., 2007; Ugrinova et al., 2007), but did not affect the association of αSat 17 with fibrillarin (Figure 1A). We conclude that in human cells, Ki-67 is required for efficient localization of NADs to nucleoli and that NCL is not required, at least for α-satellite–nucleolar interactions.
Our results with human nucleolin depletion are somewhat different from those obtained in a recent study of nucleolin’s contribution to NAD localization in the plant Arabidopsis thaliana (Pontvianne et al., 2016). As in our experiments (Figure 1B), cells from nucleolin mutant (nuc1) plants also showed altered nucleolar morphology. However, deep sequencing of purified nucleoli revealed altered NAD profiles in nuc1 cells, including decreased nucleolar association of telomeric heterochromatin, coinciding with a 30% loss of telomere length on all chromosomes. It is possible that there are altered NAD associations in nucleolin-depleted human cells that we could not have detected using an α-satellite probe specific for chromosome 17. Alternatively, there is precedence for species-specific interactions mediated by nucleolin. For example, depletion of nucleolin increases centromere–nucleolar interactions in Arabidopsis (Pontvianne et al., 2016) but decreases such interactions in Drosophila (Padeken et al., 2013).
Of interest, decreased telomere lengths were also observed in Arabidopsis fas1 and fas2 mutants (Pontvianne et al., 2016), which lack the homologues of the human p150 and p60 subunits of CAF-1. Another study found that fas1 and fas2 mutants also display decreased copy number of the 45S rRNA genes, along with increased nucleolar association of the remaining 45S rRNA genes (Pontvianne et al., 2013). Note that human p150 and Arabidopsis Fas1 share 29.8% amino acid identity, but Fas1 completely lacks the p150N domain, the N-terminal 310 amino acids of p150 previously shown to regulate human nucleolar structure (Smith et al., 2014). Together these data suggest that p150/Fas1 regulates nucleolar structure in both plants and mammals but also that there are marked mechanistic differences in how this regulation occurs in different organisms. We also note that a higher percentage of the genome is nucleolar associated in human cells (Dillinger et al., 2016) than in plant cells (Pontvianne et al., 2016).
A subset of p150 colocalizes with Ki-67 foci during mitosis and early G1 phases
Ki-67 has three distinct nuclear localization patterns, which appear at distinct cell-cycle periods (see later discussion of Figure 5A). First, Ki-67 localizes to the nucleolus during interphase (Verheijen et al., 1989a; Traut et al., 2002; Cheutin et al., 2003), and a subset colocalizes with p150 at the perinucleolar region (Smith et al., 2014).
FIGURE 5:
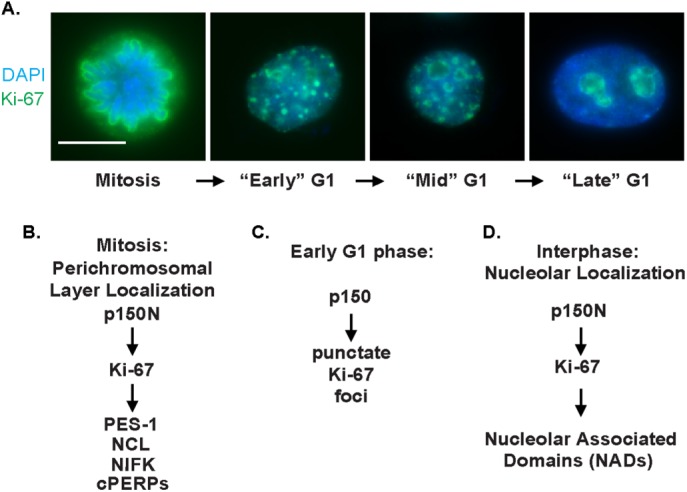
p150 regulates Ki-67 localization throughout the cell cycle: a summary of dependence relationships. (A) HeLa S3 cells were stained with DAPI to visualize DNA (blue) and antibodies against Ki-67 (green) to demonstrate Ki-67 staining patterns throughout the cell cycle. (B) Hierarchy of known proteins controlling localization to the perichromosomal layer during mitosis. Proteins dependent on Ki-67 include pescadillo (PES-1; Sobecki et al., 2016), as well as NCL, NIFK, and the cPERPs (Booth et al., 2014). (C) p150 is required for Ki-67 localization to punctate foci during early G1 (Figure 3). (D) p150N is required for localization of the Ki-67 protein and NAD loci to nucleoli during interphase (Smith et al., 2014).
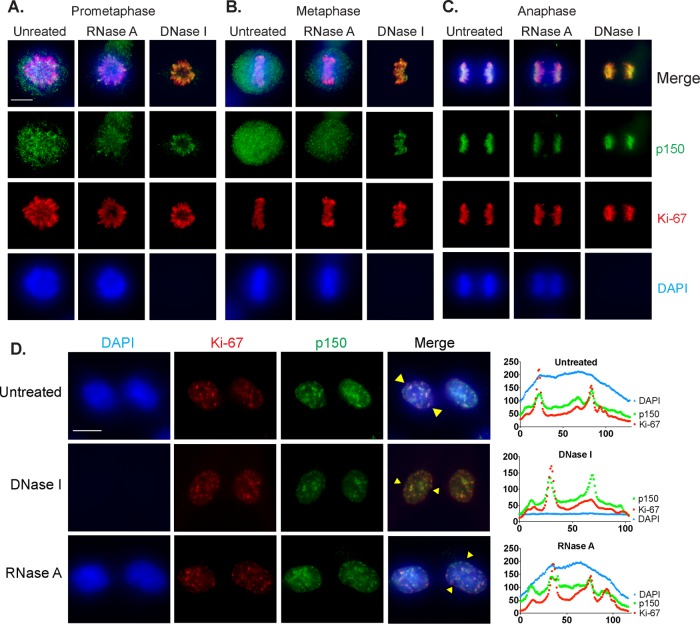
Second, Ki-67 localizes to the PCL during mitosis (Gerdes et al., 1983; Gerdes et al., 1984; Verheijen et al., 1989b). In contrast to the chromosome-associated state of mitotic Ki-67, mitotic phosphorylation evicts the majority of CAF-1 from mitotic chromosomes and inhibits its nucleosome assembly activity (Matsumoto-Taniura et al., 1996; Marheineke and Krude, 1998; Murzina et al., 1999; Keller and Krude, 2000). However, mass spectrometric analyses suggested that a mitotic chromosome–associated subset of p150 existed (Ohta et al. 2010), but its relationship to the PCL was not directly tested. To analyze this, we took advantage of the fact that Ki-67 localization to the PCL remains visibly unchanged even after extraction with 2 M NaCl and treatment with either RNase A or DNase I (Sheval and Polyakov, 2008), indicating that the structural stability of the PCL is independent of the chromatin it surrounds. We performed immunofluorescence on RPE1-hTERT cells that were digested with either RNase A or DNase I or mock digested and then high-salt extracted (Supplemental Figure S1). As previously reported, Ki-67 remained on the PCL throughout mitosis even after DNA or RNA digestion, as shown for prophase (Figure 2A), metaphase (Figure 2B), and anaphase (Figure 2C). As expected, p150, as well as Ki67, could be easily detected on anaphase chromosomes presumably because at that stage, the mitotic phosphorylations of CAF-1 are removed, triggering bulk reassociation with chromatin and reactivation of nucleosome assembly activity (Keller and Krude, 2000). In contrast, during prophase or metaphase, a large amount of p150 evicted into the nucleoplasm made it difficult to determine whether p150 localized to the PCL. The combination of high-salt extraction and DNase I digestion, however, revealed that a subset of p150 localized to the PCL during all phases of mitosis.
FIGURE 2:
A subset of p150 localizes to the PCL during mitosis and to Ki-67 foci during early G1 of interphase. RPE1-hTERT cells were either untreated or digested with RNase A or DNase I as indicated and then high-salt extracted. Cells were stained with DAPI to detect DNA (blue) and with antibodies recognizing p150 (green) and Ki-67 (red). Cells from prophase (A), metaphase (B), and anaphase (C). Note that DNase I–treated cells lack DAPI staining. Scale bar, 10 μm. (D) p150 colocalizes with Ki-67 foci during early G1. RPE1-hTERT cells were treated and stained as indicated. Pairs of recently divided cells featuring hundreds of Ki-67 foci characteristic of early G1 are shown. Line scans (right) of individual cells (yellow triangles in merged images) were used to assess colocalization of p150 with Ki-67 signal. Scale bar, 10 μm.
The third Ki-67 localization pattern occurs after cytokinesis and early in G1 phase. At this stage, Ki-67 localizes to hundreds of distinct foci (Isola et al., 1990; du Manoir et al., 1991), which colocalize with heterochromatic satellite repeats (Bridger et al., 1998). These foci are transient and appear to be intermediates in the process of reformation of nucleoli as cells transition from mitosis to interphase (Saiwaki et al., 2005). Because a subset of p150 is in the mitotic PCL (Figure 2, A–C), we sought to determine whether p150 also colocalized with early G1 Ki-67 foci. To do this, we performed immunofluorescence on early G1 RPE1-hTERT cells digested with nucleases or mock digested as previously described (Figure 2D). In all three conditions, a subset of p150 foci in early G1 cells colocalized with Ki-67 foci, as indicated by line scan analyses. These data suggest that a subset of p150 travels together with Ki67 during exit from mitosis, as PCL components transit toward reforming nucleoli.
p150 regulates Ki-67 accumulation on the perichromosomal layer
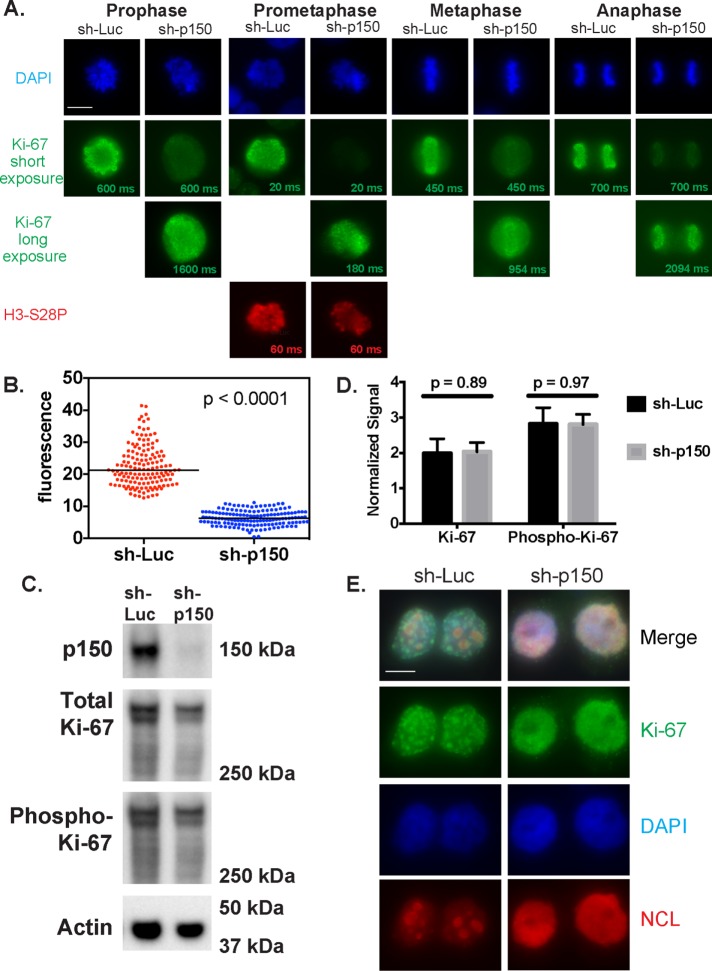
Our previous work demonstrated that p150 is required for normal steady-state accumulation of Ki-67 in the nucleolus during interphase (Smith et al., 2014). Because Ki-67 is essential for the formation of the PCL in mitotic cells (Booth et al., 2014; Sobecki et al., 2016), we tested whether p150 also regulated Ki-67 localization during mitosis. Via immunofluorescence, we examined Ki-67 distribution in HeLa S3 cells expressing an inducible short hairpin RNA (shRNA) directed against either luciferase (Luc) or p150 (Figure 3A). As expected, we found that Ki-67 robustly associated with the PCL during all phases of mitosis in the negative control cells expressing sh-Luc. In contrast, cells expressing sh-p150 displayed less Ki-67 staining on the PCL, as demonstrated in the exposure times matched with the sh-Luc samples. When exposure times were increased, Ki-67 was detected on the PCL, indicating that the PCL was not entirely disassembled upon p150 depletion. When the Ki-67 fluorescence was quantified in prometaphase cells from three biological replicates, cells expressing sh-p150 displayed, on average, a 3.5-fold decrease in fluorescence intensity compared with the control cells expressing sh-Luc (Figure 3B). To test whether this effect of p150 depletion could result from global masking of epitopes on the PCL, we costained some samples with an antibody recognizing the mitotic histone modification H3-S28ph (Goto et al., 1999, 2002). As shown in the prometaphase cell in Figure 3A, this antibody stained cells expressing sh-Luc or sh-p150 equally well, suggesting that p150 regulates Ki-67 accumulation on the PCL rather than affecting overall epitope accessibility on mitotic chromosomes.
FIGURE 3:
p150 regulates Ki-67 localization during mitosis and early G1 phase. (A) HeLa S3 cells from the indicated cell cycle stages expressed an shRNA directed against luciferase (sh-Luc) or p150 (sh-p150) for 72 h and were stained with DAPI to visualize DNA (blue) and antibodies against Ki-67 (green). Exposure times for Ki67 are indicated on each image; in the sh-p150–expressing cells, different exposure times are shown to illustrate reduced Ki-67 accumulation on the PCL. As a positive control for antibody accessibility, cells in prometaphase were also stained with antibodies recognizing the mitotic marker histone H3-S28-phosphate (red). Scale bar, 10 μm. (B) Quantified corrected total cellular fluorescence of cells from three biological replicate experiments of cells expressing sh-luc (red, N = 150) or sh-p150 (blue, N = 147). (C) Immunoblot analysis of extracts from shRNA-expressing cells described in A arrested in mitosis (12 h in 100 ng/ml nocodazole, followed by shaking off mitotic cells). Blots were probed with antibodies recognizing p150 (top), Ki-67 (upper middle), phospho-Ki-67 (lower middle), and actin (loading control; bottom). Numbers on the right indicate migration of marker proteins in kilodaltons. (D) Quantification of the Ki-67 and phospho-Ki-67 blots from C normalized to actin signal (N = 3). Quantification was performed using the Bio-Rad ChemiDoc system. (E) HeLa S3 cells that expressed an shRNA directed against luciferase (sh-Luc) or p150 (sh-p150) for 72 h were synchronized in mitosis (12 h in 100 ng/ml, nocodazole followed by shaking off), and released into drug-free medium for 2 h to enrich for early G1 cells. Cells were stained with DAPI to visualize DNA (blue) and antibodies against Ki-67 (green) and NCL (red). Scale bar, 10 μm.
To explore how p150 may be regulating Ki-67 localization, we tested whether p150 is required for maintaining steady-state levels of Ki-67. Our previous work demonstrated that Ki-67 steady-state levels were not affected upon p150 depletion in asynchronous cells (Smith et al., 2014). To determine whether p150 regulates the steady-state protein levels of Ki-67 during mitosis, we collected extracts from mitotic-arrested cells expressing sh-Luc or sh-p150 (Supplemental Figure S2) and analyzed them by immunoblotting (Smith et al., 2014; Figure 3, C and D). Total levels of Ki-67 in mitotic samples normalized to actin signals were not significantly changed upon p150 depletion (Figure 3, C and D), indicating that p150 is not required for maintaining steady-state levels of Ki-67 during mitosis. At the beginning of mitosis, Ki-67 localization to the PCL occurs in conjunction with hyperphosphorylation of the Ki-67 protein (MacCallum and Hall, 1999; Endl and Gerdes, 2000; Takagi et al., 2014). These phosphorylations are important for PCL localization, as treatment of mitotic cells with protein kinase inhibitors results in dephosphorylation of Ki-67 and relocalization of Ki-67 away from the PCL to distinct nuclear foci (MacCallum and Hall, 1999). Therefore we tested whether p150 regulates Ki-67’s mitotic phosphorylation status by reprobing the immunoblots of mitotically arrested cell extracts with antibodies that specifically recognize Ki-67 phosphorylated on Cdk consensus sites within the Ki-67 internal repeat structure (Takagi et al., 2014). However, we detected no statistically significant changes upon p150 depletion (Figure 3, C and D). Therefore p150 does not appear to regulate the steady-state levels or phosphorylation of Ki-67 during mitosis.
p150 regulates the formation of Ki-67 foci in early G1 phase
Because p150 regulates the localization of Ki-67 to the nucleolus during interphase (Smith et al., 2014) and to the PCL during mitosis (Figure 3A), we wanted to examine whether p150 is also required for the formation of the punctate Ki-67 foci seen at the beginning of G1 phase. To answer this question, we induced expression of sh-Luc or sh-p150 for 60 h and then synchronized cells by adding 100 ng/ml nocodazole for the final 12 h of shRNA expression. After vigorously shaking mitotic cells off the plate, we washed cells three times in phosphate-buffered saline (PBS) and released them into nocodazole-free medium. Based on fluorescence-activated cell sorting of a time course after release (Supplemental Figure S3), G1 cells were collected at 120 min postrelease and stained with antibodies directed against Ki-67 and NCL (Figure 3E). In the sh-Luc samples, hundreds of punctate Ki-67 foci were visible, and NCL was detected within the newly reformed nucleoli. In contrast, sh-p150 samples featured disrupted NCL localization and dispersal of Ki-67 foci. Together our data suggest that the contribution of p150N to interphase Ki-67 and NCL localization (Smith et al., 2014) results from events that begin in M and/or G1 phase. In this manner, p150 regulates Ki-67 localization throughout the cell cycle (see later discussion of Figure 5).
The SIM within the N-terminus of p150 is required for Ki-67 localization to the PCL
To map p150 domains required for regulating Ki-67 accumulation on the PCL, we used previously published HeLa cell lines stably expressing V5-epitope–tagged, shRNA-resistant, p150-derived transgenes (Smith et al., 2014). These V5-transgene cell lines were acutely depleted of endogenously encoded p150 via lentiviral expression of sh-p150 for 72 h before immunofluorescence analysis of Ki-67.
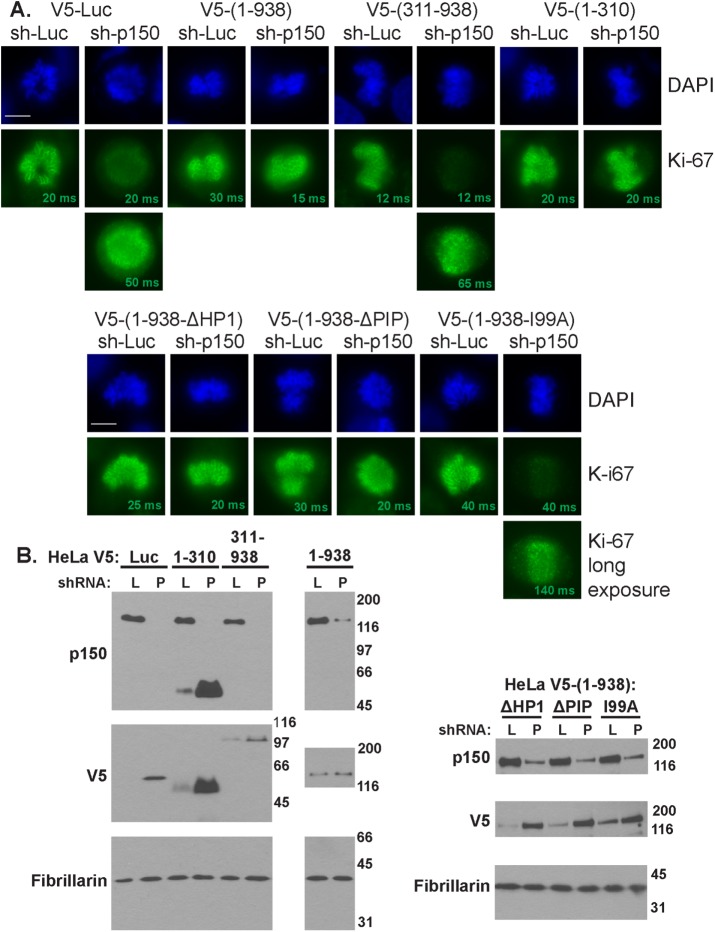
The C-terminal two-thirds of p150 serves as the scaffold for binding the other two subunits of the CAF-1 complex and thereby is essential for CAF-1’s nucleosome assembly activity (Kaufman et al., 1995; Takami et al., 2007). In contrast, p150’s N-terminus is dispensable for nucleosome assembly (Kaufman et al., 1995) but is necessary and sufficient to maintain Ki-67 localization to the nucleolus during interphase (Smith et al., 2014). We found that depletion of p150 in HeLa cells expressing either luciferase or a p150 transgene encoding the C-terminus (amino acids 311–938) displayed reduced Ki-67 localization to the PCL (Figure 4A). In contrast, cells expressing either full-length p150 (amino acids 1–938) or only the N-terminus (p150N, amino acids 1–310) maintained normal amounts of Ki-67 on the PCL (Figure 4A). Therefore the p150 N-terminus regulates mitotic Ki-67 abundance in a manner independent of the chromatin assembly activity of the CAF-1 complex.
FIGURE 4:
The SIM within the N-terminal domain of p150 is required to maintain Ki-67 localization to the PCL. (A) HeLa cells in prometaphase expressing the indicated shRNA-resistant p150 transgene were infected with lentiviruses encoding the indicated shRNAs (sh-Luc or sh-p150) for 72 h and then stained with DAPI to visualize DNA (blue) and antibodies against Ki-67 (green). Note that upon expression of sh-p150, cells expressing luciferase (V5-Luc), the p150 C-terminus (V5-(p150-311-938)), or p150 with a I99A point mutation in the SIM motif required longer exposure times to detect Ki-67 on the PCL. Scale bar, 10 μm. (B) Immunoblots of cell extracts described in A expressing either sh-Luc (L) or ph-p150 (P). Blots were probed with antibodies recognizing p150 (top), V5 transgenes (middle), and fibrillarin (loading control; bottom). Note the depletion of endogenous p150 in sh-p150 lanes and that V5-tagged transgene expression often increased in these lanes, as we had previously (Smith et al., 2014).
p150N contains several known interaction motifs, including a noncanonical PCNA-interacting peptide (PIP; Moggs et al., 2000; Rolef Ben-Shahar et al., 2009), a heterochromatin protein 1–binding domain (Murzina et al., 1999), and a sumoylation-interacting motif (SIM; Uwada et al., 2010; Sun and Hunter, 2012). Of note, the SIM in p150N is required for normal Ki-67 localization to the nucleolus during interphase (Smith et al., 2014). We therefore determined whether transgenes encoding full-length p150 with mutations in foregoing three motifs described supported normal Ki-67 accumulation during mitosis. The cell lines expressing p150 transgenes with mutations in the PIP and HP1 domains maintained normal Ki-67 levels on the PCL. However, cells expressing a p150 transgene with a single amino acid mutation (I99A) that disrupts the SUMO-binding activity of the SIM (Uwada et al., 2010) displayed reduced levels of Ki-67 on the PCL (Figure 4A). Together our data indicate that that the SIM motif within the N-terminus of p150 is required for the normal accumulation of Ki67 on the PCL during mitosis.
Together these data and our previous studies suggest a hierarchical relationship between p150N and Ki-67 in regulating nuclear architecture across the cell cycle (Figure 5, B–D). p150 and Ki-67 both colocalize during mitosis and interphase (Figure 2; Smith et al., 2014), regulate NAD localization (Figure 1; Booth et al., 2014; Smith et al., 2014; Sobecki et al., 2016), and have established roles in regulating heterochromatin (Quivy et al., 2008; Sobecki et al., 2016). It is noteworthy that p150 is required for the formation of punctate Ki-67 foci in early G1 of the cell cycle, as previous studies demonstrated that these foci colocalize with heterochromatic satellite repeats typically enriched within NADs (Bridger et al., 1998). Because ablation of these foci results in mislocalization of NAD elements, we hypothesize that the function of these foci may be to guide heterochromatin-rich NADs back to the nucleolar periphery. However, additional evidence, such as the deep sequencing of the DNA elements contained within these foci, is required to further support this hypothesis.
p150N appears to function upstream of Ki-67 in this hierarchy, as p150N is required for efficient Ki-67 association with the PCL during mitosis (Figure 4) and with the nucleolus during interphase (Smith et al., 2014). p150N is dispensable for nucleosome assembly by CAF-1, indicating that p150’s role in regulating Ki-67 localization is independent of histone deposition. During both mitosis and interphase, the SIM within p150N is required for Ki-67 localization (Figure 4; Smith et al., 2014), suggesting that this action involves an as-yet-unidentified sumoylated protein. These data suggest that future studies should explore the contributions of the p150N domain and its putative sumoylated binding partners to the relationship between Ki-67 and p150 in regulating mitotic and interphase nuclear structure.
MATERIALS AND METHODS
Cell culture
HeLa S3 cells containing the Trex CMV/TO promoters driving either sh-Luc or sh-p150 expression (Campeau et al., 2009) were maintained in RPMI medium with 5% tetracycline-free fetal bovine serum (FBS), 2 mM L-glutamine, and antibiotic/antimycotic solution (Life Technologies). shRNA expression was induced by supplementing media with 2 μg/ml doxycycline for 72 h before fixation or processing. Mitotic cells were enriched by adding 100 ng/ml nocodazole for 12 h (hours 60–72 of the doxycycline treatment) and then freed from the cell culture flask surface by vigorous shaking. To enrich for early G1 cells, shaken mitotic cells were washed three times with PBS and plated onto polylysine-coated coverslips in drug-free medium, and samples were taken at specified time points. HeLa cells continuously expressing the V5-tagged p150 transgenes (Smith et al., 2014) were maintained in DMEM and supplemented with 10% FBS, 2 mM L-glutamine, and antibiotic/antimycotic solution (Life Technologies, Carlsbad, CA). RPE1-hTERT cells (a generous gift from Judith Sharp, Department of Molecular Biology, Massachusetts General Hospital) were maintained in 50:50 DMEM-F12 medium supplemented with 10% FBS, 7.5% sodium bicarbonate, 2 mM L-glutamine, and antibiotic/antimycotic solution (Life Technologies).
Depletion reagents
For lentiviral depletion (Smith et al., 2014), cells were infected at a multiplicity of infection of 7.5 with 6 μg/ml Polybrene for 72 h. Lentivirus reagents were synthesized as previously described (Campeau et al., 2009; Smith et al., 2014). Endonuclease-prepared siRNA (esiRNA) reagents were generated as previously described (Fazzio et al., 2008; Smith et al., 2014), using primers listed in Table 1 to generate double-stranded RNAs, which were then digested using RNase III. For esiRNA transfection, 500 ng of siRNAs were transfected in 1 ml Opti-MEM (Life Technologies) with 6 μl of Oligofectamine (Life Technologies). After 6.5 h, 2.5 ml of medium was added on top of the transfection cocktail, and cells were processed after 72 h. For synthetic siRNA transfection, 10 μl of 5 μM scramble (AM4611; Ambion) or Ki-67 (4392420-s8796; Ambion) siRNAs were diluted in 400 μl of Opti-MEM (Life Technologies) with 5 μl RNAiMAX (Invitrogen) and incubated at room temperature for 10 min. The siRNA cocktail was then slowly added to six-well dishes containing 800 μl of Opti-MEM, and 6 h later, 2.5 ml of appropriate medium (lacking antibiotics) was added. Note that the esiRNA directed against Ki-67 targets the Ki-67 repeat region within exon 13, whereas the synthetic siRNA targets nucleotides 559–577 (CGUCGUGUCUCAAGAUCUAtt) within exon 6.
TABLE 1:
esiRNAs.
| Target | Forward primer | Reverse primer |
|---|---|---|
| Luciferase | gcgtaatacgactcactataggAACAATTGCTTTTACAGATGC | gcgtaatacgactcactataggAGGCAGACCAGTAGATCC |
| NCL | gcgtaatacgactcactataggGCGACGAAGATGATGAAGATGA | gcgtaatacgactcactataggGTGAGTTCCAACGCTTTCTCC |
| Ki-67 | gcgtaatacgactcactataggGTGCTGCCGGTTAAGTTCTCT | gcgtaatacgactcactataggGCTCCAACAAGCACAAAGCAA |
Immunofluorescence and immuno-FISH
Cells were plated on polylysine (Sigma-Aldrich, St. Louis, MO)–coated coverslips for 24 h before manipulation. For immunofluorescence experiments, fixation and permeabilization were performed in six-well tissue culture dishes to minimize displacement of mitotic cells. After aspiration of media, cells were immediately fixed with 4% PFA/PBS for 10 min at room temperature. Cells were then gently washed with ice-cold PBS and permeabilized with 0.5% Triton TX-100/PBS on ice for 5 min. Cells were then washed twice with room temperature PBS and then incubated in blocking buffer (1% BSA/PBS) for 5–30 min in a 37°C humid chamber. After blocking, cells were incubated with primary antibody in blocking buffer for 2 h in a 37°C humid chamber (Table 2). Coverslips were then transferred to Coplin jars and washed three times with PBS on a rotating platform for 10 min at room temperature. After washing, cells were incubated with secondary antibody in blocking buffer for 1 h in a 37°C humid chamber. Coverslips were then transferred to Coplin jars and washed three times with PBS on a rotating platform for 10 min at room temperature. After the final washing step, cells were transferred to a Coplin jar containing 4′,6-diamidino-2-phenylindole (DAPI; 50 ng/ml) in PBS for 2 min. Coverslips were mounted with Vectashield (Vector Labs, Peterborough, UK), and images were taken on a Zeiss Axioplan2 microscope with a 63× objective. Corrected total cell fluorescence (CTCF) was quantified using the ImageJ “measure” feature and background subtracted from an area of the image without a cell. Images were all captured using the same exposure time across three biological replicates, and the unpaired Student’s t test was used to generate p values. Immuno-FISH was performed as previously reported (Smith et al., 2014). Association percentages for each of the three biological replicates were transformed into arcsine units, and the unpaired Student’s t test was used (with Welch’s correction) to generate p values. p < 0.05 was considered statistically significant. All statistical analysis was performed using GraphPad Prism 6.
TABLE 2:
Antibodies.
| Epitope | Species | Manufacturer or provider | Catalog number | Immunofluorescence dilution | Western blot dilution |
|---|---|---|---|---|---|
| Fibrillarin | Rabbit | Abcam, Cambridge, UK | Ab5821 | 1:500 | 1:2000 |
| H3S28ph | Mouse (clone HTA28) | Sigma-Aldrich | H9908 | 1:100 | |
| Ki-67 | Rabbit | Abcam | Ab66155 | 1:500 | 1:2000 |
| NCL | Mouse (clone 364-5) | Abcam | Ab136649 | 1:250 | 1:1000 |
| p150 | Rabbit | Campeau et al. (2009) | 1:2000 | ||
| p150 | Mouse (ss1) | Smith and Stillman (1991) | 1:500 | ||
| p150 | Mouse (ss48) | Smith and Stillman (1991) | 1:500 | ||
| Phospho–Ki-67 | Rabbit | Takagi et al. (2014) | 1:5000 | ||
| V5 | Mouse | Thermo | R960-25 | 1:1000 | 1:2000 |
Nuclease digestion
RPE1-hTERT cells were used for these experiments rather than HeLa cells because they are sufficiently adherent during detergent permeabilization/nuclease digestion/high-salt extraction. Nuclease digestion was performed as previously reported (Sheval and Polyakov, 2008), with some modification. Cells were plated onto polylysine-coated coverslips in six-well dishes at least 24 h before manipulation, and all subsequent steps were performed with solutions containing 100 μM phenylmethylsulfonyl fluoride. Cells were washed once with digestion buffer (10 mM Tris-HCl, pH 7.6, 5 mM MgCl2, 1 mM CuSO4) and then incubated in 1% Triton TX-100/digestion buffer for 10 min at 4°C. Cells were then gently washed with digestion buffer and incubated in 100 μg/ml DNase I/digestion buffer, 100 μg/ml RNase A/digestion buffer, or mock digested (“untreated”) in digestion buffer in a 37°C humid chamber for 30 min. Proteins were then extracted by incubating for 10 min in extraction buffer (2 M NaCl, 10 mM ethylenediaminetetraacetic acid [EDTA], 20 mM Tris-HCl, pH 7.6) at 4°C. Cells on coverslips were immediately fixed as described, or RNA samples were processed using TRIzol (Invitrogen). Briefly, 1 ml of TRIzol was added directly to the plate, and cells were homogenized by pipetting. After incubation for 5 min at room temperature, samples were stored at −80°C until further processing. After thawing, 200 μl of chloroform was added, and samples were shaken vigorously for 15 s before incubation at room temperature for 2 min. Samples were then centrifuged at 13,000 × g for 15 min at 4°C. The aqueous phase was transferred to a fresh tube, and one volume of isopropanol was added and mixed well by vigorous shaking. Samples were then incubated at room temperature for 10 min before centrifugation at 13,000 × g for 15 min at 4°C. Samples were then washed with 75% ethanol and resuspended with nuclease-free water. Ten percent of recovered samples were run on a 1% agarose gel to compare RNA digestion efficiency.
Early G1 cells were selected by choosing pairs of cells that appeared significantly smaller than surrounding cells and featured hundreds of Ki-67 foci (Isola et al., 1990; du Manoir et al., 1991; Croft et al., 1999). Line scans were performed using the RGB Profiles Tool plug-in for ImageJ.
Western blotting
A 15-μg amount of protein was loaded and run through either Tris-HCl polyacrylamide gradient gel (20–5%) for asynchronous samples or a NuPAGE Novex 3–8% Tris-Acetate Protein Gel (ThermoFisher Scientific, Waltham, MA) for mitotically arrested samples. Protein samples were then transferred to a polyvinylidene fluoride membrane at 20 V for 17 h at 4°C to maximize the transfer of high–molecular weight Ki-67. Chemiluminescence was acquired using the Bio-Rad ChemiDoc system and quantified using Bio-Rad Image Lab V 5.2.1.
Acknowledgments
We thank Michael Brodsky for the generous use of the AxioPlan microscope, Masatoshi Takagi and Naoko Imamoto for the anti–phospho-Ki-67 antibody, Judith Sharp for RPE1 cells, and Tom Fazzio for fruitful discussions. This work was supported by National Institutes of Health Grants R01 GM55712 and U01 DA040588 to P.D.K.
Abbreviations used:
- CAF-1
chromatin assembly factor 1
- esiRNA
endoribonuclease-prepared siRNA
- FISH
fluorescence in-situ hybridization
- H3K27me3
histone H3, lysine 27 trimethylation
- H3K9me3
histone H3, lysine 9 trimethylation
- H4K20me3
histone H4, lysine 20 trimethylation
- Luc
luciferase
- NAD
nucleolar-associated domain
- NCL
nucleolin
- p150N
N-terminal domain of CAF-1 subunit p150
- PCL
perichromosomal layer
- PIP
PCNA-interacting peptide
- Scr
scramble sequence
- SIM
sumoylation-interacting motif
- siRNA
small interefering RNA.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-09-0659) on November 2, 2016.
REFERENCES
- Baldeyron C, Soria G, Roche D, Cook AJL, Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 2011;193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth DG, Takagi M, Sanchez-Pulido L, Petfalski E, Vargiu G, Samejima K, Imamoto N, Ponting CP, Tollervey D, Earnshaw WC, et al. Ki-67 is a PP1-interacting protein that organises the mitotic chromosome periphery. Elife. 2014;3:e01641. doi: 10.7554/eLife.01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger JM, Kill IR, Lichter P. Association of pKi-67 with satellite DNA of the human genome in early G1 cells. Chromosome Res. 1998;6:13–24. doi: 10.1023/a:1009210206855. [DOI] [PubMed] [Google Scholar]
- Bugler B, Caizergues-Ferrer M, Bouche G, Bourbon H, Amalric F. Detection and localization of a class of proteins immunologically related to a 100-kDa nucleolar protein. Eur J Biochem. 1982;128:475–480. doi: 10.1111/j.1432-1033.1982.tb06989.x. [DOI] [PubMed] [Google Scholar]
- Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, O’Donohue M-F, Beorchia A, Klein C, Kaplan H, Ploton D. Three-dimensional organization of pKi-67: a comparative fluorescence and electron tomography study using FluoroNanogold. J Histochem Cytochem. 2003;51:1411–1423. doi: 10.1177/002215540305101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S, Blaukopf C, Politi AZ, Müller-Reichert T, Neumann B, Poser I, Ellenberg J, Hyman AA, Gerlich DW. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature. 2016;535:308–312. doi: 10.1038/nature18610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillinger S, Straub T, Nemeth A. Nucleolus association of chromosomal domains is largely maintained in cellular senescence despite massive nuclear reorganisation. bioRxiv. 2016:054908. doi: 10.1371/journal.pone.0178821. doi: http://dx.doi.org/10.1101/054908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohke K, Miyazaki S, Tanaka K, Urano T, Grewal SIS, Murakami Y. Fission yeast chromatin assembly factor 1 assists in the replication-coupled maintenance of heterochromatin. Genes Cells. 2008;13:1027–1043. doi: 10.1111/j.1365-2443.2008.01225.x. [DOI] [PubMed] [Google Scholar]
- du Manoir S, Guillaud P, Camus E, Seigneurin D, Brugal G. Ki-67 labeling in postmitotic cells defines different Ki-67 pathways within the 2c compartment. Cytometry. 1991;12:455–463. doi: 10.1002/cyto.990120511. [DOI] [PubMed] [Google Scholar]
- Endl E, Gerdes J. Posttranslational modifications of the Ki-67 protein coincide with two major checkpoints during mitosis. J Cell Physiol. 2000;182:371–380. doi: 10.1002/(SICI)1097-4652(200003)182:3<371::AID-JCP8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–1715. [PubMed] [Google Scholar]
- Gerdes J, Schwab U, Lemke H, Stein H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31:13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Goto H, Tomono Y, Ajiro K, Kosako H, Fujita M, Sakurai M, Okawa K, Iwamatsu A, Okigaki T, Takahashi T, et al. Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J Biol Chem. 1999;274:25543–25549. doi: 10.1074/jbc.274.36.25543. [DOI] [PubMed] [Google Scholar]
- Goto H, Yasui Y, Nigg EA, Inagaki M. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells. 2002;7:11–17. doi: 10.1046/j.1356-9597.2001.00498.x. [DOI] [PubMed] [Google Scholar]
- Houlard M, Berlivet S, Probst AV, Quivy J-P, Héry P, Almouzni G, Gérard M. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2006;2:e181. doi: 10.1371/journal.pgen.0020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Yu Z, Zhang S, Liang X, Chen J, Li C, Ma J, Jiao R. Drosophila CAF-1 regulates HP1-mediated epigenetic silencing and pericentric heterochromatin stability. J Cell Sci. 2010;123:2853–2861. doi: 10.1242/jcs.063610. [DOI] [PubMed] [Google Scholar]
- Isola J, Helin H, Kallioniemi OP. Immunoelectron-microscopic localization of a proliferation-associated antigen Ki-67 in MCF-7 cells. Histochem J. 1990;22:498–506. doi: 10.1007/BF01007235. [DOI] [PubMed] [Google Scholar]
- Kametaka A, Takagi M, Hayakawa T, Haraguchi T, Hiraoka Y, Yoneda Y. Interaction of the chromatin compaction-inducing domain (LR domain) of Ki-67 antigen with HP1 proteins. Genes Cells. 2002;7:1231–1242. doi: 10.1046/j.1365-2443.2002.00596.x. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- Keller C, Krude T. Requirement of cyclin/Cdk2 and protein phosphatase 1 activity for chromatin assembly factor 1-dependent chromatin assembly during DNA synthesis. J Biol Chem. 2000;275:35512–35521. doi: 10.1074/jbc.M003073200. [DOI] [PubMed] [Google Scholar]
- Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- Luo Y, Ren F, Liu Y, Shi Z, Tan Z, Xiong H, Dang Y, Chen G. Clinicopathological and prognostic significance of high Ki-67 labeling index in hepatocellular carcinoma patients: a meta-analysis. Int J Clin Exp Med. 2015;8:10235–10247. [PMC free article] [PubMed] [Google Scholar]
- Ma N, Matsunaga S, Takata H, Ono-Maniwa R, Uchiyama S, Fukui K. Nucleolin functions in nucleolus formation and chromosome congression. J Cell Sci. 2007;120:2091–2105. doi: 10.1242/jcs.008771. [DOI] [PubMed] [Google Scholar]
- MacCallum DE, Hall PA. Biochemical characterization of pKi67 with the identification of a mitotic-specific form associated with hyperphosphorylation and altered DNA binding. Exp Cell Res. 1999;252:186–198. doi: 10.1006/excr.1999.4600. [DOI] [PubMed] [Google Scholar]
- Marheineke K, Krude T. Nucleosome assembly activity and intracellular localization of human CAF-1 changes during the cell division cycle. J Biol Chem. 1998;273:15279–15286. doi: 10.1074/jbc.273.24.15279. [DOI] [PubMed] [Google Scholar]
- Matheson TD, Kaufman PD. Grabbing the genome by the NADs. Chromosoma. 2015;125:361–371. doi: 10.1007/s00412-015-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Taniura N, Pirollet F, Monroe R, Gerace L, Westendorf JM. Identification of novel M phase phosphoproteins by expression cloning. Mol Biol Cell. 1996;7:1455–1469. doi: 10.1091/mbc.7.9.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moggs JG, Grandi P, Quivy JP, Jónsson ZO, Hübscher U, Becker PB, Almouzni G. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol. 2000;20:1206–1218. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- Németh A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Péterfia B, Solovei I, Cremer T, Dopazo J, Längst G. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padeken J, Heun P. Nucleolus and nuclear periphery: Velcro for heterochromatin. Curr Opin Cell Biol. 2014;28C:54–60. doi: 10.1016/j.ceb.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Padeken J, Mendiburo MJ, Chlamydas S, Schwarz H-J, Kremmer E, Heun P. The nucleoplasmin homolog NLP mediates centromere clustering and anchoring to the nucleolus. Mol Cell. 2013;50:236–249. doi: 10.1016/j.molcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Pezzilli R, Partelli S, Cannizzaro R, Pagano N, Crippa S, Pagnanelli M, Falconi M. Ki-67 prognostic and therapeutic decision driven marker for pancreatic neuroendocrine neoplasms (PNENs): a systematic review. Adv Med Sci. 2016;61:147–153. doi: 10.1016/j.advms.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Politz JCR, Scalzo D, Groudine M. The redundancy of the mammalian heterochromatic compartment. Curr Opin Genet Dev. 2016;37:1–8. doi: 10.1016/j.gde.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Theocharis SE, Klijanienko J, Savignoni A, Asselain B, Vielh P, Almouzni G. Chromatin assembly factor-1, a marker of clinical value to distinguish quiescent from proliferating cells. Cancer Res. 2004;64:2371–2381. doi: 10.1158/0008-5472.can-03-2893. [DOI] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Chandrasekhara C, Mozgová I, Hassel C, Pontes OMF, Tucker S, Mokroš P, Muchová V, Fajkus J, et al. Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev. 2013;27:1545–1550. doi: 10.1101/gad.221648.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Carpentier M-C, Durut N, Pavlištová V, Jaške K, Schorˇová Š, Parrinello H, Rohmer M, Pikaard CS, Fojtová M, et al. Identification of nucleolus-associated chromatin domains reveals a role for the nucleolus in 3D organization of the A. thaliana genome. Cell Rep. 2016;16:1574–1587. doi: 10.1016/j.celrep.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo J-S, Kang G, Sohn JH. Ki-67 labeling index can be used as a prognostic marker in gastrointestinal stromal tumor: a systematic review and meta-analysis. Int J Biol Markers. 2016;31:e204-10. doi: 10.5301/jbm.5000183. [DOI] [PubMed] [Google Scholar]
- Quivy J-P, Gérard A, Cook AJL, Roche D, Almouzni G. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol. 2008;15:972–979. doi: 10.1038/nsmb.1470. [DOI] [PubMed] [Google Scholar]
- Quivy J-P, Roche D, Kirschner D, Tagami H, Nakatani Y, Almouzni G. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 2004;23:3516–3526. doi: 10.1038/sj.emboj.7600362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese BE, Bachman KE, Baylin SB, Rountree MR. The methyl-CpG binding protein MBD1 interacts with the p150 subunit of chromatin assembly factor 1. Mol Cell Biol. 2003;23:3226–3236. doi: 10.1128/MCB.23.9.3226-3236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards-Taylor S, Ewings SM, Jaynes E, Tilley C, Ellis SG, Armstrong T, Pearce N, Cave J. The assessment of Ki-67 as a prognostic marker in neuroendocrine tumours: a systematic review and meta-analysis. J Clin Pathol. 2016;69:612–618. doi: 10.1136/jclinpath-2015-203340. [DOI] [PubMed] [Google Scholar]
- Rickards B, Flint SJ, Cole MD, LeRoy G. Nucleolin is required for RNA polymerase I transcription in vivo. Mol Cell Biol. 2007;27:937–948. doi: 10.1128/MCB.01584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger B, Moisand A, Amalric F, Bouvet P. Repression of RNA polymerase I transcription by nucleolin is independent of the RNA sequence that is transcribed. J Biol Chem. 2002;277:10209–10219. doi: 10.1074/jbc.M106412200. [DOI] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T, Castillo AG, Osborne MJ, Borden KLB, Kornblatt J, Verreault A. Two fundamentally distinct PCNA interaction peptides contribute to chromatin assembly factor 1 function. Mol Cell Biol. 2009;29:6353–6365. doi: 10.1128/MCB.01051-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiwaki T, Kotera I, Sasaki M, Takagi M, Yoneda Y. In vivo dynamics and kinetics of pKi-67: transition from a mobile to an immobile form at the onset of anaphase. Exp Cell Res. 2005;308:123–134. doi: 10.1016/j.yexcr.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Sheval EV, Polyakov VY. The peripheral chromosome scaffold, a novel structural component of mitotic chromosomes. Cell Biol Int. 2008;32:708–712. doi: 10.1016/j.cellbi.2008.01.290. [DOI] [PubMed] [Google Scholar]
- Smith CL, Matheson TD, Trombly DJ, Sun X, Campeau E, Han X, Yates JR 3rd, Kaufman PD. A separable domain of the p150 subunit of human chromatin assembly factor-1 promotes protein and chromosome associations with nucleoli. Mol Biol Cell. 2014;25:2866–2881. doi: 10.1091/mbc.E14-05-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- Smith S, Stillman B. Immunological characterization of chromatin assembly factor I, a human cell factor required for chromatin assembly during DNA replication in vitro. J Biol Chem. 1991;266:12041–12047. [PubMed] [Google Scholar]
- Sobecki M, Mrouj K, Camasses A, Parisis N, Nicolas E, Llères D, Gerbe F, Prieto S, Krasinska L, David A, et al. The cell proliferation antigen Ki-67 organises heterochromatin. Elife. 2016;5:e13722. doi: 10.7554/eLife.13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Hunter T. Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. J Biol Chem. 2012;287:42071–42083. doi: 10.1074/jbc.M112.410985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Matsuoka Y, Kurihara T, Yoneda Y. Chmadrin: a novel Ki-67 antigen-related perichromosomal protein possibly implicated in higher order chromatin structure. J Cell Sci. 1999;112:2463–2472. doi: 10.1242/jcs.112.15.2463. [DOI] [PubMed] [Google Scholar]
- Takagi M, Natsume T, Kanemaki MT, Imamoto N. Perichromosomal protein Ki67 supports mitotic chromosome architecture. Genes Cells. 2016;21:1113-1124. doi: 10.1111/gtc.12420. [DOI] [PubMed] [Google Scholar]
- Takagi M, Nishiyama Y, Taguchi A, Imamoto N. Ki67 antigen contributes to the timely accumulation of protein phosphatase 1γ on anaphase chromosomes. J Biol Chem. 2014;289:22877–22887. doi: 10.1074/jbc.M114.556647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami Y, Ono T, Fukagawa T, Shibahara K, Nakayama T. Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells. Mol Biol Cell. 2007;18:129–141. doi: 10.1091/mbc.E06-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut W, Endl E, Scholzen T, Gerdes J, Winking H. The temporal and spatial distribution of the proliferation associated Ki-67 protein during female and male meiosis. Chromosoma. 2002;111:156–164. doi: 10.1007/s00412-002-0202-8. [DOI] [PubMed] [Google Scholar]
- Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, Bouvet P. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol. 2007;8:66. doi: 10.1186/1471-2199-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwada J, Tanaka N, Yamaguchi Y, Uchimura Y, Shibahara K, Nakao M, Saitoh H. The p150 subunit of CAF-1 causes association of SUMO2/3 with the DNA replication foci. Biochem Biophys Res Commun. 2010;391:407–413. doi: 10.1016/j.bbrc.2009.11.071. [DOI] [PubMed] [Google Scholar]
- Van Hooser AA, Yuh P, Heald R. The perichromosomal layer. Chromosoma. 2005;114:377–388. doi: 10.1007/s00412-005-0021-9. [DOI] [PubMed] [Google Scholar]
- van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21:3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijen R, Kuijpers HJ, Schlingemann RO, Boehmer AL, van Driel R, Brakenhoff GJ, Ramaekers FC. Ki-67 detects a nuclear matrix-associated proliferation-related antigen. I. Intracellular localization during interphase. J Cell Sci. 1989a;92:123–130. doi: 10.1242/jcs.92.1.123. [DOI] [PubMed] [Google Scholar]
- Verheijen R, Kuijpers HJ, van Driel R, Beck JL, van Dierendonck JH, Brakenhoff GJ, Ramaekers FC. Ki-67 detects a nuclear matrix-associated proliferation-related antigen. II. Localization in mitotic cells and association with chromosomes. J Cell Sci. 1989b;92:531–540. doi: 10.1242/jcs.92.4.531. [DOI] [PubMed] [Google Scholar]