Optogenetic activation of the adenylate cyclase enzyme in Saccharomyces cerevisiae, paired with computational modeling, enables study of the dynamic quantitative properties of the cAMP/PKA signaling network. The ability to deliver such precise perturbation reveals fundamental dynamical features of PKA signaling, including the time scales of feedback.
Abstract
In eukaryotes, protein kinase A (PKA) is a master regulator of cell proliferation and survival. The activity of PKA is subject to elaborate control and exhibits complex time dynamics. To probe the quantitative attributes of PKA dynamics in the yeast Saccharomyces cerevisiae, we developed an optogenetic strategy that uses a photoactivatable adenylate cyclase to achieve real-time regulation of cAMP and the PKA pathway. We capitalize on the precise and rapid control afforded by this optogenetic tool, together with quantitative computational modeling, to study the properties of feedback in the PKA signaling network and dissect the nonintuitive dynamic effects that ensue from perturbing its components. Our analyses reveal that negative feedback channeled through the Ras1/2 GTPase is delayed, pinpointing its time scale and its contribution to the dynamic features of the cAMP/PKA signaling network.
INTRODUCTION
The second messenger cyclic-AMP (cAMP) is a ubiquitous signaling molecule whose synthesis by adenylate cyclase and degradation by phosphodiesterases (PDEs) occur in all branches of life. In eukaryotes, protein kinase A (PKA) is the most conserved cAMP-responsive protein. Binding of cAMP to the regulatory subunit of PKA frees its catalytic units to phosphorylate hundreds of targets regulating a vast swath of metabolism and cellular physiology. cAMP often exhibits pulsatile or oscillatory dynamics. In Dictyostelium, for example, waves of cAMP coordinate colony growth and differentiation (Tyson and Murray, 1989), and in humans, oscillating cAMP levels regulate insulin secretion in pancreatic beta cells (Holz et al., 2008).
In budding yeast, cAMP levels are regulated by extracellular glucose and a range of growth- and stress-related signals. Changes in these environmental variables alter cAMP levels, modulating activity of the PKA complex, which in turn fans out to regulate a wide range of cellular processes, estimated to involve at least one-third of the genome (Zaman et al., 2009). PKA has been shown to directly phosphorylate more than two dozen proteins, including the mitochondrial protein import machinery, P-body components, autophagy proteins, glycolysis machinery, and a large number of transcription factors (Ptacek et al., 2005). In conditions of plentiful resources, cAMP levels are high, and PKA promotes rapid fermentative growth by enhancing glycolysis and ribosomal production. In stressful conditions, a decrease in cAMP levels causes a drop in PKA activity, resulting in the inhibition of ribosomal and growth-related programs and the activation of stress-responsive factors such as the transcription factor Msn2. This low-PKA-activity state is in many cases transient, as negative feedback loops embedded in the PKA network and involving the small G-protein Ras cause PKA activity to rebound even as stressful conditions continue (Nikawa et al., 1987a).
PKA exerts much of its influence through regulation of the nuclear localization and activity of several transcription regulators, most notably the stress-responsive Msn2 and its homologue, Msn4. Decreased phosphorylation by PKA of these stress-responsive transcription factors leads to their nuclear localization (Smith et al., 1998). Observation of the PKA-regulated subcellular localization of Msn2 in yeast cells suggests that PKA activity is highly dynamic, with rapid pulses of activity occurring in “bursts” of Msn2 localization on the minute time scale (Cai et al., 2008).
Although substantial progress has been made in probing the downstream consequences of dynamic Msn2 pulsing (Hao and O’Shea, 2011; Hansen and O’Shea, 2013, 2015), our understanding of how PKA generates these dynamics is incomplete, partly because upstream tools to perturb the PKA system in vivo are typically slow (e.g., mutations) or nonspecific (e.g., stress). A tool that provides a rapid, specific, and reversible perturbation of PKA signaling is therefore needed. In this work, we develop the recently discovered bacterial photoactivatable adenylate cyclase (bPAC; Iseki et al., 2002; Stierl et al., 2011) as a quantitative perturbation tool to investigate PKA dynamics in Saccharomyces cerevisiae. We demonstrate that by expressing this bacterial protein in yeast cells, we can achieve high-resolution temporal control of PKA activity. Using precise optogenetic stimuli, we develop and test a model of PKA signaling and uncover important aspects of its dynamics.
RESULTS
Rapid in vivo regulation of PKA activity by a bPAC
Previous reports demonstrated that PKA exhibits complex dynamics that occurs on fast time scales, on the order of minutes (Görner et al., 1998; Garmendia-Torres et al., 2007; Cai et al., 2008). Therefore, quantitative studies of this system require perturbations with a time resolution of seconds. To achieve such rapid perturbation, we expressed in budding yeast cells a recently characterized bPAC, originally from the soil bacterium Beggiatoa (Ryu et al., 2010; Stierl et al., 2011), that contains a light-sensitive BLUF (blue light receptor using FAD) domain linked to an adenylate cyclase domain. This construct allows for regulation of cAMP levels in living cells by illumination with blue light (<500 nm), which transiently alters the conformation of the light-sensitive BLUF domain, rendering the associated cyclase domain competent to catalyze the conversion of ATP to cAMP (Figure 1A). To take advantage of this genetically encoded tool, we assembled a custom-built system that allows us to coordinate the imaging of budding yeast in a fluorescence microscope with illumination by a blue-light LED. This system is capable of providing graded illumination for 1–40 μW/mm2. Although substantially brighter than ambient light (∼4 μW/mm2), these blue-light intensities do not trigger a stress response in budding yeast (Supplemental Figure S1). Furthermore, bPAC has a fast spontaneous reversion to the inactive state (<30 s; Supplemental Figure S2; Stierl et al. 2011), enabling of bPAC activity at high time resolution regulation through control of light intensity. Using this infrastructure, we were able to apply controlled amounts of blue light to cells expressing the bPAC construct and quantify the resulting PKA activity in real time using a localization-based fluorescent Msn2 reporter (Supplemental Figure S2). Msn2 nuclear localization is modulated by PKA via phosphorylation of PKA motifs (RRxS) in the Msn2 nuclear import (nuclear localization sequence [NLS]) and export sequences (Görner et al., 1998), making the nuclear enrichment of Msn2 an accurate measure of PKA activity (Hao and O’Shea, 2011).
FIGURE 1:
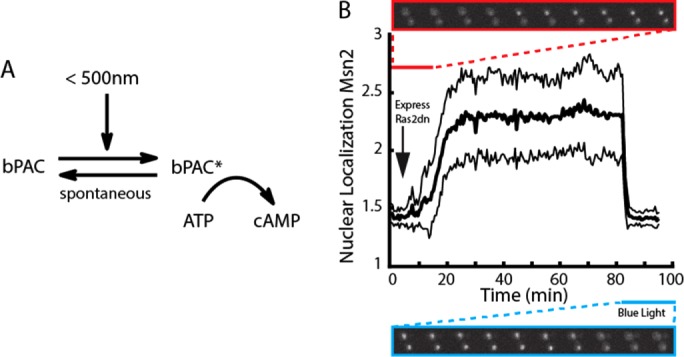
Expression of a bPAC in budding yeast allows for real-time light-gated control of PKA signaling. (A) The bPAC protein switches from an inactive to an active conformation in response to light. (B) Nuclear localization of Msn2 after inhibition of the PKA pathway by expression of a dominant-negative Ras2 allele (S24N; red inset) and subsequent blue light activation of bPAC expressed in the same strain (blue inset). Activation of the dominant-negative allele results in rapid nuclear localization of Msn2 into the nucleus, and this is synchronously reversed by bPAC activation. Nuclear localization is defined as the ratio of nuclear to cytoplasmic Msn2 (see also Materials and Methods). Time trace (thick black line) shows average of 65 cells with SDs (thin black lines).
To test the ability of bPAC to regulate PKA in vivo, we built a yeast strain harboring both a hormone-inducible dominant-negative allele of Ras2S24N (Zaman et al., 2009; Stewart-Ornstein et al., 2013) and a constitutively expressed bPAC. We used the translocation of mCherry-tagged Msn2 in and out of the nucleus as a measure of PKA activity (Hao and O’Shea, 2011; quantification of nuclear localization is discussed in Materials and Methods). Expression of Ras2S24N inhibits cAMP production by sequestering the Ras guanine exchange factor CDC25, preventing the formation of Ras-GTP and hence activation of the endogenous adenylate cyclase. As a result, when this allele is induced, we expect cAMP levels to drop, PKA to become inactive, and Msn2-mCherry to be localized to the nucleus. Consistent with this expectation, we observed complete localization of Msn2 to the nuclei of cells within 10 min of the induction of Ras2S24N expression (Figure 1B and Supplemental Movie S1). Activation of bPAC should provide an orthogonal non–Ras-dependent source of cAMP, which would activate PKA and cause Msn2-mCherry to be exported from the nucleus. Indeed, upon illumination with blue light, there was a rapid and synchronous exit of Msn2 from the nucleus. These data are consistent with a sharp increase in PKA activity upon illumination induced by cAMP production from the activated bPAC and show that bPAC can produce sufficient cAMP to compensate for the loss of Ras-dependent endogenous adenylate cyclase activity.
The dynamics of PKA signaling induced by optogenetic stimulation is explained by a computational model
Using the precise perturbation afforded by the bPAC construct, we next explored how PKA dynamics is shaped by its signaling network. To do so, we exposed wild-type cells expressing bPAC to a 3-min pulse of light (Figure 2A). As discussed earlier, in resting cells, Msn2 localization is predominantly cytoplasmic due to the PKA phosphorylation of the NLS, with occasional stochastic nuclear localization in individual cells. Consequently, blue light–mediated activation of bPAC and the subsequent increase in cAMP had only the slight and expected effect of inducing uniform Msn2 cytoplasmic localization. Of interest, however, soon after the blue light was turned off, we observed a sharp transient (lasting ∼5 min) increase in Msn2 nuclear localization above the initial localization state (Figure 2B and Supplemental Movie S2). This pulse of Msn2 nuclear localization after bPAC shutoff is surprising since we expected Msn2 to return monotonically to its prestimulus steady state. The observed behavior is a hallmark of an underdamped system, whose signature is a transient overshoot (transient PKA inactivation) before return to steady state (fully active PKA) upon change in input, as compared with an overdamped system, which exhibits monotonic return to this steady state. Such unexpected behavior can be generated by a negative feedback loop with a delay, a hypothesis that we wanted to further pursue.
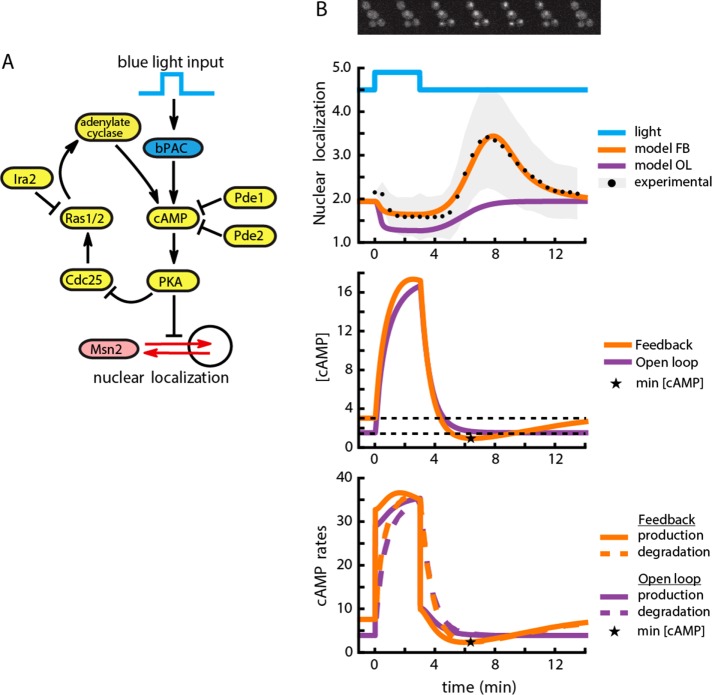
FIGURE 2:
A computational model of the cAMP-PKA circuit explains the response to bPAC stimulation. (A) Diagram of the components involved in the model and their interactions. Included are production of cAMP through bPAC stimulation and endogenous cyclase activity, Msn2 nuclear localization, autoregulatory negative feedback on PKA through Cdc25 and Ras2, and the effect of Pde1 and Pde2 on cAMP degradation. (B) Top, nuclear localization of Msn2 as a function of time (black dots are the mean from a population of 379 cells; shaded gray error bars indicate SD) after a single pulse of blue light (40 μW/mm2). The orange trace is the output of the computational model containing a negative feedback loop for a single representative parameter set. The purple trace is a representative output of a model not containing the feedback. Middle, concentration of cAMP as a function of time for the model with (orange) or without negative feedback (purple). The negative feedback model predicts a cAMP undershoot (minimum value denoted by a star), whereas the open loop model (purple) monotonically approaches the steady-state value (black dotted). The model undershoot is more pronounced than the experimentally observed undershoot because of the detection limit of the experiment. Bottom, cAMP production and degradation rates as a function of time. The undershoot in cAMP concentration is generated by a delayed production of cAMP. The cAMP minimum (star) is reached when the rate of cAMP production is balanced by the rate of degradation.
Feedback loops are believed to decorate the architecture of the PKA signaling network. To investigate the possibility that feedback is responsible for the underdamped behavior of the PKA system after bPAC-mediated rapid alteration of cAMP levels, we built two mathematical models of the PKA network. The first is a negative feedback model consisting of PKA, its core regulatory and activation components (Cdc25, Ras, and cAMP), the exogenous optogenetic stimulation, and Msn2 nuclear shuttling kinetics. The negative feedback in this system has been identified genetically and is generally considered to be implemented through regulation of the Ras1/2 GTPase proteins (Colombo et al., 2004; Jian et al., 2010). The second model is an identical open loop variant that lacks the negative feedback component, as the dependence of Cdc25 levels on PKA activity is removed (Supplemental Note S1). With this open-loop model, we wanted to test whether the Msn2 pulse of activity could be simply generated by the interaction of Msn2 with PKA without the need to invoke feedback or whether it is necessarily generated by negative feedback in PKA signaling.
In contrast to prior work (Garmendia-Torres et al., 2007; Cazzaniga et al., 2008), we did not seek to build a model that reflects all known biochemical interactions surrounding PKA signaling. Instead, by capturing only the known essential interactions in the PKA system, we aimed to build a simple model that could recapitulate the pulse of Msn2 translocation after a bPAC pulse. Because the parameters for the interactions that are captured in the model have not been measured independently, we simulated both open- and closed-loop models for >107 log-uniformly sampled parameter sets. We then selected the 5.5 × 104 parameter sets that minimized the mean square error between the model output and the wild-type experimental data of Figure 2B and optimized these sets by the Nelder–Mead algorithm to improve the fit (Nelder and Mead, 1965). We found 1.4 × 104 parameter sets for the negative feedback model that generated a good fit to the data (Figure 2B and Supplemental Data Set S1). At the same time, we could not identify any parameters for the open-loop model that could recapitulate qualitatively or quantitatively the transient pulse of Msn2 (Supplemental Figure S3 and Supplemental Data Set S2). Instead, for all parameters sampled, the open-loop model produced an overdamped Msn2 nuclear residence and depletion profile that decreased upon bPAC activation and monotonically increased to prestimulus steady state upon bPAC shutoff.
Examining the parameter sets for the feedback model that could recapitulate the Msn2 pulse data revealed a common feature they all shared: a feedback-induced delay for the activation of the endogenous adenylate cyclase after bPAC shutoff.
In simple terms, bPAC activation injects a large concentration of cAMP into the system with two consequences. First, the endogenous cAMP production is repressed by the PKA feedback on Ras1/2 activity. In addition, as cAMP concentration increases, the PDE-mediated degradation of cAMP also increases due to mass action effects. When bPAC is shut off, this cAMP degradation initially continues at a high rate. This degradation is not immediately counteracted by the endogenous production of cAMP, as Ras1/2 activity is delayed by the engaged PKA feedback. As a result of such imbalance between degradation and production dynamics, cAMP levels transiently drop below bPAC preinduction levels (Figure 2B). The corresponding drop in PKA activity consequently generates a pulse of Msn2 nuclear translocation. Msn2 localization reaches its cytoplasmic steady state when the degradation and production of cAMP equilibrate. Supporting the idea that the pulse of Msn2 nuclear localization is a result of feedback-induced delay, we observed in the model that the pulse occurs precisely when one of the components in the feedback loop traversing Ras1/2 to PKA is rate limiting (Supplemental Figure S4). Moreover, the lack of a cAMP undershoot after bPAC shutoff for the open-loop model is in agreement with the idea that the feedback-induced delay of cAMP production produces the cAMP undershoot and subsequent Msn2 pulse. Taken together, our data suggest that the observed pulse after bPAC shutoff is likely to be a structural feature of the negative feedback surrounding PKA.
Quantitative features of Msn2 nuclear pulse after bPAC shutoff depend on the components of the PKA signaling network
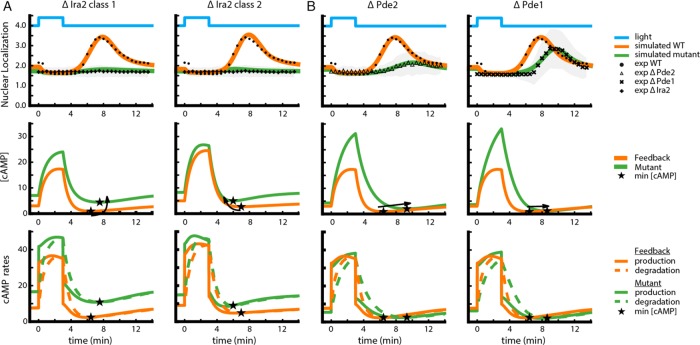
An implication of feedback in generating the transient Msn2 pulse after bPAC shutoff is that the attributes of this pulse, such as its peak height and time to peak, can be predictably modified by perturbations of different components that impinge on this feedback. To explore this idea, we built bPAC strains in which one of the following three negative regulators of PKA was deleted: Ira2, a GTPase-activating protein (GAP) that inhibits Ras1/2 and hence decreases PKA activity; and low- and high-affinity PDEs Pde1 and Pde2, respectively, which also decrease PKA activity by degrading cAMP. We then delivered a pulse of blue light input to these strains and measured the time dynamics of Msn2 nuclear localization. Deletion of Ira2, Pde1, and Pde2 generated qualitative and quantitative differences in the system’s response to the bPAC input. Compared to WT, Δira2 showed no transient Msn2 pulse upon bPAC shutoff, whereas both Δpde1 and Δpde2 displayed a delayed and attenuated Msn2 pulse (Figure 3 and Supplemental Figure S5).
FIGURE 3:
Computational modeling is used to dissect the behavior of the Δira2, Δpde1, and Δpde2 mutants. (A) Model predicted nuclear localization of Msn2 (top), concentration of cAMP (middle), and cAMP production/degradation rates (bottom) as a function of time for Δira2. The plots are shown for two representative parameter sets, one of each of the class 1 and class 2 parameter regimes that could explain this mutant. Top, experimental nuclear localization in response to 40-μW/mm2 blue light input; symbols with error bars in gray. Arrows indicate the change in minimum cAMP concentration (star) due to the deletion. (B) Same plots as in A, for the Δpde1 and Δpde2 mutants. Parameter sets used to generate these plots are sets 7 and 30 of Supplemental Dataset S3.
To capitalize on the model as a guide for our intuition in understanding these differences, we used the parameter sets of the feedback model that fit the wild-type (WT) data as seeds to perform a second stage of fitting to experimental data from all four strains (WT and three mutants; Supplemental Note S1). Of the original data set, only ∼300 parameter sets from the original ensemble produced a good fit to all experimental data (Supplemental Data Set S3), suggesting that the mutant data effectively constrained the model parameters. For any one mutant, simulated trajectories of nonfitted variables (e.g., active Ras) were quantitatively different based on the particular parameter set used. However, these trajectories had common qualitative features that provided plausible explanations for the particular Msn2 phenotype observed in the various mutants.
For Δira2, two nonexclusive parameter categories were at the root of the suppression of the Msn2 translocation pulse (Figure 3A and Supplemental Figure S6). In the first category, accumulation of Ras in the model due to the absence of the negative regulator IRA2 caused a large increase in cAMP concentration both at steady state and during bPAC application (Figure 3A, left). After bPAC shutoff, the degradation of cAMP still proceeds at a fast rate and may cause cAMP levels to transiently drop below their (elevated) steady-state value. However, for the particular parameter sets corresponding to this mutant, even the lowest cAMP level achieved is still sufficient to keep PKA activity above the threshold that needs to be traversed in order for Msn2 to translocate to the nucleus (Supplemental Figure S7). In the second category of parameters also reproducing the Δira2 data, the lack of an Msn2 pulse resulted from altered feedback. Here the pulse of Msn2 nuclear translocation was abrogated by the fact that, due to higher Ras activity in this mutant, cAMP degradation was more quickly balanced by endogenous cAMP production after bPAC shutoff (Figure 3A, right).
The model also clarified the phenotypes of Δpde1 and Δpde2, which both show a delayed and attenuated Msn2 nuclear translocation pulse. In both mutants, degradation of cAMP is reduced, resulting in rapid accumulation of cAMP to levels exceeding those of the wild type upon bPAC stimulation. After bPAC shutoff, impaired degradation of this excess cAMP keeps PKA activity high for an extended period of time, therefore maintaining Msn2 cytoplasmic localization during that time and explaining the delay. Levels of cAMP eventually decline below a level capable of keeping PKA active, and a pulse of Msn2 nuclear translocation ensues. The presence of the pulse is still a result of the feedback-induced delay in endogenous cAMP production, but its attenuated magnitude is a manifestation of the combined effect of a slower cAMP degradation (slower rise time for the pulse) and a greater ability of cAMP production to balance this degradation (Figure 3B). The different quantitative phenotypes of Δpde1 versus Δpde2 are ascribed by the model to different affinity of these enzymes to cAMP, resulting in a differential effect of their deletion on cAMP degradation rate. This differential binding of Pde1 and Pde2 to cAMP is well documented experimentally (Sass et al., 1986; Nikawa et al., 1987b). In addition, the model indicates that at steady state, cAMP levels for these mutants is similar to that of the wild type, a nonintuitive result of the feedback ensuring that decreased cAMP degradation is homeostatically counteracted by decreased Ras.
The observation that the dynamics of the Msn2 nuclear pulse is profoundly affected by perturbations of the PKA network, supported by our modeling results, lends further support to the notion that this pulse is predominantly generated by the PKA signaling network itself.
bPAC enables frequency- and amplitude-modulated control of the PKA signaling network
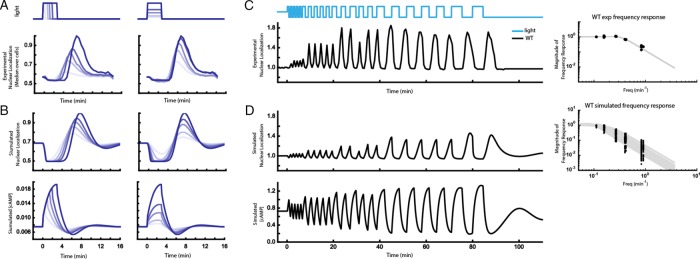
To further explore the feedback model with the constrained parameters, we subjected it to light pulses of increasing amplitudes and durations, inputs for which the model was not fit (Figure 4A). With increasing light duration at a fixed intensity, the model predicted a corresponding graded increase in peak nuclear localization of Msn2 (Figure 4B). A similar conclusion holds for increasing light intensity for a constant duration (Figures 4B and Supplemental Figure S8). This is because in this model, the cAMP response is dependent on the integral of the total light pulse within this intensity/duration regime (Figure 4B, bottom). Hence both light stimulus amplitude and duration can achieve a continuous range of values for the peak of the Msn2 pulse. Experiments performed with bPAC pulses of different intensities and durations agree with this result (Figure 4A), supporting the basic model structure.
FIGURE 4:
High-precision control of cAMP using bPAC allows for systematic characterization of the PKA system using pulse width–, amplitude-, or frequency-modulated input signals. (A) Peak nuclear localization of Msn2-mCherry increases with blue light duration (left) and amplitude (right) in cells expressing bPAC. Experimental WT data collected at 30-s intervals for light durations of 0.5, 1, 1.5, 2, and 3 min with amplitude fixed at 40 μW/mm2 (left) and light amplitudes of 8, 10, 16, 20, and 40 μW/mm2 with duration fixed at 3 min (right). (See also Supplemental Figure S8.) Plots show median values over ∼900 cells normalized to the maximum observed median value for comparison of responses across different light doses. (B) Top, the model predicts that nuclear localization shows a trend similar to the experimental data of A when light amplitude or duration increases (parameter set 0 from Supplemental Data Set S3; see also Supplemental Figure S7). Data normalized by maximum value. Bottom row, maximum simulated cAMP concentration increases with both light duration and amplitude. (C) Left, mean Msn2-mCherry nuclear localization after blue light illumination at different pulse frequencies. Right, peak mean Msn2 nuclear localization at each frequency (black dots) with fit to second-order transfer function (gray lines). (D) Left, model prediction of the frequency sweep experiment in C for a representative set of parameters (parameter set 17 in Supplemental Data Set S3). Right, peak Msn2 nuclear localization at each frequency for representative parameter sets (black dots), with fits to second-order transfer function (gray lines).
Finally, we asked whether the model can recapitulate the filtering properties of the PKA system, which can be determined by assessing the system’s output to different bPAC input pulse trains of different frequencies (Toettcher et al., 2013). Such a frequency response analysis provides a quantitative picture of the dominant time scales of PKA signaling. We therefore used the optogenetic bPAC input to apply light pulses at six distinct frequencies spanning 0.83 to 0.11 min−1, with five repeats of each frequency (except for 0.11 min−1, which only had two repeats). Experimental data revealed a low-pass filter with a cutoff frequency of 0.33 min−1 and quality factor of 0.88 (Figure 4C and Supplemental Movie S3), suggesting that the response time scale of the PKA system is <1/0.33 ≈ 3 min and that inputs with substantially higher frequency content are significantly attenuated.
We then stimulated the computational model with a bPAC input consisting of a train of pulses with decreasing frequencies and compared these simulations to the experimental results (Figure 4D, top). We repeated this exercise for all of the parameter sets that recapitulated the data in previous sections. The maximum magnitude of Msn2 nuclear localization varied among parameter sets. However, all parameter sets consistently predicted that the system has the characteristics of a second-order low-pass filter with a median cutoff frequency of 0.14 min−1 with interquartile range of 0.12–0.15 min−1 and a quality factor of 0.86 with interquartile range of 0.77–0.93 (see Supplemental Note S1 for the conventions used in the transfer function and Supplemental Figure S9 for the phase response). The cutoff frequency arises because at high pulsing frequencies, shutoff of bPAC is now closely followed by another bPAC activation pulse, such that cAMP concentration now cannot fall below the preinduction level before the next light pulse is applied (Figure 4D, bottom).
The quantitative difference between the computational and experimental cutoff frequency is not surprising. Ultimately, our model was built to be the most parsimonious representation that captures the phenomenology and overall characteristics of the system and trained on a limited set of data. Specifically, since the model was trained on 3-min light pulses, it cannot predict accurately the rates of cAMP production/degradation necessary to fit shorter light pulses and hence cannot quantitatively fit data obtained at high input frequencies. Therefore, while this model reveals the main dynamic characteristics of the PKA signaling network, featuring delayed feedback, more detailed representations and experimentation are needed to accurately capture the true quantitative complexity of the PKA system.
DISCUSSION
Growth-regulatory signaling pathways such as the PKA pathway link perception of the environment with cell proliferation. To reliably relay the environmental state, these pathways often show rapid dynamics. Rapid dynamics of PKA was previously documented in response to environmental perturbations (Garmendia-Torres et al., 2007; Hao and O’Shea, 2011). Fast and minimally pleiotropic perturbation tools are therefore required to explore these dynamics and evaluate their functional roles. Optogenetic perturbations are increasingly identified as powerful tools to carry out these studies. Although optogenetic technology has seen widespread use in neuroscience, these tools were only recently introduced in the study of a wider breadth of molecular biology. In this work, we capitalize on a naturally occurring bPAC to study PKA signaling in budding yeast. We show that when coupled with real-time reporters and computer-controlled illumination, bPAC constitutes a powerful general tool for administering precise and specific perturbations to this system to probe its quantitative properties. Using this tool, we were able to study the system-level characteristics of the PKA signaling pathway.
Specifically, our studies using bPAC indicated that strong negative feedback channeled through Ras1/2 in the PKA signaling network in budding yeast causes a delay in Ras1/2-mediated endogenous production of cAMP, and we pinpointed its relevant timescale. Using these data, we were able to build a quantitative computational model that generated a rigorous predictive understanding of the role of feedback in generating PKA activity dynamics, which may be important in regulating downstream signaling and gene expression.
Using a combination of precise optogenetic perturbation and quantitative modeling, this study enabled identification of dynamic properties of the PKA signaling network that would otherwise be difficult to dissect using slower and more pleiotropic methods such as knockouts and overexpression. More generally, our analyses revealed two salient principles. First, the presence of feedback in a system generates nonintuitive dynamic effects upon perturbation of components both within (such as Ira2) and outside the feedback loop (such as Pde1/2). Second, different regulatory components with the same overall qualitative description (e.g., both Pde1/2 and Ira2 negatively affect PKA activity) can generate vastly different qualitative and quantitative phenotypes.
Overall it is tempting to hypothesize that these regulators could be used as gateways for different environmental inputs into this system. In this scheme, different inputs can affect and perturb different regulators. Because perturbation of such regulators induces distinct dynamical phenotypes, this scheme could be the basis for encoding the identity of inputs into different dynamical patterns of the pathway. Dynamic encoding of inputs has been proposed as a strategy for implementing response specificity in a signaling pathway that propagates multiple environmental inputs (Hao and O’Shea, 2011). Although current research has focused on identifying specific molecular implementations of such a strategy, in the future, it will be interesting to investigate the dynamically malleable platform of feedback as a possible basis for generating dynamic encoding in signaling pathways.
MATERIALS AND METHODS
Yeast strains and constructs
All yeast strains used for these experiments are derived from W303A-1 in which the ade2 marker was reverted to ADE2+ to reduce the autofluorescence. Msn2-mCherry was integrated at the trp1 locus, and the bPAC construct was integrated into the leu2 locus. Overexpression construct for RAS2(S24N) was integrated into the TRP1 locus of a LEU2+ MATalpha strain that contained an estradiol-inducible construct. All strains were constructed using standard yeast protocols and LioAc/PEG transformation.
The RAS2(S24N) expression vector was constructed by amplification of the RAS2 gene from yeast genomic DNA and cloned downstream of a prGAL1 promoter in a TRP1-marked single integration plasmid. Site-directed mutagenesis was performed to create the dominant-negative allele of RAS2(S24N) using a standard QuikChange protocol and the pfuTurbo enzyme mix (Stratagene, La Jolla, CA). The bPAC gene was synthesized with yeast-optimized codons by Integrated DNA Technologies (Coralville, IA) and cloned downstream of a prNOP7 promoter in the LEU2-marked single integration vector.
Microscopy, image acquisition, and analysis
Cells expressing Msn2-mCherry or related constructs were plated in SD complete medium onto concanavalin A–coated 96-well glass-bottom plates, allowed to settle, and then washed twice with fresh medium and 100 μl of fresh medium added.
Samples were imaged on a Nikon Ti inverted scope with arc-lamp illumination using red fluorescent protein (560/40 nm excitation, 630/75 nm emission; Chroma, Bellows Falls, VT) and yellow fluorescent protein (510/10 nm excitation, 542/27 nm emission; Semrock, Rochester, NY) filters. Blue light illumination was provided by a 465-nm LED driven by a USB-controlled power source (MIGHTex, Pleasanton, CA) mounted on the bright-field condenser. Imaging and illumination were controlled and coordinated by custom Matlab (MathWorks, Natick, MA) software interfaced with the μmanager software suite (Edelstein et al., 2010).
Images were processed and analyzed with ImageJ and custom-built Matlab scripts. Nuclear localization was computed by dividing the average intensity of the brightest 10% of pixels in the cell by the median intensity of the cell.
Deterministic model
An ordinary differential equation (ODE) model of the PKA regulatory network, consisting of mostly Michaelis–Menten interactions, was constructed with five state variables and 24 parameters. Latin hypercube sampling and Nelder–Mead optimization of parameters (Nelder and Mead, 1965) were done to obtain model fits. Additional details of the computational methods are described in Supplemental Note S1.
Supplementary Material
Acknowledgments
We thank Charles Biddle-Snead, David Pincus, Ben Heineike, Michael Chevalier, and the Weissman and El-Samad labs for scientific discussion and comments on the manuscript. This work was funded by the University of California, San Francisco, Center for Systems and Synthetic Biology (National Institute of General Medical Sciences P50 GM081879) and the Paul G. Allen Family Foundation (H.E.S.), the Howard Hughes Medical Institute (J.S.W.), a Natural Sciences and Engineering Research Council of Canada postgraduate scholarship (J.S.-O.), and a National Science Foundation graduate research fellowship (S.C.).
Abbreviations used:
- bPAC
bacterial photoactivated adenylyl cyclase
- BLUF
blue-light using FAD
- cAMP
cyclic-AMP
- LED
light-emitting diode
- ODE
ordinary differential equation
- PKA
protein kinase A.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-06-0354) on November 9, 2016.
REFERENCES
- Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:485–490. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzaniga P, Pescini D, Besozzi D, Mauri G, Colombo S, Martegani E. Modeling and stochastic simulation of the Ras/cAMP/PKA pathway in the yeast Saccharomyces cerevisiae evidences a key regulatory function for intracellular guanine nucleotides pools. J Biotechnol. 2008;133:377–385. doi: 10.1016/j.jbiotec.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Colombo S, Ronchetti D, Thevelein JM, Winderickx J, Martegani E. Activation state of the Ras2 protein and glucose-induced signaling in Saccharomyces cerevisiae. J Biol Chem. 2004;279:46715–46722. doi: 10.1074/jbc.M405136200. [DOI] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μManager. Curr Protoc Mol Biol. 2010:14.20.1–14.20.17. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmendia-Torres C, Goldbeter A, Jacquet M. Nucleocytoplasmic oscillations of the yeast transcription factor Msn2: evidence for periodic PKA activation. Curr Biol. 2007;17:1044–1049. doi: 10.1016/j.cub.2007.05.032. [DOI] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schüller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, O’Shea EK. Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Mol Syst Biol. 2013;9:704. doi: 10.1038/msb.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, O’Shea EK. Limits on information transduction through amplitude and frequency regulation of transcription factor activity. Elife. 2015;4:e06559. doi: 10.7554/eLife.06559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, O’Shea EK. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat Struct Mol Biol. 2011;19:31–39. doi: 10.1038/nsmb.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz GG, Heart E, Leech CA. Synchronizing Ca2+and cAMP oscillations in pancreatic β-cells: a role for glucose metabolism and GLP-1 receptors. Am J Physiol Cell Physiol. 2008;294:C4–C6. doi: 10.1152/ajpcell.00522.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseki M, Matsunaga S, Murakami A, Ohno K, Shiga K, Yoshida K, Sugai M, Takahashi T, Hori T, Watanabe M. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature. 2002;415:1047–1051. doi: 10.1038/4151047a. [DOI] [PubMed] [Google Scholar]
- Jian D, Aili Z, Xiaojia B, Huansheng Z, Yun H. Feedback regulation of Ras2 guanine nucleotide exchange factor (Ras2-GEF) activity of Cdc25p by Cdc25p phosphorylation in the yeast Saccharomyces cerevisiae. FEBS Lett. 2010;584:4745–4750. doi: 10.1016/j.febslet.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Nelder JA, Mead R. A simplex method for function minimization. Comput J. 1965;7:308–313. [Google Scholar]
- Nikawa J, Cameron S, Toda T, Ferguson KM, Wigler M. Rigorous feedback control of cAMP levels in Saccharomyces cerevisiae. Genes Dev. 1987a;1:931–937. doi: 10.1101/gad.1.9.931. [correction published in Genes Dev (1987), 1, 1351] [DOI] [PubMed] [Google Scholar]
- Nikawa J, Sass P, Wigler M. Cloning and characterization of the low-affinity cyclic AMP phosphodiesterase gene of Saccharomyces cerevisiae. Mol Cell Biol. 1987b;7:3629. doi: 10.1128/mcb.7.10.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Ryu MH, Moskvin OV, Siltberg-Liberles J, Gomelsky M. Natural and engineered photoactivated nucleotidyl cyclases for optogenetic applications. J Biol Chem. 2010;285:41501–41508. doi: 10.1074/jbc.M110.177600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass P, Field J, Nikawa J, Toda T, Wigler M. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1986;83:9303–9307. doi: 10.1073/pnas.83.24.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Ward MP, Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Ornstein J, Nelson C, DeRisi J, Weissman JS, El-Samad H. Msn2 coordinates a stoichiometric gene expression program. Curr Biol. 2013;23:233623–233645. doi: 10.1016/j.cub.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierl M, Stumpf P, Udwari D, Gueta R, Hagedorn R, Losi A, Gärtner W, Petereit L, Efetova M, Schwarzel M, et al. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J Biol Chem. 2011;286:1181–1188. doi: 10.1074/jbc.M110.185496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the ras/erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JJ, Murray JD. Cyclic AMP waves during aggregation of Dictyostelium amoebae. Development. 1989;106:421–426. doi: 10.1242/dev.106.3.421. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Schneper L, Slonim N, Broach JR. Glucose regulates transcription in yeast through a network of signaling pathways. Mol Syst Biol. 2009;5:245. doi: 10.1038/msb.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.