Condensin II interacts with human CENP-A chaperone HJURP and is present at centromeres in early G1. Condensin II, but not condensin I, is required for efficient CENP-A deposition in human cells. HJURP-induced chromatin decondensation at de novo centromeres is counteracted by the activity of condensin II.
Abstract
Centromeric chromatin is required for kinetochore assembly during mitosis and accurate chromosome segregation. A unique nucleosome containing the histone H3–specific variant CENP-A is the defining feature of centromeric chromatin. In humans, CENP-A nucleosome deposition occurs in early G1 just after mitotic exit at the time when the CENP-A deposition machinery localizes to centromeres. The mechanism by which CENP-A is deposited onto an existing, condensed chromatin template is not understood. Here we identify the selective association of the CENP-A chaperone HJURP with the condensin II complex and not condensin I. We show CAPH2 is present at centromeres during early G1 at the time when CENP-A deposition is occurring. CAPH2 localization to early G1 centromeres is dependent on HJURP. The CENP-A chaperone and assembly factor HJURP induces decondensation of a noncentromeric LacO array, and this decondensation is modulated by the condensin II complex. We show that condensin II function at the centromere is required for new CENP-A deposition in human cells. These data demonstrate that HJURP selectively recruits the condensin II chromatin-remodeling complex to facilitate CENP-A deposition in human cells.
INTRODUCTION
Centromere protein A (CENP-A) is a specialized histone H3 variant that is specifically present in nucleosomes at centromeric chromatin and is believed to epigenetically define centromeric chromatin. New CENP-A deposition into centromeric chromatin is uncoupled from DNA replication in humans and most metazoans. New CENP-A is loaded at the centromere by its chaperone Holliday junction recognition protein (HJURP) in early G1 just after the cell exits mitosis (Jansen et al., 2007; Dunleavy et al., 2009; Foltz et al., 2009). Replication-independent CENP-A deposition presents the additional challenge of assembling new CENP-A nucleosomes into a chromatin template that is already assembled with existing nucleosomes and presumably condensed chromatin. H3.3 nucleosomes may be selectively removed in this process to make room for new CENP-A at the centromeres (Dunleavy et al., 2011).
HJURP recognizes the CENP-A histone H3 variant through a set of specific amino acids within the centromere-targeting domain of CENP-A (Black et al., 2007; Foltz et al., 2009; Bassett et al., 2012). HJURP binds CENP-A via HJURP’s N-terminal Scm3 domain, which is conserved from yeast to vertebrates (Sanchez-Pulido et al., 2009; Shuaib et al., 2010; Cho and Harrison, 2011; Feng et al., 2011; Hu et al., 2011; Zhou et al., 2011). The recruitment of HJURP to chromatin is sufficient to determine the site of new CENP-A nucleosome assembly in human cells (Barnhart et al., 2011). At the centromere, the selective recruitment of HJURP depends on the missegregation protein 18 (Mis18) complex (Barnhart et al., 2011; Bernad et al., 2011). Mis18 binding to the HJURP centromere-targeting domain is believed to directly couple new CENP-A deposition to the existing CENP-A at the centromere (Zasadzinska et al., 2013; Wang et al., 2014; Nardi et al., 2016). In addition to binding HJURP, the Mis18 complex may also influence histone acetylation at centromeric repeats (Hayashi et al., 2004; Fujita et al., 2007), thus priming centromeric chromatin in a way that makes it compliant for HJURP recruitment and CENP-A deposition. Mis18α has also been shown to interact with DNMT3A and DNMT3B and to enhance DNA methylation within centromeric chromatin and CENP-A assembly (Gopalakrishnan et al., 2009; Kim et al., 2012). Thus the Mis18 complex may function to prepare centromeric chromatin for accessibility by the CENP-A deposition machinery.
The condensin complexes have been extensively characterized for their fundamental roles in inducing chromatin condensation during mitosis, but they are also involved in a variety of nuclear functions in the interphase nucleus (Hirano, 2012a; Piazza et al., 2013). Two condensin complexes are present in metazoans. The condensin I and II complexes share the large structural maintenance of chromosomes (SMC) subunits SMC2 and SMC4, which are chromosomal ATPases (Hirano and Hirano, 2006). Condensin I and II each contain three complex-specific subunits—CAPD2, CAPG, and CAPH; and CAPD3, CAPG2, CAPH2, respectively (Figure 1A; Hirano, 2012b). Together, the condensin complex proteins form a ring-like structure (Schleiffer et al., 2003). Condensin II is largely responsible for the axial shortening of mitotic chromosomes, and it works alongside condensin I, which is responsible for lateral compaction of mitotic chromosomes (Ono et al., 2003). Condensin II is localized in the nucleus throughout the cell cycle, whereas condensin I gains access only upon nuclear envelope breakdown in prometaphase. Condensin I gains access to the nucleus after nuclear envelope breakdown and is loaded onto mitotic chromosomes after phosphorylation events by CDK1, Aurora B, and Polo-like kinases (Kimura et al., 1998; Giet and Glover, 2001; St-Pierre et al., 2009). Condensin II also undergoes multiple phosphorylation events by mitotic kinases Mps1, CDK1, and Plk1 that contribute to its activity (Abe et al., 2011; Kagami et al., 2014). The interaction of condensin I with mitotic chromosomes is more dynamic than that of condensin II, suggesting that these two complexes interact with chromatin differently (Gerlich et al., 2006).
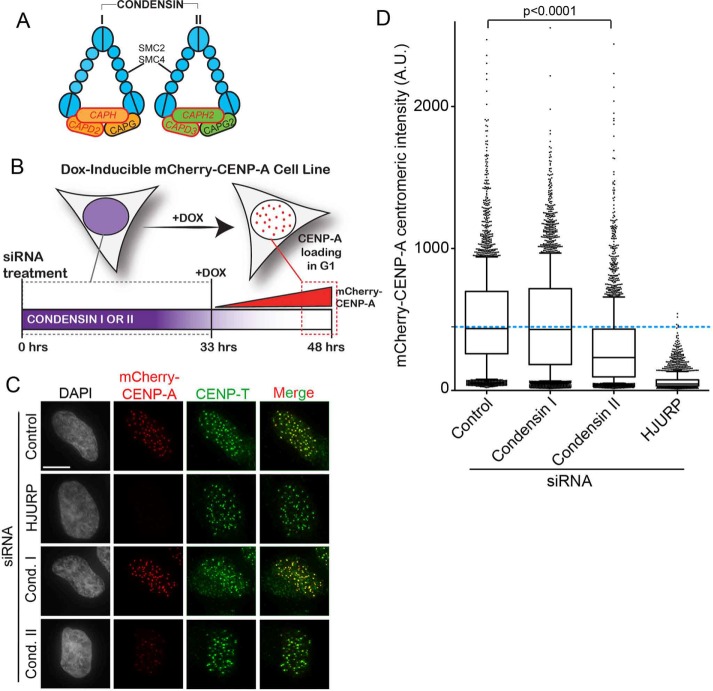
FIGURE 1:
Condensin II depletion results in CENP-A deposition defect in human cells. (A) Schematics of human condensin I and II protein complexes. (B) Schematic of inducible mCherry-CENP-A cell line. Cells were treated for 48 h with negative control, HJURP, CAPH, and CAPD2 (condensin I siRNA) or CAPH2 and CAPD3 siRNA (condensin II siRNA), and then mCherry-CENP-A expression was induced by Dox addition for the last 15 h. (C) Representative images of HeLa-TRex cells expressing inducible mCherry-CENP-A after siRNA to indicated targets. Cells were preextracted and fixed, then immunostained with CENP-T antibody to mark centromeres. mCherry-CENP-A signal is scaled evenly between conditions. Scale bar, 5 μm. (D) Quantification of centromeric mCherry-CENP-A intensity from experiment in C; ≥5200 centromeres/condition, three biological replicates. Bar represents the mean, and whiskers mark the SD. Blue dashed line marks the mean mCherry-CENP-A intensity in negative control siRNA–treated cells. p < 0.0001 as compared with negative control by Kruskal–Wallis test.
In the context of centromeric chromatin, depleting the common condensin subunits SMC2 and SMC4 results in a reduction in CENP-A/Cse4 loading in yeast and human cells (Yong-Gonzalez et al., 2007; Samoshkin et al., 2009). Depleting specifically the condensin II complex in Xenopus egg extracts also results in decreased CENP-A loading (Bernad et al., 2011). This suggests that specifically the condensin II complex may have a unique contribution at centromeric chromatin. Whether the condensin II complex is selectively involved in CENP-A deposition and how the complex is specifically linked to centromeric chromatin in human cells are not known.
Here we demonstrate a role for condensin II in human CENP-A deposition. Condensin II is present at early G1 centromeres in human cells and interacts with the CENP-A chaperone HJURP. This early G1 centromeric enrichment of the condensin II complex depends on HJURP. Complementary to this, depletion of the condensin II complex reduces HJURP recruitment to centromeres. We also find that the CENP-A chaperone HJURP induces chromatin decondensation when targeted to a LacO/TRE array in U2OS cells. The same region of HJURP that induces this decondensation is sufficient to specifically recruit the condensin II complex, and condensin II modulates the extent of decondensation. Our data identify a new step in the epigenetic deposition of CENP-A requiring HJURP and condensin II presence at early G1 centromeres for successful deposition to occur.
RESULTS
Condensin II depletion results in a CENP-A deposition defect in human cells
Studies in human tissue culture cells have implicated condensin involvement in the CENP-A deposition pathway but have not delineated the roles of condensin I versus condensin II (Samoshkin et al., 2009). Previous studies in Xenopus egg extracts demonstrated that depleting condensin II reduces new CENP-A loading at centromeres (Bernad et al., 2011). We therefore asked whether specifically the condensin II complex has a role in human CENP-A deposition.
To determine the role of condensin II in new CENP-A deposition in human cells, we depleted the condensin I–specific complex members CAPH and CAPD2 or the condensin II–specific complex members CAPH2 and CAPD3 (Figure 1, A and B) in HeLa-TRex cells expressing inducible mCherry-CENP-A. HJURP depletion was used as a positive control and served as a benchmark for the degree to which CENP-A deposition was decreased. Cells were treated with small interfering RNA (siRNA) for 48 h and then additionally treated with doxycycline (Dox) to induce Cherry-CENP-A expression for the last 15 h (Figure 1B). This experimental setup allowed us to specifically investigate the fate of the newly expressed mCherry-CENP-A under conditions in which the condensin complexes had already been depleted. As expected, HJURP depletion resulted in a complete lack of new mCherry-CENP-A deposition at centromeres, consistent with previous reports and validating the use of the inducible mCherry cell line to assess new CENP-A deposition (Figure 1, C and D, and Supplemental Figure S1A; Foltz et al., 2009).
siRNA against either the condensin I or the condensin II subunits significantly reduced mRNA levels compared with negative control–treated cells (Supplemental Figure S1B). Depleting the condensin II complex by targeting both the CAPH2 and CAPD3 subunits resulted in a significant decrease of the level of newly assembled mCherry-CENP-A at centromeres. Upon condensin II depletion, the average mCherry-CENP-A centromeric intensity was reduced to ∼65% of the negative control–treated intensity (Figure 1D). In contrast, depleting the condensin I complex resulted in mCherry-CENP-A levels that were 87% of those for the negative control–treated cells (Figure 1D). siRNA treatment against condensin I had a mild effect on condensin II mRNA levels and may explain the modest effect of condensin I on CENP-A assembly (Supplemental Figure S1B). The effect of condensin II siRNA on condensin I levels is mild, whereas the effect on CENP-A deposition is significant. The converse is not true; that is, direct siRNA targeting condensin I has a major effect on condensin I levels but does not significantly affect CENP-A deposition. Therefore we conclude that, as is the case in Xenopus, the condensin II complex is important for efficient human CENP-A deposition. However, we cannot completely rule out a minor role for the condensin I complex in CENP-A deposition. Compared with HJURP depletion, the reduction of new CENP-A observed in the condensin II siRNA was intermediate, suggesting that condensin II may facilitate CENP-A deposition but not be absolutely essential.
Condensin II is present at centromeres in early G1 and requires HJURP for recruitment
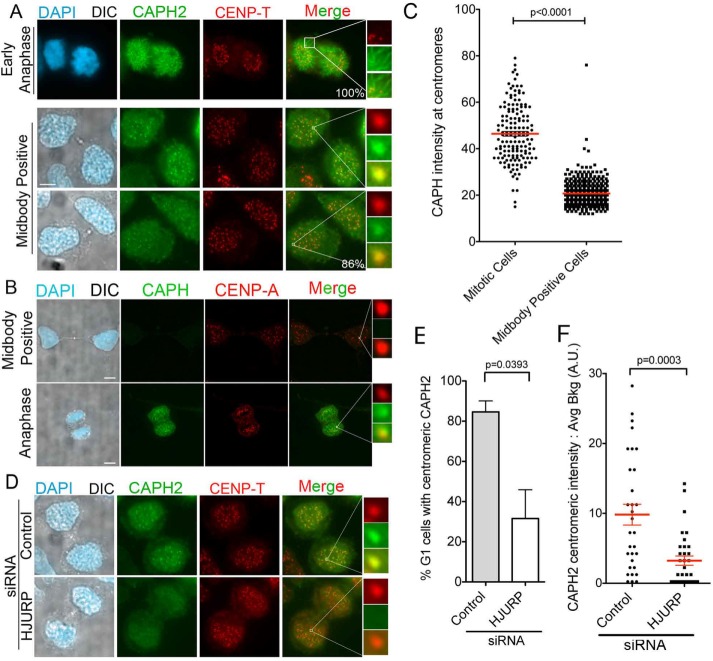
Although previous work implicated condensin function in centromere assembly, support for the condensin II complex acting directly at centromeres was lacking, and the effect could therefore be indirect. If the condensin II complex is specifically required to assist in the process of CENP-A deposition, we would expect it to localize to centromeres in early G1, when CENP-A deposition occurs in human cells (Jansen et al., 2007; Dunleavy et al., 2009; Foltz et al., 2009). To assess whether this is the case, we used a HeLa line that expresses a Dox-inducible CAPH2–green fluorescent protein (GFP). As expected, CAPH2-GFP localized along the arms of chromosomes during mitosis (Figure 2A, top; Ono et al., 2004). We then analyzed the CAPH2-GFP localization pattern in early G1 cell pairs, identified by the presence of a midbody between the two recently divided cells. Strikingly, CAPH2-GFP enriched specifically at centromeric loci in 86% of these early G1 cells, marked by the colocalization between the GFP signal and CENP-T (Figure 2A). This indicates that condensin II is present at centromeres during the time frame when CENP-A deposition is occurring. In contrast, although CAPH-GFP was expressed to a lower level than CAPH2, CAPH-GFP was present at mitotic centromeres but absent at early G1 midbody–positive centromeres (Figure 2, B and C). This further supports a unique role for condensin II and not condensin I during the early G1 CENP-A deposition window.
FIGURE 2:
CAPH2 is present at centromeres in an HJURP-dependent manner in early G1 cells. (A) Representative images of HeLa-TRex cells expressing Tet-inducible CAPH2-GFP. Cells were fixed and then stained with antibody to CENP-T to mark centromeres. Top, CAPH2-GFP localization pattern in early anaphase mitotic cell. Bottom, CAPH2-GFP localization pattern in early G1, midbody-positive cells. Percentages are localization percentages in population of 100 cells. Two biological replicates. Scale bar, 5 μm. (B) Representative images of HeLa-TRex cells stably expressing Tet-inducible CAPH-GFP. Cells were fixed and then stained with an antibody to CENP-A to mark centromeres. Top, CAPH-GFP localization pattern in midbody-positive cells. Bottom, localization in early anaphase mitotic cell. Scale bar, 5 μm. (C) Quantification of experiment in B; 150 centromeres measured per condition. p < 0.0001 by Mann–Whitney test. (D) Representative images of CAPH2-GFP Tet-inducible cells treated with HJURP or negative control siRNA for 48 h with Dox added to induce CAPH2-GFP expression for the last 15 h. Cells were fixed and then stained with antibody to CENP-T to mark centromeres. (E) Quantification in experiment in D. p = 0.0393 by two-tailed t test. Two biological replicates. (F) CAPH2 intensity measurements at centromeres after 48 h of control or HJURP siRNA treatment. Red line indicates mean, and whiskers mark SE; >25 centromeres/condition. p = 0.0003 by Mann–Whitney test.
Because the condensin II complex is present at G1 centromeres and contributes to CENP-A deposition, we asked whether HJURP is responsible for early G1 enrichment of condensin II at human centromeres. To answer this question, we depleted HJURP from the CAPH2-GFP stable line for 48 h (Supplemental Figure S1C) and then induced CAPH2-GFP expression for 12 h and analyzed early G1 cells for CAPH2-GFP centromeric localization. We observed a 50% decrease in the percentage of midbody-positive cells with CAPH2-GFP centromeric localization upon HJURP depletion (Figure 2, D and E). The intensity of CAPH2-GFP at these midbody-positive G1 centromeres was also statistically reduced with HJURP compared with control siRNA treatment (Figure 2F).
HJURP interacts with the condensin II complex
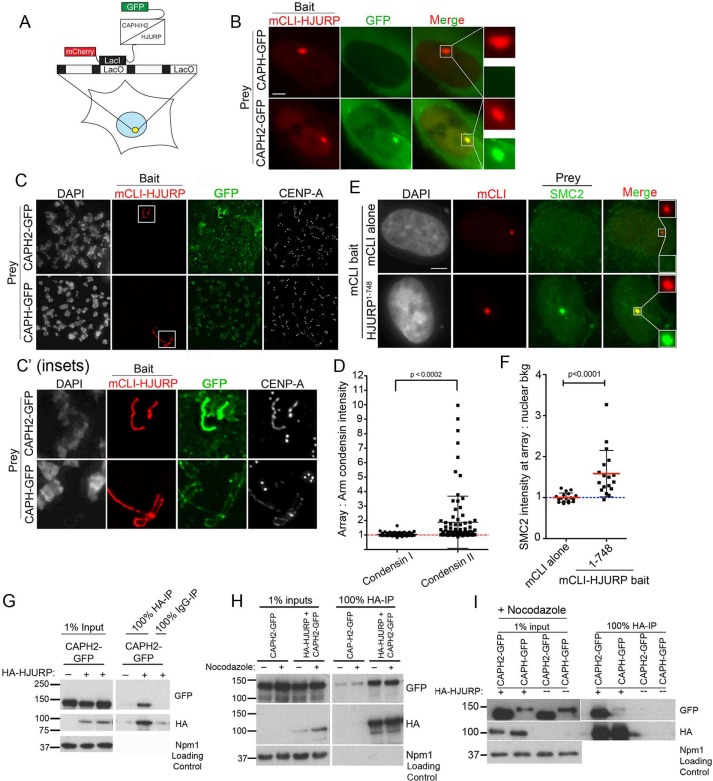
HJURP recruitment to a noncentromeric locus is sufficient to determine the site of CENP-A nucleosome assembly and create a de novo centromere (Barnhart et al., 2011; Bergmann et al., 2011). Because we observe an HJURP-dependent recruitment of condensin II to the centromere (Figure 2), and the condensin II complex affects CENP-A deposition (Figure 1), we asked whether HJURP was sufficient to recruit condensin II to the de novo centromeres. U2OS cells containing a stably integrated LacO array were cotransfected with mCherry-LacI-HJURP1-748 (full-length HJURP) and either CAPH-GFP (condensin I subunit) or CAPH2-GFP (condensin II subunit; Figure 3A). Using live-cell imaging, we looked for condensin recruitment to the HJURP-positive arrays. We observed strong recruitment of CAPH2-GFP to the HJURP arrays. Although CAPH-GFP levels were lower than those of CAPH2-GFP, we readily detected GFP signal in the cytoplasm but observed no recruitment of CAPH-GFP to the LacO array (Figure 3B). Because condensin I is excluded from the nucleus except during mitosis, we wanted to verify that the specific CAPH2 recruitment was also true in mitotic cells, in which both the condensin I and the condensin II complexes have access to the chromatin. To investigate this, we arrested U2OS-LacO cells cotransfected with mCherry-LacI-HJURP1-748 and either CAPH-GFP or CAPH2-GFP in mitosis with nocodazole and then prepared them as metaphase spreads. We found that mCherry-LacI-HJURP1-748 was again specifically able to recruit the condensin II subunit, CAPH2-GFP, and not the condensin I subunit, CAPH-GFP (Figure 3, C, C′, and D). When targeted to mitotic chromatin, we note and will address that HJURP causes chromatin decondensation at the LacO array, indicated by the lengthened array shape (Figure 3, C and C′). To quantify the specific recruitment of CAPH2-GFP, we measured its intensity at the array and set it as a ratio against the arm staining. CAPH2 was significantly enriched at the LacO array compared with the condensin I CAPH subunit (Figure 3D). CAPH-GFP was expressed at lower levels than CAPH2-GFP but strongly decorated the arms of the mitotic chromosomes. However, CAPH-GFP was not enriched at the LacO array when HJURP was targeted there, underpinning our findings that HJURP interacts preferentially with condensin II and not condensin I (Figure 3D). To determine whether the presence of CAPH2 indicated that the full condensin II complex was recruited by HJURP, we examined recruitment of the non–complex-specific condensin subunits SMC2/4 to the mCherry-LacI-HJURP1-748–positive arrays. Endogenous SMC2 was robustly recruited to mCherry-LacI-HJURP1-748–positive arrays and was completely absent at control mCherry-LacI–alone arrays (Figure 3, E and F), suggesting that a functional condensin II complex is assembling at the array.
FIGURE 3:
HJURP interacts with the condensin II complex. (A) Schematic of U2OS-LacO cells with mCherry-LacI-HJURP1-748 (mCLI-HJURP) and CAPH or CAPH2-GFP coexpressed. (B) Representative live-cell images of U2OS-LacO cells expressing mCLI-HJURP as bait and CAPH or CAPH2-GFP as prey protein. Scale bar, 5 μm. (C) Representative images of mitotic spreads of U2OS-LacO cells transfected for 48 h with mCLI-HJURP and CAPH or CAPH2-GFP. Spreads were fixed and stained with an antibody to CENP-A. (C′) Insets of boxed regions in C. (D) Quantification of experiment in C, represented as a ratio of GFP intensity at mCLI-HJURP positive arrays to GFP intensity on the adjacent chromosome arm; 30 arrays, two biological replicates. p < 0.0002 by two-tailed t test. (E) Representative images of U2OS-LacO cells transfected with mCLI-HJURP as bait for 48 h and then stained with antibody for endogenous SMC2. mCLI and SMC2 intensities are scaled equally. Scale bar, 5 μm. (F) Quantification of experiment in E. SMC2 intensity was measured at the array as a ratio over nuclear background signal to show enrichment. Blue dotted line represents a ratio of 1, or no enrichment. Red lines mark the mean, and whiskers are the SD; 20 cells, two biological replicates. p < 0.0001 by Mann–Whitney test. (G) HA immunoprecipitation from HEK cells transfected for 24 h with HA-HJURP with or without CAPH or CAPH2-GFP. CAPH2-GFP alone was used as a negative control. Npm1 is shown as an input loading control. Three biological replicates. (H) HA immunoprecipitation from HEK cells transfected for 48 h with HA-HJURP plus CAPH2-GFP in the presence or absence of nocodazole for the last 15 h of transfection. CAPH2-GFP alone was used as a negative control. Npm1 is shown as an input loading control. Two biological replicates. (I) HA-IP from HEK cells transfected for 48 h with HA-HJURP and CAPH2-GFP or CAPH-GFP. Cells were arrested in nocodazole for the final 12 h of transfection. Blots were probed with antibodies to GFP, HA, and Npm1 as a loading control for the input lanes.
HA-HJURP was also able to efficiently immunoprecipitate CAPH2-GFP from randomly cycling (Figure 3G) and mitotic cell extracts (Figure 3H). To address whether HA-HJURP could also immunoprecipitate CAPH-GFP, we performed immunoprecipitations from mitotic cell extracts to best equalize the expression of CAPH2 and CAPH. Similar amounts of HA-HJURP pulled down greater amounts of CAPH2-GFP than CAPH-GFP (Figure 3I). We conclude that the human CENP-A chaperone HJURP interacts preferentially with the condensin II complex to facilitate its recruitment during G1 when new CENP-A deposition is occurring at the centromere. However, because CAPH2 and CAPH accumulate to different levels in the cells, even during mitosis, we cannot completely rule out the ability of CAPH to interact with HJURP, at least in the case when HJURP is not bound to chromatin.
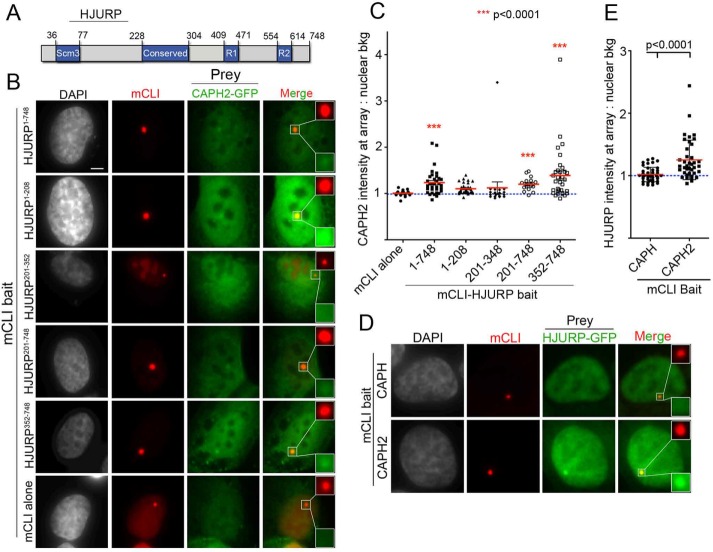
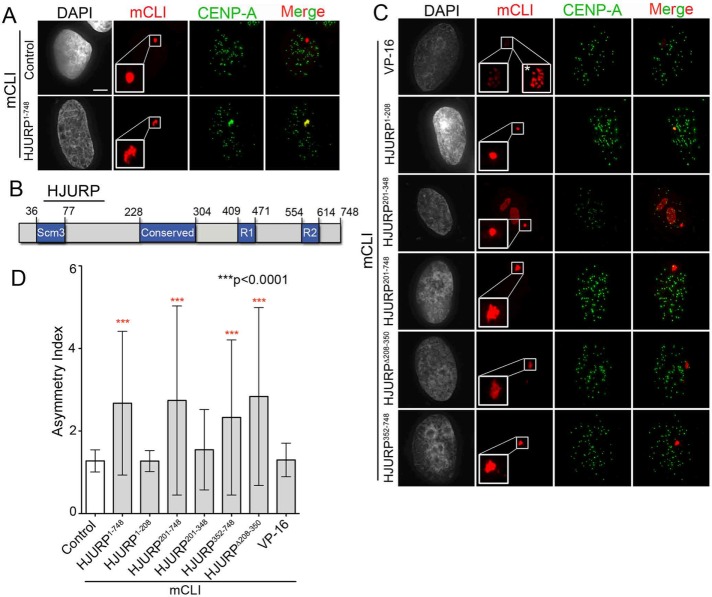
We next wanted to determine which region within HJURP is required to interact with the condensin II complex. Full-length (HJURP1-748) and truncations of HJURP (Figure 4A) were targeted to the LacO array for 48 h in cells cotransfected with CAPH2-GFP. We observed that fragments of HJURP including amino acids 201–748 were sufficient to recruit the condensin II subunit (Figure 4, B and C). The minimal region of HJURP required to recruit statistically significant amounts of CAPH2-GFP was mCherry-LacI-HJURP352-748 (Figure 4, B and C). We additionally demonstrated in a reciprocal experiment that tethering mCherry-LacI-CAPH2 to the LacO array was sufficient to recruit HJURP-GFP (Figure 4, D and E).
FIGURE 4:
HJURP interacts with condensin II via its C-terminal amino acids 352–748. (A) Schematic of HJURP and its known domains. Numbers are amino acids. (B) Representative images of U2OS-LacO cells cotransfected for 48 h with indicated mCLI-HJURP fragments and CAPH2-GFP. Cells were fixed without preextraction. Scale bar, 5 μm. (C) Quantification of experiment in B. Graph displays intensity of CAPH2-GFP at individual mCLI-HJURP arrays as a ratio over the average nuclear background. Red bar marks the mean, and whiskers mark the SD for each condition; ≥30 arrays/condition. ***p < 0.0001 when compared with mCLI by Kruskal–Wallis test followed by Dunn’s multiple comparison test. Blue dotted line marks an intensity ratio of 1, indicating no array enrichment. (D) Representative images of U2OS-LacO cells cotransfected for 48 h with mCLI-CAPH or CAPH2 as bait and HJURP-GFP as prey. Cells were fixed without preextraction. (E) Quantification of experiment in D. Graph displays intensity of HJURP-GFP at individual mCLI-CAPH or CAPH2 arrays as a ratio over the average nuclear background. Red bar marks the mean, and whiskers mark the SD for each condition; ≥20 arrays/condition, two biological replicates. p < 0.0001 by Mann–Whitney test. Blue dotted line marks an intensity ratio of 1, indicating no array enrichment.
Condensin II depletion reduces HJURP centromeric recruitment
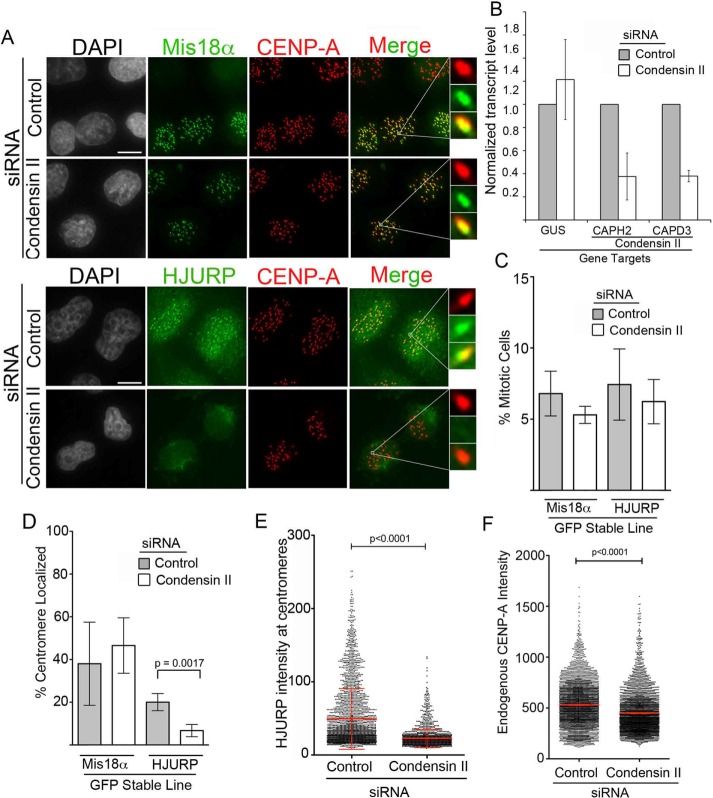
We tested whether condensin II function influences the ability of HJURP to access centromeric chromatin in early G1. On condensin II depletion, the percentage of HJURP-GFP centromere–localized cells was significantly reduced to 6.7% relative to 20% of cells in control siRNA (Figure 5, A and D). In contrast to HJURP-GFP, the percentage of Mis18α-GFP centromere–positive cells was unchanged (Figure 5, A and D). Similarly to parental HeLa cell lines, when we targeted the condensin II complex by siRNA (subunits CAPH2 and CAPD3) in cells stably expressing Mis18α-GFP or HJURP-GFP, the transcript levels of both subunits were depleted to ∼30% of their endogenous levels (Figure 5B). This reduction in HJURP centromere recruitment was not due to a mitotic arrest resulting in fewer cells entering into G1 because the mitotic index percentage was uniform among all conditions (Figure 5C and Supplemental Figure S2). Similarly to the decrease in percentage of cells showing recruitment, the intensity of HJURP-GFP at centromeres was significantly reduced with condensin II siRNA (Figure 5E). Consistent with a reduction in HJURP, endogenous CENP-A levels were also reduced after condensin II siRNA in the HeLa HJURP-GFP stable lines (Figure 5F). Therefore we cannot rule out the possibility that reduction of CENP-A deposition due to condensin II depletion leads to fewer binding sites for HJURP in the following cell cycle, although if this were the case, we would expect a loss of Mis18 as well. These results indicate that interaction between HJURP and the condensin II complex is required for HJURP’s proper centromeric recruitment and stability once it reaches the centromere in early G1.
FIGURE 5:
Condensin II depletion reduces HJURP centromeric recruitment. (A) Representative images of Mis18α-GFP HeLa or HJURP-LAP HeLa-TRex stable lines treated with negative control siRNA or CAPH2 plus CAPD3 (condensin II) siRNA for 48 h. Insets highlight centromere-localized examples for Mis18α-GFP and HJURP-GFP. Cells were fixed and then stained with an antibody to endogenous CENP-A. Scale bar, 5 μm. (B) Quantitative PCR analysis of CAPH2 and CAPD3 transcript levels relative to negative control siRNA after 48-h siRNA treatment. GUS was used as an endogenous control transcript. Error bars represent SD. Three biological replicates. (C) Graph displaying percentage of mitotic cells from experiments in A. Error bars represent SD. p = 0.5079 by Kruskal–Wallis test followed by Dunn’s multiple comparison test. Three biological replicates. (D) Quantification of experiment in A. Graph displays the percentage out of ≥100 cells of either Mis18α-GFP or HJURP-GFP centromere-localized cells after negative control or CAPH2 plus CAPD3 (condensin II) siRNA. Error bars represent SD. p = 0.0017 by two-tailed t test. Three biological replicates. (E) Graph of HJURP-LAP intensities of individual centromeres in HJURP-LAP stable line after 48 h of control or condensin II (CAPH2 and CAPD3) siRNA; >2000 centromeres measured per condition. p < 0.0001 by Mann–Whitney test. (F) Endogenous centromere CENP-A intensities in LAP-HJURP HeLa-TRex stable lines treated for 48 h with control or condensin II (CAPH2+CAPD3) siRNA; >3500 centromeres/condition, three biological replicates. Red line represents mean. p < 0.0001 by Mann–Whitney test.
HJURP alters chromatin compaction
Targeting the CENP-A chaperone HJURP to a noncentromeric, LacO/TRE array induced CENP-A chromatin establishment and de novo kinetochore formation at that site (Barnhart et al., 2011). In addition to recruiting and depositing CENP-A, we observed that HJURP affected the chromatin compaction state at the LacO/TRE array as compared with targeting LacI alone (Figures 3C and 6A). Because the LacO repeats are a linear array, we expect that the unfolding of the LacO locus would result in an asymmetric extension of the locus. Therefore the chromatin compaction status at the array was quantified by taking the ratio of the longest axis of each array to its shortest axis (asymmetry axis). Targeting full-length mCherry-LacI-HJURP1-748 expanded the LacO array to an asymmetry index of 2.67, whereas the mCherry-LacI alone showed an asymmetry index close to 1, as expected (Figure 6, A and D). Full-length HJURP recruitment showed a similar effect on array area (Supplemental Figure S3). This observation is not limited to interphase because extension of the array was also seen during mitosis. We noted that the tip of chromosome 1, where the LacO/TRE array is integrated, was visibly extended compared with the remainder of the mitotic chromosome when mCherry-LacI-HJURP1-748 was targeted in mitotic spreads (Figure 3C).
FIGURE 6:
HJURP induces chromatin decondensation at LacO array via its C-terminal amino acids 352–748. (A) Representative images of U2OS-LacO cells transfected for 48 h with mCLI or mCLI-HJURP. Cells were preextracted and then fixed. Centromeres are marked using a monoclonal antibody to CENP-A. Scale bar, 5 μm. (B) Schematic of HJURP domains. Numbers are amino acids. (C) Representative images of indicated fragments of mCLI-HJURP transfected into U2OS-LacO cells for 48 h. mCLI-VP-16 was used as a positive control for chromatin morphology change. Inset in mCLI-VP-16 image with asterisk was scaled differently to better display differences in array condensation. Cells were prepared as in A. (D) Quantification of array decondensation represented in A and B. The decondensation is quantified as the asymmetry index (ratio of the longest length of each array to its widest width); ≥30 arrays/condition, two biological replicates. Asterisks represent conditions statistically different as compared with control (mCLI alone), p ≤ 0.0001, by Kruskal–Wallis test followed by Dunn’s multiple comparison test.
To investigate what aspects of HJURP contribute to this expansion, we targeted fragments containing separate portions of HJURP to the LacO/TRE array (Figure 6, B and C). Previously we showed that the Scm3 domain–containing fragment of HJURP, HJURP1-208, was sufficient to deposit CENP-A nucleosomes. Targeting of the HJURP1-208 fragment did not result in extension of the LacO array; however, a fragment of HJURP that excluded the CENP-A–binding Scm3 domain (HJURP201-748) showed similar extension of the array to full-length HJURP1-748 (Figure 6, C and D). This demonstrates that CENP-A deposition is not required for the HJURP-induced expansion of the array, but it does require the condensin II–binding domain HJURP352-748 (Figures 6, C and D, and 4,B and C, and Supplemental Figure S3).
Targeting the activator protein VP-16 to LacO arrays has been shown to induce a chromatin morphology change (Rafalska-Metcalf et al., 2010). VP-16 induces a characteristic rosette shape of local decondensation associated with transcription initiation (Tumbar et al., 1999). We therefore used this as a positive control for chromatin condensation change in our assay (Figure 6, C and D). Although VP-16 did produce a localized change in chromatin morphology at the LacO array as previously observed, HJURP dramatically extended the LacO arrays instead of just causing a local decondensation event like VP-16. We additionally verified that condensin II recruitment was not a general response to chromatin condensation changes by targeting mCherry-LacI-VP-16. Although it changed the chromatin compaction at the array, mCherry-LacI-VP-16 was not sufficient to recruit condensin II, indicating that condensin II recruitment by HJURP is specific and not a general response to any type of chromatin condensation change (Supplemental Figure S4).
Condensin II tempers HJURP chromatin compaction capabilities
Given that the same region of HJURP (HJURP352-748) is responsible for both condensin II recruitment and chromatin expansion, we investigated whether condensin II depletion influences array expansion in any way. We observed endogenous SMC2 recruited by HJURP to the LacO array (Figure 3E), suggesting that the endogenous condensin II complex may be functioning there. If the purpose of condensin II complex recruitment is to counteract the chromatin decondensation induced by HJURP, then depleting the condensin II complex should enhance the ability of HJURP to decondense chromatin at the array. To test this, we transfected U2OS-LacO cells with mCherry-LacI-HJURP1-748, followed by 48 h of either negative control siRNA or siRNA to two condensin II complex subunits, CAPH2 and CAPD3 (Figure 7, A and B). Consistent with our hypothesis, we observed an increase in the asymmetry index and area of the mCherry-LacI-HJURP1-748 arrays with condensin II siRNA treatment (Figure 7, A and C, and Supplemental Figure S5). The increase in asymmetry index and area in the population as a whole was modest but was statistically significantly different from treatment with negative control siRNA (Figure 7C and Supplemental Figure S5). The modest effect may be due to the fact that the condensin complex is highly abundant in the cell and is difficult to deplete entirely (Figure 7B). mCherry-LacI-HJURP1-748 arrays from different replicates displayed highly decondensed character after condensin II depletion, some with lengths between 20 and 30 times their widths (Figure 7, A and C). However, depletion of condensin II did not significantly alter the volume of the endogenous G1 centromere (Supplemental Figure S6), suggesting that changes in centromere compaction orchestrated by condensin II may not directly translate into expansion of the centromere, and other factors may act to constrain the locus.
FIGURE 7:
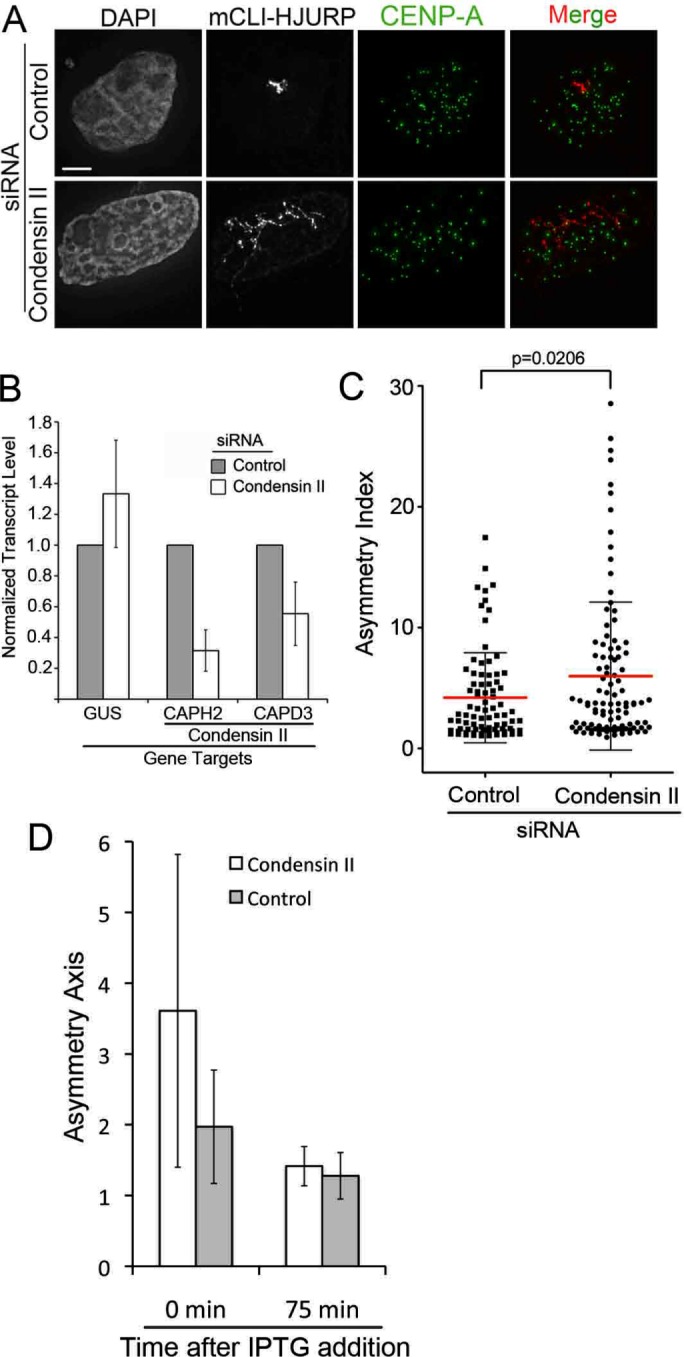
Condensin II tempers HJURP chromatin decompaction capabilities. (A) Representative images of U2OS-LacO cells transfected with mCLI-HJURP and then treated with either negative control siRNA or CAPH2 plus CAPD3 (condensin II) siRNA for 48 h. Cells were preextracted and then fixed and stained with a CENP-A antibody. Scale bar, 5 μm. (B) Quantitative PCR analysis of CAPH2 and CAPD3 transcript levels relative to negative control siRNA after 48-h siRNA treatment. GUS was used as an endogenous control transcript. Error bars represent SD. Three biological replicates. (C) Asymmetry index measurements of experiments in A. Bar represents the mean, and whiskers mark the SD. p = 0.0206 by two-tailed t test; 30 arrays/condition, three biological replicates. (D) Graph of average asymmetry axis values from U2OS-LacO cells expressing mCLI-HJURP and GFP-TetR after 48 h of either control or condensin II siRNA. Cells were treated with 5 mM IPTG to remove mCLI-HJURP from the LacO array for the indicated time points; 20 arrays per time point per condition. Corresponding representative images are in Supplemental Figure S7.
Finally, we tested whether the expansion of the array induced by mCherry-LacI-HJURP at the LacO array was reversible. We treated mCherry-LacI-HJURP and GFP-TetR (independent array marker)–transfected LacO containing cells with control or condensin II (CAPH2 and CAPD3) siRNA for 48 h. The size of the array was examined after 75 min of isopropyl-β-d-thiogalactoside (IPTG) treatment to remove LacI-HJURP from the array. We observed that the chromatin at the array was able to return from its extended shape to a simple round shape in both the control and condensin II siRNA–treated cells (Figure 7D and Supplemental Figure S7). We conclude that the extension induced by HJURP when it is targeted to chromatin is a reversible and actively maintained state that can be influenced by condensin II recruitment.
DISCUSSION
Here we demonstrate an interaction between the condensin II complex and HJURP in human cells that supports a specific requirement for condensin II in CENP-A deposition. We identify a pool of condensin II present at early G1 centromeres during the CENP-A deposition window in human cells. In addition, we show that the presence of HJURP can induce compaction changes to chromatin in vivo. The activity of condensin II complex appears to work in opposition to the intrinsic decondensation activity of HJURP. Our data support a model in which the condensin II complex works alongside HJURP to facilitate new CENP-A deposition (Figure 8).
FIGURE 8:
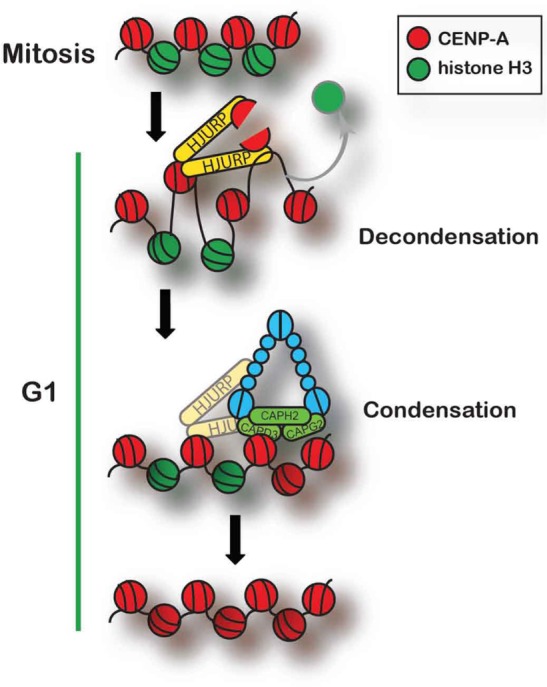
Model of condensin II and HJURP at the centromere during CENP-A deposition. HJURP brings new CENP-A with it to the centromere. Once at centromeric chromatin, HJURP decondenses the chromatin there and loads new CENP-A into the chromatin. Condensin II is locally enriched with HJURP and recondenses the chromatin after HJURP deposits CENP-A, leading to stable new CENP-A assembly. Condensin II is also involved in stably recruiting or keeping HJURP at the centromere in early G1, and so they are drawn as interacting at that time.
HJURP and the Mis18 complex are the major determinants of CENP-A assembly, providing a connection between the existing centromere, new CENP-A deposition, and the nucleosome assembly activity necessary to form the new CENP-A nucleosomes (Fujita et al., 2007; Dunleavy et al., 2009; Foltz et al., 2009). However, because deposition of new CENP-A occurs into existing chromatin in human cells, the process of new CENP-A deposition may also require additional factors to remodel or remove existing H3.3 nucleosomes that are believed to occupy centromeric loci in late G2 and early mitosis (Dunleavy et al., 2011). Condensin II activity is able to induce positive supercoiling in DNA, which will destabilize canonical nucleosomes and may facilitate the removal of H3 nucleosomes and provide access of HJURP to DNA for CENP-A deposition (Kimura et al., 1999; Hirano and Hirano, 2006). In addition, the facilitates chromatin transcription (FACT) complex—nucleophosmin 1, retinoblastoma binding protein 7 (RBBP7; also known as RbAp46), and retinoblastoma binding protein 4 (RBBP4, also known as RbAp48), which have all been implicated in chromatin remodeling—have also been shown to interact with CENP-A or its deposition machinery. However, the exact roles of these proteins in centromere assembly are not well understood (Hayashi et al., 2004; Foltz et al., 2006, 2009; Okada et al., 2006; Fujita et al., 2007; Dunleavy et al., 2009; Shuaib et al., 2010; Mellone et al., 2011).
HJURP and CENP-A were previously implicated in the repair of DNA double-strand breaks (DSBs; Kato et al., 2007; Zeitlin et al., 2009). DNA damage repair is similar to human CENP-A deposition in that it requires the proteins involved in repair to act on previously chromatinized DNA templates. HJURP has been shown to bind synthetic Holliday junctions, but the role of HJURP in DNA damage is unclear (Kato et al., 2007). The condensin II complex is also required for efficient DSB repair (Wood et al., 2008). It is possible that the decondensation phenomenon we characterized here and the interaction of HJURP with condensin II may also be important for the poorly understood role of HJURP in DNA damage.
In humans, as well as in several other species, the condensin II complex is specifically enriched at centromeric, CENP-A–containing chromatin during mitosis, whereas the condensin I complex is absent from these regions (Stear and Roth, 2002; Ono et al., 2004; Savvidou et al., 2005; Shintomi and Hirano, 2011). This enrichment of condensin II at the centromere in human cells requires Aurora B (Ono et al., 2004). During early G1, we observed that the condensin II complex is specifically enriched at centromeric chromatin (Figure 2). It is not clear whether mitotic and G1 pools of condensin II are the same or condensin II is recruited to centromeres de novo during G1. We observed a decrease in HJURP recruitment when condensin II is reduced by siRNA (Figure 5). It may be that the activity of the mitotic pool of condensin II is required for full access of HJURP to centromeric DNA. From our experiments, it is not clear whether the reduction of HJURP is due to less HJURP recruitment to the centromere or reduced residence time of the protein; both would explain the reduction in the percentage of cells showing HJURP recruitment and reduced intensity of HJURP at centromeres in early G1 (Figure 5).
We observed axial expansion of the LacO array after targeting HJURP. In chicken cells, tethering of HJURP leads to an expansion of CENP-A beyond the LacO array (Perpelescu et al., 2015). Although the axial distortion of the array we observe is reversible, we expect that the expanded area of CENP-A deposition may not be. Therefore we believe that these two phenomena are distinct events driven by HJURP. However, we do not know whether condensin function or the expansion induced by HJURP may facilitate expanded deposition of CENP-A.
The expansion did not require the deposition of CENP-A nucleosomes (Figure 6). The similarity between vertebrate HJURP and yeast Scm3 proteins is limited to the Scm3 domain (Sanchez-Pulido et al., 2009). The Scm3 domain is responsible for CENP-A binding and is sufficient to direct the deposition of new CENP-A nucleosomes (Shuaib et al., 2010; Barnhart et al., 2011; Cho and Harrison, 2011; Hu et al., 2011; Zhou et al., 2011; Bassett et al., 2012). HJURP amino acids 352–748, which were sufficient to induce chromatin expansion and also recruit the condensin II complex, contain the HCTD domains of human HJURP, one of which contains the centromere-targeting domain of HJURP (Figures 4 and 6; Zasadzinska et al., 2013). These domains are absent from yeast Scm3. This may reflect that fact that new CENP-A nucleosome addition to the centromere in Saccharomyces cerevisiae and Schizosaccharomyces pombe is coincident with DNA replication and does not necessitate removal of H3 nucleosomes or remodeling of the existing chromatin to achieve CENP-A deposition. Therefore we propose that HJURP-induced decondensation of chromatin and recruitment of the condensin II complex may be unique to vertebrate systems. We propose that these processes have been necessitated by the uncoupling of CENP-A deposition from DNA replication and therefore require remodeling of the centromeric chromatin template in addition to the accrual of new CENP-A to the centromere.
MATERIALS AND METHODS
siRNA, Western blotting, and quantitative PCR
U2OS-LacO or HeLa-TRex cell lines stably expressing Mis18α-GFP, HJURP-LAP, or mCherry-CENP-A were plated at 1 × 105 cells in six-well plates on polylysine-coated coverslips if used for immunofluorescence (IF). If applicable, cells were transfected with Lipofectamine 2000 as described and then treated with siRNA after 8 h of transfection. Transfection medium was left in the well. For siRNA treatment alone without transfection, 24 h after plating, cells were treated with siRNA. Concentrations in well (3-ml total volume in six-well plate format) and product information: 50 nM CAPD3 custom Stealth siRNA from Life Technologies (Carlsbad, CA) (5′ CAA GCC UCU GUU AAC UUG AAU UCC U 3′), 33 nM custom Stealth siRNA from Life Technologies CAPH2 (5′ UUC CAG AGA UGA AAU CAA GGG CCU G 3′), 20 nM HJURP Silencer Select siRNA from Life Technologies (siRNA ID s30814), or equal amount of Negative Control #2 Silencer Select siRNA from Life Technologies (4390846). RNAiMAX was used as lipofection reagent. After 24 h, one-third of the plating volume DMEM with 10% heat-inactivated fetal bovine serum (FBS) plus 5% penicillin/streptomycin was added. For CAPH2 plus CAPD3 depletion, existing medium in the well was removed, and a second siRNA treatment was done at 48 h after plating. For HJURP depletion Western blot analysis, cells were harvested 48 h after siRNA treatment with phosphate-buffered saline (PBS) plus 3 mM ethylenediaminetetraacetic acid (EDTA) and counted and whole-cell lysates were made in SDS–PAGE sample buffer. Lysates from 1 × 105 cells/lane were separated on 10% SDS–PAGE gel and transferred to nitrocellulose. Blots were incubated in primary anti-HJURP (3399) or anti-tubulin (AA2) antibody overnight at 4ºC and in secondary antibodies (Jackson Laboratories) for 1 h at room temperature. For CAPH2 and CAPD3 depletion analysis, cells were harvested using PBS plus 3 mM EDTA and were washed once with PBS. RNA was extracted using Qiagen (Hilden, Germany) RNeasy Mini Kit (74104). cDNA library was prepared using 1 μg of RNA as input for an iScript cDNA synthesis kit (1708890; Bio-Rad). Quantitative PCR with primers to GUS, CAPH2, or CAPD3 from 1 μl of cDNA template was performed (Sybr Green iQ SYBR Green Supermix; 170-8880).
Cell culture, transfections, and immunocytochemistry
HeLa or U2OS-LacO cells were plated to polylysine-coated coverslips at 1 × 105 cells/well in six-well plates, 0.6 × 105 cells/well in 24-well plates, or 1 × 106 cells for 10-cm2 plates. Cells were transfected in Opti-MEM 24 h later with 0.2–0.25 μg of plasmid DNA (24-well plate), 1 μg (six-well plate), or 5.8 μg (10-cm2 dish) using 0.4 μl (24-well plate), 2 μl (six-well plate), or 11.6 μl (10-cm2 dish) of Lipofectamine 2000 (11668-027; Life Technologies). Cells were left in Opti-MEM plus transfection complexes for 9 h, and then medium was changed to DMEM plus 10% FBS and 5% penicillin/streptomycin. HEK293T cells were transfected in serum-free, penicillin/streptomycin–free DMEM using 4 μg of DNA and 30 μl of Polyfect (Qiagen 301105) per 6-cm2 dish.
Live-cell imaging was conducted on the U2OS-LacO cell line at 37°C in Leibovitz’s L-15 medium including 10% FBS after 48 h of transfection. Images were collected at 6-min intervals on a DeltaVision Microscope (Pittsburgh, PA) equipped with a Weatherstation environmental chamber maintained at 37°C.
For fixation, U2OS-LacO and HeLa-TRex cells were preextracted with 0.1% Triton-X in PHEM (PIPES [piperazine-N,N’-bis], HEPES [4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid], EGTA [ethylene glycol-bis[β-aminoethyl ether]-N,N,N’,N’-tetraacetic acid], magnesium sulfate) buffer for 3 min, fixed with 4% paraformaldehyde in PBS for 10 min, and then quenched by addition of 100 mM Tris-HCl, pH 7.5, for another 5 min at room temperature. Experiments that were not preextracted are indicated in figure legends. Cells were blocked in 2% FBS and 2% bovine serum albumin in 0.1% Triton plus PBS. Centromeres were visualized with a rabbit polyclonal anti–CENP-T antibody 1:2000 (3408) or monoclonal CENP-A at 1:1000 dilution (ab13939; Abcam, Cambridge, UK). DNA was stained with 4′,6-diamidino-2-phenylindole (0.2 mg/ml). Donkey-anti-rabbit Cy5-conjugated (111175003; Jackson Laboratories, Bar Harbor, ME) or Cy-3 or fluorescein isothiocyanate–conjugated goat anti-mouse or secondary antibodies were used for detection, and coverslips were mounted with 4 μl of Prolong Gold (Life Technologies).
All micrograph images were collected using a 60 or 100× oil-immersion Olympus (Tokyo, Japan) objective lens (numerical aperture 1.40) on a DeltaVision deconvolution microscope using a Photometrics (Tucson, AZ) CoolSNAP HQ2 camera. Acquisition software used was SoftWoRx (Applied Precision). Images were deconvolved and are presented as stacked images. All representative images within cell lines in figures were collected with identical exposure times and scaled equally for Figures 2–7. Figure 1 mCherry-CENP-A levels were collected with identical exposure times and scaled equally. CENP-T levels in Figure 1 were scaled independently for visualization. Intensities in live-cell and fixed images were analyzed using ImageJ. The CRaQ plug-in was used to measure >5000 centromeres for the mCherry-CENP-A deposition experiment, as well as for the HJURP-GFP and endogenous CENP-A intensity at centromeres with and without condensin II knockdown (Bodor et al., 2012). Details of each analysis method are provided in the appropriate figure legends.
Mitotic chromosome spreads
U2OS-LacO cells were arrested overnight in 0.1 μg/ml nocodazole in DMEM GlutaMAX medium 32 h after transfection. Mitotic cells were harvested using a transfer pipette to blow cells off the plate. Cells were spun down, washed in 1× PBS, and resuspended at 1 × 106 cells/ml in a hypotonic solution (20 mM HEPES, pH 7.0, 1 mM MgCl2, 0.2 mM CaCl2, 20 mM KCl, LPC, and 0.5 μg/ml nocodazole/Colcemid). After 10 min in the hypotonic solution, cells were spun onto glass slides using a cytospin at 2000 rpm for 4 min (30,000 cells/slide), immediately hydrated with 1× PBS, and then fixed and immunostained as described. The anti-mouse CENP-A antibody was used at a 1:1000 dilution (13939; Abcam).
Immunoprecipitations
HEK293T cells were plated to 90% confluency in 6-cm2 dishes. After 24 h, cells were transfected using Polyfect (Qiagen) as described. For mitotically arrested populations, 0.1 μg/ml nocodazole was added to the medium for 12 h. After 24 h, cells were harvested on ice using PBS plus 3 mM EDTA. Cells were spun down and then washed once with PBS. Cells were lysed for 10 min on ice in 1 ml of RIPA buffer with occasional vortexing (150 mM NaCl, 1% NP-40, 0.3% deoxycholate, 0.15% SDS, 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 10% glycerol, 1× Roche (Basel, Switzerland) protease inhibitors, 200 μM NaV, 0.5 mM phenylmethylsulfonyl fluoride, 5 mM NaF, 50 mM β-glycerophosphate). Lysates were sonicated by 2 × 10 cycles of 30 s on/30 s off, using a Biorupter. Lysates were spun down at maximum speed, and then the full 1-ml supernatant was precleared with 10 μl of Protein A agarose beads for 1 h on ice. Beads were spun out, supernatants were transferred to a fresh tube, and 1 μl of hemagglutinin antibody (HA.11 161312; Covance) or 0.2 μl of immunoglobulin G was added overnight at 4°C with rotation. The next day, 8 μl of Protein A Dynabeads (10001D; Life Technologies) was added for 1 h on ice. Beads were washed once with radioimmunoprecipitation (RIPA) buffer and three times with PBST. Beads were resuspended in sample buffer.
Inducible CENP-A, CAPH2, and CAPH cell lines
HeLa-TRex cells containing a Dox-inducible mCherry-CENP-A or CAPH/H2-GFP integration were plated at 1 × 105 cells/well in a six-well plate, and cells were plated on polylysine-coated coverslips for IF wells. Cells were kept in serum-free medium during this protocol (following the initial plating step) to ensure that inducible constructs were not expressed until Dox was added. After 24 h, cells were treated with siRNA plus RNAiMAX. See siRNA treatment described earlier for details. After 32 h of siRNA treatment (8 h after a second round of siRNA, if applicable), Dox was added at a concentration of 1 μg/ml to induce the expression of the mCherry-CENP-A or CAPH/H2-GFP. siRNA was left in the well to ensure that depletion continued during CENP-A or CAPH/H2 loading. Cells were induced with Dox overnight and then fixed and stained.
Antibodies
We used the following antibodies: CENP-A mouse monoclonal (13939; Abcam), 1:1000, IF; CENP-T rabbit polyclonal (3408), 1:2000, IF; GFP rabbit polyclonal (3404), 1:1000, immunoblotting (IB); HA mouse monoclonal (HA.11 161312; Covance, Princeton, NJ), 1:1000, IB; Npm1 mouse monoclonal, 1:10,000, IB; and HJURP rabbit polyclonal (3399), 1:1000, IF.
Supplementary Material
Acknowledgments
We thank members of the Foltz, Stukenberg, and Burke (University of Virginia) labs, especially Ewelina Zasadzinska and Sathyan Mattada, for experimental advice and feedback. We thank Toru Hirota for condensin II constructs. M.C.B.D. was supported by a Farrow Fellowship from the University of Virginia Cancer Center, and D.R.F. is funded by National Institutes of Health Grant GM111907 and a Research Scholar Award from the American Cancer Society. P.T.S. was supported by National Institutes of Health Grant R01 GM063045.
Abbreviations used:
- CENP-A
centromere protein A
- HJURP
Holliday junction recognition protein
- Mis18
missegregation protein 18.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-12-0843) on November 2, 2016.
REFERENCES
- Abe S, Nagasaka K, Hirayama Y, Kozuka-Hata H, Oyama M, Aoyagi Y, Obuse C, Hirota T. The initial phase of chromosome condensation requires Cdk1-mediated phosphorylation of the CAP-D3 subunit of condensin II. Genes Dev. 2011;25:863–874. doi: 10.1101/gad.2016411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, Denizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, Foltz DR, Black BE. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev Cell. 2012;22:749–762. doi: 10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R, Sanchez P, Rivera T, Rodriguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, Losada A. Xenopus HJURP and condensin II are required for CENP-A assembly. J Cell Biol. 2011;192:569–582. doi: 10.1083/jcb.201005136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Bodor DL, Rodriguez MG, Moreno N, Jansen LE. Analysis of protein turnover by quantitative SNAP-based pulse-chase imaging. Curr Protoc Cell Biol. 2012. Chapter 8, Unit 8.8. [DOI] [PubMed]
- Cho US, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci USA. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Almouzni G, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Feng H, Zhou Z, Zhou BR, Bai Y. Structure of the budding yeast Saccharomyces cerevisiae centromeric histones Cse4-H4 complexed with the chaperone Scm3. Proc Natl Acad Sci USA. 2011;108:E596. doi: 10.1073/pnas.1109548108. author reply, E597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Gerlich D, Hirota T, Koch B, Peters JM, Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol. 2006;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Giet R, Glover DM. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, Sullivan BA, Trazzi S, Della Valle G, Robertson KD. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet. 2009;18:3178–3193. doi: 10.1093/hmg/ddp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Hirano M, Hirano T. Opening closed arms: long-distance activation of SMC ATPase by hinge-DNA interactions. Mol Cell. 2006;21:175–186. doi: 10.1016/j.molcel.2005.11.026. [DOI] [PubMed] [Google Scholar]
- Hirano T. Chromosome territories meet a condensin. PLoS Genet. 2012a;8:e1002939. doi: 10.1371/journal.pgen.1002939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012b;26:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami Y, Nihira K, Wada S, Ono M, Honda M, Yoshida K. Mps1 phosphorylation of condensin II controls chromosome condensation at the onset of mitosis. J Cell Biol. 2014;205:781–790. doi: 10.1083/jcb.201308172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67:8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- Kim IS, Lee M, Park KC, Jeon Y, Park JH, Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH, Kim KI. Roles of Mis18alpha in epigenetic regulation of centromeric chromatin and CENP-A loading. Mol Cell. 2012;46:260–273. doi: 10.1016/j.molcel.2012.03.021. [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano M, Kobayashi R, Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- Kimura K, Rybenkov VV, Crisona NJ, Hirano T, Cozzarelli NR. 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi IK, Zasadzinska E, Stellfox ME, Knippler CM, Foltz DR. Licensing of centromeric chromatin assembly through the Mis18alpha-Mis18beta heterotetramer. Mol Cell. 2016;61:774–787. doi: 10.1016/j.molcel.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Ono T, Fang Y, Spector DL, Hirano T. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell. 2004;15:3296–3308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- Perpelescu M, Hori T, Toyoda A, Misu S, Monma N, Ikeo K, Obuse C, Fujiyama A, Fukagawa T. HJURP is involved in the expansion of centromeric chromatin. Mol Biol Cell. 2015;26:2742–2754. doi: 10.1091/mbc.E15-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza I, Haering CH, Rutkowska A. Condensin: crafting the chromosome landscape. Chromosoma. 2013;122:175–190. doi: 10.1007/s00412-013-0405-1. [DOI] [PubMed] [Google Scholar]
- Rafalska-Metcalf IU, Powers SL, Joo LM, LeRoy G, Janicki SM. Single cell analysis of transcriptional activation dynamics. PLoS One. 2010;5:e10272. doi: 10.1371/journal.pone.0010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samoshkin A, Arnaoutov A, Jansen LE, Ouspenski I, Dye L, Karpova T, McNally J, Dasso M, Cleveland DW, Strunnikov A. Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation. PLoS One. 2009;4:e6831. doi: 10.1371/journal.pone.0006831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvidou E, Cobbe N, Steffensen S, Cotterill S, Heck MM. Drosophila CAP-D2 is required for condensin complex stability and resolution of sister chromatids. J Cell Sci. 2005;118:2529–2543. doi: 10.1242/jcs.02392. [DOI] [PubMed] [Google Scholar]
- Schleiffer A, Kaitna S, Maurer-Stroh S, Glotzer M, Nasmyth K, Eisenhaber F. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell. 2003;11:571–575. doi: 10.1016/s1097-2765(03)00108-4. [DOI] [PubMed] [Google Scholar]
- Shintomi K, Hirano T. The relative ratio of condensin I to II determines chromosome shapes. Genes Dev. 2011;25:1464–1469. doi: 10.1101/gad.2060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stear JH, Roth MB. Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment. Genes Dev. 2002;16:1498–1508. doi: 10.1101/gad.989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Douziech M, Bazile F, Pascariu M, Bonneil E, Sauve V, Ratsima H, D’Amours D. Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity. Mol Cell. 2009;34:416–426. doi: 10.1016/j.molcel.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Dou Z, Chen L, Jiang H, Fu C, Fu G, Liu D, Zhang J, Zhu T, et al. Mitotic regulator Mis18beta interacts with and specifies the centromeric assembly of molecular chaperone HJURP. J Biol Chem. 2014;289:8326–8336. doi: 10.1074/jbc.M113.529958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JL, Liang Y, Li K, Chen J. Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J Biol Chem. 2008;283:29586–29592. doi: 10.1074/jbc.M804080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong-Gonzalez V, Wang BD, Butylin P, Ouspenski I, Strunnikov A. Condensin function at centromere chromatin facilitates proper kinetochore tension and ensures correct mitotic segregation of sister chromatids. Genes Cells. 2007;12:1075–1090. doi: 10.1111/j.1365-2443.2007.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasadzinska E, Barnhart-Dailey MC, Kuich PH, Foltz DR. Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J. 2013;32:2113–2124. doi: 10.1038/emboj.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin SG, Baker NM, Chapados BR, Soutoglou E, Wang JY, Berns MW, Cleveland DW. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci USA. 2009;106:15762–15767. doi: 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, Bai Y. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.