Abstract
Objective
This study investigated the correlation between choline/creatine (Cho/Cr) ratios determined by multivoxel proton magnetic resonance spectroscopy (1H-MRS) and the distribution of cancer stem-like cells (CSLCs) in high-grade gliomas.
Patients and methods
Sixteen patients with high-grade gliomas were recruited and underwent 1H-MRS examination before surgery to identify distinct tumor regions with variable Cho/Cr ratios. Using intraoperative neuronavigation, tumor tissues were accurately sampled from regions with high and low Cho/Cr ratios within each tumor. The distribution of CSLCs in samples from glioma tissue regions with different Cho/Cr ratios was quantified by neurosphere culture, immunohistochemistry, and Western blot.
Results
The mean neurosphere formation rate in tissues with high Cho/Cr ratios was significantly increased compared with that in low Cho/Cr ratio tissues (13.94±5.94 per 100 cells vs 8.04±3.99 per 100 cells, P<0.001). Immunohistochemistry indicated that tissues with high Cho/Cr ratios had elevated expression of CD133, nestin, and CD15, relative to low Cho/Cr ratio tissue samples (23.6%±3.8% vs 18.3%±3.3%, 25.2%±4.5% vs 19.8%±2.8%, 24.5%±3.8% vs 17.8%±2.2%, respectively; all P<0.001). Western blot demonstrated that relative CD133 and nestin protein expression in high Cho/Cr ratio regions was significantly higher than that in low Cho/Cr ratio tissue samples (0.50±0.17 vs 0.30±0.08, 0.45±0.13 vs 0.27±0.07, respectively; both P<0.001). The protein expression levels of CD133 and nestin were highly correlated with Cho/Cr ratios (r=0.897 and r=0.861, respectively).
Conclusion
Cho/Cr ratios correlate with the distribution of CSLCs in high-grade gliomas, and this may assist in identifying foci enriched with CSLCs and thus improve the management of high-grade gliomas.
Keywords: high-grade glioma, 1H-MRS, cancer stem-like cells, choline, creatine, Cho/Cr
Introduction
Gliomas are the most prevalent primary brain tumors and are of various histological types and grades, according to the World Health Organization (WHO) classification criteria.1 Among them, grade III anaplastic astrocytoma and grade IV glioblastoma are the most common, with the latter comprising 15% of all intracranial neoplasms and 60%–75% of astrocytic tumors.2 Conventional treatment for high-grade gliomas (grades III and IV) involves neurosurgery, chemotherapy, and radiotherapy; however, therapy is almost inevitably followed by therapeutic resistance and recurrence, which are due largely to the propagation of residual glioma cancer stem cells (CSCs).3,4 CSCs isolated from high-grade gliomas are capable of self-renewal and formation of neurospheres in serum-free medium.5 Cancer stem cells within the same tumor are also recognized to exhibit a high degree of cellular heterogeneity, and this has been demonstrated at both functional (neurosphere culture) and molecular levels6 to contribute to tumor progression and resistance to treatment. Although great efforts have been made to characterize glioma CSCs, such studies are rare and have yet to be translated into clinical practice to efficiently remove these cells. It is therefore imperative to develop a method of identifying areas enriched with CSCs within gliomas to maximize surgical resection, concomitant with targeted radiotherapy and chemotherapy.
High-grade gliomas, and in particular glioblastomas, display unique metabolic features compared with normal brain tissues and low-grade gliomas.7 In addition to glycolysis, tricarboxylic cycle, pentose phosphate pathway, and amino acid metabolism, choline (Cho) metabolism is dysregulated within tumors by cell-intrinsic and cell-extrinsic factors.8 Given that metabolic heterogeneity has been demonstrated in various human tumors,9–11 it is likely that gliomas with heterogeneous CSCs will similarly exhibit a mixed pattern of Cho metabolism. In this study, we aimed to identify sub-regions of gliomas enriched with CSCs based on altered Cho levels. In our study, we used the stem cell markers, CD133, nestin, and CD15, to identify cells with stem-like qualities, along with neurosphere culture assay; however, since the best way to definitively prove the presence of CSCs is by serial xenograft experiments, we refer to the cells isolated in this study as “cancer stem-like cells (CSLCs)”.
Magnetic resonance spectroscopy (MRS) is a noninvasive imaging method used to characterize the metabolic activity of brain tumors.12 By obtaining two-dimensional (2D) or three-dimensional (3D) arrays of spectra covering multiple endogenous metabolites, the technique is able to define the molecular signature of a glioma, in addition to obtaining anatomical information. For example, the concentrations of Cho (a marker of membrane turnover and cell density) and creatine (Cr, levels of which are related to cellular energy state) can be measured in normal and tumor tissues; they appear to be distinctively distributed in high-grade brain tumors, which have significantly higher Cho/Cr ratios than benign lesions.13,14 Although proton MRS (1H-MRS) imaging of Cho and other metabolites has been used for decades in the diagnosis and staging of gliomas,15 grading of tumors,16,17 and following treatment,18,19 the relationship between biochemical metabolic heterogeneity within gliomas and CSC niches has yet to be established. A recent study reported that 1H-MRS integrated with the use of a neuronavigation system can facilitate intraoperative planning and maximal resection, based on the metabolic activities of gliomas, leading to a better patient prognosis.20 We hypothesized that 1H-MRS metabolic profiles within gliomas could assist in the management of these tumors and improve patient prognosis, by facilitating the location of CSC niches.
In this study, 1H-MRS imaging of Cho/Cr distribution and conventional magnetic resonance imaging (MRI) were integrated into our neuronavigational system to obtain tissue samples from distinct regions within the same gliomas. CSLCs from these samples were isolated using a neurosphere culture assay and identified in vivo using the glioma stem cell markers CD133, nestin, and CD15. Our study demonstrated a strong correlation between increased Cho/Cr ratios and enrichment of CSLCs in high-grade gliomas. MRS may provide critical intraoperative information to enable targeting of CSC-enriched foci of gliomas and improve the outcome of radical resection and adjuvant therapies.
Patients and methods
Patients
In the period between August 2012 and March 2015, 16 patients with high-grade gliomas treated at General Hospital of Ningxia Medical University were recruited to the study. There were 10 male and 6 female patients aged 21–72 years (mean, 50.5 years). All patients enrolled in the study met the following criteria: 1) histopathologically proven high-grade gliomas (WHO grade III, n=9; grade IV, n=7), 2) had received no radio- or chemotherapies prior to the study, and 3) no history of cranial trauma, cerebral infection, or cerebrovascular events.
This study was approved by the ethics and review board at the General Hospital of Ningxia Medical University. All patients received and signed consent forms before participating in the study.
MRI and 1H-MRS
Conventional MRI and 1H-MRS scans were performed at 3.0 T using a Siemens Magnetom Verio 3.0 T MRI scanner (Siemens Medical Solutions, Erlangen, Germany). The protocol for MRI included T1-weighted imaging (repetition time [TR], 1.90 ms; echo time [TE], 2.93 ms; matrix size, 256×215; slice thickness, 1 mm; field of view [FOV], 250×219 mm2; acquisition averages 1) and T2-weighted fluid-attenuated inversion recovery (T2-FLAIR) sequence (TR, 9.00 ms; TE, 96 ms; TI, 2.5 ms; slice thickness, 2 mm; matrix size, 256×160).
Regular brain MRI and contrast-enhanced scans were followed by 1H-MRS imaging.
MRS imaging was acquired for the gliomas (including areas of necrosis and edema) and contralateral normal brain tissues using 3D multivoxel pointed-resolved spectroscopy sequence (PRESS; TR, 1.70 ms; TE, 135 ms; slice thickness, 15 mm; phase encoding, 16×16×16; FOV, 120×120 mm2). The voxel size was 10×10×10 mm3.
Reconstruction and postprocessing of 1H-MRS data
MRS datasets were processed by Fourier transformation and reconstruction of metabolic maps, including N-acetylaspartate (NAA), Cho, and Cr, and were performed using automation software (Syngo MultiModality Workplace; Siemens Medical Solutions). From the maps of the spatial distribution of Cho and Cr, two locations within each glioma were predetermined as having the highest and lowest Cho/Cr ratios for stereotactic sampling. Cases where the foci of peak and nadir Cho/Cr ratios were located outside of the glioma, or within functional areas, were excluded from the study.
Neurosurgery and glioma sampling
MRS data were processed and transferred to the surgical navigation computer workstation, iPlan Cranial 3.0 (BrainLab, Germany). Registration of the two locations within the glioma with predetermined Cho/Cr ratios was carried out as previously described.20 Tumor samples were obtained by experienced neurosurgeons following standard procedures.21 Specimens from each location were divided and immediately sent for neurosphere culture and histopathological evaluation.
Neurosphere culture
Glioma tissue samples from each site were briefly rinsed and finely minced in 0.25% trypsin/EDTA (Gibco, NY, USA). After incubation at 37°C, samples were dissociated by pipetting up and down thoroughly. The dissociated samples were washed with DMEM/F12 medium (Hyclone, UT, USA) containing 10% fetal bovine serum (TransGen Biotech, Beijing, People’s Republic of China). The suspension was filtered to single cells and spun briefly. Cells were resuspended in serum-free medium, counted using a hemacytometer, and plated at 100 cells/well in 96-well plates. Five replicate wells were cultured for each sample. Culture medium was free of serum and contained 20 ng/mL epidermal growth factor (EGF; Peprotech, NJ, USA), 20 ng/mL basic fibroblast growth factor (bFGF; Peprotech), and 1× B27 (Gibco). Cells were maintained at 37°C in an incubator with an atmosphere of 95% air and 5% CO2.
Histology and immunohistochemistry
Glioma specimens for histopathology were fixed in 4% formalin for 24 h. After being embedded in paraffin, tissue blocks were sectioned (5 μm), and slides were stained with hematoxylin and eosin for evaluation of tumor type and grading. Immunohistochemistry with DAB staining was performed on adjacent slides. Primary antibodies used were rabbit anti-CD133 (1:50; Cat #18470-1-AP; Proteintech, Wuhan, People’s Republic of China), rabbit antinestin (1:100; Cat #AP2020D; Abgent, Jiangsu, People’s Republic of China), and rabbit anti-CD15 (1:100; Cat #19497-1-AP; Proteintech). Secondary antibody was goat antirabbit (1:500). A horseradish-peroxidase-based Vectastain ABC Kit (Vector Laboratories, California, LA, USA) was used to visualize antibody staining. Slides were counterstained with hematoxylin and mounted in aqueous media. Images were captured by standard light microscopy. The percentages of CD133+, nestin+, and CD15+ cells were counted and calculated from five random nonoverlapping 100× high power fields.
Western blot
Whole cell lysates were extracted from fresh glioma tissues using a commercial protein extraction kit (KeyGEN Biotech, Jiangsu, People’s Republic of China). Protein concentration was measured by BCA assay (Bio-Rad, California, LA, USA). Equal amounts of protein (35 μg) were separated by 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Proteins transferred to polyvinylidene difluoride membranes were incubated with primary antibodies, including rabbit anti-CD133 (1:500), anti-GAPDH (1:8,000; Cat #10494-1-AP; Proteintech), and antinestin (1:1,500). Chemiluminescence-exposed films were developed, scanned, and band intensities determined using Gel-Pro Analyzer software (Media Cybernetics, MD, USA) to quantify protein expression levels. GAPDH was used as an internal control to determine the relative levels of CD133 and nestin.
Statistical analysis
Data were analyzed using SPSS 20.0 (IBM Corporation, Armonk, NY, USA) and were presented as the mean ± standard deviation. We used Student’s t-test to evaluate the difference between samples from two regions of gliomas. The association between Cho/Cr ratios and enrichment with CSLCs was determined by Pearson’s correlation analysis. P<0.05 was considered statistically significant.
Results
Brain MRI
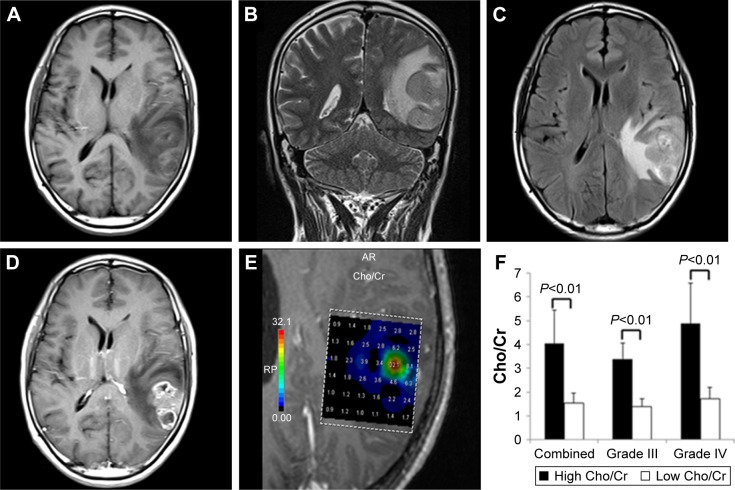
Typical high-grade gliomas were recognized as irregular and heterogeneous lesions that were isointense or hypointense on T1-weighted images (WIs; Figure 1A) and were often hyperintense masses on T2-WI and T2-FLAIR images (Figure 1B and C). Postcontrast images (Figure 1D) showed a thickened ring of enhancement, representing a combination of vasogenic edema and tumor infiltration.
Figure 1.
Conventional MRI and 1H-MRS images of a typical glioblastoma.
Notes: (A) An isointense/hypointense lesion visible on axial T1-WI. (B) T2-WI and (C) T2-FLAIR showed a hyperintense mass with remarkable infiltration. (D) Contrast-enhanced image demonstrated clear enhancement of the tumor. (E) Cho/Cr spectra from multiple-voxel regions inside the tumor. (F) Differences in Cho/Cr ratios between high and low metabolic regions in combined high-grade gliomas and grade III and grade IV gliomas separately; all were highly significantly different (P<0.01).
Abbreviations: MRI, magnetic resonance imaging; 1H-MRS, proton magnetic resonance spectroscopy; WI, weighted image; T2-FLAIR, T2-weighted fluid-attenuated inversion recovery; Cho, choline; Cr, creatine.
1H-MRS scan and glioma sampling
Multivoxel 1H-MRS imaging was routinely performed after traditional MRI scans to visualize the metabolic spectra of gliomas (Figure 1E). Compared to contralateral normal brain tissue, high-grade gliomas showed significantly elevated Cho peaks, lower NAA levels, and mildly reduced levels of Cr (data not shown; Figure 1E). Cho/Cr ratios from distinct regions within the same gliomas varied significantly (Figure 1F). Mean Cho/Cr ratios in high metabolic areas were 4.03±0.67, which were significantly higher than those in low metabolic areas (1.53±0.34; P<0.01). Separately, mean Cho/Cr ratios in high metabolic regions of grade III gliomas were 3.38±0.67 and those in grade IV tumors were 4.87±1.71, both of which were significantly higher than those from low metabolic regions in corresponding grade tumors (grade III 1.38±0.34 and grade IV 1.72±0.49; both P<0.01; Figure 1F).
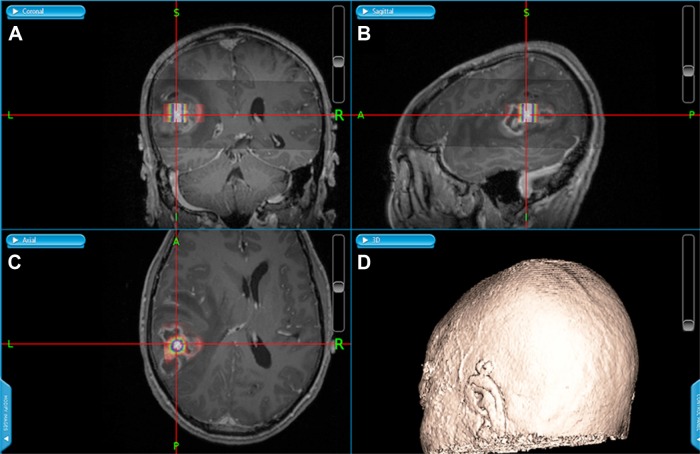
Varied levels of Cho and Cr within tumors may reflect clonal heterogeneity, which is characteristic of high-grade gliomas, and especially glioblastomas. We hypothesized that these regions may correspond to rich and poor sources of tumor-initiation niches of glioma CSCs. For each glioma, the two locations with the highest and lowest Cho/Cr ratios were determined on the 1H-MRS maps (Figure 1E). MRS maps and brain MRIs were coregistered and integrated into the neuronavigational system. Under the guidance of the system (Figure 2), glioma samples were obtained from regions with contrasting Cho/Cr ratios for stem cell assays and histopathological evaluation.
Figure 2.
Neuronavigational system-guided stereotactic biopsy and tumor resection.
Notes: (A) Coronal, (B) sagittal, and (C) axial sections of the tumor. (D) 3D rendition of the patient.
Abbreviation: 3D, three-dimensional.
Glioma neurosphere culture
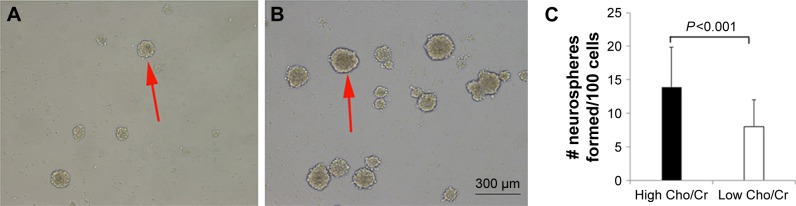
Single-cell suspensions were prepared from fresh glioma specimens. Cells were diluted and cultured in serum-free medium. After 3–4 days of culture, small neurospheres started to form. They grew to 150–200 μm in diameter after 6–8 days (Figure 3A and B). The mean number of neurospheres cultured from samples from high Cho/Cr ratio areas for all cases (13.94±5.94) was significantly higher than that from low Cho/Cr ratio area samples (8.04±3.99, P<0.001) (Figure 3C), indicating that there were more CSLCs in high Cho/Cr ratio regions. Moreover, significantly increased numbers of neurospheres were also formed from high Cho/Cr ratio regions of grade III and grade IV gliomas separately (data not shown).
Figure 3.
Neurospheres formed from glioma tissues with distinct Cho/Cr metabolic activities.
Notes: (A) Neurospheres formed from low Cho/Cr ratio tissues were fewer and smaller in size. (B) Neurospheres formed from high Cho/Cr ratio regions grew more quickly and were more numerous. (C) There was a highly significant difference between the mean numbers of neurospheres formed from distinct regions within same gliomas (P<0.001). Red arrows show the neurospheres cultured from high-grade glioma samples.
Abbreviations: Cho, choline; Cr, creatine.
Identification of CD133+, nestin+, and CD15+ CSLCs in gliomas
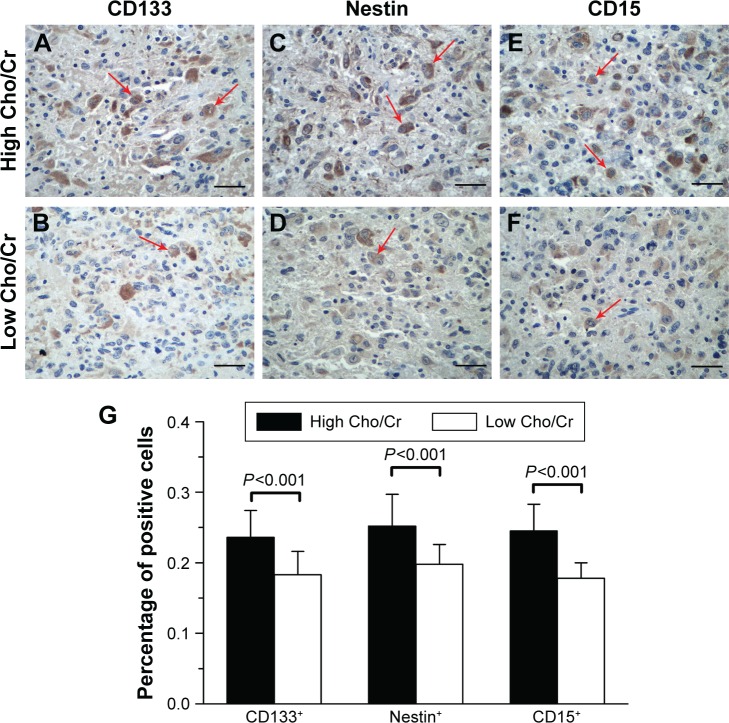
To quantify CSLCs from gliomas in vivo, we used the neural/tumor stem cell markers CD133, nestin, and CD1522 to examine glioma tissues with distinct Cho/Cr metabolic profiles. CD133 strongly labeled niches in gliomas, with staining mostly concentrated on cell membranes and cytoplasm (Figure 4A and B). CD133+ cells were found in aggregates of diverse size. Nestin showed immunoreactivity in the cytoplasm and labeled positive cells evenly (Figure 4C and D). A small fraction of glioma cells also displayed nuclear distribution of nestin, in addition to diffuse cytoplasmic staining. Nestin+ cells were clustered in cords and islands. Another cancer stem cell marker, CD15, was expressed in cell cytoplasm and membranes (Figure 4E and F). Quantification of CD133+, nestin+, and CD15+ cells showed that there was a significantly increased percentage of CSLCs (23.6%±3.8% CD133+, 25.2%±4.5% nestin+, and 24.5%±3.8% CD15+) in the areas with high Cho/Cr ratios, compared with low Cho/Cr ratio areas (18.3%±3.3% CD133+, 19.8%±2.8% nestin+, and 17.8%±2.2% CD15+; all P<0.001; Figure 4G).
Figure 4.
Cancer stem-like cells within gliomas varied in number in distinct regions of gliomas.
Notes: Tissues with high Cho/Cr ratios, stained with the stem cell markers, (A) CD133, (C) nestin, and (E) CD15. Tissues with low Cho/Cr ratios, stained with the stem cell markers, (B) CD133, (D) nestin, and (F) CD15. (G) There were highly significant differences in the percentage of CD133+, nestin+, and CD15+ cells between high and low Cho metabolic regions. Red arrows show the CD133+, nestin+, or CD15+ cells in high-grade glioma samples. Magnification, 100×. Scale bar, 200 μm.
Abbreviations: Cho, choline; Cr, creatine.
Correlation between Cho/Cr ratios and CD133/nestin expression
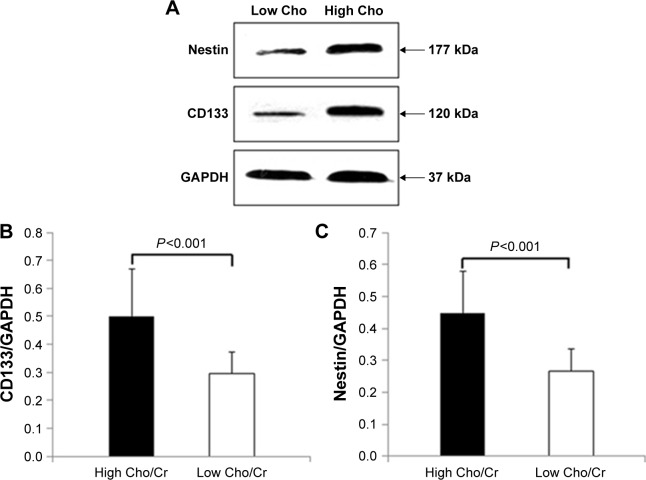
To confirm our immunohistochemistry findings, we extracted proteins from both high and low Cho/Cr ratio areas of gliomas. The relative protein levels of CD133 and nestin were significantly elevated in high Cho/Cr ratio areas (CD133/GAPDH, 0.50±0.17; nestin/GAPDH, 0.45±0.13) compared with those in low Cho/Cr ratio areas (CD133/GAPDH, 0.30±0.08; nestin/GAPDH, 0.27±0.07; both P<0.001; Figure 5). The relative amounts of CD133 and nestin correlated with Cho/Cr ratios. Pearson’s correlation analysis suggested that the positive relationships between CD133 and nestin and Cho/Cr were highly significant (CD133 r=0.897, nestin r=0.861; both P<0.01). Overall, Cho/Cr ratios determined by 1H-MRS appeared to be strongly associated with enrichment of CSLCs in high-grade gliomas.
Figure 5.
Expression of CD133 and nestin correlated with Cho/Cr ratios in gliomas.
Notes: (A) Western blot incubated with nestin and CD133 antibodies showed stronger expression of stem cell markers in high Cho/Cr ratio regions than those in low Cho/Cr ratio regions. There were highly significant differences in (B) CD133 and (C) nestin expression levels between high and low Cho/Cr ratio regions.
Abbreviations: Cho, choline; Cr, creatine.
Discussion
The challenges of managing high-grade gliomas include the extent of resection (EOR), tumor progression, and recurrence from residual glioma CSCs.23 The current practice of maximal surgical resection improves survival rates of patients with gliomas;24 however, given their characteristic of infiltrative growth, it is almost impossible to achieve 100% resection of gliomas and surrounding abnormalities, particularly when the tumor is located close to functional brain areas. Postoperative residual glioma stem cells survive aggressive chemo-and radiotherapies and contribute to tumor recurrence.3,4,25 Currently, clinical trials targeting glioma CSCs are ongoing.26 Our goal in this study was to identify CSC-enriched regions within gliomas to maximize the EOR and retain as few residual CSCs as possible to improve the treatment of high-grade gliomas and patient survival rates.
We used 1H-MRS as a noninvasive method to study the metabolic activity of gliomas in vivo. Distinct regions within the same gliomas, which displayed remarkably different metabolic activities, and especially Cho/Cr ratios, were observed. CSLCs isolated from these regions varied in number and capacity to form neurospheres in culture. There was a strong correlation between elevated Cho/Cr ratios and enrichment of CSLCs in high-grade gliomas. Thus, 1H-MRS Cho/Cr metabolic profiles may be useful as biomarkers to identify “hot zones” enriched with CSLCs within gliomas both pre- and intraoperatively. The information acquired by 1H-MRS could be crucial for both neurosurgeons in planning EOR and neuro-oncologists in delivering targeted focal radiation. A recent report of MRS-guided resection of gliomas demonstrated that the technique allows metabolic delineation of tumor margins and results in better patient prognosis,20 which may be attributable to greater elimination of glioma CSCs, corroborating the findings of our study.
MRS has been used since the 1980s to provide functional information about brain tumors and other neurological lesions.15,27 The advantage of multivoxel 1H-MRS is that high-resolution metabolic maps of whole brains can be reconstructed from 3D arrays of spectra. Multivoxel MRS combined with conventional MRI can provide information about the spatial extent and functional metabolic heterogeneity of gliomas and surrounding areas.12,28 Thus, 1H-MRS has been widely used for diagnosing gliomas, tumor grading, and tracking the response to therapy at subcellular level.29 Choline, NAA, and creatine are the most common metabolites visualized by 1H-MRS imaging to investigate gliomas. NAA is a derivative of the neurotransmitter aspartate, and its metabolic profile indicates neuronal viability. Creatine is a marker of cellular energy state, frequently applied as a standard to normalize and interpret other metabolite peaks (eg, Cho/Cr and NAA/Cr). Choline is an important component of phospholipids in cell membranes, and its accumulation is caused by several Cho metabolic enzymes, which are dysregulated by multiple oncogenic signaling pathways during malignant transformation.8 Choline is also the source of the methyl group,30 which is essential for epigenetic regulation, such as methylation of the O6-methylguanine-DNA methyltransferase (MGMT) gene promoter31 and the BMP pathway32 in glioblastoma. Elevated Cho peaks in 1H-MRS arise from signals from the trimethyl groups of Cho compounds and correlate with histological findings of cellular density and proliferation in glioma.33,34 Increased Cho/Cr ratios and decreased NAA/Cr ratios are associated with high-grade gliomas.16,35
In our study, we observed heterogeneous distributions of Cho/Cr within tumors, consistent with clonal diversity and the complex hierarchical cellular structure in gliomas.6 Histopathology and stem cell assays further confirmed our hypothesis that the regions of higher Cho/Cr ratios harbored increased numbers of CSLCs.6 The mechanism by which aggregation of CSLCs leads to local elevation of choline requires further investigation. One possibility is that increased choline levels reflect accelerated membrane turnover and anabolic metabolism in the CSLC-enriched areas. Another option is that oncogenic pathways within CSLCs and the surrounding hypoxic microenvironment activate choline metabolic transporters and enzymes,8 leading to increased uptake of choline and cell autonomous accumulation of choline compounds.
Although there is no consensus on the gold standard for identifying glioma stem cells, neurosphere culture and staining of CSC markers are the most commonly used assays. Neurosphere culture tests for self-renewal and proliferation of CSCs. We observed increased numbers of large neurospheres formed from tumor tissues exhibiting high Cho/Cr ratios. These neurospheres were able to form at limiting dilutions and passages, suggesting that there are more self-renewing CSLCs in high Cho/Cr ratio regions. Significantly lower numbers of neurospheres grew from tissue samples with low Cho/Cr ratios, and the neurospheres were smaller in size, likely due to the presence of fewer self-renewing CSLCs with lower proliferative potentials. These CSLCs could be less aggressive clones that have accumulated fewer mutations compared to the more malignant isolates from high Cho/Cr ratio regions. Our current ongoing studies, including the genomic sequencing of CSCs and xenograft transplantations into animal models, will provide information to help resolve these issues.
To complement the neurosphere assay, we examined glioma tissues by staining for glioma CSC markers. CD133 is a cell surface glycoprotein used for enriching stem cells. Staining for CD133 may vary depending on the glycosylation state of the protein, technical differences, and clones of antibodies, which recognize diverse epitopes.36 Although there is still debate regarding the validity of its use in defining stemness, the combined use of CD133 and nestin can identify at least a subset of glioma CSCs, which are associated with survival rates of glioma patients.37 In human glioblastomas, Son et al38 have found that CD15 is an enrichment marker of stem cells in CD133− negative tumors. It is often used as a marker of glioma stem cell now.39 In our study, we observed higher numbers of CD133+, nestin+, and CD15+ cells in the foci with high Cho/Cr ratios relative to those in low Cho/Cr foci. The expression levels of CD133 and nestin were consistently elevated in high Cho/Cr ratio regions and correlated closely with Cho/Cr ratios; therefore, the Cho/Cr ratio acquired by 1H-MRS may be a useful in vivo clinical biomarker that can identify glioma foci enriched with CSLCs.
Conclusion
Our study investigated 1H-MRS Cho/Cr metabolic profiles in an attempt to improve the management of high-grade gliomas. This is the first report, linking imaging of tumor metabolism to the enrichment of CSLCs within gliomas. The patterns of metabolites determined by 1H-MRS imaging could be useful in planning surgical resection and delivery of focal radiation, targeting the source of tumor-initiating CSCs. Follow-up of these patients will help to validate the strategy of identifying CSLC-enriched niches by 1H-MRS imaging of Cho/Cr.
Acknowledgments
The authors thank our colleagues Tao Zhang, Xinghao Duan, and Xvlei Hu for their cooperation with data collection. We thank our colleague Linna Zhang for data analysis. We also thank our colleague Michael Rallo for critical reading and comments on the manuscript. This work was supported by the National Key Technology R&D Program of China (grant no 2014BAI04B05) and the Natural Science Foundation of Ningxia (grant no NZ16127).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2014;16(suppl 4):iv1–iv63. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39(3):193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 6.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark PM, Mai WX, Cloughesy TF, Nathanson DA. Emerging approaches for targeting metabolic vulnerabilities in malignant glioma. Curr Neurol Neurosci Rep. 2016;16(2):17. doi: 10.1007/s11910-015-0613-6. [DOI] [PubMed] [Google Scholar]
- 8.Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer. 2011;11(12):835–848. doi: 10.1038/nrc3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caro P, Kishan AU, Norberg E, et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell. 2012;22(4):547–560. doi: 10.1016/j.ccr.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hensley CT, Faubert B, Yuan Q, et al. Metabolic heterogeneity in human lung tumors. Cell. 2016;164(4):681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terunuma A, Putluri N, Mishra P, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest. 2014;124(1):398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Bisbal MC, Celda B. Proton magnetic resonance spectroscopy imaging in the study of human brain cancer. Q J Nucl Med Mol Imaging. 2009;53(6):618–630. [PubMed] [Google Scholar]
- 13.Hourani R, Brant LJ, Rizk T, Weingart JD, Barker PB, Horska A. Can proton MR spectroscopic and perfusion imaging differentiate between neoplastic and nonneoplastic brain lesions in adults? AJNR Am J Neuroradiol. 2008;29(2):366–372. doi: 10.3174/ajnr.A0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Usenius JP, Vainio P, Hernesniemi J, Kauppinen RA. Choline-containing compounds in human astrocytomas studied by 1H NMR spectroscopy in vivo and in vitro. J Neurochem. 1994;63(4):1538–1543. doi: 10.1046/j.1471-4159.1994.63041538.x. [DOI] [PubMed] [Google Scholar]
- 15.Bruhn H, Frahm J, Gyngell ML, et al. Noninvasive differentiation of tumors with use of localized H-1 MR spectroscopy in vivo: initial experience in patients with cerebral tumors. Radiology. 1989;172(2):541–548. doi: 10.1148/radiology.172.2.2748837. [DOI] [PubMed] [Google Scholar]
- 16.Zeng Q, Liu H, Zhang K, Li C, Zhou G. Noninvasive evaluation of cerebral glioma grade by using multivoxel 3D proton MR spectroscopy. Magn Reson Imaging. 2011;29(1):25–31. doi: 10.1016/j.mri.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Zhang H, Zhang J, et al. The diagnostic performance of magnetic resonance spectroscopy in differentiating high-from low-grade gliomas: a systematic review and meta-analysis. Eur Radiol. 2016;26(8):2670–2684. doi: 10.1007/s00330-015-4046-z. [DOI] [PubMed] [Google Scholar]
- 18.Fink JR, Carr RB, Matsusue E, et al. Comparison of 3 Tesla proton MR spectroscopy, MR perfusion and MR diffusion for distinguishing glioma recurrence from posttreatment effects. J Magn Reson Imaging. 2012;35(1):56–63. doi: 10.1002/jmri.22801. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Zeng Q, Liu Y, Li C, Feng D, Wang J. Assessment of the intrinsic radiosensitivity of glioma cells and monitoring of metabolite ratio changes after irradiation by 14.7-T high-resolution (1)H MRS. NMR Biomed. 2014;27(5):547–552. doi: 10.1002/nbm.3091. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Zhuang DX, Yao CJ, et al. Metabolic approach for tumor delineation in glioma surgery: 3D MR spectroscopy image-guided resection. J Neurosurg. 2016;124(6):1585–1593. doi: 10.3171/2015.6.JNS142651. [DOI] [PubMed] [Google Scholar]
- 21.Najbauer J, Zhao J, He H, et al. Neuronal transcription factors induce conversion of human glioma cells to neurons and inhibit tumorigenesis. PLoS One. 2012;7(7):e41506. doi: 10.1371/journal.pone.0041506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brescia P, Richichi C, Pelicci G. Current strategies for identification of glioma stem cells: adequate or unsatisfactory? J Oncol. 2012;2012:376894. doi: 10.1155/2012/376894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis HP, Greenslade M, Powell B, Spiteri I, Sottoriva A, Kurian KM. Current challenges in glioblastoma: intratumour heterogeneity, residual disease, and models to predict disease recurrence. Front Oncol. 2015;5:251. doi: 10.3389/fonc.2015.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bloch O, Han SJ, Cha S, et al. Impact of extent of resection for recurrent glioblastoma on overall survival: clinical article. J Neurosurg. 2012;117(6):1032–1038. doi: 10.3171/2012.9.JNS12504. [DOI] [PubMed] [Google Scholar]
- 25.Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohka F, Natsume A, Wakabayashi T. Current trends in targeted therapies for glioblastoma multiforme. Neurol Res Int. 2012;2012:878425. doi: 10.1155/2012/878425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin A, Ross BD, Harris K, Wong W. Efficacy of proton magnetic resonance spectroscopy in neurological diagnosis and neurotherapeutic decision making. NeuroRx. 2005;2(2):197–214. doi: 10.1602/neurorx.2.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson SJ. Multivoxel magnetic resonance spectroscopy of brain tumors. Mol Cancer Ther. 2003;2(5):497–507. [PubMed] [Google Scholar]
- 29.Verma A, Kumar I, Verma N, Aggarwal P, Ojha R. Magnetic resonance spectroscopy – revisiting the biochemical and molecular milieu of brain tumors. BBA Clin. 2016;5:170–178. doi: 10.1016/j.bbacli.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niculescu MD, Zeisel SH. Diet, methyl donors and DNA methylation: interactions between dietary folate, methionine and choline. J Nutr. 2002;132(8 suppl):2333S–2335S. doi: 10.1093/jn/132.8.2333S. [DOI] [PubMed] [Google Scholar]
- 31.Wick W, Weller M, van den Bent M, et al. MGMT testing – the challenges for biomarker-based glioma treatment. Nat Rev Neurol. 2014;10(7):372–385. doi: 10.1038/nrneurol.2014.100. [DOI] [PubMed] [Google Scholar]
- 32.Lee J, Son MJ, Woolard K, et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13(1):69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKnight TR, Lamborn KR, Love TD, et al. Correlation of magnetic resonance spectroscopic and growth characteristics within Grades II and III gliomas. J Neurosurg. 2007;106(4):660–666. doi: 10.3171/jns.2007.106.4.660. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu H, Kumabe T, Shirane R, Yoshimoto T. Correlation between choline level measured by proton MR spectroscopy and Ki-67 labeling index in gliomas. AJNR Am J Neuroradiol. 2000;21(4):659–665. [PMC free article] [PubMed] [Google Scholar]
- 35.Caivano R, Lotumolo A, Rabasco P, et al. 3 Tesla magnetic resonance spectroscopy: cerebral gliomas vs. metastatic brain tumors. Our experience and review of the literature. Int J Neurosci. 2013;123(8):537–543. doi: 10.3109/00207454.2013.774395. [DOI] [PubMed] [Google Scholar]
- 36.Hermansen SK, Christensen KG, Jensen SS, Kristensen BW. Inconsistent immunohistochemical expression patterns of four different CD133 antibody clones in glioblastoma. J Histochem Cytochem. 2011;59(4):391–407. doi: 10.1369/0022155411400867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang M, Song T, Yang L, et al. Nestin and CD133: valuable stem cell-specific markers for determining clinical outcome of glioma patients. J Exp Clin Cancer Res. 2008;27:85. doi: 10.1186/1756-9966-27-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4(5):440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamuro S, Okamoto Y, Sano E, et al. Characterization of glioma stem-like cells from human glioblastomas. Int J Oncol. 2015;47(1):91–96. doi: 10.3892/ijo.2015.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]