Abstract
For the past three decades, methods for culturing mouse embryos ex vivo have been optimized in order to improve embryo viability and physiology throughout critical stages of embryogenesis. Combining advances made in the production of transgenic animals and in the development of different varieties of fluorescent proteins (FPs), time-lapse imaging is becoming more and more popular in the analysis of dynamic events during mouse development. Targeting FPs to specific cell types or subcellular compartments has enabled researchers to study cell proliferation, apoptosis, migration, and changes in cell morphology in living mouse embryos in real time. Here we provide a guide for time-lapse imaging of early stages of mouse embryo development.
1. Introduction
During development, cell–cell interactions, divisions, and coordinated movements underlie the changes in shape known as morphogenesis. How cells and tissues form and rearrange during morphogenesis has fascinated scientists for generations, yet the ability to directly observe such profound and fundamental changes has only recently been attained. Before the 1950s (when video cameras were combined with microscopes), knowledge about morphogenetic events was stored only in the minds of the observers and could only be communicated in drawings (Inoue and Gliksman, 2003). In more recent times, video microscopy and continued advances in technology have provided new tools for cell and developmental biologists to discern closer and clearer views of morphogenesis in action. The advent of confocal and multiphoton microscopes in the 1980s and 1990s together with the development of fluorescent proteins (FPs) that can be used to vitally label cells of interest (also emerging in the 1990s) has transformed what we can directly visualize in a living embryo, and provides us with exciting new insights into both cellular and subcellular dynamics during development. Moreover, advances in digital media now make it possible not only to record the dynamic events of development but to publish and share movies with the world with the click of a button.
Despite advances in imaging technology, probe development, and digital media, there are still limitations imposed by the samples themselves as microscopic analysis of the embryo requires that the organism can be grown on a microscope stage. Thus, most imaging studies in vertebrate embryos have been carried out using zebrafish, avian (chick and quail), and Xenopus embryos which can be readily maintained in culture. Imaging mammalian model systems, such as mouse or rat embryos, has been significantly more difficult as uterine implantation, which supports growth and development of the embryo, does not permit direct visualization of embryonic development at all stages. To observe rodent embryos in utero, approaches such as MRI and ultrasound have been used (Dickinson, 2006). These methods have excellent depth penetration but with relatively poor resolution (50–200 μm) and relatively few specific labels exist to visualize cells or subcellular details of interest. Fluorescence microscopy methods offer cellular and subcellular resolution and a rapidly growing list of promoters and enhancers to label cells with FPs, but fluorescence microscopy cannot be performed through the thick and light scattering uterine wall. However, it is possible to maintain mouse and rat embryos in culture during pre- and early postimplantation stages, prior to forming a maternal–placental connection (Copp and Cockroft, 1990; Hsu, 1979; Jones et al., 2002; New and Cockroft, 1979; Tam, 1998; Wiley et al., 1978). We and others have adapted these protocols for static culture on the microscope stage to allow time-lapse imaging of mouse embryo development (Aulehla et al., 2008; Fraser et al., 2005; Kwon et al., 2008; Plusa et al., 2008; Srinivas et al., 2004). In this chapter, we review the methods that have been used successfully to enable direct imaging of mouse embryos. We have highlighted some of the key concerns and criteria for successful embryo maintenance in static culture on the microscope stage and have also included insight into imaging and image analysis to aid researchers in obtaining the most from live imaging experiments.
2. Overview of Key Parameters
For imaging mouse embryonic development, optimization of three important criteria have been shown to be essential—culture conditions, cell/tissue labeling, and imaging tools.
2.1. Culture conditions
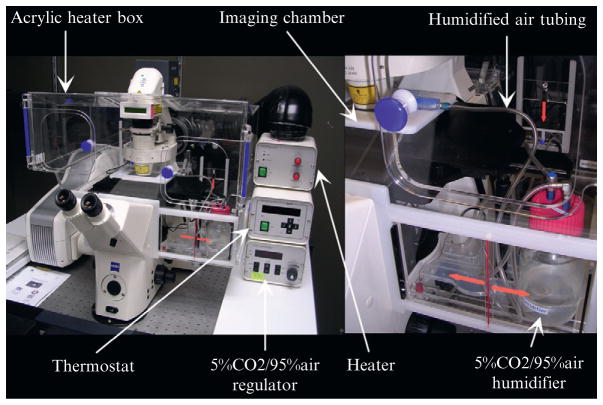
To grow embryos ex vivo in static culture, culture media must be carefully maintained at the appropriate temperature, pH, and humidity and must be supplied with the right balance of gasses. Carefully controlling these parameters allow mouse embryos to survive and develop comparably to embryos grown in utero. Mouse embryos can be cultured successfully during two different stages of development—preimplantation stages up until the early somite stages (Hsu, 1979; Sadler and New, 1981; Sherbahn et al., 1996; Tam, 1998) and prestreak stages up until embryonic day 10.5 (E10.5) (Hsu, 1979; Jones et al., 2002). The protocols discussed here have been optimized for E6.5–E10.5 embryos. Embryos at these stages are sensitive to changes in environmental conditions and should be imaged using an environmental stage unit which can be used to control these parameters such as the one shown in Fig. 19.1. Detailed culture conditions are given below.
Figure 19.1.
Zeiss LSM 5 LIVE confocal laser scanning microscope setup. The microscope is equipped with an acrylic heater box which surrounds the microscope stage. Directly on the microscope stage is the imaging chamber which consists of a lower platform in which the culture dish rests on, and a top platform that encloses the imaging chamber and has a clear acrylic portion to allow light to pass through for bright-field imaging. The lower platform has an open circular area at the bottom that allows the objective to collect light. Two thermosensors are placed in the imaging chamber and within the heater box to control the thermostat. The thermostat regulates the temperature (37°C) by controlling the heater, which is attached to the back of the heater box. For humidified air, 5% CO2/95% air flows through the regulator and humidifier, and it is sent through tubing to the imaging chamber.
2.2. Cell/tissue labeling
Over the past 10 years, there have been tremendous advances in vital cell labeling with improvements both in dyes as well as the development of bright and stable FPs. While much can be said about fluorescent labels and further details are provided below, the best labels are bright and nontoxic. Bright fluorescent molecules or proteins are generally those that have both a high extinction coefficient and a high quantum yield. This means that the label readily absorbs photons and emits photons as the excited electron relaxes back to the ground state. Fluorochromes with a poor quantum yield exhibit nonradiative decay which can result in local heating or increases in free radicals. Also, dim fluorochromes require higher power illumination which can damage the tissue directly. The fluorescent-labeling strategy should be optimized as much as possible to reduce excess illumination. The choice of fluorescent label will also depend somewhat on the imaging system being used, so take care to match these choices with the illumination wavelengths and emission filters that are available (see below).
2.3. Imaging tools
The protocols that have been established to grow mouse embryos on the microscope stage can be used in conjunction with a number of imaging strategies or microscopies (for instance, using optical coherence tomography; Larina et al., 2008, 2009), but fluorescence microscopy offers the most utility for imaging cellular and subcellular events with the widest range of available cell labels. Both wide-field fluorescence microscopy and confocal laser scanning microscopy (CLSM) have been used for time-lapse microscopy of developing mouse embryos, but for many applications confocal microscopy can produce considerably better images with enhanced resolution and signal-to-noise over wide-field fluorescence approaches. Most modern confocal microscopes have time-lapse imaging capabilities built into their acquisition software for automated collection and offer considerable flexibility in scan strategies to enable efficient and sensitive collection of fluorescence signal while minimizing damage. Movies generated from these experiments can be further processed with image analysis tools to quantify changes in cell proliferation, apoptosis, morphology, and migration.
There are many examples in which live imaging of cultured mouse embryos or embryonic tissue explants, in combination with transgenic reporters, has revealed insights into the mechanisms behind various cellular and developmental events. For example, by labeling precursors to specific cell types in early development, a more accurate understanding of the specific morphogenetic events driving germ layer formation has been determined (Burtscher and Lickert, 2009; Kwon et al., 2008; Plusa et al., 2008; Srinivas et al., 2004). Imaging has revealed new information about notochord formation (Yamanaka et al., 2007), peripheral nerve outgrowth (Brachmann et al., 2007), neurogenesis from basal neuroepithelium (Haubensak et al., 2004), ureteric bud branching (Srinivas et al., 1999; Watanabe and Costantini, 2004), and FP fusions have provided insights into specific cellular events such as cell migration and motion (Anderson et al., 2000; Druckenbrod and Epstein, 2005; Jones et al., 2002, 2004; Molyneaux et al., 2001; Young et al., 2004), cell mitosis and G1 to S-phase transitions (Fraser et al., 2005; Sakaue-Sawano et al., 2008), nodal cilia dynamics (Nonaka et al., 1998; Okada et al., 1999), and cell death in primitive endoderm-fated cells (Hadjantonakis and Papaioannou, 2004; Plusa et al., 2008).
This chapter details an approach for live imaging the mouse embryonic yolk sac. However, similar culturing and imaging methods can be adapted to study other embryonic structures. Here, we will discuss culturing conditions, dissection strategies, use of FP transgenics, confocal fluorescence microscopy, time-lapse imaging, and image analyses.
3. Whole Embryo Culture
3.1. Whole embryo culture staging
Although there are well-described roller bottle protocols for culturing postimplantation mouse embryos (Downs and Gardner, 1995; Lawson et al., 1986; New and Cockroft, 1979; Sadler and New, 1981), static culture is required for microscopy. For early postimplantation stage embryos (starting at E6.5) grown in static culture, cultures can begin between early streak formation to E9.5 ( Jones et al., 2002; Nagy, 2003; Tam, 1998). They can be grown for 18–24 h until E9.5, and with limited success to E10.5 ( Jones et al., 2002). By E10.5 there is limited diffusion through thicker, more complex embryos and the lack of placental support prevents extensive normal development. For imaging dynamic events in later stage embryos, explant culture has been used successfully to image salivary glands, lungs, kidneys, ovaries, testes, and heart (epicardium) (Coveney et al., 2008; Nel-Themaat et al., 2009; Rhee et al., 2009; Sakai and Onodera, 2008; Watanabe and Costantini, 2004) but with the caveat that the tissues are not perfused by blood flow as in live, intact embryos.
To verify success of the static cultures, developmental timing can be compared to embryos grown in vivo at similar time points by assessing morphological changes. For instance, for embryo cultures starting at E8.5, the formation of the neural plate, appearance of the first somite, formation of a linear heart tube, appearance of the first heart beat, commencement of head fold closure, initiation of heart looping, fusion of head folds, and the onset and completion of axial rotation or turning are the various developmental changes that can be observed ( Jones et al., 2002). In embryo cultures starting at E9.5, changes in embryo size and maturation of head features can be compared. In addition, heart rate and the rate of somite formation can be used as metrics of continuing development ( Jones et al., 2002; Nagy, 2003) and more sophisticated approaches to measure blood flow ( Jones et al., 2004) can provide detailed information about cardiovascular physiology.
3.2. Culture conditions
For whole embryo culture of postimplantation embryos, two types of media are prepared fresh on the day of use in a sterile tissue culture hood (see Section 3.4): dissection media and culture media. Dissection media is comprised of DMEM/F12 (Invitrogen, Cat. # 11330) which contains a mixture of salts, buffers, amino acids, nutrients, vitamins, and pH indicator. To prepare dissection media, 90% (v/v) DMEM/F12 is supplemented with 10% (v/v) fetal bovine serum (Gibco) and penicillin (1 unit/ml)/streptomycin (1 μg/ml) antibiotics. For culturing embryos at E7.5 and beyond, culture media is comprised of a 1:1 ratio of DMEM/F12 to homemade rat serum and with penicillin (1 unit/ml)/streptomycin (1 μg/ml) antibiotics (see Section 3.3); however, we have also had success in using a 6:1 ratio of dissection media to homemade rat serum. It is important to note that the homemade rat serum (on the day of use) should be incubated for a minimum of 1 h at 37°C (5% CO2/95% air, for E7.5–E9.5 cultures) with an open cap in a sterile 50 ml Falcon® tube (BD Biosciences), to allow for excess ether to evaporate (too much ether can reduce cardiac contractility and blood flow). After incubating the rat serum, the culture media is prepared and placed through a 0.45 μm syringe filter (Nalgene®) for sterilization. Both dissection media and culture media are warmed to 37°C prior to use.
An important parameter for a successful whole embryo culture is maintenance of the pH of the media. For bicarbonate or HEPES-based buffers, exposure to 5% CO2 can maintain the pH of the media to ~7.2, the appropriate physiological pH. Culture media should be exposed to CO2 by placing the media in a gas incubator (which is fed 5% CO2/95% air) in a 50 ml Falcon® tube with the cap partially unscrewed. If pH levels are not sufficiently maintained, extra HEPES buffer can be added to the culture media to bring the final concentration of HEPES from 7.5 to 15 mM.
Evaporation can also affect the overall health of the embryo. In postimplantation embryos with an intact yolk sac, the yolk sac becomes wrinkled upon excessive evaporation which can subsequently impede blood flow throughout the yolk sac and embryo ( Jones et al., 2002). Thus, media must be kept in a humidified environment. In static culture, embryos can be kept in a gas incubator with humidifier pan or when being imaged they can be kept in an imaging chamber that is fed 5% CO2/95% air at a low flow rate sent through a bubbler to keep the air humidified. Evaporation rates are dependent upon the percent humidity present in the laboratory environment. Some regions of the country are drier than others. In these cases, extra measures can also be taken to prevent excess evaporation including the use of a layer of sterile mineral oil on top of the culture media, silicon grease to seal the edges of the culture dish, or the addition of Teflon® tape to seal the edges of the imaging chamber ( Jones et al., 2002).
Temperature must also be maintained at 37°C. Thus, the incubator can be kept at this temperature and the confocal microscope stage must have an enclosure that maintains this temperature. Keeping both the temperature of the stage and the objective can help to prevent drift caused by changes in the glass so an environmental chamber that encases the stage and nosepiece is recommended. Many microscope systems offer environmental control chambers (such as the Zeiss XL systems) that are custom fitted to that microscope base and that surrounds a portion of the microscope (Fig. 19.1). The box has a thermostat that can detect the temperature both within the box and the imaging chamber, and the amount of heat is adjusted to maintain the temperature. Alternatively, a homemade box can be made out of cardboard, insulating material, a heater, and a temperature regulated power outlet ( Jones et al., 2002, 2005a). Embryos can be very sensitive to fluctuations in temperature which can sometimes affect the timing of development and rate of cardiac contraction; thus, appropriate temperature regulation is critical (Nishii and Shibata, 2006).
For culturing late-stage embryos (E9.5–E11.5), growth of the embryo can limit O2 diffusion rates. Thus, the concentration of O2 should be adjusted depending on the developmental stage at the beginning of a culture. For example, cultures starting at E9.5 should be cultured in 20% O2; whereas cultures starting at E10.5 should be cultured in 95% O2 (Nagy, 2003).
Overall, dissection conditions should be as sterile as possible. Media is prepared using sterile containers and pipette tips in a sterile tissue culture hood. Dissection tools are cleaned with distilled water and ethanol. The tools should never be placed in contact with detergents or fixative, as this can adversely affect the health of the embryos.
3.3. Preparation of rat serum
For our culturing purposes, we have utilized many different types of commercially available rat serum from several different companies, but this was met with limited success, as normal cardiovascular physiology was impaired ( Jones et al., 2002). Thus, homemade rat serum is prepared essentially according to previously established protocols (Fraser et al., 2005; Hogan, 1994; Jones et al., 2005a,b). This protocol requires two people to perform. To collect rat serum, adult male Sprague-Dawley rats at around 12 weeks of age weighing approximately 300 g (Charles River) are anesthetized (as approved by animal protocols) by exposure to ether. Once rats are unresponsive, they are laid down in a supine position, and the Rat’s nose and mouth is placed inside a 50-ml Falcon® tube containing an ether-soaked paper towel to maintain anesthetization. The abdomen is prepared by spraying with 70% ethanol and a V-shaped incision is made in the peritoneum to open up the abdominal cavity. The internal organs are moved aside and excess fascia is wiped away with a Kimwipe®, leaving the dorsal aorta exposed. A beveled butterfly needle (BD Biosciences) is inserted into the dorsal aorta. Directly after insertion (when blood is present in the needle), the opposite end of the needle is inserted into a Vacutainer® blood collection tube with anticoagulants (BD Biosciences). When blood flows into the collection tube, the tube is inverted to mix the anticoagulants with the blood. The collection tube is then placed on ice, and rats are euthanized as per animal protocols (decapitation using a guillotine). Carcasses are placed in a fume hood for several hours or overnight to allow ether to evaporate before disposing and should be stored in an explosion proof freezer prior to disposal. Note: working with ether is dangerous and all safe practices should be observed according to the regulations of your home institution.
Once blood collection is complete, the blood is centrifuged in the collection tubes at 1300×g for 20 min to separate the blood cells from the serum. After centrifugation, the sera (top layer) quality is evaluated. Low-quality sera with a pinkish hue, as compared to a high-quality and less-turbid/clearer sera, is discarded because the pinkish color represents components from lysed red blood cells which impede embryo growth when present in media. Only high-quality sera is collected and transferred to a 15ml Falcon® tube and centrifuged again at 1300×g for 10 min. The serum is pooled into a 50 ml Falcon® tube and is heat inactivated by placing in a 56°C water bath for 30 min in a tissue culture hood with the lid partially unscrewed to allow the ether to evaporate. After incubation, the sera is sterile filtered through a 0.45 μm syringe filter (Nalgene®), and in a sterile hood dispensed into microcentrifuge tubes which then are stored at −80°C for up to 1 year. Typically, we collect sera from about 30 rats, which produce about 100 ml of rat serum.
3.4. Dissection and isolation of postimplantation mouse embryos
To isolate E8.5 embryos, timed matings are performed. The presence of a vaginal plug in the morning after mating signifies a potential pregnancy. The resulting embryos are E0.5 on the afternoon of the vaginal plug. Eight days later, embryos are harvested from them others. Dissections are performed on a dissection stereomicroscope surrounded by a homemade heater box regulated to 37°C to avoid interruptions in cardiac activity. The heater box is made from insulated cardboard and is heated by a space heater connected to a thermostat (Fisher Biosciences) ( Jones et al., 2005a,b). Embryos are isolated by humanely sacrificing mothers (using euthanization procedures approved by animal protocols). A V-shaped incision is made in the peritoneum, starting from the posterior and working anteriorly, to expose the abdominal cavity. The uterus is removed by cutting the uterus at the uterotubal/ovary junction for each horn of the uterus. The uterus is subsequently cut at the cervix and it is then placed in dissection media in a 35 mm culture dish, and further dissected in the heated box. A sterile transfer pipette is used to wash away some of the blood with dissection media in order to more clearly see the uterus. Using dissection scissors, incisions are made perpendicular to the uterus to separate each individual embryo still surrounded by the uterus (Fig. 19.2). The embryos with the uterus are then transferred (by forceps or transfer pipette) to a new 35 mm culture dish with fresh dissection media. To prevent excess nutrient expenditure, it is critical that each embryo has at least 1 ml of dissection or culture media. Typically, we place three embryos per culture dish in about 3–4 ml of media. Embryos that are not immediately being dissected are kept in dissection media and moved to the incubator until ready to dissect. Using two pairs of forceps, remove the rest of the uterus by cutting away the uterine tissue that surrounds the decidua. The embryo proper is surrounded by visceral yolk sac, parietal yolk sac, and the decidua. The decidua is shaped with the distal region being more pointed and the proximal region being wider at the ectoplacental cone (Fig. 19.2). At the base of the ectoplacental cone, carefully use both forceps to remove the decidua by cutting around this base, and slightly pulling off the decidua proximally. Then, remove the parietal endoderm cutting in the same manner. Finally, remove the clear and thin membrane, Reichert’s membrane, from the yolk sac to allow for better nutrient or media exchange. For future immobilization of an intact embryo during an imaging session, keep a portion of the ectoplacental cone attached to the embryo (see Section 5.2 and Fig. 19.2). Embryos that are freshly dissected are transferred to fresh dissection media using a sterile transfer pipette cut off at the base, and then moved to the incubator. After all embryos are dissected, healthy embryos with strong heart beats are chosen and transferred to a 35 mm culture dish with a 10 mm glass bottom microwell (Mat-Tek®) containing 3 ml of culture media, or they can be transferred to Lab-Tek™ culture chambers (Lab-Tek™ II chambered coverglass) with 2 ml per chamber. A maximum of three embryos per dish/chamber can be cultured. Embryos are then allowed to recover in a 37°C tissue culture incubator for 15–30 min before they are imaged.
Figure 19.2.
Example for dissecting E8.25–E8.75 embryos for static culture (bottom right embryo ~16 somites). Uteri are removed and placed in dissection medium in a culture dish. The embryos are dissected out by first cutting perpendicular to the uterus between implanted embryos using dissection scissors. Using forceps, the uterus is removed from the decidua, and the distal portion of the uterus is removed by holding one portion of the decidua with one pair of forceps, and gently pulling of the other part of the decidua away from the embryo with another pair of forceps. This is done around the embryo, and what remains is the embryo attached to the ectoplacental cone. The parietal endoderm and Reichert’s membrane are removed, and a portion of the ectoplacental cone can be pared down and placed adjacent to the sticky sides of the Mat-Tek® culture dish glass bottom microwell to prevent drift.
4. Labeling Cells of Interest
As mentioned above, there are two categories of fluorescent labels that are used in embryos for time-lapse analysis, dyes and FPs. Molecular Probes (Invitrogen) is an excellent source of dyes and dye conjugates that can be used to label tissues within embryos. Some examples of these probes that have been used for vital imaging are fluorescent dextrans for labeling blood flow or iontophoretic injection into single cells for lineage tracing, lipophilic dyes (i.e., DiI and DiO) for labeling clusters of cells for tracking migration and movement or axonal connections, Cell Tracker dyes which are taken up into the cytoplasm marking cells of interest, BODIPY-ceramide which labels cell membranes and can be used to outline tissue structures and organization, and SYTO dyes (cell-permeant cyanine nucleic acid stains) which label nuclei in order to follow mitosis, apoptosis, or single cell migration. In cases where the investigator wishes to label a cluster of cells within a tissue, vital lipophilic fluorescent dyes such as carbocyanine lipophilic dye (DiI/DiO) can be used. While application of exogenous dyes can be very effective and convenient, these need to be injected or applied, requiring additional manipulation of the embryo which can lead to impaired viability or abnormal development and suffer from the drawback of having limited specific control over the cells that are labeled.
Genetically encoded markers such as FPs provide bright, stable markers and have a number of advantages over traditional chemical dyes. FPs can be introduced as stable elements in the genome either via viruses or by the production of transgenic mice. In transgenic mice, FPs can be driven by particular promoter sequences that have either part of a gene expression construct introduced via pronuclear injection or by “knocking in” the FP into a specific loci (Gordon and Ruddle, 1981; Hadjantonakis et al., 2003; Megason et al., 2006). While the knock-in approach has the advantage that specific promoter elements need not be defined, only a single FP gene per locus is expressed which can result in weak expression and low fluorescence signal unless the targeted gene is expressed at high levels. This is less of a problem with transgenes introduced via pronuclear injection since multiple copies of the transgene integrate as an array into the genome. The mutation and optimization of FPs has now resulted in dozens of available colors with many bright, stable choices for live cell imaging (Davidson and Campbell, 2009; Shaner et al., 2005). Currently, there are 29 common FPs that are excitable and can emit light in the visible spectrum, where at least three spectra can be easily separated with standard filter sets and laser sources (Nowotschin et al., 2009).
FPs also offer the advantage of being able to direct the fluorescence to subcellular compartments such as the cell membrane, cytoplasm, and nucleus; thus permitting direct visualization of membrane dynamics and cell migration, cell division, or changes in cell shape (Hadjantonakis et al., 2003; Nowotschin and Hadjantonakis, 2009a; Passamaneck et al., 2006; Rizzo et al., 2009). For example, myristoylation (myr) or glycosylphosphotidylinosital (gpi) FP fusions can be used to localize FPs at the cell membrane (Hadjantonakis et al., 2003; Nowotschin and Hadjantonakis, 2009b). Histone H2B::FP fusions can target FPs to the cell nucleus. Lack of these tags causes FPs to distribute throughout the cell including the cytoplasm and the nucleus. The use of these tags not only provides information about overall morphology of subcellular, cellular, and tissue structures, but it also allows researchers to visualize many dynamic processes such as cell membrane dynamics, mitosis, apoptosis, migration, nuclear import/export, and vesicular trafficking. In addition, concentrating FPs within specific domains within the cell can produce a brighter signal, allowing for less illumination light to be used to image fluorescence. There is a great deal of interest in both continuing to improve the properties of FPs as well as to adapt FPs for new purposes, as in the development of photactivateable/photoconvertible FPs and sensors for intracellular signaling pathways as well as in facilitating very high resolution microscopy (Davidson and Campbell, 2009; Nowotschin and Hadjantonakis, 2009b). The combination of different cell-specific promoters driving expression of FPs proves to be very popular markers for live cell imaging in many systems, especially the mouse.
5. Time-Lapse Imaging of Early Mouse Embryos
5.1. Confocal fluorescence microscopy setup
As discussed above, CLSM is a popular tool for in vivo time-lapse imaging of embryos. However, since these systems can use fairly powerful lasers, care must be taken to limit the exposure of embryos to excessive laser illumination which can cause cell damage and cell death. The amount of light exposure can be reduced by altering several aspects: scanning time intervals, transmission of the laser to the sample, optimizing the balance of illumination versus efficiency of detection specific filters, use of objectives or zooming features, and the number of optical slices or images acquired. Here are some key issues to remember when optimizing these conditions.
5.1.1. Start by knowing the excitation and emission spectrum of your FP or dye
The wavelength of the excitation laser should be as close to the excitation peak as possible and the emission filters that you choose should encompass as much of the emission spectra as possible. For instance, a long-pass 505 filter will allow you to collect more emission signal than a 505–530 band-pass filter which means you will be able to use a lower laser power setting and expose the specimen to less light for the same amount of emission signal. Thus, using single fluorochromes or well-separated pairs is advantageous over labels that have close or overlapping spectra. Narrower emission filters wastes valuable photons since they are blocked from the detector. To avoid using narrow band-pass filters, another option is to collect different color signals sequentially to avoid overlap. For instance, excite the green dye and collect the signal with a long-pass or broad band-pass filter, then excite the red dye in the second image and collect with a long-pass filter. In many cases this will produce the same amount of emission signal for less overall laser power.
5.1.2. Maximize the gain
Make sure when you are adjusting the power of the laser that you do this with the gain set to the maximum. Many people make the mistake of optimizing the gain and leaving the laser power at some default level. Set the laser at zero and the gain at its maximum, then bring up the laser power in small increments until you get the signal level that you need. If there is still considerable noise from the detector, lower the gain gradually, paying close attention to how much laser power needs to be increased to compensate.
5.1.3. Embrace the ugly
The settings that are the safest and most informative for your imaging studies may not always yield cover photo ready images but may provide plenty of information. Averaging or slow scanning can both reduce noise and improve the signal-to-noise ratio, but if viability is a concern, consider scanning faster without averaging. Faster scan times not only enable faster processes to be tracked but also minimizes laser exposure to the sample resulting in less damage. However, these benefits come with increased noise. Longer pixel dwell times and averaging reduce noise, but require longer or repeated laser exposure and can damage tissue, so beauty has its price. Consider aiming to make the images pretty enough for what you need to answer your question of interest.
5.1.4. Open the pinhole
For many live imaging experiments using thick samples, Z-stacks are necessary to follow cells migrating along the Z-axis or to image other changes in 3-D. That said, it is not always the case that thin optical sections are needed. Unless the highest possible resolution is important to the experiment (such as in imaging submicron subcellular domains), the pinhole can be opened up to allow for thicker optical sections and more signal to reach the detector and this may be sufficient to follow cell translocations, mitosis, etc. Also, opening the pinhole even a little will reduce the number of optical sections that will need to be acquired in the Z-axis which will reduce the amount of scans used to generate 3-D datasets and reduce the chance of damage to the embryo. Many confocal microscopes provide an estimation of the size of the optical slice thickness and this will change from lens to lens so think about how much resolution you really need. Moreover, if resolution is very important, you may also want to consider using deconvolution after you have acquired the data to make further improvements without endangering the sample. Take some stacks through a sample and try out different pinhole settings to evaluate these settings for your particular question.
5.1.5. Choose the best objective
Although most people think about the magnification needed to image a particular sample, the numerical aperture (NA) is actually most important. This number relates to the axial and lateral resolution that can be achieved as well as the amount of light that can be collected. Higher values are more desirable but higher NA lenses usually also have a shorter working distance (w.d.) which may limit the depth along the Z-axis that can be imaged. Also, if fluorescence imaging is being used, choose a lens that is optimized for fluorescence transmission, not for DIC or phase contrast (PH). Lenses that have DIC or PH on the lens itself are designed specifically for these types of contrast and are optimized for aberration correction and preservation of polarization but at the expense of fluorescence transmission. These lenses can contain more glass elements which limit transmission. Before you start, you should ask your local microscope representative or core director about the best lenses available for live imaging. Many manufacturers now offer lenses that are optimized for live imaging with improved transmission, longer working distances for imaging thicker specimens, temperature stability for use at 37°C and for immersion into biological media. The right lens can make all the difference, so compare those that are available to you to find the best lens for your sample.
5.2. Transferring embryos to the microscope stage
Embryos are dissected following appropriate procedures (see Section 3.4). For imaging, embryos are placed in culture media in either Mat-Tek® culture dishes or Lab-Tek™ culture chambers. After allowing the embryos to recover in the incubator for a minimum of 15 min, they are quickly removed and placed in the imaging chamber. It is important that the imaging chamber be equilibrated to the appropriate temperature, gaseous phase and humidity for at least 1 h prior to imaging. To prevent drift of the embryos on the XY and Z positions, embryos with an intact yolk sac can be stabilized by placing the ectoplacental cone adjacent to the sticky edge of the glass microwell. If using the Lab-Tek™ culture chambers, a human hair can be tied around the ectoplacental cone in a manner which can prop up the embryo in place at the bottom of the dish ( Jones et al., 2005a). Alternatively, embryos can also be positioned using a holding pipette attached to a micromanipulator. In some cases, drift can still occur. This drift can be in both the XY plane (often caused by microcurrents in the media) and in the Z plane (often caused by the expansion of the yolk sac or growth of the embryo that occurs during the culturing period). To adjust for this drift, periodic evaluation of the culture should be performed, and appropriate readjustments of the XY and Z planes should be made if necessary. Time-lapse images that are readjusted in the middle of an imaging session can be realigned by adjusting for drift computationally using the Imaris software program (see Section 6).
The appropriate amount of light, magnification, Z-stack number and scanning intervals are empirically chosen as to not disrupt tissue viability. To assess tissue viability, there are three major parameters that should be performed to assess health of the embryo being imaged. First, cells labeled with a nuclear-localized FP can be used to assess whether abnormal apoptosis occurs by visualizing nuclear fragmentation. For example, time-lapse imaging of Flk1-H2B::eYFP labeled vessels can clearly show nuclear fragmentation events which can take anywhere between 0.5 and 1.5 h to observe before the fragments are cleared (Fig. 19.3). The duration of this process ensures that all apoptotic events can be captured by the 5–10 min scan time intervals. Cell death can also be assessed in nonlabeled embryos after the imaging session to assess viability. Second, normal cardiovascular physiology can be assessed by performing periodic evaluations of blood flow (visual inspection of blood flow through the eye piece or by performing bright-field imaging to detect streaks of dark cells within vessels that represent moving blood cells) during the imaging session. Also, normal yolk sac vessel remodeling in embryos cultured from E8.5–E9.5 is a good sign of normal cardiovascular physiology. Lastly, cultured embryos should form similar morphological structures, at similar time points, to embryos grown in utero as shown in ( Jones et al., 2002). These structures can thus be used as hallmarks to compare health of the embryo being imaged.
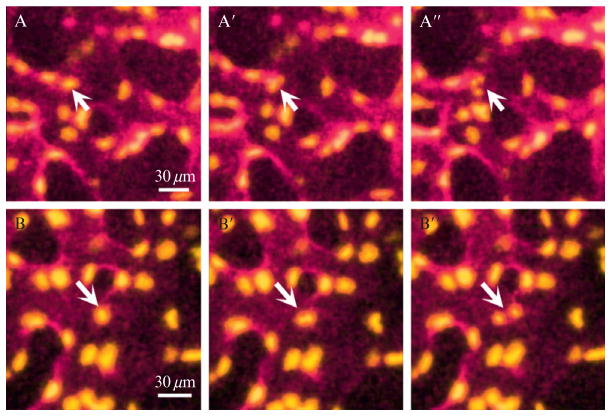
Figure 19.3.
Time-lapse movie of the developing Yolk sac vessels (cultures starting at E8.5). Yolk sac vessels of intact Flk1-myr::mCherrytg/tg (to visualize endothelial cell membrane); Flk1-H2B::eYFPtg/tg (to visualize endothelial cell nuclei) embryos are imaged on the LSM 5 LIVE confocal with a 25× objective (NA of 0.45). Images are taken every 6 min at three Z-planes (~30 μM) each with the 488 nm (0.3% power) and 561 nm (2.0% power) lasers to image eYFP and mCherry, respectively. Endothelial cell apoptosis (panels A-A″) and mitosis (panels B-B″) can be captured in movies by observing fragmentation of nuclei and division of a single nucleus into two nuclei, respectively. Beginning of chromatin condensation is observed in A′ (6 minutes after A), and nuclear fragmentation is observed in A″ (1 hour after A). Beginning of mitosis is observed in B′ (12 minutes after B) and formation of two separate nuclei is apparent in B″ (18 minutes after B).
6. Image Analysis
There are many different parameters that can be analyzed for different types of morphogenetic events. The rate of mitosis (Fig. 19.3), apoptosis, cell/tissue morphology changes, and cell migration rate/directionality of movement are among the parameters that can be quantified by computational means. Tools for image analysis are available from several sources. Many convenient tools can be found within the confocal software. For instance, in the Zeiss LSM software there are many tools such as those for adding scale bars and time stamps, creating 3-D or 4-D reconstructions, analyzing signal colocalization, applying look up tables, cropping an image series, joining different image series and measuring changes in signal intensity. In addition to the LSM software, the Bitplane Imaris software has additional tools for image analysis and is used routinely by our lab for tracking cells, 3-D reconstructions and image postprocessing for denoising and drift. Other tools are also available from MatLab, Volocity, Slidebook, and other programs as well as for free in ImageJ. In this section, we will highlight some of the routine methods that we use for image analysis and data analysis but note that there are many available tools for many applications. Talk to your local microscope experts or company representatives if you are looking for a particular image analysis tool.
6.1. Image processing for a typical 3-D, time-lapse sequence
To begin processing the time-lapse images, entire movies are first reconstructed by performing an extended-depth view (projection) in the Zeiss LSM software. This projects the 3-D data from optical slices into a single image, so the resulting movie will be a simple time sequence. Sequential movies are combined together by concatenating them in order using the concatenation macro in the macros menu of the Zeiss LSM software. Image contrast and brightness can be adjusted using this software; however, it is recommended to do this in the Imaris software program where there are more tools for denoising. To use Imaris, we input the LSM-derived concatenated image sequences into Imaris, and open the image as a Surpass scene using the volume function. Imaris has a plug-in to recognize LSM images from Zeiss confocals to make this relatively easy. If needed, the image sequence is further processed by correcting the levels of each individual color (e.g., eYFP and mCherry). If the signal-to-noise ratio is low, Gaussian filters are then applied to denoise the images. Care is always taken in postprocessing to be sure that important features of the images are preserved and additional features are not created. Files are ultimately saved as Audio Video Interleaved (AVI) movies, and playback time is adjusted depending on the speed of morphogenesis and on the particular events to be displayed.
6.2. Adjusting for specimen drift using Imaris
Specimen drift is common in time-lapse imaging sessions. In order to correct for drift in the XY plane using Imaris, the user assigns several reference points on the specimen that can be followed in each image over time. The reference points represent a center of mass which the software will use to stabilize the structure. In our samples containing Flk1-H2B:: eYFP labeled nuclei, we use this signal as our reference spots but other markers or regions of reproducible contrast can be used. Nuclei are identified by selecting “objects” and “adding new spots.” The appropriate channel (YFP) is selected, and the diameter of the spots to be detected is set (~10 μm for endothelial cell nuclei). To account for most nuclei, the threshold is appropriately set until all or most nuclei are detected and this is confirmed by visual inspection from the operator. The next step is to track spots by selecting the Tracking folder and choosing the “Autoregressive motion gap close 3 function.” The motion of each spot is then tracked throughout the movie, and information is gathered in the spot track group 1 folder. The last step is to depress the “correct for drift” icon in the Tracks folder (make sure the spot track group 1 folder is selected). This produces an image series that has been corrected for drift. Regions that are not common to all the corrected images are cropped so there is some sacrifice of the field of view encountered by drift correction. Also, structures that leave the plane of the imaging view during the middle of the time-lapse movie cannot be compared and are thus subsequently cropped off.
6.3. Tracking cell migration using Imaris
Another benefit of using nuclear markers is to analyze cell migration. The ability to easily identify each nucleus of an individual cell makes it possible to easily track the movement of cells with these markers. In instances where labeled cells invade a nonlabeled region, cytoplasmic or membrane-localized markers can be easily distinguished. However, in similarly labeled tissues, uniform expression of a cytoplasmic- or membrane-localized FP makes it difficult to distinguish between neighboring cells. Thus, nuclear-localized FPs can be used to define the nucleus of each cell in order to discern the migratory abilities of individual cells in similarly labeled tissues.
There are several parameters that can be quantified from a migrating cell. For instance, the overall speed, distance traveled, displacement, and directionality of cell migration can be determined. In the Imaris software program, each individual cell can be traced by tracking spots (as shown above). Spots are tracked by Imaris by plotting the location of a nucleus at a particular XY coordinate from one time point to the next. The program can take the sum of the distance traveled at all time points (total distance traveled), can compare the distance traveled at the beginning to the end of the time-lapse session (distance displaced), and determine the average speed of migration. Tracking information can be displayed in a Microsoft® Excel spreadsheet by depressing the “tracking folder” and then “statistics” tabs. After collecting the data, results can be averaged to determine the overall affect of migration in the sample of interest.
7. Summary
Here we have provided some details and examples into the procedures used by our lab to study cell dynamics in early mouse embryos using time-lapse confocal microscopy to guide new scientists toward the use of imaging to study mouse embryogenesis. We are grateful to all the investigators that have established protocols that we have modified for imaging strategies in our lab and we encourage more people to adapt and extend these protocols to generate new approaches to image developmental events.
References
- Anderson R, Copeland TK, Scholer H, Heasman J, Wylie C. The onset of germ cell migration in the mouse embryo. Mech Dev. 2000;91:61–68. doi: 10.1016/s0925-4773(99)00271-3. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Wiegraebe W, Baubet V, Wahl MB, Deng C, Taketo M, Lewandoski M, Pourquie O. A beta-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat Cell Biol. 2008;10:186–193. doi: 10.1038/ncb1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann I, Jakubick VC, Shaked M, Unsicker K, Tucker KL. A simple slice culture system for the imaging of nerve development in embryonic mouse. Dev Dyn. 2007;236:3514–3523. doi: 10.1002/dvdy.21386. [DOI] [PubMed] [Google Scholar]
- Burtscher I, Lickert H. Foxa2 regulates polarity and epithelialization in the endoderm germ layer of the mouse embryo. Development. 2009;136:1029–1038. doi: 10.1242/dev.028415. [DOI] [PubMed] [Google Scholar]
- Copp AJ, Cockroft DL. Postimplantation Mammalian Embryos: A Practical Approach. IRL Press; Oxford, UK; New York, USA: 1990. [Google Scholar]
- Coveney D, Cool J, Oliver T, Capel B. Four-dimensional analysis of vascularization during primary development of an organ, the gonad. Proc Natl Acad Sci USA. 2008;105:7212–7217. doi: 10.1073/pnas.0707674105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson MW, Campbell RE. Engineered fluorescent proteins: Innovations and applications. Nat Methods. 2009;6:713–717. doi: 10.1038/nmeth1009-713. [DOI] [PubMed] [Google Scholar]
- Dickinson ME. Multimodal imaging of mouse development: Tools for the postgenomic era. Dev Dyn. 2006;235:2386–2400. doi: 10.1002/dvdy.20889. [DOI] [PubMed] [Google Scholar]
- Downs KM, Gardner RL. An investigation into early placental ontogeny: Allantoic attachment to the chorion is selective and developmentally regulated. Development. 1995;121:407–416. doi: 10.1242/dev.121.2.407. [DOI] [PubMed] [Google Scholar]
- Druckenbrod NR, Epstein ML. The pattern of neural crest advance in the cecum and colon. Dev Biol. 2005;287:125–133. doi: 10.1016/j.ydbio.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Fraser ST, Hadjantonakis AK, Sahr KE, Willey S, Kelly OG, Jones EA, Dickinson ME, Baron MH. Using a histone yellow fluorescent protein fusion for tagging and tracking endothelial cells in ES cells and mice. Genesis. 2005;42:162–171. doi: 10.1002/gene.20139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JW, Ruddle FH. Integration and stable germ line transmission of genes injected into mouse pronuclei. Science. 1981;214:1244–1246. doi: 10.1126/science.6272397. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis AK, Papaioannou VE. Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol. 2004;4:33. doi: 10.1186/1472-6750-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis AK, Dickinson ME, Fraser SE, Papaioannou VE. Technicolour transgenics: Imaging tools for functional genomics in the mouse. Nat Rev Genet. 2003;4:613–625. doi: 10.1038/nrg1126. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: A major site of neurogenesis. Proc Natl Acad Sci USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. [Google Scholar]
- Hsu YC. In vitro development of individually cultured whole mouse embryos from blastocyst to early somite stage. Dev Biol. 1979;68:453–461. doi: 10.1016/0012-1606(79)90217-3. [DOI] [PubMed] [Google Scholar]
- Inoue T, Gliksman N. Techniques for optimizing microscopy and analysis through digital image processing. Methods Cell Biol. 2003;72:243–270. doi: 10.1016/s0091-679x(03)72012-3. [DOI] [PubMed] [Google Scholar]
- Jones EA, Crotty D, Kulesa PM, Waters CW, Baron MH, Fraser SE, Dickinson ME. Dynamic in vivo imaging of postimplantation mammalian embryos using whole embryo culture. Genesis. 2002;34:228–235. doi: 10.1002/gene.10162. [DOI] [PubMed] [Google Scholar]
- Jones EA, Baron MH, Fraser SE, Dickinson ME. Measuring hemodynamic changes during mammalian development. Am J Physiol Heart Circ Physiol. 2004;287:H1561–H1569. doi: 10.1152/ajpheart.00081.2004. [DOI] [PubMed] [Google Scholar]
- Jones EA, Baron MH, Fraser SE, Dickinson ME. Dynamic in vivo imaging of mammalian hematovascular development using whole embryo culture. Methods Mol Med. 2005a;105:381–394. doi: 10.1385/1-59259-826-9:381. [DOI] [PubMed] [Google Scholar]
- Jones EA, Hadjantonakis AK, Dickinson ME. Imaging in Neuroscience and Development: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2005b. Imaging mouse embryonic development. [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis AK. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev Cell. 2008;15:509–520. doi: 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larina IV, Sudheendran N, Ghosn M, Jiang J, Cable A, Larin KV, Dickinson ME. Live imaging of blood flow in mammalian embryos using Doppler swept-source optical coherence tomography. J Biomed Opt. 2008;13:060506. doi: 10.1117/1.3046716. [DOI] [PubMed] [Google Scholar]
- Larina IV, Ivers S, Syed S, Dickinson ME, Larin KV. Hemodynamic measurements from individual blood cells in early mammalian embryos with Doppler swept source OCT. Opt Lett. 2009;34:986–988. doi: 10.1364/ol.34.000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Meneses JJ, Pedersen RA. Cell fate and cell lineage in the endoderm of the presomite mouse embryo, studied with an intracellular tracer. Dev Biol. 1986;115:325–339. doi: 10.1016/0012-1606(86)90253-8. [DOI] [PubMed] [Google Scholar]
- Megason S, Amsterdam A, Hopkins N, Lin S. Uses of GFP in transgenic vertebrates. Methods Biochem Anal. 2006;47:285–303. doi: 10.1002/0471739499.ch13. [DOI] [PubMed] [Google Scholar]
- Molyneaux KA, Stallock J, Schaible K, Wylie C. Time-lapse analysis of living mouse germ cell migration. Dev Biol. 2001;240:488–498. doi: 10.1006/dbio.2001.0436. [DOI] [PubMed] [Google Scholar]
- Nagy A. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. [Google Scholar]
- Nel-Themaat L, Vadakkan TJ, Wang Y, Dickinson ME, Akiyama H, Behringer RR. Morphometric analysis of testis cord formation in Sox9-EGFP mice. Dev Dyn. 2009;238:1100–1110. doi: 10.1002/dvdy.21954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New DA, Cockroft DL. A rotating bottle culture method with continuous replacement of the gas phase. Experientia. 1979;35:138–140. doi: 10.1007/BF01917926. [DOI] [PubMed] [Google Scholar]
- Nishii K, Shibata Y. Mode and determination of the initial contraction stage in the mouse embryo heart. Anat Embryol (Berl) 2006;211:95–100. doi: 10.1007/s00429-005-0065-x. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Nowotschin S, Hadjantonakis AK. Photomodulatable fluorescent proteins for imaging cell dynamics and cell fate. Organogenesis. 2009a;5:135–144. doi: 10.4161/org.5.4.10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotschin S, Hadjantonakis AK. Use of KikGR a photoconvertible green-to-red fluorescent protein for cell labeling and lineage analysis in ES cells and mouse embryos. BMC Dev Biol. 2009b;9:49. doi: 10.1186/1471-213X-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotschin S, Eakin GS, Hadjantonakis AK. Live-imaging fluorescent proteins in mouse embryos: Multi-dimensional, multi-spectral perspectives. Trends Biotechnol. 2009;27:266–276. doi: 10.1016/j.tibtech.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Nonaka S, Tanaka Y, Saijoh Y, Hamada H, Hirokawa N. Abnormal nodal flow precedes situs in versus in iv and inv mice. Mol Cell. 1999;4:459–468. doi: 10.1016/s1097-2765(00)80197-5. [DOI] [PubMed] [Google Scholar]
- Passamaneck YJ, Di Gregorio A, Papaioannou VE, Hadjantonakis AK. Live imaging of fluorescent proteins in chordate embryos: From ascidians to mice. Microsc Res Tech. 2006;69:160–167. doi: 10.1002/jemt.20284. [DOI] [PubMed] [Google Scholar]
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135:3081–3091. doi: 10.1242/dev.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee DY, Zhao XQ, Francis RJ, Huang GY, Mably JD, Lo CW. Connexin 43 regulates epicardial cell polarity and migration in coronary vascular development. Development. 2009;136:3185–3193. doi: 10.1242/dev.032334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MA, Davidson MW, Piston DW. Fluorescent protein tracking and detection: Applications using fluorescent proteins in living cells. CSH Protoc. 2009;2009 doi: 10.1101/pdb.top64. pdb top 64. [DOI] [PubMed] [Google Scholar]
- Sadler TW, New DA. Culture of mouse embryos during neurulation. J Embryol Exp Morphol. 1981;66:109–116. [PubMed] [Google Scholar]
- Sakai T, Onodera T. Embryonic organ culture. Curr ProtocCell Biol. 2008;Chapter 19(Unit 19):8. doi: 10.1002/0471143030.cb1908s41. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, Imamura T, Ogawa M, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Sherbahn R, Frasor J, Radwanska E, Binor Z, Wood-Molo M, Hibner M, Mack S, Rawlins RG. Comparison of mouse embryo development in open and microdrop co-culture systems. Hum Reprod. 1996;11:2223–2229. doi: 10.1093/oxfordjournals.humrep.a019081. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Goldberg MR, Watanabe T, D’Agati V, al-Awqati Q, Costantini F. Expression of green fluorescent protein in the ureteric bud of transgenic mice: A new tool for the analysis of ureteric bud morphogenesis. Dev Genet. 1999;24:241–251. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<241::AID-DVG7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Rodriguez T, Clements M, Smith JC, Beddington RS. Active cell migration drives the unilateral movements of the anterior visceral endoderm. Development. 2004;131:1157–1164. doi: 10.1242/dev.01005. [DOI] [PubMed] [Google Scholar]
- Tam PP. Postimplantation mouse development: Whole embryo culture and micro-manipulation. Int J Dev Biol. 1998;42:895–902. [PubMed] [Google Scholar]
- Watanabe T, Costantini F. Real-time analysis of ureteric bud branching morphogenesis in vitro. Dev Biol. 2004;271:98–108. doi: 10.1016/j.ydbio.2004.03.025. [DOI] [PubMed] [Google Scholar]
- Wiley LM, Spindle AI, Pedersen RA. Morphology of isolated mouse inner cell masses developing in vitro. Dev Biol. 1978;63:1–10. doi: 10.1016/0012-1606(78)90108-2. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Tamplin OJ, Beckers A, Gossler A, Rossant J. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev Cell. 2007;13:884–896. doi: 10.1016/j.devcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Young HM, Bergner AJ, Anderson RB, Enomoto H, Milbrandt J, Newgreen DF, Whitington PM. Dynamics of neural crest-derived cell migration in the embryonic mouse gut. Dev Biol. 2004;270:455–473. doi: 10.1016/j.ydbio.2004.03.015. [DOI] [PubMed] [Google Scholar]