Abstract
Cardiomyocytes from human pluripotent stem cells (hPSCs-CMs) could revolutionise biomedicine. Global burden of heart failure will soon reach USD $90bn, while unexpected cardiotoxicity underlies 28% of drug withdrawals. Advances in hPSC isolation, Cas9/CRISPR genome engineering and hPSC-CM differentiation have improved patient care, progressed drugs to clinic and opened a new era in safety pharmacology. Nevertheless, predictive cardiotoxicity using hPSC-CMs contrasts from failure to almost total success. Since this likely relates to cell immaturity, efforts are underway to use biochemical and biophysical cues to improve many of the ~ 30 structural and functional properties of hPSC-CMs towards those seen in adult CMs. Other developments needed for widespread hPSC-CM utility include subtype specification, cost reduction of large scale differentiation and elimination of the phenotyping bottleneck. This review will consider these factors in the evolution of hPSC-CM technologies, as well as their integration into high content industrial platforms that assess structure, mitochondrial function, electrophysiology, calcium transients and contractility. This article is part of a Special Issue entitled: Cardiomyocyte Biology: Integration of Developmental and Environmental Cues in the Heart edited by Marcus Schaub and Hughes Abriel.
Keywords: Human embryonic stem cells, Human induced pluripotent stem cells, Cas9/CRISPR genome editing, Cardiomyocytes, Drug screening, Disease modelling, Maturation factors, Muscular thin films, Engineered heart tissue, Automated scalability, High content platforms, Calcium imaging, Electrophysiology, Mitochondria, Contractility
Highlights
-
•
hPSC-CM drug screening, disease modelling & Cas9/CRISPR engineering becoming routine.
-
•
hPSC-CMs used to refine patient treatment & assist in progressing drugs to clinic.
-
•
Transplantation into heart failure pigs and primates now translated to humans.
-
•
Barriers to progression include cost of goods, subtype specification & maturation.
-
•
2D and 3D hPSC-CM high content phenotyping platforms evolving rapidly.
1. Introduction
Human embryonic stem cells (hESCs) were first isolated from blastocyst stage embryos in 1998 [1], with later demonstration of human induced pluripotent stem cell (hiPSC) reprogramming from somatic cells by just four genetic factors [2]. The ability to culture these human pluripotent stem cell (hPSC) populations long-term and yet induce their differentiation into a wide variety of cell types potentially offers a new era in biomedicine, particularly for understanding human development, drug screening, disease modelling and cell therapy to replace lost or damaged tissues.
The socioeconomic drivers for development of new technologies to address these biomedical areas are strong. The UK Department for Innovation, Research & Skills [3] concluded that 80% of healthcare costs go towards treating the late stages of illnesses, which in the future could be cured early or better managed by regenerative medicine and cell replacement approaches. The global burden of heart failure is currently USD $45bn, with a forecast of USD $90bn by 2030. These sobering statistics have prompted more than a decade of autologous stem cell trials to treat heart failure, predominantly using cell populations derived the bone marrow. However, the efficacy of such treatments has been called into doubt with the realisation that only trials containing flaws (e.g. design or reporting errors) showed positive outcomes, while error-free trials showed no benefit [4].
The pharmaceutical industry faces equivalent challenges. Average drug development duration is 10–15 years with costs as high as USD $11bn [5]. Between 1980 and 2009, ~ 1 in 7 licenced drugs deemed efficacious in Phase III trials had to be withdrawn from the market. The main reasons included unanticipated side-effects, such as cardiotoxicity, hepatotoxicity and gastro-intestinal issues [6]. Unexpected cardiotoxicity was implicated in 28% of drug withdrawals in the USA [7]. For the heart, side effects can damage the structural integrity and survival of cardiomyocytes (CMs), as is the case with the anti-inflammatory drug, Vioxx [8], and many anti-cancer drugs, such as doxorubicin [9]. Beat regularity and duration (QT prolongation or shortening) can also be affected, potentially leading to polymorphic ventricular tachyarrhythmia, seizures and sudden death. In 2010 this was the reason for the US Food and Drug Administration (FDA) requesting withdrawal of propoxyphene, an opioid pain reliever marketed by Xanodyne Pharmaceuticals [10], and of sibutramine, a weight loss agent marketed by Abbott Laboratories [11]. In the worst cases, adverse side effects lead to fatalities, as was the case with the serotonin agonist, cisparide, which caused 125 deaths before its use ceased [12].
1.1. Current safety assessment platforms are suboptimal
Underlying poor predictivity of cardiotoxicity are suboptimal safety assessment platforms. While human primary CMs would be the in vitro model of choice, their large-scale use is hindered by limited availability, poor consistency, almost non-existent proliferation and de-differentiation in culture [13]. Consequently, various other models are used. Provisional safety screens often involve aneuploid tumour cell lines (e.g. CHO or HEK cells) genetically engineered to over-express an ion channel of choice. While high throughput, these cells do not replicate the complexity of the working CM and can lead to false negatives or positive. Thus, the multi-channel blocking drug, verapamil, is considered safe and “QT-neutral” on account of dual blocking of potassium IKr and calcium Ica,L channels but can be flagged as potentially harmful in the single ion channel assays [14]. Ex vivo systems, such as ventricular wedge preparations [15] and Purkinje fibres [16], have been extensively used in physiological and pharmacological studies, but low-throughput and inter-species differences are limitations.
Species differences are particularly highlighted in the mouse [13]. While this species benefits from genetic tractability via gene targeting, the beat rate of the mouse heart is ~ 10 times faster than human (500 bpm vs 60 bpm) and has an electrocardiogram duration 5–10 times shorter (450 ms vs 50-100 ms). Increases in heart rate are associated with increased force of contraction in humans but decreased force in mice [17]. Whereas repolarisation of the mouse CMs is driven primarily by Ito, IK,slow1, IK,slow2, ISS ion channels, this role is achieved by the potassium channels, IKr and IKr in human cells [18]. There are species differences in the role of the regulatory molecule, phospholamban, while expression of structural genes also varies. In humans, expression of alpha and beta myosin heavy chains (α −/β-MHC) locates to the atria and ventricles, respectively, but in the mouse αMHC is expressed in both locations. There are also differences in developmental progression and location of the myosin light chains, MLC2a and MLC2v. The surface marker, SIRPA, is expressed on human but not mouse CMs. Such differences mean that mice are at least 10 × more tolerant to 37% of drugs than humans. Issues extend to rats and dogs, which tolerate 4.5- to 100-fold the concentration of various chemotherapeutic agents than humans (e.g. ThioTEPA, Myleran, Actinomycin-D, Mitomycin C, Mithramycin, Fludarabine) [19].
Reducing drug attrition by 5% in Phase 1 clinical development could reduce drug development costs by 5.5–7.1% [20] equating to savings of about USD $100 m. Thus, there has been considerable effort invested in finding additional tools for safety assessment, which include hPSC-CMs.
1.2. Evolution of hPSC-CM differentiation
With the issues above, it was a certain degree of excitement that, in 2000, Joseph Itskovitz-Eldor's team demonstrated contracting structures containing CMs could be produced by spontaneous differentiation of hESCs via three-dimensional embryoid bodies [21]. Subsequent research has shown that CMs derived from both hESC and hiPSC display many of the structural and functional features associated with heart cells (for review [13]). This promoted development and evaluation of three general strategies to improve differentiation efficiency: 3-dimensional aggregates known as embryoid bodies; co-cultures with an inducer END-2 cell line; 2-dimensional monolayers (reviewed in [22]). Initially, these approaches produced purities of < 50% hPSC-CMs and additional enrichment was needed to go beyond 90% purity. Genetic selection strategies were developed first. These employed random integration into the hESC genome of expression cassettes that coupled cardiac specific promoters (e.g. MYH6 encoding αMHC) with puromycin antibiotic resistance [23]. Gene targeting allowed refinement by precise positioning of the NeoR gene downstream of MHY6 [24]; this approach is still used for commercial production by Cellular Dynamics International (CDI) since it enables selection of CMs at mass scale.
Purity of hPSC-CMs has also been improved by FACS sorting for cells expressing markers associated with cardiac progenitors (CD15; SSEA1 [25]) or CMs (e.g. VCAM1, SIRPA [26]) and for cells incubated with molecular beacons targeting mRNAs that encode cardiac troponin and MHCs [27]. Sorting has been achieved following staining with the mitochondrial dye, TMRM, on the basis that highly metabolic CMs will have the highest numbers of mitochondria and hence have high level of fluorescence [28]. However, these approaches are neither economically nor practically viable for large scale production of hPSC-CMs due to slow sort speeds (maximum 70,000 cells/s) and poor survival after coupling dissociation with FACS. This approach would require over 2 weeks to harvest 10 billion hPSC-CMs, which is the number estimated from primate studies to be required for transplantation to restore function into the infarcted human heart of a single patient [29].
In an effort to develop mass enrichment strategies that do not require bespoke genetic modification approaches for each hPSC line, Tohyama et al. [30] exploited the metabolic differences between CMs and the contaminating (predominantly) fibroblast populations. While fibroblasts rely on glucose-based metabolism, hPSC-CMs can utilise both lactate and glucose. Purities of 99% were achieved by incubating hPSC-CMs in medium that contained 4 mM lactate but lacked glucose and this approach is becoming popular. Microfluidic approaches has also been used to separate cells on the basis of size [31] or electrophysiological signals [32] but the considerable heterogeneity in differentiated hPSC-CM cultures means these physical approaches will require further validation.
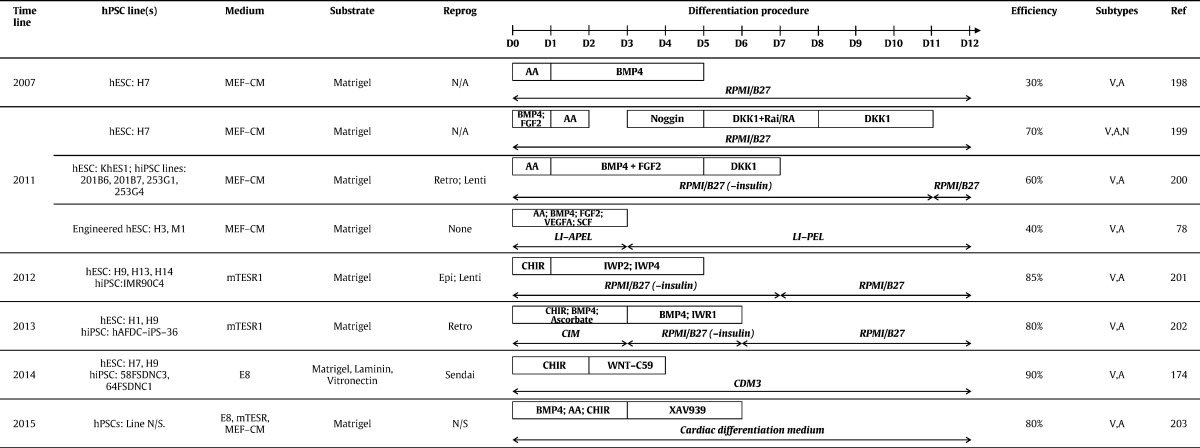
While these enrichment approaches have been useful as an interim measure, efforts to develop differentiation protocols that produce hPSC-CMs at high purity have reached fruition in the last 3–4 years. Monolayer protocols (see Table 1) are popular due to ease of handling and large scale production of ~ 7 billion hPSC-CMs [29]. Clues on how cardiogenesis proceeds in vivo in a coordinated, stepwise manner have been derived from various model species. In mammals, progression from gastrulation, through formation of the primitive streak and epiblast towards development of the linear heart tube shows the importance of signalling via pathways involving transforming growth factor beta (TGF-β; including bone morphogenetic protein [BMP] and Activin A), basic fibroblast growth factor (bFGF; FGF2) and Wingless (WNT). Translating these developmental signals to hPSCs in vitro has been empirical and iterative but has defined the concentrations and timings of growth factors and/or small molecules that activate or inhibit relevant cardiogenic pathways (Table 1). Commercial media, such as mTeSR and Essential 8 (E8), increase reproducibility of culture and differentiation by eliminating need for steps involving serum, serum replacements or conditioned medium. Relative to recombinant growth factors, use of chemically synthesised small molecules reduces variability and cost. Thus, monolayer cardiogenic protocols now routinely employ defined medium with the GSK3β inhibitor, CHIR99021, during the first 1–3 days of differentiation process, followed by inhibition of WNT using IWR1, IWR4, C59 or XAV939 (Table 1).
Table 1.
Methods for monolayer differentiation of hPSC-CMs. Abbreviations: MEF-CM, mouse embryonic fibroblast-conditioned medium; E8, Essential 8 medium; Reprog, Reprogramming method; N/A, not applicable; Retro, retrovirus; Lenti, lentivirus; Epi, Epsiomal; AA, activin A; V, ventricular; A, atrial; N, nodal (or pacemaker).
Although efficiencies of > 80% are now regularly reported for hPSC-CM differentiation (Table 1), unpublished anecdotal evidence suggests reproducibility and robustness of protocols needs to be improved so there is greater consistency between lines and laboratories. Also, protocols yield a mixed population of CM subtypes, including ventricular-, atrial- and pacemaker-like cells. Cultures containing a single subtype are preferred; for example, ventricular cells are needed to evaluate drugs that have Torsades de Pointe liabilities or for transplantation after myocardial infarction. In this regard, there have been recent exciting developments. Birket and colleagues [33] combined a complex but elegant double transgenic approach, wherein an NKX2.5-GFP targeted hESC line was further transfected with an inducible MYC expression construct. In the presence of insulin-like growth factor-1 (IGF-1) and a hedgehog pathway agonist, cardiovascular progenitor cells could be isolated and proliferated for over 40 population doublings. Moreover, modulating exogenous BMP, FGF, WNT and RA signalling led to multi-lineage differentiation, as well as directed specification to pacemaker and ventricular cells. This report was remarkable because it not only showed long-term proliferation of hPSC-derived cardiac progenitors (in 11 other reports using mouse and human PSCs, maximum expansion was 4-fold [34]), but it was the first robust demonstration of subtype specification. In an alternative approach, modulation of retinoic acid signalling during hESC differentiation was used to generate atrial- and ventricular-like CMs. These CM subtypes were used to show that the multi-ion channel blocker, vernakalant, and Kv1.5 blocker, XEN-D0101, caused a reduction in early repolarization only in the atrial cells [35], providing a novel preclinical test platform for these drug classes.
1.3. Genotypes are now readily captured using hiPSC reprogramming
Improvements in CM differentiation have been paralleled by advances in hiPSC production methods. The original landmark papers by Shinya Yamanaka's team and the reports thereafter described low efficiency (< 0.1%) production of hiPSCs via integrating retroviruses [2] in undefined medium on mitotically-inactivated mouse embryonic fibroblasts. Technologies have evolved so that academic and commercial labs now produce hiPSC lines at efficiencies of ~ 4.4% using non-integrating approaches in defined medium on recombinant matrices [36] Indeed, large scale banking schemes including Human Induced Pluripotent Stem Cells Initiative (HIPSCI), StemBANCC/IMI, California Institute for Regenerative Medicine and New York Stem Cell Foundation will create hiPSC lines from 7000 normal or diseased skin biopsy donors using Sendai-virus, episomes or mRNA with a combination of SOX2, c-MYC, OCT4, KLF4 and/or LIN28 [37]. Nevertheless, each integration-free method has pros and cons and there is not yet consensus on which reprogramming method is best. Episomal plasmids have lower reprogramming efficiencies and the potential for residual plasmid integration; Sendai-virus require higher biosafety containment levels and are relatively costly; mRNA reprogramming is labour intensive, requiring repeated (daily) transduction and costly Pluriton medium. In addition, there are licencing cost implications and restrictions to be considered for commercial use for Sendai-virus and mRNA approaches.
1.4. hPSC-CMs are becoming valuable for in vitro and in vivo biomedical application
The relative ease of efficient reprogramming and directed cardiogenesis has accelerated progress towards biomedical application. This is helped by hiPSCs largely eliminating ethical or legal restrictions that prohibited use of hESCs in many companies and countries. For predictive cardiotoxicity, many reports show hPSC-CMs are effective in safety screening. In the 13 years from 2000 to 2013, pharmacological responses of hPSC-CMs to only 60 different compounds had been demonstrated [13]. These numbers are now being exceeded by single studies; one report assessed impact of 131 compounds of hPSC-CM function [38].
Accuracy of the assays is also improving and gaining interest from the pharmaceutical industry. Using hPSC-CMs, AstraZeneca showed 70% specificity and 87% sensitivity for a 51 compound screen [39], while a study commissioned by J&J recorded an accuracy of 90% following blind testing of electrical toxicity in 20 compounds [40]. Work from GlaxoSmithKline cross-compared pharmacological responses of hPSC-CMs and animal models, concluding that the human cells offered a reliable and cost-effective surrogate to preclinical in vitro testing [41]. Direct comparison between CMs isolated from hPSCs or dog and rabbit hearts showed the human cells more accurately predicted moxifloxacin-induced cardiotoxicity [42]. Studies have extended to screening antivirals as a treatment for B3-strain of coxsackievirus, a major causative agent for viral myocarditis [43]. Notably, hPSC-CMs were used to show that toxicity was reduced when the anti-cancer drug, doxorubicin, was delivered via a HER2-targeted liposomal pathway; this assisted the decision to advance to Phase I testing [44]. Such studies have led the CIPA initiative (Comprehensive In Vitro Proarrhythmia Assay) to propose integration of hPSC-CMs into the ICH (International Conference on Harmonisation) S7a/b and E14 guidelines by the end of 2015. These guidelines have been the mainstay over the last decade of preclinical assessment of cardiac electrophysiology for new drugs [45].
Patient-specific hiPSC-CMs are being used increasingly to evaluate altered phenotype and drug rescue of various channelopathies affecting the heart, including long QT syndrome (LQTS)-1 [46], [47], [48], − 2 [49], [50], [51], [52], [53], − 3 [54], − 8 [55], LQTS3/Brugada overlap [56] and catecholaminergic polymorphic ventricular tachycardia (CPVT) [57], [58], [59]. Disorders that affect structure, contractility and survival have also been modelled, such as Duchenne muscular dystrophy (DMD) [60], dilated cardiomyopathy [61], [62], hypertrophic cardiomyopathy (HCM) [63], [64], Leopard Syndrome [65], Barth Syndrome [66], [67] and arrhythmogenic right ventricular cardiomyopathy (ARVC) [68], [69], [70], [71]. These have been used to understand disease mechanisms and evaluate novel therapeutics. Thus, dantrolene abolished isoprenaline-induced arrhythmias in CPVT1 hiPSC-CMs [58], while trichostatin A was shown to prevent hypertrophy in HCM hiPSC-CMs [64]. Tests for efficacy of genetic intervention include oligonucleotide-mediated exon skipping and allele-specific RNAi to correct DMD [72] and LQTS2 [53] hiPSC-CMs, respectively. Most notably, the inability to manage effectively treatment of an individual with complex LQTS was addressed by deriving hiPSC-CMs and performing multi-parameter in vitro drug testing until a suitable combinatorial regime was identified. This treatment was used in the clinic to improve the patient's care [73] showing feasibility of personalised medicine.
Nevertheless, while the examples above show the potential offered by hiPSC-CMs, there are several reports of deficiencies relative to their hESC derived counterparts. Thus, Foldes and colleagues [74] described robust hypertrophic responses to phenylephrine in hESC-CMs but not hiPSC-CMs. This was irrespective of the reprogramming or differentiation method used. Indeed, a hESC line was differentiated to fibroblasts, which were reprogrammed to hiPSC. When this hiPSC line and the parental hESC line were differentiated to CMs, only the cells derived from hESCs showed hypertrophy, despite the cells sharing the same genotype. Similar issues have been reported for improper reprogramming and disease modelling in hiPSC from patients with Fragile-X relative to hESCs derived from pre-implantation genetic diagnosis embryos [75].
Beyond their in vitro use, hPSC-CMs are also being evaluated for treatment of damaged of diseased heart. Pilot studies using hPSC-CM engraftment into mouse, rat, guinea-pig and pig models of myocardial infarction were escalated to pigtail macaque non-human primates in mid-2014 [29]. In the primate studies, 1 billion cryopreserved hPSC-CMs were transplanted in a complex pro-survival cocktail to the infarct site of each of 7 animals. Transplanted cells led to extensive remuscularisation, accompanied by host vasculature perfusion, electromechanical junction formation between graft & host, and synchronous calcium transients. Nonetheless, there were two cautionary notes. While the hPSC-CMs constituted a graft size of up to 5.3% of the left ventricular mass, survival of transplanted cells was less than 10% (< 108 of 109 cells) despite the powerful pro-survival cocktail. Secondly, although the macaques remained free of distress, continuous electrocardiogram recordings showed that all animals receiving hESC-CMs developed ventricular arrhythmias. There are similarities but also differences between these primate studies and results from transplantation into swine. In a study in pigs [76], the tri-lineage differentiation potential of hiPSCs was exploited to produce CMs, endothelial cells and smooth muscle cells. A total of 6 million cells (2 million of each lineage) were complexed with a 3D epicardial fibrin patch loaded with microspheres to allow prolonged release of the pro-survival factor, insulin-like growth factor 1 (IGF-1). The complex was then transplanted into a porcine model of myocardial infarction. Similar to the primate study, over a 4 week period cell survival was around 9%, although without the fibrin patch was reduced to 3–4%. Surprisingly, given the low cell numbers transplanted (160-fold less than the primate study), there were improvements in myocardial wall stress, metabolism and contractile performance. However, distinct to macaques, development of ventricular arrhythmias was not reported in the pigs; whether this important difference is down to the animal model, cell types, numbers or preparation method, inclusion of different survival factors or transplant route are now all questions that need to be addressed. Moreover, these reports have not included methods to improve vasculature to the grafted cells and this will be a consideration for the future.
The preclinical studies have led to the first clinical trial for the heart using hPSC derivatives. Menasché and co-workers [77] sought to direct differentiation from hESCs and then use immunomagnetic sorting to isolate ISL-1 +/SSEA-1 + cardiac progenitor cells. These were embedded into a fibrin scaffold, which was surgically delivered onto the infarct area in a 68-year-old patient suffering from severe heart failure. At the 3 month follow-up stage, the patient showed no complications, such as arrhythmias, tumour formation or immunosuppression-related adverse events, but was symptomatically improved, wherein echocardiographically showed the damaged region of the heart regained contractility. The progress of this patient, and those who follow, will be keenly awaited. Nevertheless, nearly a year on it is not clear whether any further patients have been recruited to this trial, even though the expected start and end dates are 2013 and 2017 to treat a total of 6 patients. This perhaps highlights the challenges of coordinating complex processes of large scale cardiac progenitor cell production, surgical procedures and immunosuppression regimens with highly selective patient inclusion criteria. Thus, for inclusion, patients must display severe left ventricular systolic dysfunction with left ventricular ejection fraction (LVEF) ≤ 35% as assessed by echocardiography or scintigraphy and have an echocardiography history of myocardial infarction with a residual akinesia involving more than 2 of 16 contiguous segments. They will show New York Heart Association (NYHA) Class III or IV, despite optimal standard of care including diuretics and angiotensin receptor blockers and, if possible, beta blockers and aldosterone blockers, as well as previous implantation of an automatic internal defibrillator associated to ventricular resynchronization [78]. If the current Phase I trial continues to provide optimism as a new treatment route for patients, the issues of bioprocess, surgery and patient selection will need to be reviewed carefully. Indeed, it will only be through larger trials that true improvements in the patient's heart function can be attributed to cell transplantation rather than natural recovery or impact of past treatment (e.g. bypass surgery, drug treatment, ventricular resynchronization).
1.5. Genome editing marks a new era for in vitro genotype modelling
Until hiPSC approaches provided a route to capturing a wide range of patient-relevant genotypes, reliance had been on establishing hESC lines from pre-implantation genetic diagnosis (PGD) embryos [79] or by gene targeting [80]. However, PGD is available for only a limited number of genetic conditions, few scientists have access to these facilities and the use of embryos (even those that harbour detrimental genetic lesions) is ethically sensitive in many countries. Similarly, gene targeting by homologous recombination was initially achieved in a few laboratories to create knockouts (e.g. HPRT1 to model of the metabolic disorder, Lesch Nyhan syndrome [80]) or reporter constructs downstream of developmentally important genes, such as NKX2.5 [81]. In rare cases, creation of isogenic pairs was used to study role of mutations in genes such as KCNH2, which underlies the sudden cardiac death condition of LQTS2 [82]. Further progress was stymied because of low recombination frequency (1 in 106–109 cells) in most mammalian cells, which made the generation of isogenic models almost unachievable because this often requires biallelic targeting. However, progress in genome editing tools now allows rapid engineering of the genotypes available in hPSCs. If there is not the need for patient history to draw in vitro-in vivo correlations, the speed, flexibility, ease and low cost of gene targeting will be used in preference to hiPSC reprogramming to capture specific genotypes; indeed, in our own laboratory, this is the situation for some diseases.
For gene targeting, it has been known for 25 years that introduction of specific double strand breaks at the target locus can improve efficiency. In human cells, complexing the Fok1 endonuclease to a pair of zinc finger nucleases (ZFNs) [83] allowed double strand breaks at a model GFP locus in 293 T cells [84] and endogenous PIG-A locus in hPSCs [84] resulting in targeting efficiency improvements of 200- to 2000-fold. However, the complex design and construction for each ZFN attracted a high commercial cost of USD $25,000. The advent of transcription activator-like effector nucleases (TALENs) used the same principle as ZFNs, relying on a dimeric protein-based DNA binding domain coupled to endonuclease. Construction kits, such as GoldenGate [85] and FLASH assembly [86], meant individual labs could produce their own TALEN vectors and hence reduce costs by 20-fold relative to ZFNs. Moreover, TALENs showed greater specificity in hPSCs, with less off-target activity and toxicity in comparison to ZFNs [87].
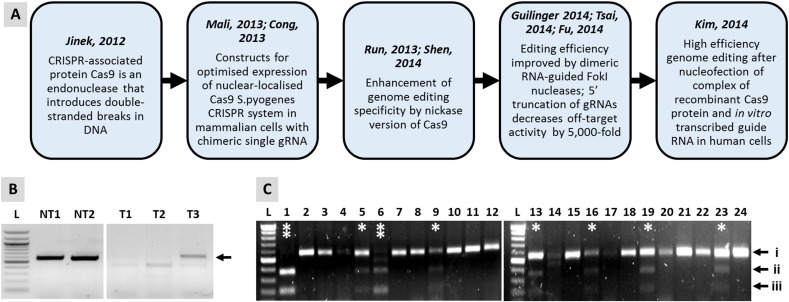
The real breakthrough came with the development of the Cas9/CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat) system (Fig. 1) [88] which is derived from various strains of bacteria and is often described as their immune system. The adapted version of this system relies on 100 base site-specific guide RNA (gRNA) to direct the Cas9 endonuclease to the target site, which eliminates the need for time-consuming production of DNA binding-endonuclease fusion proteins [89]. Such approaches have been used to perform correction methods using exon skipping, frameshifting and exon knock-in into hiPSC lines carrying mutations in DMD, which underlie severe muscle degenerative disease [90].
Fig. 1.
Genome editing in hPSCs with the Cas9/CRISPR system. Panel (A) shows a timeline of the key events involved in adaptation of the Cas9/CRISPR system for use in mammalian cells. In (B), non-targeted, parental undifferentiated hESCs (NT1) or derived CMs (NT2) express KCNH2 RNA (arrow), which encodes components of the Ikr channel. These hESCs were targeted with a construct that induced a G1681A SNP conversion in the coding region of the KCNH2 locus, whilst concurrently introducing a FRT-flanked blasticidin resistance cassette into the neighbouring intron. The selection cassette was to facilitate selection of targeted clones and did not interfere with any known KCNH2 regulatory elements. However, correct expression of the RNA was compromised to levels that were barely detectable in targeted hESC-CMs (T1) and undifferentiated hESCs (T2). Only when the selection cassette was removed by Flp recombinase was expression of the correct transcript size restored (T3). Panel (C) shows oligonucleotide-mediated gene editing in the absence of drug selection in hESCs at the ADRB2 locus, which encodes the b2-adrenoceptor. Transfection of hESCs was by Amaxa 4D nucleofection to introduce recombinant Cas9 protein complexed with an in vitro transcribed gRNA and a single-stranded 110 base DNA oligonucleotide carrying an XbaI restriction site plus TAA stop codon. This enables functional knock out of targeted allele(s) and detection of targeting events by RFLP (restriction fragment length polymorphism) analysis of clones. Incubation of PCR products with XbaI enzyme show non-targeted clones as a non-digested band (i); heterozygote clones as three bands,indicated by single * showing non-targeted (band i) and targeted (digested bands ii/iii) alleles; or homozygote cells as two bands, indicated by double ** showing both alleles targeted (digested bands ii/iii). Efficiency of targeting was 8/24 (33%) clones, of which 2/8 (25%) were homozygote. L = 100 bp Ladder.
There have been several further refinements in the Cas9/CRISPR system (Fig. 1). Use of dual guide RNA/Cas9-Nickase (D10A) reduces off-target activity to an almost undetectable level [91]. Moreover, pre-synthesised in vitro transcribed gRNAs can be complexed with recombinant Cas9 protein and transfected by nucleofection, providing a rapid route to gene knockout or small changes in sequence, including substitutions [92]. Thus, in the absence of a homology domain, Cas9-induced DNA cleavage leads to non-homologous end joining and causes insertions or deletions, known as ‘indels’, that can cause gene knockouts [93]. To induce base substitutions, small changes or deletions of up to 100 kbases, the conventional ~ 5–15 kb targeting vector can be replaced with a ~ 100–120 nucleotide single-stranded DNA oligo [94]. With this approach, Kim et al. [92] showed up to 79% targeting efficiency in CCR5 locus in human leukaemia K562 cell line, BJ fibroblasts and H9 hESCs.
The recombinant Cas9 approach has other advantages. Since it is active immediately upon entry to the nucleus but continues to function for only 24 h, off-target effects and toxicities are low, while targeting frequency is high and eliminates the need for drug selection. Our own work corroborates these findings (Fig. 1). We used Amaxa 4D nucleofection to deliver a combination of gRNA, recombinant Cas9 protein and 110-nucleotide single-stranded DNA oligo template into ReBl-PAT hiPSCs, which resulted in a targeting frequency at the β2-adrenoceptor locus of 33%, of which 25% of clones were biallelic. Such efficiencies can be expected to rise further with the finding that small molecules can enhance targeting by up to 9-fold in hPSCs [95], which will ultimately lead to panels of isogenic pairs in hPSCs to study disease mechanisms and therapies.
The high efficiency of this oligo-based method of Cas9/CRISPR targeting will mark a step-change in gene editing. By avoiding the need for a selection cassette eliminates the need for residual transgenic DNA sequences to reside in the genome. Even by placing the selection cassette in the intron, we have shown this can interfere with the expression of the KCNH2 endogenous gene in undifferentiated hPSCs and derived CMs (Fig. 1). Only by excising the selection cassette with Flp recombinase was endogenous gene expression restored (Fig. 1). Nevertheless, this strategy still leaves residual FRT recombination sites in the genome. An alternative route to producing isogenic footprint-free hiPSC models was used to achieve biallellic targeted correction of AATD (α1-antitrypsin deficiency). After using ZFNs to target a construct containing a drug selection cassette and the sequences to correct the polymorphic mutation, the selection cassette was seamlessly excised via transposase acting on flanking PiggyBac transposons [96]. However, this two-step approach requires more time and more population doublings, which increases the likelihood of genome instability in the targeted hPSCs.
Advances in the Cas9/CRISPR system mean that there is now exponential uptake of the technology, with over 20 publications a week appearing in PubMed. Commercial providers are now offering the reagents to target every gene in the human and mouse genome, and projects are underway within the Wellcome Trust Sanger Institute to produce panels of mouse ESC lines in which every gene in the genome has been knocked out. Notably, the Cas9/CRISPR system was used in a somewhat abortive and controversial [97] attempt to target the β-globin gene in human embryos. However, the efficiency of targeting was low (14%), edited embryos were mosaic and the level of off-target events was high due to homology with delta-globin gene [98]. Inevitably, the debate will continue on the relative efficiencies, fidelities and off-target characteristics of each nuclease-based platform. However, unpublished data indicate that the Cas9/CRISPR elicits gene editing at up to 10-fold greater frequency than TALENs, whilst off-target events are comparable between the systems when constructs or gRNAs are produced to current best practise. It can be expected that these next generation genome engineering approaches will start to provide the tools required to decipher the phenotypic impact of polymorphism data arising from GWAS (genome-wide association studies).
1.6. Current and future technology challenges for hPSC-CMs
In the last 3 years, there have been considerable advances in the methods to produce hiPSCs and genome engineer hPSCs. Concurrently, improved protocols for differentiation have yielded novel approaches in safety assessment, modelling disease, developing patient-specific therapies and transplanting cardiac progenitor cells into the diseased heart. Nevertheless, the technologies are still very much in the development phase. The next sections consider some of the challenges facing the field of hPSC-CM biology, along with current progress.
2. Immaturity of hPSC-CMs
Reports from academia and industry show that hPSC-CMs display phenotypes consistent with a variety of disease- or drug-induced states. Nevertheless, it is now well established that current differentiation protocols produce cells with an immature phenotype consistent with mid-gestation of human foetal heart development [13]. The consensus viewpoint in the field is that each improvement made to hPSC-CM maturity will further increase utility. Table 2 provides a summary of parameters collated from the literature (for specific details see [13], [99], [100], [101], [102], [103], [104]) and/or generated by our own lab reflecting the differences between hPSC-CMs and adult CMs, providing a benchmark to measure effectiveness of prospective maturation approaches.
Table 2.
Comparison of characteristic between adult-CMs and hPSC-CMs, showing the latter lack maturity.
| Adult-CM | hPSC-CM | ||
|---|---|---|---|
| Structure | Structure | Rod-shaped | Round or polygonal |
| Alignment | Longitudinally aligned | Chaotically organised | |
| Nucleation | ~ 30% cells bi- or poly-nuclear | Very limited bi-nucleation | |
| Sarcomere organisation | Highly organised | Disorganised | |
| Aspect ratio | 5–9.5:1 | 2–3:1 | |
| Banding | Z-discs, I-, H-, A- and M-bands | Mainly Z-discs and I-bands | |
| Sarcomere length | 2.2 μm | 1.6 μm | |
| SR | Sarcoplasmic reticulum | Well developed | Mixed response: caffeine, Thapsigargin & ryanodine |
| SR proteins | e.g. CSQ, PLN, RYR2, SERCA/ATP2A2 | Expression lower than adult | |
| T-Tubules | Yes | No | |
| Expr. | Gene expression | MYH7 (β-MHC) > MYH6 (αMHC) TNNI3 (cTnI) > TNNI1 (foetal ssTnI) MYL2 (MLC2v) > MYL7 (MLC2a) Titin isoform N2B predominates ADRA1A (α-adrenoceptor) expressed |
MYH6 (αMHC) > MYH7 (βMHC) TNNI1 (foetal ssTnI) > TNNI3 (cTnI) MYL2:MYL7 ratio not determined Titin isoform N2BA predominates ADRA1A (α-adrenoceptor) not expressed |
| Energy & force | Metabolism | Mainly fatty acids | Glucose and lactate but can use fatty acids |
| Energy production | Mainly oxidative phosphorylation | Mainly oxidative phosphorylation | |
| Mitochondria | Throughout cell; occupies 20–40% of cell volume | Near nuclei; numbers increase during differentiation | |
| Beating | Quiescent | Many cells spontaneous | |
| Force | 40 to 80 mN/mm2 (muscle strips) ~ μN range (single cells) |
0.08–4 mN/mm2 (3D constructs) ~ 200 nN (single cells) |
|
| Conductn | Capacitance | 150 pF | 20–50 pF |
| Resting mem potential | − 80 to − 90 mV | − 20 to − 60 mV | |
| Upstroke velocity | 150–350 V/s | 10–50 V/s | |
| Conduction velocity | 60 cm/s | 10–20 cm/s | |
| Location of gap junctions | Intercalated discs | Circumference of cells | |
| Ion channel density (pA/pF) | INa | − 196 | − 100 to − 244 |
| ICaL | − 4.3 to − 10.2 | − 2.2 to − 10 | |
| Ito | 2.3 to 10.6 | 2.5 to 13.7 | |
| IKs | 0.18 to 0.58 | Most publications 0.3 to 0.7 | |
| IKr | 0.5 | 0.4 to 0.8 | |
| IK1 | − 12 | 0 to − 3.4 | |
| INCX | 2.5 to 3 | 3.6 to 7.9 (inward mode) | |
| Ca2 + kinetics | APD90 | 260 ms | 300–700 ms |
| Cycle Length | 0.8–1 s | 0.8–2 s | |
| T-rise | 2.5 ms | 3.5-10 ms | |
| Triangulation | 45 ms | 45-120 ms | |
Structurally (Table 2), hPSC-CMs are round or multi-angular, small cells with a single nucleus and show chaotic alignment. They have disorganised and short sarcomeres (1.6 μm), an aspect ratio of 2 or 3:1 and no T-tubules. Most commonly, only Z-discs and I-bands can be detected during microscopic analysis. Conversely, adult CMs are highly organised, large rod-shaped poly-nuclear cells with sarcomeres of 2.2 μm and show longitudinal alignment. They have an aspect ratio of 5 to 9.5:1 and prominent T-tubules. Microscopic analysis shows Z-discs, and I-, H-, A- and M-bands.
Differences continue into gene expression and cell function (Table 2). A recent report that showed gene expression in hPSC-CMs was similar to that seen in the first trimester foetal heart [99]. Moreover, MYH6 (αMHC), TNNI1 (foetal ssTnI) and Titin isoform N2BA predominate in hPSC-CMs, whereas it is MYH7 [βMHC], TNNI3 [cTnI] and Titin isoform N2B in adult CMs. Low levels of calsequestrin, phospholambam, ryanodine receptor and SERCA are reported for hPSC-CMs. This is corroborated by relatively little sarcoplasmic reticulum (SR) and mixed responses to caffeine, Thapsigargin & ryanodine. While in both hPSC-CMs and adult CMs oxidative phosphorylation predominates and fatty acids can be used as an energy substrate, hPSC-CMs rely more heavily on glucose and lactate. In adult CMs, mitochondria are located throughout the cell and occupy 20–40% of the cell volume, which contrasts to hPSC-CMs, where numbers are lower and location is perinuclear.
Electrical immaturity of hPSC-CMs is evident from spontaneous beating, since mature adult ventricular CMs are quiescent. This belies high expression of the pacemaker current, If, and low expression of inwardly rectifying potassium current, IK1, which stabilises the resting membrane potential to around -85 mV in adult cells; the value is − 20 to -60 mV in hPSC-CMs (Table 2). Density of IKs potassium and INa sodium channels is highly heterogeneous and can be lower than in adult. Indeed, the presence of the IKs channel in hPSC-CMs, or the right tools to detect it, has recently been called into question [105]. This is of particular concern since CIPA [45] will require these currents in addition to IKr and ICaL. Collectively, these currents usually provide a capacitance of 30-50 pF versus ~ 150 pF in adult CMs and upstroke velocity of 10-50 V/s versus 150-350 V/s. Conduction velocity is also slower in hPSC-CMs (10–20 cm/s versus 60 cm/s) on account of gap junctions being located around the cell circumference rather than at the intercalated discs.
2.1. Improving maturity of hPSC-CMs
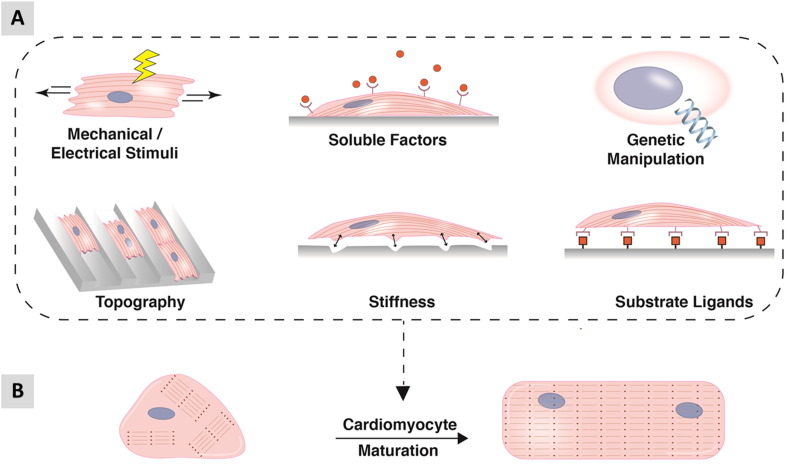
The immaturity associated with hPSC-CMs is delaying progress in the field. The observation that hPSC-CMs undergo structural and functional maturation when transplanted into the working myocardium of model species shows the cells can mature when placed in an appropriate environment [106]. Consequently, many investigators are evaluating physical, chemical, genetic and environmental inducers to facilitate maturation (Fig. 2). Current protocols typically produce beating clusters of CMs by day 6–12 of differentiation and gene expression continues to mature over time. While some reports indicate expression of specific pathways (e.g. β2-adrenoceptor [62]) or global transcriptome [107] largely stabilises between 4 and 8 weeks of differentiation, others have shown improvements in ratiometric markers (e.g. ssTnI:cTnI isoform conversion) continue to occur for periods of up to nearly a year [102]. Extended time in culture has also been shown to improve ultrastructure of the sarcomere, calcium handling and ion channel expression [13].
Fig. 2.
Schematic of in vitro maturation strategies for hPSC-CMs. (A) Methods include biophysical stimuli such as mechanical cues, electrical stimulation, optimising substrate stiffness and topography. Biochemical cues can be presented as soluble factors or substrate ligands within biological or synthetic matrices. Genetic manipulation such as forced expression of missing ion channels has also been adopted as a maturation strategy. (B) The aim of these strategies is to drive the polygonal morphology and disorganised myofibril banding of immature CMs towards a more mature state indicated by rod shaped morphology and parallel myofibrils (see also Table 2).
Although it is not feasible to use extended culture for routine biomedical application, it does provide a useful developmental tool. Thus, hESC-CMs maintained in culture for a year showed molecular signatures similar to those seen for in vivo-derived mature cardiac tissues [108]. The data identified let-7 as the most highly up-regulated microRNA in the culture-matured hESC-CMs. The authors went on to show overexpression of let-7 family members enhanced cell size, sarcomere length, force of contraction, and respiratory capacity in hESC-CMs. It was also suggested that the mechanism of let-7-driven maturation may be via down-regulation of the phosphoinositide 3 kinase (PI3K)/AKT protein kinase/insulin pathway and an up-regulation of fatty acid metabolism [108]. Similar approaches to over-express components of the CM machinery, such as miR-1, calsequestrin or Kir2.1 have also facilitated maturation of hPSC-CMs [13].
2.2. Maturation: medium additives
Adding supplements to the culture medium is potentially a straightforward way to modulate hPSC-CM maturation. Table 3 shows factors that have undergone some level of testing in mouse and human cells to evaluate their ability to induce CM maturity. Triiodothyronine (T3) is essential for normal cardiac development and during the perinatal period, it regulates isoform switching of several myocardial proteins, including MHC and titin. Incubation of hPSC-CMs with T3 for 1–2 weeks led to changes consistent with maturation, including 11-fold up-regulation of αMHC, lower proliferation rates (but not increased bi-nucleation), 1.5-fold increase in twitch force (to ~ 12nN/cell), higher calcium-derived maximal upstroke and decay velocities enhanced oxygen consumption rates [109].
Table 3.
Factors with potential for facilitating maturation hPSC-CMs.
| Factor | Known function | Model system | Effect | Ref |
|---|---|---|---|---|
| Insulin | Regulates glucose uptake and postnatal cardiac growth | ARVD hiPSC-CM | Induction of adult-like metabolism in model of adult onset disease | [70] |
| IBMX | Induction of adipogenesis | |||
| Dexamethasone | Induction of adipogenesis | |||
| Corticosterone | Structural and functional maturation of the foetal heart in vivo | Mouse foetal cardiomyocytes | Improve contractility, Z-disc assembly, mature myofibrils and mitochondrial capacity | [207] |
| PPARα | Regulator of fatty acid metabolism in adult CMs | ARVD hiPSC-CM | Co-activation of PPARα and PPARγ promoted lipogenesis, apoptosis & channel deregulation | [110] |
| PGC-1α (PPARγ coactivator 1α) | Promotes cardiac mitochondrial biogenesis | hESC-CM | Controlling PGC-1α and reactive oxygen species implied in recapitulating mature phenotypes | [208] |
| Klf15 | Glucocorticoid receptor target that interacts with PPARα to regulate cardiac lipid metabolism | Cardiac progenitors from mouse hearts | Cells with plakoglobin mutation showed increased Klf15, CEBPα, Wnt5b | [209] |
| 13-HODE | Component of oxidised low-density lipoprotein via PPARγ | ARVD hiPSC-CM | Induced lipogenesis and apoptosis in model of adult onset disease | [70] |
| Rosiglitazone | PPARγ activator that increases adiponectin in CMs | |||
| Indomethacin | Mediates agonists for PPARγ to regulate adipogenesis | |||
| Insulin-like growth factor | IGF1 receptor induces heart growth via the PI3K pathway | mESC | Insulin or IGF1/2 during early differentiation increased mesodermal cell proliferation | [210] |
| T3 | Thyroid hormone essential for optimal heart development | hiPSC-CM | T3 drives maturation | [109] |
| EPA | Fish oil that affects developmental bioenergetics | mESC | Increases in gene expression associated with cardiac development | [211] |
Abbreviations: IBMX, 3-isobutyl-1-methylxanthine; ARVD, arrhythmogenic right ventricular dysplasia; PPARα, peroxisome proliferator-activated receptor α; 13-HODE, 13-hydroxyocta-decadienoic acid; T3, Tri-iodo-L-thyronine; EPA, eicosapentaenoic acid.
An alternative approach was taken by Wen and colleagues to model the late onset disorder, arrhythmogenic right ventricular dysplasia (ARVD) [110]. A combination of insulin, dexamethasone (a glucocorticoid) and IBMX (3-isobutyl-1-methil-xanthine; a phosphodiesterase inhibitor) was used to drive metabolic maturation by increasing fatty acid synthesis and triggering activation of PPARα, which led to enhanced mitochondrial oxidative phosphorylation. Further addition of the PPARγ activators, rosiglitazone and indomethacin, to the medium caused abnormal PPARγ activation in ARVD hiPSC-CMs. This unveiled the pathological phenotypes associated with this condition, which include exaggerated lipogenesis, apoptosis, Na+ channel down-regulation and defective intracellular Ca2 + handling. However, it is unlikely that medium additives alone will induce complete maturation of CMs, hence the effect of modulating other components of the cell environment are being investigated.
2.3. Maturation: biophysical cues
Unlike the hESC-CMs, primary CMs in the atria and ventricles of adult human heart do not exhibit spontaneous beating. Instead, they are innervated by the autonomic nervous system via nodal CMs, which determines pace and contractility. To mimic this excitation–contraction coupling, Radisic and colleagues applied extrinsic electrical field stimulation to neonatal rat ventricular myocytes (NRVM) [111]. Compared to non-stimulated cells, field stimulation induced elongated morphology concurrent with increased sarcomere volume and numbers of mitochondria, intercalated discs, gap junctions and contractility. This work has been translated to hPSC-CMs [112], wherein three dimensional (3D) configurations have been subjected to electrical field stimulation of increasing frequency (see 3D engineering section below).
Other investigators have assessed the impact of mechanical cues to mimic the intra- and extra-cellular stresses that CMs experience. This can be achieved by altering cyclic stretch, mechanical load and substrate stiffness. Thus, Mihica and co-workers [113] reported hESC-CMs seeded onto gelatin-based scaffolds and stressed with cyclical stretching showed several hallmarks of maturation, namely, cell elongation, increased expression of gap junction proteins and ion channels, while imaging confirmed shorter calcium cycle durations. Implantation of the hPSC-CM constructs under the epicardium of ischemic rat hearts demonstrated enhanced survival and engraftment in the stretched constructs.
Modulating substrate stiffness provides an alternative route to varying the level of load experienced by the CMs. The rationale is that substrate stiffness of the myocardium changes dynamically during development. In the mouse, the elastic modulus increases from 12kPa in the embryonic heart to 39kPa in the neonate [114]. In the human heart, the modulus is 10kPa at the start of diastole but increases to 500kPa at the end [115]. While these dynamic ranges are known to exist, there is considerable variation in the literature as to what the ‘correct’ range of elastic moduli to translate from in vivo to in vitro. When cultured on substrates that mimic the elasticity of the developing myocardium (i.e. 1-11kPa, values for rat and quail), CMs from chicken embryos produced contractile force and developed actomyosin striations [116]. In contrast, CMs cultured on harder substrates (34kPa) designed to mimic post-infarct fibrotic scar tissue cells overstrained themselves, lack striated myofibrils and stop beating [116]. For neonatal rat ventricular CMs cultured on hydrogels of 90kPa elastic modulus, there was a high level of sarcomeric content and microtubule polymerisation relative to cells cultured on 13kPa hydrogels [117]. The same study also showed that peak systolic force was generated from CMs seeded to micro-patterned shapes of ~ 7:1 aspect ratio/13kPa substrates but ~ 2:1 aspect ratio/90kPa substrates [117]. In another report, neonatal rat ventricular CMs cultured on collagen-coated polyacrylamide gels with an elastic modulus 10kPa showed enhanced maturation, as evidenced by increases in sarcomere alignment, mechanical force, improved calcium transients and sarcoplasmic calcium stores relative to cells on substrates with higher elastic moduli [118]. Most recently, single hPSC-CMs were cultured on 10kPa polyacrylamide substrates patterned with Matrigel in 2,000μm2 rectangles of aspect ratio between 5:1 and 7:1. The key findings were that translation of sarcomere shortening to mechanical output was highest in 7:1, while increased substrate stiffness or applied overstretch perturbed myofibril structure and mechanical output in 7:1 hPSC-CMs [119]. It is possible these discrepancies reflect the different approaches used to measure elastic modulus or differences in the cell types, as well as their isolation and culture methods. Irrespective of the reasons, the diversity of data makes it difficult to pinpoint conclusions and a careful analysis of the impact of elastic modulus on hPSC-CM function is needed.
2.4. Maturation: chemical cues from the substrate
The substrate chemistry and structure can have a significant influence on the maturity of hPSC-CMs. It is known that different extracellular matrices can influence structure and cell behaviour, with phenylephrine-induced maturation absent when neonatal rat ventricular CMs were cultured on gelatin but present on fibronectin or laminin [120]. This has prompted investigations into the impact of synthetic polymers on hPSC-CMs. A library of combinatorial polymers was used to identify a mixture of 4% polyethylene glycol:96% carboxylated PCL as enabling the greatest level of contractility and mitochondrial function. This was concurrent with increases in expression of MLC2v and integrin α7, as well as a modest level of isoform switch from foetal ssTnI to the postnatal cTnI [121]. Patel and co-workers [122] screened almost 700 polymers for their utility as growth substrates for hPSC-CMs. These were refined down to identify chemically-defined methacrylate co-polymers (isobornyl and tert-butylamino-ethyl) on which hPSC-CMs exhibited a 6-fold faster upstroke velocity and significantly longer sarcomeres relative to gelatin controls. This copolymer also enhanced detection of the anti-cancer drug, doxorubicin, by up to 10-fold when myofibril disruption was used as the parameter for cardiotoxicity.
Combining substrates and enhanced maturation medium has also been investigated. Single hPSC- or second trimester human foetal-CMs were seeded to gelatin patterned lines on an acrylamide substrate loaded with fluorescent beads, which allowed measurement of contraction force [123]. While hPSC-CMs showed distinctly lower contraction stress than the foetal counterparts (~ 0.25mN vs ~ 0.4mN/mm2), incubation with a proprietary commercial medium containing T3 promoted contraction force to beyond that seen in the foetal cells (~ 0.5mN vs ~ 0.4mN/mm2). Concurrently, there was evidence of improved electrophysiology (upstroke velocities, action potential amplitudes, resting membrane potentials), sarcomeric organisation and cardiac-specific gene expression.
Nevertheless, while these improvements are encouraging, the data showed that the hPSC-CMs mirror the late-stage foetus rather than the adult myocardium.
2.5. Development of muscular thin films (MTFs)
Notwithstanding the variables above, relative to unpatterned substrates, hPSC-CMs seeded onto fibronectin-coated micro-grooved polydimethylsiloxane (PDMS) scaffolds (~ 1.8 MPa) showed cellular alignment, sarcomeric organisation, enhanced calcium properties and heightened responses to caffeine, suggesting improved cycling [124]. Micro-patterned PDMS can also be incorporated into muscular thin films (MTFs), which have tunable stiffness and flexibility to mimic healthy as well as diseased myocardium conditions [125]. Shortening of CMs during synchronous contraction causes the MTF to flex and adopt a pseudo 3D conformation, thereby enabling the force of contraction to be calculated [126]. The MTF platform has been used to evaluate function of various cell types. Feinberg and co-workers [127] investigated the impact of architectures comprising isotropic (ISO) monolayers, anisotropic (ANISO) monolayers and 20 μm wide 20 μm spaced lines (LINES) on neonatal rat ventricular CMs. Relative to the ISO configuration, ANISO and LINES showed uniaxial alignment, enhanced calcium handling and conduction velocity, and a 10-fold increase in peak systolic stress [127]. Treatment of human umbilical arterial vascular smooth muscle cells as an anisotropic monolayer on MTFs showed application of 50 nM endothelin-1 increased basal contractile stress from ~ 17kPa to ~ 22kPa [128]. The same study went on to show MTFs seeded with neonatal rat ventricular CMs generated a peak systole stress of ~ 9kPa, similar to contractility measurements performed on papillary muscle from adult rats [128].
The MTF platform has been extended to modelling of the mitochondrial myopathy, Barth syndrome, which is caused by mutations in the X-linked gene, Tafazzin (TAZ). Patient-derived hiPSCs with mutations in TAZ were differentiated to CMs and cultured as self-organising laminar, anisotropic MTF myocardium constructs for “heart-on-a-chip” analyses [129]. Control CMs showed better sarcomeric alignment than the disease samples. During electrical field stimulation from 1 to 5 Hz in galactose-containing medium, control hiPSC-CM MTFs produced a twitch stress of 250 Pa relative to the significantly weaker values of 100 Pa in the diseased tissues. This recapitulated the Barth Syndrome myopathic phenotype and provides a basis for further investigation of the mechanisms that underlie the condition in the engineered tissue. The development of MTFs that have electrodes incorporated for electrical field stimulation and micro-fluidic channels for drug loading will allow the platform to be used for cost-effective and scalable for pharmacological testing [126].
Nevertheless, 2D systems and MTFs lack the structure of the adult heart. In part, this may be due to improper cell attachment to the substrate. Bidirectional translation of mechanical forces between the contractile apparatus and ECM is governed by integrins [130]. While there are similarities in integrin expression between adult-CM and hPSC-CMs, the adult cells typically attach to surfaces with their distal cellular regions (costameres integrin rich area) [131], whereas hPSC-CMs form integrin attachments along their basal surface. In addition, sarcomeres in adult-CMs are aligned in perpendicular direction from the cell axis, which results in higher probability of actin-myosin cross-bridge formation and hence greater contractile force. Collectively, these differences lead to different mechanotransduction mechanism and contractility patterns between adult- and hPSC-CMs. It may be that nano-patterning the substrate could promote better adhesion and alignment in hPSC-CMs and will need to be tested.
2.6. Engineering heart tissues in three dimensions
In parallel to the development of the various 2D and MTF systems available, 3D approaches to incorporate hPSC-CMs have been investigated. The 3D systems that have been validated to a higher level include engineered heart tissues (EHTs [132]), cardiac microtissues (CMTs or microtissue gauges; μTUGs [133]) and cardiac biowires [112]. EHTs and CMTs rely on casting cell-hydrogel mixtures in moulds featuring elastic anchors that guide cardiac tissue organisation in an aligned conformation. This enables the CMs to perform contractile work against the anchors, thus developing an auxotonic tension that resembles physiological conditions. EHT fabrication involves encapsulating CMs in fibrin gels between two silicon posts, whereas CMTs are based on CM-seeded fibrin/collagen gels tethered to PDMS cantilevers. Functional evaluation of CM contractility can be monitored indirectly in the EHTs by video-optically analysing the deflection of the silicon posts with known mechanical properties. In CMTs, direct force reporting is via a microelectromechanical sensor coupled to the cantilevers. Importantly, CMs in these systems can be loaded with calcium- or voltage-sensitive dyes, enabling functional analysis of calcium transients and electrophysiology [134]. Cardiac biowires consist of a cell-laden collagen gel around a surgical suture placed in a PDMS mould.
These 3D systems each have pros and cons. EHTs and biowires are produced in centimetre-scale that require 0.5–1 million CMs per unit. For EHTs, format is 24-well requiring 12–24 million hPSC-CMs, which is costly at current commercial rates of USD$1000/million cells. In contrast, a million cells are sufficient to produce 100–200 microscale CMTs and so 96-well formats are feasible. Measurements in EHTs are possible over several weeks, potentially enabling acute and chronic drug effects to be monitored, whereas long-term analysis in CMTs is more difficult as they are harder to handle. Dynamic load of the silicon posts in EHT and cantilevers in CMTs can be varied to mimic heart failure. Biowires do not permit measurement of contractility, which is a major disadvantage.
The 3D configuration has been shown to enhance CM maturation. CMs in EHTs align along the force lines between the silicon posts, with 3.4-fold improved longitudinal orientation relative to embryoid body-CMs. Sarcomeres in the EHTs become evenly distributed both around the nuclei and in the periphery of the cells, although connexin-43 is still expressed along the sarcolemma (unlike adult CMs that express this gap junctional protein in the intercalated discs). Elevated expression of adult isoforms of sarcomeric genes (e.g. MYH7 encoding β-MHC) also occurs. EHTs display key responses to physiological and pharmacological stimulation, such as increased contractile forces at higher extracellular Ca2 + concentrations and upon treatment with β-adrenergic agonists [135]. CMTs developed with hESC-CMs promote cell alignment and expression of mature CM markers such as BNP.
These improvements in maturation appear to be further enhanced by incorporating electrical stimulation, even if not always as anticipated. Beat rate is ~ 3 Hz in human foetal hearts beat but ~ 1 Hz in adults. This suggests that reduction in rate might correlate with maturation but the opposite was observed in hPSC-CMs. Thus, increasing stimulation frequency from 1 to 6 Hz in hPSC-CM biowires caused maturation, as evidenced by improvements in structure and function [112]. It may be that forcing mechanical stress by pacing is more important than the electrical stimulus per se. Nevertheless, stimulated biowires had myofibrils with a higher degree of ultrastructural organisation (aligned Z-discs displaying up to two I-bands per disc; organised sarcomeres showing up to 0.4-H zones per sarcomere) and enhanced expression of cardiac contractile proteins, such as sarcomeric α-actinin, actin and cTnT. Conduction velocity increased (~ 15 vs ~ 10 cm/s) and there was greater sarcoplasmic reticulum maturity, with caffeine treatment resulting in higher cytosolic Ca2 + transients. Electrical stimulation of biowires induced higher IKr (~ 0.81 vs 0.52pA/pF) and IK1 (~ 1.53 vs 0.94pA/pF) currents [112]. Similar data have been created for hPSC-CM derived EHTs, wherein electrical stimulation improved Ca2 + transients, contraction force and response to isoprenaline [134].
Nevertheless, 3D platforms have not produced fully mature CMs and are absent for properties including (i) formation of T-tubules, (ii) expression of the full array of sarcomeric proteins (including α-sarcomeric protein and myosin-binding protein C), (iii) physiological potassium ion channel densities, and (iv) contraction forces. For example, infarcted heart muscle has a twitch force of 40–80 mN/mm2, which is ~ 30- or 600-fold greater that EHTs comprising rat-CMs (2-4mN/mm2) or hPSC-CMs (0.08–0.12mN/mm2) [136]. Whether these parameters can be improved by combining electrical pacing, medium supplementation with adrenergic agonists, thyroid hormones and growth factors and/or co-culture with supporting cell types (e.g. cardiac fibroblasts [137]) requires further investigation.
As well as use in vitro, the utility of EHTs in correcting myocardial function deficit in animal models of heart failure has been tested. Most studies have used allogeneic transplantation of rat EHTs or xeno-grafting of human EHTs into myocardial infarcted immunosuppressed rats. By 4–12 weeks post transplantation, ~ 25–30% of grafted cells survive intervention [138], [139] and show electrical integration without arrhythmias [140]. Of note, EHT-grafted hearts showed maximum conduction velocities similar to non-infarcted rat myocardium (VT = 0.19 m/s vs VT = 0.16 m/s, respectively) and host-derived angiogenesis with a ~ 2.8-fold increase in vascular density in the EHT-borderzone region [139]. Grafted hearts showed slowing of disease progression, evidenced by improved fractional shortening and lower maximum left ventricular volume [140]. Nonetheless, recovery of the infarcted heart after EHT transplantation was not to ‘healthy’ levels and additional challenges to overcome include: i) reducing immunogenicity of the graft by developing defined, xeno/serum-free culture and EHT fabrication conditions; ii) scaling graft size to include the cell quantities (≥ 1010 cells) needed for the human heart; iii) increasing graft complexity to include not only cardiomyocytes, but also smooth muscle cells and cardiac fibroblasts. Further inclusion of endothelial cells should help to overcome the important issues of vascularization, which could reduce the levels of cell death seen in the transplanted EHTs [138]. Integration of pre-formed vascular structures [141] or of additional extracellular matrix (e.g. collagen + Matrigel) has already shown progress to improved vasculargenesis of EHT-derived grafts in animal models of heart disease [142] and will be an area to explore further in the future.
In summary, different physical, chemical, genetic and environmental factors have been shown to mature hPSC-CMs in 2D and 3D configurations. The varying success may be due to biology or to differences in technique. Recent work by Du and colleagues showed that non-invasive optical mapping of action potential morphology in hiPSC-CMs seeded as confluent monolayers or as sparse cultures did not predict cardiac chamber specificity but, instead, was dependent on cell density [143]. Thus, establishing experimental standards will ensure greater comparability between reports.
3. Towards industrial scalability of hPSC-CM platforms
Most of the studies described above have been carried out by individual academic or industrial labs using less than 5 hPSC lines in conjunction with low throughput technology to test low numbers of parameters and so require only a few million hPSC-CMs. For these technologies to be used as widespread commercial tools, there will be a need to upscale culture and differentiation. Analysis of a hundred compounds over 6 x ½ log doses for 10 replicates using existing 96-well calcium imaging systems [144] or 24-well multiplexed EHTs [135] would require approximately 500 million or 3 billion hPSC-CMs, respectively. For in vivo use, recent studies have shown that transplanting 1 billion hPSC-CMs into the infarcted hearts of pigtailed macaques led to substantial remuscularisation [29] but to achieve the same in the larger human heart would require at least 10 billion CMs. Current commercial rates for 1 million hPSC-CMs costs are USD 1000, meaning at the in vitro and in vivo scales above permitted budgets of most companies or healthcare providers would be exceeded.
3.1. Scale-up of hPSC culture and CM differentiation
The three core requirements for adherent culture of hPSCs are medium, matrix and passaging method. The labour intensive nature of mechanical dissection of individual colonies was never compatible with commercial upscaling and soon gave way to bulk passaging methods [145]. Initially, clump passaging methods used collagenase, dispase or cell scraping, but have largely been replaced by small clump passaging with EDTA and by Accutase and TrypLE enzymes that produce single cells or small clusters (2–5 cells). The latter enzymatic approaches are compatible with single cell cloning needed for genome editing technologies and automated cell counting for integration of hPSCs into robotic culture platforms.
Similar progress has been achieved with culture medium. Poorly defined serum, serum replacements and conditioned medium were superseded by more refined medium such as StemPro, Nutristem, mTeSR and TeSR2 celiz [146], some of which were available as “xeno-free” formulation and potentially compatible with clinical-grade Good Manufacturing Practice (GMP). However, these media often showed considerable batch to batch variability. Development of Essential 8 (E8) has addressed this issue and is becoming the defined medium of choice for many laboratories. It can also be coupled with Sendai-virus and Essential 6 medium to enable efficient integration-free reprogramming of somatic cells into hiPSCs and is compatible with high efficiency monolayer differentiation of hPSCs to CMs (see Table 1). Nevertheless, the high cost of commercially-produced E8 (~ USD $450 per litre) means many research labs are using the published formulation to make their own medium for around USD $70 per litre.
Perhaps the most challenging part of the culture system to define is the matrix. Very few labs now rely on the early methods of using mitotically-inactivated mouse or human feeder cells to support hPSC cultures. However, use of cell derived (e.g. Matrigel, Geltrex) or recombinant (e.g. laminin, collagen, fibronectin, vitronectin, E-cadherin) are frequently used but are expensive, variable and/or labile [145]. Developments in the field have included formulation of humanised versions of the proteins (e.g. CellStart), peptide-polymer conjugates (e.g. Synthemax) and plasma-treated polystyrene. Recently, Celiz and co-workers [145] employed a high throughput materials discovery approach by microarray screening of 909 unique polymers to identify the first synthetic polymeric substrate that achieved both hPSC expansion in commercially-available StemPro and mTeSR media and subsequent multi-lineage differentiation, including to CMs. Nevertheless, the compatibility of the polymer substrate with E8 was not shown and the largest culture format was 6-well so further development is needed.
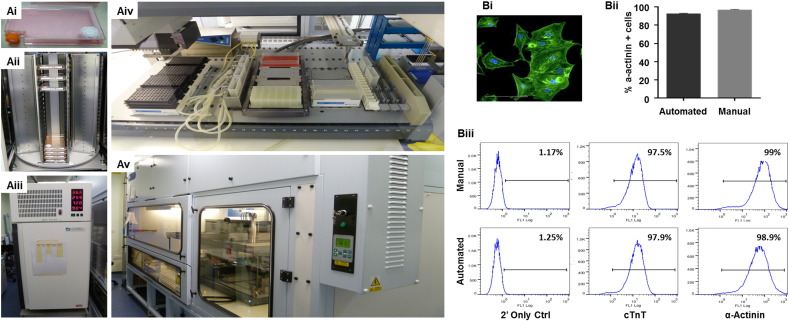
In parallel with the development of improved culture systems, there have been various attempts to evaluate the compatibility of culture protocols on automated liquid handling platforms for robotic scale-up of hPSCs and their differentiated derivatives. Automated systems have been shown to improve the consistency, quality and failure rates often reported from manual handling of cell cultures [147]. The CompacT SelecT system was used to demonstrate feasibility of producing ~ 2.5 x 109 undifferentiated hESCs, although this early report used mouse embryonic fibroblast conditioned medium coupled with Matrigel [148]. The Biomek FXP liquid handler workstation has been used to automate the cardiomyogenic differentiation of mouse ESC in a 384 well plate format [149]. However, in a tour de force of automation Paull and co-workers [150] development a modular system that enabled 1008 mRNA reprogramming events from human adult and control fibroblasts to be processed in batches of 48 samples per run. This resulted in 221 successes, as judged by presence of nascent TRA-1-60 + hiPSC colonies. The automated system was used to induce spontaneous or directed differentiation of several of the lines into lineages including midbrain-type dopaminergic neurons, hepatocytes, metanephric mesenchyme and oligodendrocytes. CMs were also produced at efficiencies of ~ 40–65%, similar to cultures handled manually. In our own lab, we have demonstrated the feasibility of automating large scale production of hPSC-CMs in 90cm2 Roboflask™ format on a Tecan Evoware Liquid Handling Platform (Fig. 3). This custom built system has a capacity of ~ 100 x 90cm2 Roboflasks™, giving a potential maximum batch production yield of ~3 x 109 hPSC-CMs in a fully defined and reproducible manner.
Fig. 3.
Fully automated Tecan Evoware Liquid Handling platform for hPSCs expansion and differentiation. Panel (A) shows the anatomy of the automated platform.
hPSCs cultured in 92 cm2 Roboflasks™ (Corning®) (Ai) in tower stacks (Aii) within integrated 37 °C/5% CO2 automated incubators (LiCONiC Instruments) (Aiii). All media exchanges, cell dissociations, hPSC counting and reseedings are fully automated on a liquid handling deck (Aiv) within a custom build class 2 cabinet (Bigneat Ltd) (Av). In (B), automated hPSC-CM differentiation in monolayer yields populations that stain positive for α-actinin (Bi), with comparable purities between cultures produced by manual or automated processes, as judged by automated image analysis to a-actinin (Bii) or flow cytometry for cTnT or a-actinin (Biii).
An alternative route to scaling hPSCs and differentiated lineages is the use of suspension bioreactors. Undifferentiated hPSCs have been upscaled across 10 passages as multi-cellular aggregates in stirred tank bioreactors in suspension [151], including in mTeSR or E8 medium [152], whilst retaining normal karyotype, expression of pluripotency-associated markers and multi-lineage differentiation potential. While it has been long-established that mouse PSCs can be differentiated in stirred bioreactors to yield > 3 × 109 CMs in stirred bioreactors [153], only more recently have suspension cultures been successfully used for cardiomyogenesis in hPSCs. Thus, in 100 ml bioreactors, batch and cyclic perfusion controlled feeding strategies with induction of using the GSK3 inhibitor, CHIR99021, produced 40 million hPSC-CMs [154]. Based on electrophysiology, 85% cells were ventricular subtype, although no molecular characterisation was performed and the use of action potential morphology to assign subtype has been questioned recently du [143]. Elegant work has also showed pipeline conversion of mouse fibroblasts into iPSCs and then into iPSC-CMs in a single suspension bioreactor [155]. The challenge now is to translate the high efficiency ‘inducible secondary’ iPSC reprogramming into a technology that is compatible with human cells.
4. Towards industrial phenotyping of hPSC-CMs
For in vitro assays, such as safety assessment, there will be a need to integrate hPSC-CMs into existing or new platforms to assess cell function. In parallel with issues above of hPSC-CM maturation, cell phentopying is rapidly becoming a bottleneck. In the following sections, we consider some of the platforms that have been (or might be) used for the medium- to high-throughput measurement of structure, metabolism, electrophysiology, calcium and contractility.
4.1. High content imaging
The use of high content imaging is an industry standard for assessment of physiology of tumour cell with regards cell number, cell shape/size, proliferation, viability, membrane integrity, phagocytosis, apoptosis, cell migration, cell-cell contacts and organelle health (e.g. numbers, size, shape, activity of nucleus, mitochondria, lysosomes) [156]. Assays are typically carried out in 96-, 384- and 1536-well plates, which are compatible with various fully automated platforms [157] such as BD pathway (BD Biosciences), InCell Analyser (GE-healthcare), ImageXpress (Molecular Devices), Opera (Perkin Elmer) and Cellomics Arrayscan (ThermoFisher). For undifferentiated hPSCs, high content imaging was used to screen putative chemicals for their ability to maintain pluripotency in the absence of growth factors or to improve cell survival after passage [158], [159]. Such approaches identified a series of inhibitors of the Rho kinase pathway and pro-survival compounds such as Y27632 that are now used by many labs during routine hPSC culture. A different approach was taken to evaluate the influence of substrate chemistry on pluripotency. By forming up to 2000 microspots of 200–300 μm on a single microscope slide, 909 unique methacrylate-based polymers were assessed for hPSC attachment and OCT4 expression in 4356 individual assays. A polymer was identified that could support pluripotency by using a combination of Imstar digital imaging, Operetta confocal imaging and CellProfiler software [145].
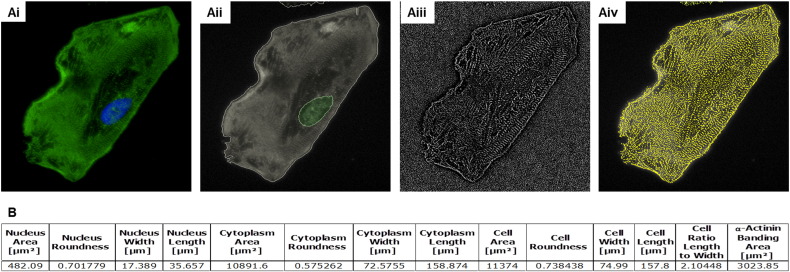
These imaging approaches have been extended to hPSC-CMs (Table 4), allowing identification of polymers that facilitate maturation [122]. Further progress in using imaging to define hPSC-CM structure was made by Pasqualini and co-workers [160]. Using commercial CDI iCell and Axiogenesis CorAt hPSC-CMs stained for the sarcomeric protein, α-actinin, a set of 11 metrics were developed to score the degree of organisation and alignment that sarcomeres acquire during myofibrillogenesis, which were used evaluate phenotypic maturity. This parallels our own work using the Operetta confocal plate reader platform to assess sarcomere number and area within hPSC-CMs (Fig. 4).
Table 4.
Phenotyping platforms for hPSC-CMs. Examples currently have medium-high content capacity or have future potential.
| Platform | hESC-CM | hiPSC-CM | Condition | Mutation | Purpose | Assay | Outcome | Refs |
|---|---|---|---|---|---|---|---|---|
| High content imaging | ||||||||
| Confocal microscope | n/a | 10 patients | HCM | βMHC: Arg663His | Hypertrophy | L-type Ca2 + block | 100 nM verapamil ↓ hypertrophy | [212] |
| ArrayScan VTi 2D Confocal opera LX |
H7, HUES7, SHEF3 | CDI iCells Axio Cor4U ReproCardio2 Patient cells |
Healthy & LQTS2 | KCNH2 c.G1681A | Hypertrophy | Phenylephrine stress | ↑ in 2D cell area of hESC-CMs but not hiPSC-CMs | [74] |
| IC 100/200 | H9 | CDI iCells | Healthy | n/a | Ca2 + & drug risk | Fluo-4AM | EC50 Verapamil, 0.26μM; BayK 8644, 11nM | [213] |
| ImageXpress Micro | n/a | CDI iCells | Healthy | n/a | Hypertrophy | Endothelin-1 + BNP | EC50 enthothelin-1, 11pM | [161] |
| ImageXpress micro | n/a | CDI iCells | Healthy | n/a | Toxicity/viability | 384-well CalceinAM, Hoechst, MitoTracker | Cardiotoxicity of a 131 modulators of Na+, K+, Ca2 + channels & adreno-, dopamine, histamine receptors | [38] |
| “Pulse” all-in-one system | n/a | CDI iCells Axio Cor4U GCaMP-engineered |
Healthy | n/a | Cardioactives on beating | Video imaging of in 24-well plates | Norepinephrine: rate ↑; cisapride, E-4031: arrhythmia, APD ↑; sotalol, quinidine: APD ↑; verapamil, nifedipine: APD ↓ |
[214] |
| “OptoDyce” | n/a | ChR2-engineered | Healthy | n/a | n/s | 96-well all-optical pacing & E-phys | Nifedipine: ↓ APD, ↓ CTD | [215] |
| Mitochondrial function | ||||||||
| ArrayScan vTi | n/a | CDI iCells | Healthy | n/a | Mitochondrial membrane potential | TMRM dye | Chelerythrine: ↓ TMRM
signal Doxorubicin: no change |
[162] |
| Seahorse XF analyser | hESC1 & hESC2 | Healthy & PomD | Pompe Disease | GAA | Bioenergetics | Extracellular flux | Up to 2-fold increase in OCR & ECAR in PomD hiPSC-CMs | [167] |
| Seahorse XF analyser | n/a | Healthy & JK#2, JK#11 | ARVD | PKP2 | Bioenergetics | Extracellular flux | Treatment with insulin, dex, IBMX, rosiglitazone, indamethacin changes glycolysis & fatty acid oxidation | [216] |
| Seahorse XF analyser | n/a | Healthy & DMD | Duchene | DMD | Bioenergetics | Extracellular flux | No difference between healthy & DMD | [166] |
| Seahorse XF analyser | n/a | hiPSC | Healthy | n/a | Bioenergetics | Extracellular flux | T3: ↑ basal & max respiration, non-mitochondrial OCR | [109] |
| Seahorse XF analyser | H7 & RUES2 | n/a | Healthy | n/a | Bioenergetics | Extracellular flux | ↑ Max respiration capacity | [108] |
| Seahorse XF analyser | n/a | Healthy & T1DMiPSC | Type 1 Diabetes | n/s | Bioenergetics | Extracellular flux | Diabetes: ↓ basal OCR and ECAR; + Glucose: ↑ ECAR, no change in OCR; +2DG: ↓ ECAR, no change in OCR |
[168] |
| Seahorse XF analyser | NKX2-5eGFP/w engineered | n/a | Healthy | n/a | Bioenergetics | Extracellular flux | ↓ Maximum respiration ↓ Basal ATP turnover |
[208] |
| Seahorse XF analyser & plate luminometer | n/a | BTH-H, BTH-C, WT1, WT2, WT3 | Barth Syndrome | TAZ | Bioenergetics | Extracellular flux | ↓ Respiration capacity in patient CMs | [129] |
| ATP production | Bioluminescent ATP | ↓ ATP generation in patient CMs | ||||||
| Calcium imaging | ||||||||
| ImageXpress Micro | n/a | CDI iCells | Healthy | n/a | Beat rates | Calcein AM | IC50 epinephrine:
50nM IC50: isoproterenol: 6nM |
[163] |
| IonOptix | n/a | EHT | Healthy | n/a | Ca2 + transients | Fura-2 | EC50 of 1.05mM Ca2 + in human EHTs | [134] |
| FDSS/μCELL | n/a | Axio Cor4U | Healthy | n/a | Calcium transients | FLIPR Calcium 5 | Ca2 + transients change with astemizole | [187] |
| FLIPR Tetra 384 | n/s | n/s | Healthy | n/a | Beat rate, peak shape and regularity | FLIPR Calcium 5 | 131 compounds: cardiac glycosides, anti-arrhythmics, α-/β-adrenoceptors, Ca2 + blockers, anti-histamines | [38] |
| Leica confocal | n/a | Healthy | n/a | Ca2 + profile; toxicity | Fluo-4AM | Ca2 + perturbations: astemizole > thioridazine > cisapride > flecainide > valdecoxib > sotalol > nadolol | [217] | |
| IonOptix & IonWizard | n/a | CDI iCells | Healthy | n/a | Ca2 + transients | Fluo-2 | 51 compounds: 70% specificity; 87% sensitivity; IC50 of 18 drug panel | [39] |
| FLIPR Tetra | FLIPR Calcium 5 | |||||||
| Automated planar patch clamp for intracellular/whole-cell/patch recordings | ||||||||
| PatchXpress 7000A | n/a | CDI iCells | Healthy | n/a | INa, ICa, IKr | Voltage clamp | IC50: TTX, 0.64 μM; nifedipine, 38nM | [24] |
| QPatch | GE Cytiva | CDI iCells | Healthy | n/a | INa, ICa, IK; AP induction | Voltage + current clamp | IC50: TTX, 10.3μM; nifedipine, 95.3nM; APA: 80Mv; APD: 400ms | [176] |
| Patchliner | n/a | Axio Cor4U | Healthy | n/a | INa, IK; AP induction | Voltage + current clamp | BayK 8644: ↑ APD & APA; TTX: blocked AP induction | [177] |
| CytoPatch™2 | n/a | Axio Cor4U | Healthy | n/a | INa, IK; AP induction; pharmacology | Voltage + current clamp | Nifedipine: ↓ APD90; cisapride: ↑ APD90; TTX: ↓ APD90 | [178] |
| Multi-electrode arrays for recordings of Extracellular Field Potentials (EFP) | ||||||||
| MEA2100 | n/a | CDI iCells | Healthy | n/a | Compounds on beat rate and amplitude | 6-well MEA | TTX, ZD7288: ↓ BR; ISO: ↑ BR + ↑ BA; Ouabain ↑ BA; Nifedipine: ↓ BA + ↑ BR; E4031/RO5657: arrhythmias suppressed by nifedipine | [218] |
| MEA1060-Inv-BC | n/a | UTA.04602.WT | Healthy | n/a | E4031 on EFP profile | 6-well MEA | E4031 ↑ FPD & ↓ amplitude of Na peak | [219] |
| MEA60 | n/a | CDI iCells | Healthy | n/a | Compounds on FPD | 6-well MEA | IKr blockers: ↑ FPDC, arrhythmias; IKs blockers: ↑ FPDC; ICa,L blockers: ↓ FPDC | [220] |
| Maestro | GE cytiva | n/a | Healthy | n/a | Compounds on FPD | 48-well MEA | 21-drug panel: hERG blockers, ↑ FPD; Na blockers, ↑ FPD; Ca blockers, ↓ FPD; Ca activators, ↑ FPD | [221] |
| Maestro | n/a | CDI iCells | Healthy | n/a | Compounds on FPD | 48-well MEA | 15-drug panel: hERG blockers, ↑ FPD; IKs blocker L768673, No effect on FPD | [181] |
| Maestro | n/a | CDI iCells | Healthy | n/a | Compounds on FPD | 48-well MEA | Verapamil, Ouabain: ↓ FPD; ranolazine, amiodarone, terfenadine, flecainide, cisapride: ↑ FPD | [180] |
| Impedance for quantification of CM contractility | ||||||||
| xCELLigence RTCA | n/a | CDI iCells | Healthy | n/a | Compounds on beating | 96-well assay beat rate & amplitude | TTX, ZD7288: ↓ BR; ISO: ↑ BR + ↑ BA; Ouabain ↑ BA; Nifedipine: ↓ BA + ↑ BR; E4031/RO5657: arrhythmias suppressed by nifedipine | [218] |
| xCELLigence RTCA | n/a | CDI iCells | Healthy | n/a | Compounds on beating | 96-well assay beat rate & amplitude | Amlodipine: ↑ BR, ↓ BA; Mibefradil: No effect; E4031: ↓ BR, arrhythmias; Zatebradine: ↓ BR | [222] |
| xCELLigence RTCA | n/a | CDI iCells | Healthy | n/a | Compounds on beating | 96-well assay beat rate & amplitude | Beat rate: 42.8 ± 0.87bpm; crizotinib, sunitinib: ↓ BR / beating cessation | [223] |
| xCELLigence RTCA | n/a | CDI iCells | Healthy | n/a | Compounds on beating | 96-well assay beat rate & amplitude | Dofetilide/E4031: ↓ BA, arrhythmias; HMR1556: no change; TTX: ↓ BR, ↓ BA; Verapamil: ↓ BA, ↑ BR | [224] |
| xCELLigence RTCA | n/a | CDI iCells | Healthy | n/a | Compounds on beating | 96-well assay beat rate & amplitude | Detected 7/9 positive inotropes (78%); 20/21 negative inotropes (95%); 14/19 negative controls had no effect | [205] |
| xCELLigence RTCA | n/a | CDI iCells | Healthy | n/a | Cardiotoxicity | 96-well assay beat rate & amplitude | 16/18 drugs with known toxicities caused changes in beat pattern | [225] |
| 2D platforms for studying cardiac contractility | ||||||||
| Atomic force microscopy | H7 | Stemcell technologies | Healthy | n/a | Beat rate, contraction force | AFM cantilever tip (single well) | Beat rate: 0.80/s (single cell) or 1.72/s (aggregates); Force: single cells, 2.37 nN; norepinephrine from 0.18 nN to 0.48nN for Ipsc-CMs and 0.097nN to 0.37 nN for H7 | [196] |
| Microposts | n/a | IMR-90 | Healthy | n/a | Contraction, relaxation, power | High-speed video microscopy (single well) | Twitch velocities 1.4-2.0 μm/s contraction; 1.2-1.6 μm/s relaxation; peak contraction: 29 fW; systolic forces: 15nN/cell | [198] |
| Impedance | n/a | CDI iCells | Healthy | n/a | Beat rate and amplitude | 96-well Impedance | 49 positive & negative inotropes for cardiac contractility: change beat rate | [205] |
| Displacement of fluorescent beads | n/s | n/s | Healthy | n/a | Contractility | 6-well microscopy | 0.26 mN/mm2 for iPSC-CMs and 0. 29 mN2 for hESCs | [123] |
| 3D platforms for studying cardiac contractility | ||||||||
| Bioartificial cardiac tissue (BCT) | n/a | n/s | Healthy | n/a | Contractility | Bioreactor | 4.4 mN/mm2 | [199] |
| Fibrin based engineered human tissues (EHTs) | hESC | clone C25 | Healthy | n/a | Contractility | 24-well video-optical | ISO: force ↑ (185%), contraction time ↓,relaxation time ↓; ISO + xiaflex: force ↓, relaxation time ↑; twitch force 0.08 mN/mm2 for Ipsc-CMs | [134], [226] |
| 3D cardiac micro tissues (CMTs) | hES2 | CD34 + precursors for iPSCs | Healthy and Ad-PLB | n/a | Contractility | Optical (photometric camera) up to 96 well | Twitch tension for hvCMTs after Ad-PLB treatment ↓ from 5.8 μN to 2.2 μN, ISO: 119% ↑ of force and twitch tension a 4.5 μN | [227] |
| Multi-parameter analysis | ||||||||
| CellOPTIQ | n/a | CDI iCells | Healthy | n/a | Action potential & T-rise | Voltage sensitive dye di-4-ANEPPS | E4031: ↓ APD50, APD75 & APD90; Nifedipine: ↓ APD75; Mexiletine ↑ Trise | [144] |
| CellOPTIQ | n/a | CDI iCells | Healthy | n/a | Action potential | Voltage sensitive dye di-4-ANEPPS | bs906: no effect on APD50 & APD90; ISO: ↑ APD50 & APD90 reversed with bs906 | [228] |
| CardioExcyte96 | GE Cytiva Cellectis clusters |
CDI iCells Axio Cor4U ReproCardio2 |
Healthy | n/a | Compounds on beat profile & EFP | CardioExcyte 96 combined impedance & EFP | Nifedipine: ↓ Amplitude/FPD; E4031: ↓ amplitude, ↑ FPD, EADs and beating cessation; S-BayK8644: ↓ amplitude | [206] |
HCM, hypertrophic cardiomyopathy; LQTS, long QT syndrome; n/a, not applicable; n/s, not specified; APD, action potential duration; CTD, calcium transient duration, OCR, oxygen consumption rate; ECAR, extracellular acidification rate; TMRM, tetramethylrhodamine, methyl ester; T3, triiodothyronine; EHT, engineered heart tissue; APA, action potential amplitude; TTX, tetrodotoxin; FPD, field potential duration; BR, beat rate; ISO, isoprenaline; BA, beat amplitude; CDI, Cellular Dynamics International; Axio, Axiogenesis.
Fig. 4.
Automated image analysis of hPSC-CMs. Panel (A) shows images captured on the Perkin Elmer Operetta confocal plate reader of hPSC-CMs stained with of α-actinin (Ai). Analysis using Harmony™ image software identifies cellular nuclei and cytoplasmic regions (Aii), which is then texture-filtered (Aiii) to allow recognition of subcellular structures (Aiv). In (B), automated processing gives quantitative metrics in a high throughput manner that can be used for rapid phenotyping of hPSC-CMs.
High content imaging is now being used to evaluate the effect of chemical insult on hPSC-CMs (Table 4). As examples, chemical induction of hypertrophy in hPSC-CMs by angiotensin II, phenylephrine, or endothelin-1 has been evaluated in 96-well or 384-well plates. The Cellomics Arrayscan was used to show that some aspects of the pathways underlying hypertrophy are defective in multiple lines of hiPSC-CMs relative to hESC-CMs [74], while the ImageXpress Micro platform was used to calculate the EC50 of enthothelin-1 as 11 pM in commercial hiPSC-CMs [161]. Imaging was also used to examine hypertrophic responses of patient-specific hiPSC-CMs from a ten-member family cohort carrying a hereditary hypertrophic cardiomyopathy missense mutation (Arg663His) in the MYH7 gene [63].
For assessment of cardiotoxicity, Mioulane and co-workers [162] triggered cell stress with the cell-permeable protein kinase C inhibitor, chelerythrine. Coupling staining cells with potentiometric dye (TMRM; tetramethylrhodamine methyl ester), caspase-3 or BOBO-1 with Cellomics Arrayscan imaging assessed mitochondrial function, apoptosis and cell death, respectively. Application notes from CDI show quantification of the cardiotoxic effect of valinomycin, etoposide and rotenone in hPSC-CMs using high content imaging of changes in mitochondrial and lysosomal physiology, DNA damage and oxidative stress (cellulardynamics.com). Unpublished data presented by GE-healthcare show how 26 anti-cancer agents changed 19 different cell morphological and functional parameters in hPSC-CMs, with analysis being carried out on 3 replicates, 2 timepoints and 7 doses in a 384-well plate format using the InCell 2000 platform. This analysis produced graphical profile sets that were associated with high, moderate, low or no drug-induced cellular toxicity. In an impressive study, Sirenko and colleagues [163] used CalceinAM, Hoechst and MitoTracker imaging on the ImageXpress Micro to evaluate cardiotoxicity of 131 modulators of Na+, K+ and Ca2 + channels, as well as adreno-, dopamine- and histamine-receptors on CDI iCell hiPSC-CMs 384-well format. This shows that using hPSC-CMs in high content imaging has now become reality.
4.2. Mitochondrial function
Mitochondria can be influenced by developmental stage, disease state and toxic insult. Hence, analysis of function in hPSC-CMs has been evaluated to gauge maturity, model disease and quantify toxicity (Table 4). As well as the imaging systems discussed above, function of mitochondria can be determined by measuring glycolysis and oxidative phosphorylation (OXPHOS), for which the preferred substrates are glucose in immature CMs and fatty acids in mature CMs, respectively. Initial platforms for evaluation were low throughput. This prompted the development of MitoXpress™, a long-decay phosphorescent oxygen probe [164], which allows estimation of IC50 values of 25 drugs/day using a 96-well format [165]. As an alternative platform, the Seahorse Extracellular Flux Analysers uses 24- or 96-well custom plates to determine in vitro oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) to assess OXPHOS, glycolysis and fatty acid oxidation. These approaches have been used to confirm a preference of hPSC-CMs for glycolysis when cultured in medium containing glucose, consistent with developmentally immature state. However, hPSC-CMs can undergo some level of maturation by culturing in galactose/fatty acid-containing medium to force transition towards OXPHOS or treating with T3 [109] or overexpressing let7 miRNA family [108].
The Seahorse platform is now becoming a common tool in evaluating disease state in hPSC-CMs (Table 4). Changes in OCR, ECAR, glycolysis, fatty acid oxidation, maximum respiration capacity and ATP generation have been studied in hPSC-CM models of Barth syndrome [67], Duchenne muscular dystrophy [166], Pompe's disease [167], ARVD [70] and type I diabetes [168]. Usually, this involves recording basal levels of activity from the hPSC-CMs and then subjecting them to a sequential treatment of electron transport chain inhibitors (e.g. oligomycin, rotenone, antimycin-A) or uncouplers (FCCP). The same approaches are now being adopted for compound screening. Enzo Life Sciences used Axiogenesis Cor.4 U hiPSC-CMs to measure oxygen consumption and glycolytic flux in real-time via oxygen and pH sensor probes in 96- or 384-well to inform on drug induced toxicity and mitochondrial function.
4.3. Electrophysiology and calcium
The use of sharp glass electrodes to access the cell interior via conventional patch clamp electrophysiology is considered the gold-standard for studying ion channel activity. No other technique offers more insight into the functionality, kinetics, gating properties and pharmacology of ion channels. However, the complexity of this low throughput, labour-intensive approach allows only 10–15 data points a day, which is insufficient to meet the demands of both academic and industrial labs. This has prompted the development of various platforms for direct or indirect measurement of electrophysiology and/or calcium flux, which are now being applied to hPSC-CMs (Table 4).
4.3.1. Planar patch clamp electrophysiology
To overcome the low throughput of conventional sharp electrodes, several ‘planar’ patch clamp platforms have been developed that can process 16 to 384 multiple recordings in parallel. While these systems differ in their technical specifications and level of automation, they increase data throughput 10- to 100-fold depending on the ion channel under investigation and the platform used [169]. By reducing the complexity of the process, they also make patch clamping accessible to more users, regardless of previous experience in electrophysiology [170]. The use of platforms including IonWorks Quattro [171], PatchXpress [172], Patchliner [173], SynchroPatch [174] and Opatch [175] has been mainstream for the analysis of recombinant cell lines overexpressing single ion channels, and they are now being trialled for use with more complex hPSC-CMs [24], [175], [176], [177] with varying degrees of success (Table 4).
The PatchXpress has been used with high purity hiPSC-CMs to simultaneously record from 16 channels [24]. The authors measured densities of INa, Ica and IKr currents in the presence or absence of their respective pharmacological inhibitors, tetrodotoxin, nifedipine and E4031. However, the study used an engineered hiPSC line with a neomycin phosphotransferse drug resistance cassette targeted downstream of the MYH6 (αMHC) locus to enrich the CMs, a strategy that is not easily adopted across multiple hPSC lines. Seal rates were relatively low at ~ 50% across 58 wells, as was the quality of the seal (~ 200 megaOhms [MΩ]). Genetically purified hiPSC-CMs have also been used on the CytoPatch2 platform [178]. Analysis in current clamp mode enabled retrieval of ventricular, atrial/nodal and S-type action potential morphologies. By achieving seal resistances over 1 GΩ, the action potential durations of ventricular-like cells were shown to be decreased by nifedipine and tetrodotoxin, but increased by cisapride [178]. In our own work (submitted CD, DR), we have used high efficiency differentiation protocols to produce CM purities of > 80% across 6 healthy or diseased hPSC lines. Processing with the Patchliner platform enabled catch rates of ~ 81%, seal rates of ~ 80% and seal qualities of up to > 2GΩ. Analysis in voltage clamp mode, allowed evaluation of INa, Ica,L and IK current densities and proof of principle for pharmacological testing with tetrodotoxin, nifedipine and E-4031. These studies provide an initial demonstration that planar patch systems can be used to investigate specific ionic currents in single hPSC-CMs.
4.3.2. Multi-electrode arrays (MEAs) and nanopillars
The invasive nature of planar patch permits only short-term (minutes to hours) recordings from single cells. However, some applications, including acute and chronic toxicity, may benefit from long-term recordings (days/weeks) from multi-cell clusters. Multi-electrode arrays (MEAs) comprise glass slides photo-etched with arrays of micro-electrodes that can record field potentials from cell types for periods from hours to days. Most analysis with MEAs has been done using low throughput single-well formats using multi-cellular clusters of hPSC-CMs in 2D monolayers or 3D structures, including evaluation of drug responses in hiPSC-CMs carrying KCNH2 (IKr) mutations that underlie LQTS2 [51]. Nevertheless, there have been some reports of multiplexed MEAs being used for evaluation of cardiotoxicity in hPSC-CMs (Table 4). By seeding hESC-CMs to 4-well MEAs, field potential duration (FPD) was calculated after challenge with escalating doses of 12 cardiac or non-cardiac drugs. This showed that the dose known to cause QT prolongation in patients corresponded with increases in FPD in hESC-CMs [179]. In a follow-up paper, this approach was extended to evaluate the effect of the IKs inhibitors, HMR1556 and JNJ303, on hPSC-CMs. However, prolongation could only be detected when the high density IKr current was reduced by pharmacology (dofetilide or sotalol) or genetics (hiPSC-CMs with a mutation in KCNH2) [40], supporting the notion that IKs is a weak current in hPSC-CMs [105]. Positive in vitro-in vivo correlations have also been made with the hPSC-CM model being used in conjunction with a 48-well MEA system. Recordings were made 0.5, 1, 2 and 4 h post exposure to compounds with a combination of IKr, QT and clinical proarrhythmia properties, including verapamil, ranolazine, flecainide, amiodarone, ouabain, cisapride and terfenadine [180].
However, a contradictory report used the Maestro MEA Platform (1- to 96-wells) to perform blinded evaluation of field potential duration, Na+ slope/amplitude, beat rate and early after-depolarizations to 15 IKr, INa or IKs blockers in hPSC-CMs (Table 4). The authors concluded that these parameters were unable to distinguish torsadogenic from benign compounds, with the suggestion that hPSC-CMs on MEAs would have a high false-positive rate in regard to proarrhythmic risk [181]. It is not clear why these discrepancies have arisen but will be important to identify to ensure consistency of analysis in the MEA platform. It is possible differences in cell seeding density or process reduces contact with the electrode, which could reduce signal strength, quality and accuracy of cellular action potentials recordings, as could the proportion of quiescent CMs in the cell preparations [182]. Additional factors that will need to be borne in mind as new generations of 96-well MEAs capable of electrical pacing are developed will include the need for light transmission, reducing plate cost and improved level of user-friendly software interface that automates data analysis.
Recent technological improvements have allowed the merger of the advantages of substrate-integrated extracellular MEAs with intracellular electrodes. These switch between extra- and intra-cellular recordings using vertical nanopillar electrodes capable of nano-scale electroporation and re-sealing events. While such systems are capable of detecting action potential waveforms of hPSC-CMs [183], the transient nature of the intracellular recordings currently limits their use for drug screening purposes.
4.3.3. Optical imaging
The challenges of direct recording of electrophysiology or use of impedance-based approaches have stimulated the development of optical mapping to measure action potentials and calcium wave propagation using voltage-sensitive dyes (VSDs) [184] like the ANEPPS dyes or genetically-encoded voltage indicators (GEVIs) [185]. These techniques allow action potential detection with high spatial and temporal resolution, and are amenable to scale-up. Fluorescence data can be easily and rapidly acquired simultaneously from several cells in a single field of view, permitting the analysis of over 440 single hESC-CMs in a day (Table 4) [186]. Furthermore, the potential to carry out sequential measurements would allow each cell to be used as its own control helping overcome the inherent heterogeneity of preparations of hPSC-CMs (Herron et al., 2012).
Optical imaging has been used to measure calcium transients or beat rates in hPSC-CMs, with high throughput platforms including FLIPR tetra, ImageXpress Micro, FDSS/μCELL and CellOPTIQ in 96- to 384-well formats (Table 4). In particular, the 96-well cellOPTIQ showed nifedipine caused a dose dependent decrease in APD75 illustrating its effect on Ca2 + channels [144], while the FDSS/μCELL was used to monitor changes in astemizole-induced calcium transients in Cor.4 U CMs in a 384-well format [187]. Using the FLIPR tetra in conjunction with a Calcium 5 kit enabled the beat rate, peak shape and regularity to be monitored in response to 131 compounds comprising cardiac glycosides, anti-arrhythmics, α −/β-adrenoceptors, Ca2 + blockers and anti-histamines [187].
However, fluorescent dyes are inherently phototoxic, which can cause temporal degradation of sample and signal quality over time, thereby limiting recording times [185]. This can be overcome by introducing genetically encoded voltage indicators (GEVIs), such as ArcLight GEVI, which has recently been used to image hiPSC-CM action potentials during drug screening [186]. In this study, ArcLight allowed the accurate prediction of arrhythmic effects, which was comparable to patch clamp measurements [186]. Unlike VSDs, GEVIs also allow specific cell populations to be targeted and provide homogenous signal intensities independent of cell uptake [186]. Nevertheless, VSDs or GEVIs provide surrogates of membrane potentials and tend to overestimate action potential duration [185], [188].
4.3.4. Contraction and force generation
Excitation–contraction coupling leads to force generation, which is the ultimate goal of the working myocardium. As such, various methods have been developed to measure contractility of CMs, including atomic force microscopy [103], flexible cantilevers [189] strain gauges [190], magnetic beads [191], optical edge detection [192], polyacrylamide gels [193], muscular thin films [194], carbon fibre deflection [195], microposts [196] and image based label-free method [197]. Each method prepares, measures and reports force in a different way (e.g. clamping, bonding, poking), which makes cross-comparison of the values produced rather challenging. Nevertheless, force generation has been measured from hPSC-CMs in presence or absence of pharmacology (Table 4). Twitch forces for hPSCs-CM range from 2-260nN for single hPSC-CMs [123], [196], [198] and 0.08–4.4 mN/mm2 for 3D hPSC-CM derived EHTs [199]. This is relatively low compared with the microN range of single adult CM [124] and 40-80mN/mm2 range of muscle human strips [200]. This reinforces the need to evaluate the effect on hPSC-CM contractility of the different maturation strategies discussed above, including incorporation of non-cardiomyocyte populations found in the heart (i.e. endothelial cells, fibroblasts, smooth muscle cells).
Translating these approaches into medium- to high-throughput is challenging and only a few examples exist. The use of impedance to report on contractility is becoming popular even though this is a surrogate measure and its accuracy needs to be established more thoroughly. The premise is that as CMs contract and relax, nano-scale changes in morphology and membrane structure alter the level of impedance between cell and underlying substrate. These characteristics are exploited by electric cell-substrate impedance sensing platforms, which incorporate gold-film electrodes into the bottom of tissue-culture plates [201], [202]. Rapid (12.9 ms update rate/plate) data acquisition frequencies [203] allow the resolution of subtle changes in CM beat profiling including arrhythmic events and the continuous monitoring of cell index is useful for cardiotoxicity profiling [204]. Of the impedance platforms, the xCELLigence has been most widely used on hPSC-CMs. This showed testing of a panel of 49 cardioactive compounds on hiPSC-CM had sensitivity, specificity and accuracy of 90%, 74%, and 82%, respectively, which compared favourably when compared to in vitro data for dog (83%, 84% and 82%) and rat (77%, 74%, and 74%) CMs [205]. The CardioExcyte96 [206] is a newer platform that measures both impedance and extracellular field potentials, allowing study of hPSC-CM beat rates, amplitudes and electrophysiology (Table 4). Nonetheless, our own experiences are that improvements are needed in the number of wells that lead to productive recordings (often as low as 15%), as well as to signal recording and data analysis, which are predominately handled as manually processes and limit throughput.
It is probable that direct measurement of contractility in 3D EHTs will provide a more accurate readout than the indirect method of impedance. Multiplexing hPSC-CM EHTs in a 24-well format has shown differences between the maximal twitch forces (0.22, 0.05, and 0.08mN) and sensitivity to external Ca2 + (EC50: 0.15, 0.39, and 1.05 mM Ca2 +) of rat, mouse, and human EHTs. Evaluating the impact of drug panels on contractility in hPSC-CM derived EHTs is now also underway, which will provide an important resource to the pharmaceutical industry [135].
5. Conclusions & future challenges
Over the last 3–5 years, hPSC-CM technologies have transformed. Challenges of hPSC culture have largely been overcome by the development of defined medium, while differentiation strategies to produce CMs at scale are being refined rapidly. New tools for producing integration-free hiPSC and engineered Cas9/CRISPR cells mean that the range of genotypes available is broader than ever. Isogenic pairs of healthy and diseased hPSCs provide sets of cell lines in which only the mutation differs in an otherwise identical genetic background. This will offer unrivalled opportunities to examine impact of polymorphisms identified from genome-wide association studies (GWAS). These advances have enticed interest from the pharmaceutical industry with the promise of replacing suboptimal drug safety assessment platforms with physiologically-relevant human cardiomyocytes. Indeed, there have been spectacular successes with hPSC-CMs being used to refine patient treatment, assist in progressing drugs to Phase I clinical trial and evaluate tens if not hundreds of drugs in medium content assays. Such observations have prompted the CIPA initiative, which will write a new chapter of safety assessment guidelines using hPSC-CMs. The progression of hPSC-derived cardiac cells to the clinic is also an important milestone.
Nevertheless, the excitement needs to be tempered with the realisation that, relative to many in vitro models, hPSC-CMs are expensive to produce and beyond the tolerated ‘price point per well’ of most Pharma screening budgets. Consistency of hPSC-CM production needs to be improved during scale up and there is a need for better definition of CM subtype following differentiation. In this regard, it is encouraging that recent protocols have been developed to specify ventricular, atrial and pacemaker cells from hPSCs, and it will now be important to see if these methods can be reproduced in other laboratories. Maturation of hPSC-CMs continues to be a considerable issue for the field. While individual biochemical and mechanical cues provide incremental improvements in maturity, there has not yet been the leap needed to produce the adult CM phenotype. Whether this will ever be achieved is unknown but it might not be needed. Each improvement will extend the utility of hPSC-CMs further and they are already proving their worth.
The glut of patient-derived or engineered hPSC lines poses a phenotyping bottleneck to understand the biology of the cardiomyocytes. Integration of these cells into medium- to high-throughput platforms is now playing catch-up and over the next 2–3 years it can be expected that single, cost-effective platforms will become available for simultaneously reporting on electrophysiology, calcium and contractility in the presence or absence of pathophysiological load to mimic the failing heart. This will be a major step towards changing the face of drug safety screening and disease modelling.
Conflict of interest
There are no conflicts of interests from any of the authors.
Transparency document
Transparency document
Acknowledgements
C.D. is supported by Engineering and Physical Sciences Research Council (EPSRC) (EP/H045384/1), Medical Research Council (MRC) (MR/L012618/1; MR/M017354/1), British Heart Foundation (BHF Centre for Regenerative Medicine and Programme Grant) (04BX14CDLG; PG/14/59/31000; RG/14/1/30588; P47352), Heart Research UK (TRP01/12) and National Centre for the Replacement, Refinement and Reduction of Animals in Research (NC3Rs) (35911-259146; NC/K000225/1).
Footnotes
This article is part of a Special Issue entitled: Cardiomyocyte Biology: Integration of Developmental and Environmental Cues in the Heart edited by Marcus Schaub and Hughes Abriel.
The Transparency document associated with this article can be found, in the online version.
References
- 1.Thomson J.A. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391) doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5) doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.GOV.UK . In: Regenerative Medicine in the UK: Taking Stock. I.S. Department for Business, editor. 2011. [Google Scholar]
- 4.Nowbar A.N. Discrepancies in autologous bone marrow stem cell trials and enhancement of ejection fraction (DAMASCENE): weighted regression and meta-analysis. BMJ [Br. Med. J.] 2014;348 doi: 10.1136/bmj.g2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herper M. 2012. The truly staggering cost of inventing new drugs. [Google Scholar]
- 6.Qureshi Z.P. Market withdrawal of new molecular entities approved in the United States from 1980 to 2009. Pharmacoepidemiol. Drug Saf. 2011;20(7):772–777. doi: 10.1002/pds.2155. [DOI] [PubMed] [Google Scholar]
- 7.Gwathmey J.K., Tsaioun K., Hajjar R.J. Cardionomics: a new integrative approach for screening cardiotoxicity of drug candidates. Expert Opin. Drug Metab. Toxicol. 2009;5(6):647–660. doi: 10.1517/17425250902932915. [DOI] [PubMed] [Google Scholar]
- 8.Braam S.R., Passier R., Mummery C.L. Cardiomyocytes from human pluripotent stem cells in regenerative medicine and drug discovery. Trends Pharmacol. Sci. 2009;30(10):536–545. doi: 10.1016/j.tips.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Pereira G.C. Drug-induced cardiac mitochondrial toxicity and protection: from doxorubicin to carvedilol. Curr. Pharm. Des. 2011;17(20):2113–2129. doi: 10.2174/138161211796904812. [DOI] [PubMed] [Google Scholar]
- 10.Hawton K. Impact of withdrawal of the analgesic co-proxamol on nonfatal self-poisoning in the UK. Crisis. J. Crisis Interv. Suicide Prev. 2011;32(2):81–87. doi: 10.1027/0227-5910/a000063. [DOI] [PubMed] [Google Scholar]
- 11.James W.P.T. Effect of sibutramine on cardiovascular outcomes in overweight and obese subjects. N. Engl. J. Med. 2010;363(10):905–917. doi: 10.1056/NEJMoa1003114. [DOI] [PubMed] [Google Scholar]
- 12.Quigley E.M.M. Cisapride: what can we learn from the rise and fall of a prokinetic? J. Dig. Dis. 2011;12(3):147–156. doi: 10.1111/j.1751-2980.2011.00491.x. [DOI] [PubMed] [Google Scholar]
- 13.Rajamohan D. Current status of drug screening and disease modelling in human pluripotent stem cells. BioEssays. 2013 doi: 10.1002/bies.201200053. (p. n/a-n/a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer T. QT-Screen: High-throughput cardiac safety pharmacology by extracellular electrophysiology on primary cardiac myocytes. Assay Drug Dev. Technol. 2004;2(5) doi: 10.1089/adt.2004.2.507. [DOI] [PubMed] [Google Scholar]
- 15.Yan G.X., Shimizu W., Antzelevitch C. Characteristics and distribution of M cells in arterially perfused canine left ventricular wedge preparations. Circulation. 1998;98(18):1921–1927. doi: 10.1161/01.cir.98.18.1921. [DOI] [PubMed] [Google Scholar]
- 16.Varro A. The role of the delayed rectifier component I-Ks in dog ventricular muscle and Purkinje fibre repolarization. J. Physiol. Lond. 2000;523(1) doi: 10.1111/j.1469-7793.2000.00067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nerbonne J.M. Studying cardiac arrhythmias in the mouse — a reasonable model for probing mechanisms? Trends Cardiovasc. Med. 2004;14(3) doi: 10.1016/j.tcm.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Salama G., London B. Mouse models of long QT syndrome. J. Physiol. Lond. 2007;578(1) doi: 10.1113/jphysiol.2006.118745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price P.S., Keenan R.E., Swartout J.C. Characterizing interspecies uncertainty using data from studies of anti-neoplastic agents in animals and humans. Toxicol. Appl. Pharmacol. 2008;233(1):64–70. doi: 10.1016/j.taap.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 20.Schachter A.D., Ramoni M.F. Clinical forecasting in drug development. Nat. Rev. Drug Discov. 2007;6(2):107–108. doi: 10.1038/nrd2246. [DOI] [PubMed] [Google Scholar]
- 21.Itskovitz-Eldor J. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol. Med. 2000;6(2):88–95. [PMC free article] [PubMed] [Google Scholar]
- 22.Burridge P.W. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10(1):16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson D. Transgenic enrichment of cardiomyocytes from human embryonic stem cells. Mol. Ther. 2007;15(11):2027–2036. doi: 10.1038/sj.mt.6300303. [DOI] [PubMed] [Google Scholar]
- 24.Ma J. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am. J. Physiol. Heart Circ. Physiol. 2011;301(5):H2006–H2017. doi: 10.1152/ajpheart.00694.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calderon D. Immune response to human embryonic stem cell-derived cardiac progenitors and adipose-derived stromal cells. J. Cell. Mol. Med. 2012;16(7):1544–1552. doi: 10.1111/j.1582-4934.2011.01435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skelton R.J. SIRPA, VCAM1 and CD34 identify discrete lineages during early human cardiovascular development. Stem Cell Res. 2014;13(1):172–179. doi: 10.1016/j.scr.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 27.Wile B.M. Molecular beacon-enabled purification of living cells by targeting cell type-specific mRNAs. Nat. Protoc. 2014;9(10):2411–2424. doi: 10.1038/nprot.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hattori F. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat. Methods. 2010;7(1):61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- 29.Chong J.J. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510(7504):273–277. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tohyama S. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12(1):127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W. Microfluidics separation reveals the stem-cell-like deformability of tumor-initiating cells. Proc. Natl. Acad. Sci. U. S. A. 2012;109(46):18707–18712. doi: 10.1073/pnas.1209893109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers F.B. Label-free electrophysiological cytometry for stem cell-derived cardiomyocyte clusters. Lab Chip. 2013;13(2):220–228. doi: 10.1039/c2lc40905d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birket M.J. Expansion and patterning of cardiovascular progenitors derived from human pluripotent stem cells. Nat. Biotechnol. 2015 doi: 10.1038/nbt.3271. [DOI] [PubMed] [Google Scholar]
- 34.Birket M.J., Mummery C.L. Pluripotent stem cell derived cardiovascular progenitors–a developmental perspective. Dev. Biol. 2015;400(2):169–179. doi: 10.1016/j.ydbio.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 35.Devalla H.D. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015;7(4):394–410. doi: 10.15252/emmm.201404757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren L. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5) doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soares F.A. International coordination of large-scale human induced pluripotent stem cell initiatives: wellcome trust and ISSCR workshops white paper. Stem Cell Rep. 2014;3(6):931–939. doi: 10.1016/j.stemcr.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sirenko O. Assessment of beating parameters in human induced pluripotent stem cells enables quantitative in vitro screening for cardiotoxicity. Toxicol. Appl. Pharmacol. 2013;273(3):500–507. doi: 10.1016/j.taap.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pointon A. Assessment of cardiomyocyte contraction in human-induced pluripotent stem cell-derived cardiomyocytes. Toxicol. Sci. 2015;144(2):227–237. doi: 10.1093/toxsci/kfu312. [DOI] [PubMed] [Google Scholar]
- 40.Braam S.R. Repolarization reserve determines drug responses in human pluripotent stem cell derived cardiomyocytes. Stem Cell Res. 2013;10(1):48–56. doi: 10.1016/j.scr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 41.Harris K. Comparison of electrophysiological data from human-induced pluripotent stem cell–derived cardiomyocytes to functional preclinical safety assays. Toxicol. Sci. 2013;134(2):412–426. doi: 10.1093/toxsci/kft113. [DOI] [PubMed] [Google Scholar]
- 42.Nalos L. Comparison of the I(Kr) blockers moxifloxacin, dofetilide and E-4031 in five screening models of pro-arrhythmia reveals lack of specificity of isolated cardiomyocytes. Br. J. Pharmacol. 2012;165(2):467–478. doi: 10.1111/j.1476-5381.2011.01558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma A. Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circ. Res. 2014;115(6):556. doi: 10.1161/CIRCRESAHA.115.303810. − + [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynolds J.G. HER2-targeted liposomal doxorubicin displays enhanced anti-tumorigenic effects without associated cardiotoxicity. Toxicol. Appl. Pharmacol. 2012;262(1):1–10. doi: 10.1016/j.taap.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Sager P.T. Rechanneling the cardiac proarrhythmia safety paradigm: A meeting report from the cardiac safety research consortium. Am. Heart J. 2014;167(3):292–300. doi: 10.1016/j.ahj.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Moretti A. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med. 2010;363(15):1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 47.Egashira T. Disease characterization using LQTS-specific induced pluripotent stem cells. 2012;95:419–429. doi: 10.1093/cvr/cvs206. [DOI] [PubMed] [Google Scholar]
- 48.Zhang M. Recessive cardiac phenotypes in induced pluripotent stem cell models of Jervell and Lange-Nielsen syndrome: disease mechanisms and pharmacological rescue. Proc. Natl. Acad. Sci. 2014;111(50):E5383–E5392. doi: 10.1073/pnas.1419553111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Itzhaki I. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471(7337):225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 50.Lahti A.L. Human disease model for long QT syndrome type 2 using iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis. Model. Mech. 2011 doi: 10.1242/dmm.008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsa E. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur. Heart J. 2011;32(8):952–962. doi: 10.1093/eurheartj/ehr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bellin M. vol. 32. 2013. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome; pp. 3161–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsa E. Allele-specific RNA interference rescues the long-QT syndrome phenotype in human-induced pluripotency stem cell cardiomyocytes. Eur. Heart J. 2014;35(16):1078–1087. doi: 10.1093/eurheartj/eht067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma D. Modeling type 3 long QT syndrome with cardiomyocytes derived from patient-specific induced pluripotent stem cells. Int. J. Cardiol. 2013;168(6):5277–5286. doi: 10.1016/j.ijcard.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 55.Yazawa M. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471(7337) doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis R.P. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation. 2012;125(25) doi: 10.1161/CIRCULATIONAHA.111.066092. [DOI] [PubMed] [Google Scholar]
- 57.Fatima A. In vitro modeling of ryanodine receptor 2 dysfunction using human induced pluripotent stem cells. Cell. Physiol. Biochem. 2011;28(4) doi: 10.1159/000335753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jung C.B. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol. Med. 2012;4(3) doi: 10.1002/emmm.201100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Itzhaki I. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. J. Am. Coll. Cardiol. 2012;60(11):990–1000. doi: 10.1016/j.jacc.2012.02.066. [DOI] [PubMed] [Google Scholar]
- 60.Lin B. Modeling and study of the mechanism of dilated cardiomyopathy using induced pluripotent stem cells derived from individuals with Duchenne muscular dystrophy. Dis. Model. Mech. 2015;8(5):457–466. doi: 10.1242/dmm.019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun N. vol. 4. 2012. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. (130ra47-130ra47) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu H. Epigenetic regulation of phosphodiesterases 2A and 3A underlies compromised β-adrenergic signaling in an iPSC model of dilated cardiomyopathy. Cell Stem Cell. 2015;17(1):89–100. doi: 10.1016/j.stem.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lan F. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12(1):101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han L. vol. 104. 2014. Study familial hypertrophic cardiomyopathy using patient-specific induced pluripotent stem cells; pp. 258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carvajal-Vergara X. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465(7299) doi: 10.1038/nature09005. (p. 808-U12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dudek J. Cardiolipin deficiency affects respiratory chain function and organization in an induced pluripotent stem cell model of Barth syndrome. Stem Cell Res. 2013;11(2):806–819. doi: 10.1016/j.scr.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 67.Wang G. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014;20(6):616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma D. vol. 34. 2013. Generation of patient-specific induced pluripotent stem cell-derived cardiomyocytes as a cellular model of arrhythmogenic right ventricular cardiomyopathy; pp. 1122–1133. [DOI] [PubMed] [Google Scholar]
- 69.Caspi O. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circ. Cardiovasc. Genet. 2013;6(6):557–568. doi: 10.1161/CIRCGENETICS.113.000188. [DOI] [PubMed] [Google Scholar]
- 70.Kim C. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494(7435):105–110. doi: 10.1038/nature11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wen J.-Y. Maturation-based model of arrhythmogenic right ventricular dysplasia using patient-specific induced pluripotent stem cells. Circ. J. 2015;79(7):1402–1408. doi: 10.1253/circj.CJ-15-0363. [DOI] [PubMed] [Google Scholar]
- 72.Dick E. Exon skipping and gene transfer restore dystrophin expression in human induced pluripotent stem cells-cardiomyocytes harboring DMD mutations. Stem Cells Dev. 2013;22(20):2714–2724. doi: 10.1089/scd.2013.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Terrenoire C. Induced pluripotent stem cells used to reveal drug actions in a long QT syndrome family with complex genetics. J. Gen. Physiol. 2013;141(1):61–72. doi: 10.1085/jgp.201210899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Foldes G. Aberrant alpha-adrenergic hypertrophic response in cardiomyocytes from human induced pluripotent cells. Stem Cell Rep. 2014;3(5):905–914. doi: 10.1016/j.stemcr.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sheridan S.D. Epigenetic Characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6(10) doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ye L. Cardiac repair in a porcine model of acute myocardial infarction with human induced pluripotent stem cell-derived cardiovascular cells. Cell Stem Cell. 2014;15(6):750–761. doi: 10.1016/j.stem.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Menasche P. Human embryonic stem cell-derived cardiac progenitors for severe heart failure treatment: first clinical case report. Eur. Heart J. 2015;36(30):2011–2017. doi: 10.1093/eurheartj/ehv189. [DOI] [PubMed] [Google Scholar]
- 78.Menasché P. 2013. Transplantation of human embryonic stem cell-derived CD15 + Isl-1 + progenitors in severe heart failure. (ClinicalTrials.gov Internet) [Google Scholar]
- 79.Mateizel I. Derivation of human embryonic stem cell lines from embryos obtained after IVF and after PGD for monogenic disorders. Hum. Reprod. 2006;21(2):503–511. doi: 10.1093/humrep/dei345. [DOI] [PubMed] [Google Scholar]
- 80.Urbach A., Schuldiner M., Benvenisty N. Modeling for Lesch–Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells. 2004;22(4):635–641. doi: 10.1634/stemcells.22-4-635. [DOI] [PubMed] [Google Scholar]
- 81.Elliott D.A. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods. 2011;8(12):1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- 82.Bellin M. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J. 2013;32(24):3161–3175. doi: 10.1038/emboj.2013.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim Y.G., Cha J., Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U. S. A. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Porteus M.H., Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300(5620):763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 85.Cermak T. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reyon D. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30(5):460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mussolino C. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39(21):9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mali P. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H.L. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Rep. 2015;4(1):143–154. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ran F.A. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cong L. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen F. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods. 2011;8(9):753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu C. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16(2):142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yusa K. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478(7369):391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lanphier E. Don't edit the human germ line. Nature. 2015;519(7544):410–411. doi: 10.1038/519410a. [DOI] [PubMed] [Google Scholar]
- 98.Liang P. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6(5):363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van den Berg C.W. Transcriptome of human foetal heart compared with cardiomyocytes from pluripotent stem cells. Development. 2015 doi: 10.1242/dev.123810. [DOI] [PubMed] [Google Scholar]
- 100.Yang X., Pabon L., Murry C.E. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2014;114(3):511–523. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Veerman C.C. Immaturity of human stem-cell-derived cardiomyocytes in culture: fatal flaw or soluble problem? Stem Cells Dev. 2015;24(9):1035–1052. doi: 10.1089/scd.2014.0533. [DOI] [PubMed] [Google Scholar]
- 102.Bedada F.B. Acquisition of a quantitative, stoichiometrically conserved ratiometric marker of maturation status in stem cell-derived cardiac myocytes. Stem Cell Rep. 2014;3(4):594–605. doi: 10.1016/j.stemcr.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Blazeski A. Cardiomyocytes derived from human induced pluripotent stem cells as models for normal and diseased cardiac electrophysiology and contractility. Prog. Biophys. Mol. Biol. 2012;110(2–3):166–177. doi: 10.1016/j.pbiomolbio.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jonsson M.K. Application of human stem cell-derived cardiomyocytes in safety pharmacology requires caution beyond hERG. J. Mol. Cell. Cardiol. 2012;52(5):998–1008. doi: 10.1016/j.yjmcc.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 105.Christ T., Horvath A., Eschenhagen T. LQT1-phenotypes in hiPSC: are we measuring the right thing? Proc. Natl. Acad. Sci. U. S. A. 2015;112(16) doi: 10.1073/pnas.1503347112. (p. (E1968-E1968)) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van Laake L.W. Human embryonic stem cell-derived cardiomyocytes survive and mature in the mouse heart and transiently improve function after myocardial infarction. Stem Cell Res. 2007;1(1):9–24. doi: 10.1016/j.scr.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 107.Piccini I. Human pluripotent stem cell-derived cardiomyocytes: genome-wide expression profiling of long-term in vitro maturation in comparison to human heart tissue. Genomic Data. 2015;4:69–72. doi: 10.1016/j.gdata.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kuppusamy K.T. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc. Natl. Acad. Sci. U. S. A. 2015;112(21):E2785–E2794. doi: 10.1073/pnas.1424042112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang X. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J. Mol. Cell. Cardiol. 2014;72:296–304. doi: 10.1016/j.yjmcc.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wen J.Y. Maturation-based model of arrhythmogenic right ventricular dysplasia using patient-specific induced pluripotent stem cells. Circ. J. 2015;79(7):1402–1408. doi: 10.1253/circj.CJ-15-0363. [DOI] [PubMed] [Google Scholar]
- 111.Radisic M. Functional assembly of engineered myocardium by electrical stimulation of cardiac myocytes cultured on scaffolds. Proc. Natl. Acad. Sci. U. S. A. 2004;101(52):18129–18134. doi: 10.1073/pnas.0407817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nunes S.S. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat. Methods. 2013;10(8):781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mihic A. The effect of cyclic stretch on maturation and 3D tissue formation of human embryonic stem cell-derived cardiomyocytes. Biomaterials. 2014;35(9):2798–2808. doi: 10.1016/j.biomaterials.2013.12.052. [DOI] [PubMed] [Google Scholar]
- 114.Jacot J.G., Martin J.C., Hunt D.L. Mechanobiology of cardiomyocyte development. J. Biomech. 2010;43(1):93–98. doi: 10.1016/j.jbiomech.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huyer L.D. Biomaterial based cardiac tissue engineering and its applications. Biomed. Mater. 2015;10(3):034004. doi: 10.1088/1748-6041/10/3/034004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Engler A.J., C.-K. C., J. C.P., R. M., T. H., S. D.W., S. J.W. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. Cell Sci. 2008;121(22):3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McCain M.L. Matrix elasticity regulates the optimal cardiac myocyte shape for contractility. Am. J. Physiol. Heart Circ. Physiol. 2014;306(11):H1525–H1539. doi: 10.1152/ajpheart.00799.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jacot J.G., McCulloch A.D., Omens J.H. Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Biophys. J. 2008;95(7):3479–3487. doi: 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ribeiro A.J.S. Contractility of single cardiomyocytes differentiated from pluripotent stem cells drepends on physiological shape and substrate stiffness. Proc. Natl. Acad. Sci. 2015 doi: 10.1073/pnas.1508073112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Taylor J.M., Rovin J.D., Parsons J.T. A role for focal adhesion kinase in phenylephrine-induced hypertrophy of rat ventricular cardiomyocytes. J. Biolumin. Chemilumin. 2000;275(25):19250–19257. doi: 10.1074/jbc.M909099199. [DOI] [PubMed] [Google Scholar]
- 121.Chun Y.W. Combinatorial polymer matrices enhance in vitro maturation of human induced pluripotent stem cell-derived cardiomyocytes. Biomaterials. 2015;67:52–64. doi: 10.1016/j.biomaterials.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Patel A.K. A defined synthetic substrate for serum-free culture of human stem cell derived cardiomyocytes with improved functional maturity identified using combinatorial materials microarrays. Biomaterials. 2015;61:257–265. doi: 10.1016/j.biomaterials.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ribeiro M.C. Functional maturation of human pluripotent stem cell derived cardiomyocytes in vitro—correlation between contraction force and electrophysiology. Biomaterials. 2015;51:138–150. doi: 10.1016/j.biomaterials.2015.01.067. [DOI] [PubMed] [Google Scholar]
- 124.Rao C. The effect of microgrooved culture substrates on calcium cycling of cardiac myocytes derived from human induced pluripotent stem cells. Biomaterials. 2013;34(10):2399–2411. doi: 10.1016/j.biomaterials.2012.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Palchesko R.N. Development of polydimethylsiloxane substrates with tunable elastic modulus to study cell mechanobiology in muscle and nerve. PLoS One. 2012;7(12):e51499. doi: 10.1371/journal.pone.0051499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Grosberg A., N. A.P., G. J.A., B. M.D., M. M.L., P. K.K. Muscle on a chip: in vitro contractility assays for smooth and striated muscle. J. Pharmacol. Toxicol. Methods. 2012;65(3):126–135. doi: 10.1016/j.vascn.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Feinberg A.W. Controlling the contractile strength of engineered cardiac muscle by hierarchal tissue architecture. Biomaterials. 2012;33(23):5732–5741. doi: 10.1016/j.biomaterials.2012.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Alford P.W. Biohybrid thin films for measuring contractility in engineered cardiovascular muscle. Biomaterials. 2010;31(13):3613–3621. doi: 10.1016/j.biomaterials.2010.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang G. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014;20(6):616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kresh J.Y., Chopra A. Intercellular and extracellular mechanotransduction in cardiac myocytes. Pflugers Arch. 2011;462(1):75–87. doi: 10.1007/s00424-011-0954-1. [DOI] [PubMed] [Google Scholar]
- 131.Borg T.K.R., K.; Lundgren E., Borg K., Obrink B. Recognition of extracellular matrix components by neonatal and adult cardiac myocytes. Dev. Biol. 1984;104:86–96. doi: 10.1016/0012-1606(84)90038-1. [DOI] [PubMed] [Google Scholar]
- 132.Schaaf S. Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology. PLoS One. 2011;6(10):e26397. doi: 10.1371/journal.pone.0026397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Boudou T. A microfabricated platform to measure and manipulate the mechanics of engineered cardiac microtissues. Tissue Eng. A. 2012;18(9–10):910–919. doi: 10.1089/ten.tea.2011.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stoehr A. Automated analysis of contractile force and Ca2 + transients in engineered heart tissue. Am. J. Physiol. Heart Circ. Physiol. 2014;306(9):H1353–H1363. doi: 10.1152/ajpheart.00705.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Eder A. Human engineered heart tissue as a model system for drug testing. Adv. Drug Deliv. Rev. 2015 doi: 10.1016/j.addr.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 136.Cao H. Electrical and mechanical strategies to enable cardiac repair and regeneration. Biomed. Eng. IEEE Rev. 2015;8:114–124. doi: 10.1109/RBME.2015.2431681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Saini H. 3D cardiac microtissues encapsulated with the co-culture of cardiomyocytes and cardiac fibroblasts. Adv. Healthcare Mater. 2015 doi: 10.1002/adhm.201500331. (p. n/a-n/a.) [DOI] [PubMed] [Google Scholar]
- 138.Riegler J. Human engineered heart muscles engraft and survive long term in a rodent myocardial infarction model. Circ. Res. 2015;117(8):720–730. doi: 10.1161/CIRCRESAHA.115.306985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang L. Derivation and high engraftment of patient-specific cardiomyocyte sheet using induced pluripotent stem cells generated from adult cardiac fibroblast. Circ. Heart Fail. 2015;8(1):156–166. doi: 10.1161/CIRCHEARTFAILURE.114.001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zimmermann W.-H. Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat. Med. 2006;12(4):452–458. doi: 10.1038/nm1394. [DOI] [PubMed] [Google Scholar]
- 141.Vollert I. In vitro perfusion of engineered heart tissue through endothelialized channels. Tissue Eng. A. 2013;20(3–4):854–863. doi: 10.1089/ten.TEA.2013.0214. [DOI] [PubMed] [Google Scholar]
- 142.Leontyev S. Transplantation of engineered heart tissue as a biological cardiac assist device for treatment of dilated cardiomyopathy. Eur. J. Heart Fail. 2013;15(1):23–35. doi: 10.1093/eurjhf/hfs200. [DOI] [PubMed] [Google Scholar]
- 143.Du D.T. Action potential morphology of human induced pluripotent stem cell-derived cardiomyocytes does not predict cardiac chamber specificity and is dependent on cell density. Biophys. J. 2015;108(1):1–4. doi: 10.1016/j.bpj.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Smith G., Craig M.A., Anson B., Hortigon-Vinagre M., Burton F., Wallis R., Ghouri I. SOT Annual Meeting. 2013. Optical measurements of electrical activity from hiPSC-derived cardiomyocytes is a robust and high-throughput method for measuring NCE-effects on the cardiac action potential. [Google Scholar]
- 145.Celiz A.D. Discovery of a novel polymer for human pluripotent stem cell expansion and multilineage differentiation. Adv. Mater. 2015 doi: 10.1002/adma.201501351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.!!! INVALID CITATION !!!
- 147.Hussain W. Reproducible culture and differentiation of mouse embryonic stem cells using an automated microwell platform. Biochem. Eng. J. 2013;77(100):246–257. doi: 10.1016/j.bej.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Thomas R.J. Automated, scalable culture of human embryonic stem cells in feeder-free conditions. Biotechnol. Bioeng. 2009;102(6):1636–1644. doi: 10.1002/bit.22187. [DOI] [PubMed] [Google Scholar]
- 149.Kowalski M.P. Controlling embryonic stem cell growth and differentiation by automation: enhanced and more reliable differentiation for drug discovery. J. Biomol. Screen. 2012;17(9):1171–1179. doi: 10.1177/1087057112452783. [DOI] [PubMed] [Google Scholar]
- 150.Paull D. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nat. Methods. 2015 doi: 10.1038/nmeth.3507. [DOI] [PubMed] [Google Scholar]
- 151.Steiner D. Derivation, propagation and controlled differentiation of human embryonic stem cells in suspension. Nat. Biotechnol. 2010;28(4):361–364. doi: 10.1038/nbt.1616. [DOI] [PubMed] [Google Scholar]
- 152.Elanzew A. A reproducible and versatile system for the dynamic expansion of human pluripotent stem cells in suspension. Biotechnol. J. 2015 doi: 10.1002/biot.201400757. [DOI] [PubMed] [Google Scholar]
- 153.Schroeder M. Differentiation and lineage selection of mouse embryonic stem cells in a stirred bench scale bioreactor with automated process control. Biotechnol. Bioeng. 2005;92(7):920–933. doi: 10.1002/bit.20668. [DOI] [PubMed] [Google Scholar]
- 154.Kempf H. Controlling expansion and cardiomyogenic differentiation of human pluripotent stem cells in scalable suspension culture. Stem Cell Rep. 2014;3(6):1132–1146. doi: 10.1016/j.stemcr.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fluri D.A. Derivation, expansion and differentiation of induced pluripotent stem cells in continuous suspension cultures. Nat. Methods. 2012;9(5):509–516. doi: 10.1038/nmeth.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zock J.M. Applications of high content screening in life science research. Comb. Chem. High Throughput Screen. 2009;12(9):870–876. doi: 10.2174/138620709789383277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zanella F., Lorens J.B., Link W. High content screening: seeing is believing. Trends Biotechnol. 2010;28(5):237–245. doi: 10.1016/j.tibtech.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 158.Desbordes S.C. High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell. 2008;2(6):602–612. doi: 10.1016/j.stem.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Barbaric I., Gokhale P.J., Andrews P.W. High-content screening of small compounds on human embryonic stem cells. Biochem. Soc. Trans. 2010;38(4):1046–1050. doi: 10.1042/BST0381046. [DOI] [PubMed] [Google Scholar]
- 160.Pasqualini F.S. Structural phenotyping of stem cell-derived cardiomyocytes. Stem Cell Rep. 2015;4(3):340–347. doi: 10.1016/j.stemcr.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Carlson C. Phenotypic screening with human iPS cell-derived cardiomyocytes: HTS-compatible assays for interrogating cardiac hypertrophy. J. Biomol. Screen. 2013;18(10):1203–1211. doi: 10.1177/1087057113500812. [DOI] [PubMed] [Google Scholar]
- 162.Mioulane M. Development of high content imaging methods for cell death detection in human pluripotent stem cell-derived cardiomyocytes. J. Cardiovasc. Transl. Res. 2012;5(5):593–604. doi: 10.1007/s12265-012-9396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sirenko O. Multiparameter in vitro assessment of compound effects on cardiomyocyte physiology using iPSC cells. J. Biomol. Screen. 2013;18(1):39–53. doi: 10.1177/1087057112457590. [DOI] [PubMed] [Google Scholar]
- 164.Will Y. Analysis of mitochondrial function using phosphorescent oxygen-sensitive probes. Nat. Protoc. 2006;1(6):2563–2572. doi: 10.1038/nprot.2006.351. [DOI] [PubMed] [Google Scholar]
- 165.Hynes J. Investigation of drug-induced mitochondrial toxicity using fluorescence-based oxygen-sensitive probes. Toxicol. Sci. 2006;92(1):186–200. doi: 10.1093/toxsci/kfj208. [DOI] [PubMed] [Google Scholar]
- 166.Guan X. Dystrophin-deficient cardiomyocytes derived from human urine: new biologic reagents for drug discovery. Stem Cell Res. 2014;12(2):467–480. doi: 10.1016/j.scr.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Huang H.P. Human Pompe disease-induced pluripotent stem cells for pathogenesis modeling, drug testing and disease marker identification. Hum. Mol. Genet. 2011;20(24):4851–4864. doi: 10.1093/hmg/ddr424. [DOI] [PubMed] [Google Scholar]
- 168.Kikuchi C. Comparison of cardiomycyte differentiation potential between type 1 diabetic donor- and non-diabetic donor-derived induced pluripotent stem cells. Cell Transplant. 2015 doi: 10.3727/096368914X685762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Brueggemann A. Planar patch clamp: advances in electrophysiology. Methods Mol. Biol. 2008;491:165–176. doi: 10.1007/978-1-59745-526-8_13. [DOI] [PubMed] [Google Scholar]
- 170.Shroder R. In: Exploring Stem Cell-Derived Cardiomyocytes with Automated Patch Clamp Techniques. Sophion, editor. 2012. [Google Scholar]
- 171.Dale T.J. Population patch clamp electrophysiology: a breakthrough technology for ion channel screening. Mol. BioSyst. 2007;3(10):714–722. doi: 10.1039/b706152h. [DOI] [PubMed] [Google Scholar]
- 172.Donovan B.T. Utility of frozen cell lines in medium throughput electrophysiology screening of hERG and NaV1.5 blockade. J. Pharmacol. Toxicol. Methods. 2011;64(3):269–276. doi: 10.1016/j.vascn.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 173.Stoelzle S. State-of-the-art automated patch clamp devices: heat activation, action potentials, and high throughput in ion channel screening. Front. Pharmacol. 2011;2 doi: 10.3389/fphar.2011.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Polonchuk L. In: Industrializing Electrophysiology: HT Automated Patch Clamp on SyncroPatch® 96 Using Instant Frozen Cells, in Patch-Clamp Methods and Protocols. Martina M., Taverna S., editors. Springer; New York: 2014. pp. 81–92. [DOI] [PubMed] [Google Scholar]
- 175.Friis, S., et al., Current clamp of stem cell derived cardiomyocytes on Qpatch. Biophys. J. 106(2): p. 764a.
- 176.Schroder R.L. Exploring stem cell-derived cardiomyocytes with automated patch clamp techniques. Biophys. J. 2012;102(3) (p. 544A-544A) [Google Scholar]
- 177.Becker N. Minimized cell usage for stem cell-derived and primary cells on an automated patch clamp system. J. Pharmacol. Toxicol. Methods. 2013;68(1):82–87. doi: 10.1016/j.vascn.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 178.Scheel O. Action potential characterization of human induced pluripotent stem cell–derived cardiomyocytes using automated patch-clamp technology. Assay Drug Dev. Technol. 2014;12(8):457–469. doi: 10.1089/adt.2014.601. [DOI] [PubMed] [Google Scholar]
- 179.Braam S.R. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 2010;4(2):107–116. doi: 10.1016/j.scr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 180.Gilchrist K.H. High-throughput cardiac safety evaluation and multi-parameter arrhythmia profiling of cardiomyocytes using microelectrode arrays. Toxicol. Appl. Pharmacol. 2015 doi: 10.1016/j.taap.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 181.Qu Y., Vargas H.M. Proarrhythmia risk assessment in human induced pluripotent stem cell-derived cardiomyocytes using the maestro MEA platform. Toxicol. Sci. 2015 doi: 10.1093/toxsci/kfv128. [DOI] [PubMed] [Google Scholar]
- 182.Spira M.E., Hai A. Multi-electrode array technologies for neuroscience and cardiology. Nat. Nanotechnol. 2013;8(2):83–94. doi: 10.1038/nnano.2012.265. [DOI] [PubMed] [Google Scholar]
- 183.Burridge P.W. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11(8):855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Salzberg B.M., Davila H.V., Cohen L.B. Optical recording of impulses in individual neurones of an invertebrate central nervous system. Nature. 1973;246(5434):508–509. doi: 10.1038/246508a0. [DOI] [PubMed] [Google Scholar]
- 185.Chang Liao M.-L. Sensing cardiac electrical activity with a cardiac myocyte targeted optogenetic voltage indicator. Circ. Res. 2015 doi: 10.1161/CIRCRESAHA.117.306143. [DOI] [PubMed] [Google Scholar]
- 186.Leyton-Mange J.S. Rapid cellular phenotyping of human pluripotent stem cell-derived cardiomyocytes using a genetically encoded fluorescent voltage sensor. Stem Cell Rep. 2014;2(2):163–170. doi: 10.1016/j.stemcr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kettenhofen, R., A Gossmann, M Bertram, B Luerman, G Rascher-Eggstein, G, The newcomer is getting up and running: human iPS cell-derived cardiomyocyte implementation in multiple cardiac safety assessment assays, in Safety Pharmacology Society Annual Meeting. 2014.
- 188.Lopez-Izquierdo A. A near-infrared fluorescent voltage-sensitive dye allows for moderate-throughput electrophysiological analyses of human induced pluripotent stem cell-derived cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2014;307(9):H1370–H1377. doi: 10.1152/ajpheart.00344.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tanaka Y. Demonstration of a PDMS-based bio-microactuator using cultured cardiomyocytes to drive polymer micropillars. Lab Chip. 2006;6(2):230–235. doi: 10.1039/b512099c. [DOI] [PubMed] [Google Scholar]
- 190.Vannier C., Chevassus H., Vassort G. Ca-dependence of isometric force kinetics in single skinned ventricular cardiomyocytes from rats. Cardiovasc. Res. 1996;32(3):580–586. [PubMed] [Google Scholar]
- 191.Yin S. Measuring single cardiac myocyte contractile force via moving a magnetic bead. Biophys. J. 2005;88(2):1489–1495. doi: 10.1529/biophysj.104.048157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Edes I.F. Rate of tension redevelopment is not modulated by sarcomere length in permeabilized human, murine, and porcine cardiomyocytes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;293(1):R20–R29. doi: 10.1152/ajpregu.00537.2006. [DOI] [PubMed] [Google Scholar]
- 193.Hazeltine L.B. Effects of substrate mechanics on contractility of cardiomyocytes generated from human pluripotent stem cells. Int. J. Cell Biol. 2012:508294. doi: 10.1155/2012/508294. (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shim J. Modeling of cardiac muscle thin films: pre-stretch, passive and active behavior. J. Biomech. 2012;45(5):832–841. doi: 10.1016/j.jbiomech.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Iribe G., Helmes M., Kohl P. Force-length relations in isolated intact cardiomyocytes subjected to dynamic changes in mechanical load. Am. J. Physiol. Heart Circ. Physiol. 2007;292(3):H1487–H1497. doi: 10.1152/ajpheart.00909.2006. [DOI] [PubMed] [Google Scholar]
- 196.Liu J. Atomic force mechanobiology of pluripotent stem cell-derived cardiomyocytes. PLoS One. 2012;7(5):e37559. doi: 10.1371/journal.pone.0037559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hayakawa T. Image-based evaluation of contraction-relaxation kinetics of human-induced pluripotent stem cell-derived cardiomyocytes: correlation and complementarity with extracellular electrophysiology. J. Mol. Cell. Cardiol. 2014;77:178–191. doi: 10.1016/j.yjmcc.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 198.Rodriguez M.L. Measuring the contractile forces of human induced pluripotent stem cell-derived cardiomyocytes with arrays of microposts. J. Biomech. Eng. 2014;136(5):051005. doi: 10.1115/1.4027145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kensah G. Murine and human pluripotent stem cell-derived cardiac bodies form contractile myocardial tissue in vitro. Eur. Heart J. 2013;34(15):1134–1146. doi: 10.1093/eurheartj/ehs349. [DOI] [PubMed] [Google Scholar]
- 200.Hasenfuss G. Energetics of isometric force development in control and volume-overload human myocardium. Comparison with animal species. Circ. Res. 1991;68(3):836–846. doi: 10.1161/01.res.68.3.836. [DOI] [PubMed] [Google Scholar]
- 201.Giaever I., Keese C.R. Monitoring fibroblast behavior in tissue culture with an applied electric field. Proc. Natl. Acad. Sci. U. S. A. 1984;81(12):3761–3764. doi: 10.1073/pnas.81.12.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Giaever I., Keese C.R. A morphological biosensor for mammalian cells. Nature. 1993;366(6455):591–592. doi: 10.1038/366591a0. [DOI] [PubMed] [Google Scholar]
- 203.Lamore S.D. Cellular impedance assays for predictive preclinical drug screening of kinase inhibitor cardiovascular toxicity. Toxicol. Sci. 2013;135(2):402–413. doi: 10.1093/toxsci/kft167. [DOI] [PubMed] [Google Scholar]
- 204.Peters M. Human stem cell-derived cardiomyocytes in cellular impedance assays: bringing cardiotoxicity screening to the front line. Cardiovasc. Toxicol. 2015;15(2):127–139. doi: 10.1007/s12012-014-9268-9. [DOI] [PubMed] [Google Scholar]
- 205.Scott C.W. An impedance-based cellular assay using human iPSC-derived cardiomyocytes to quantify modulators of cardiac contractility. Toxicol. Sci. 2014;142(2):331–338. doi: 10.1093/toxsci/kfu186. [DOI] [PubMed] [Google Scholar]
- 206.Doerr L. New easy-to-use hybrid system for extracellular potential and impedance recordings. J. Lab. Autom. 2015;20(2):175–188. doi: 10.1177/2211068214562832. [DOI] [PubMed] [Google Scholar]
- 207.Rog-Zielinska E.A. Glucocorticoids promote structural and functional maturation of foetal cardiomyocytes: a role for PGC-1alpha. Cell Death Differ. 2015;22(7):1106–1116. doi: 10.1038/cdd.2014.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Birket M.J. PGC-1α and reactive oxygen species regulate human embryonic stem cell-derived cardiomyocyte function. Stem Cell Rep. 2013;1(6):560–574. doi: 10.1016/j.stemcr.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lombardi R. Nuclear plakoglobin is essential for differentiation of cardiac progenitor cells to adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circ. Res. 2011;109(12):1342–1353. doi: 10.1161/CIRCRESAHA.111.255075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Engels M.C. Insulin-like growth factor promotes cardiac lineage induction in vitro by selective expansion of early mesoderm. Stem Cells. 2014;32(6):1493–1502. doi: 10.1002/stem.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Shabani P. Exogenous treatment with eicosapentaenoic acid supports maturation of cardiomyocytes derived from embryonic stem cells. Biochem. Biophys. Res. Commun. 2015;461(2):281–286. doi: 10.1016/j.bbrc.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 212.Lan F. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12(1):101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Cerignoli F. High throughput measurement of Ca(2)(+) dynamics for drug risk assessment in human stem cell-derived cardiomyocytes by kinetic image cytometry. J. Pharmacol. Toxicol. Methods. 2012;66(3):246–256. doi: 10.1016/j.vascn.2012.08.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Maddah M. A non-invasive platform for functional characterization of stem-cell-derived cardiomyocytes with applications in cardiotoxicity testing. Stem Cell Rep. 2015;4(4):621–631. doi: 10.1016/j.stemcr.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Klimas A. 2015. OptoDyCE: automated system for high-throughput all-optical dynamic cardiac electrophysiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kim C. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494(7435):105–110. doi: 10.1038/nature11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lewis K.J. A new system for profiling drug-induced calcium signal perturbation in human embryonic stem cell-derived cardiomyocytes. J. Biomol. Screen. 2015;20(3):330–340. doi: 10.1177/1087057114557232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Guo L. Estimating the risk of drug-induced proarrhythmia using human induced pluripotent stem cell-derived cardiomyocytes. Toxicol. Sci. 2011;123(1) doi: 10.1093/toxsci/kfr158. [DOI] [PubMed] [Google Scholar]
- 219.Pradhapan P. Cardiomyocyte MEA data analysis (CardioMDA) — a novel field potential data analysis software for pluripotent stem cell derived cardiomyocytes. PLoS One. 2013;8(9):e73637. doi: 10.1371/journal.pone.0073637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Nozaki Y. Availability of human induced pluripotent stem cell-derived cardiomyocytes in assessment of drug potential for QT prolongation. Toxicol. Appl. Pharmacol. 2014;278(1):72–77. doi: 10.1016/j.taap.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 221.Clements M., Thomas N. High-throughput multi-parameter profiling of electrophysiological drug effects in human embryonic stem cell derived cardiomyocytes using multi-electrode arrays. Toxicol. Sci. 2014;140(2):445–461. doi: 10.1093/toxsci/kfu084. [DOI] [PubMed] [Google Scholar]
- 222.Jonsson M.K.B., Wang Q.-D., Becker B. Impedance-based detection of beating rhythm and proarrhythmic effects of compounds on stem cell-derived cardiomyocytes. Assay Drug Dev. Technol. 2011;9(6):589–599. doi: 10.1089/adt.2011.0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Doherty K.R. Multi-parameter in vitro toxicity testing of crizotinib, sunitinib, erlotinib, and nilotinib in human cardiomyocytes. Toxicol. Appl. Pharmacol. 2013;272(1):245–255. doi: 10.1016/j.taap.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 224.Himmel H.M. Drug-induced functional cardiotoxicity screening in stem cell-derived human and mouse cardiomyocytes: effects of reference compounds. J. Pharmacol. Toxicol. Methods. 2013;68(1):97–111. doi: 10.1016/j.vascn.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 225.Doherty K.R. Structural and functional screening in human induced-pluripotent stem cell-derived cardiomyocytes accurately identifies cardiotoxicity of multiple drug types. Toxicol. Appl. Pharmacol. 2015;285(1):51–60. doi: 10.1016/j.taap.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 226.Hirt M.N. Functional improvement and maturation of rat and human engineered heart tissue by chronic electrical stimulation. J. Mol. Cell. Cardiol. 2014;74:151–161. doi: 10.1016/j.yjmcc.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 227.Chen G. Phospholamban as a crucial determinant of the inotropic response of human pluripotent stem cell-derived ventricular cardiomyocytes and engineered 3-dimensional tissue constructs. Circ. Arrhythm. Electrophysiol. 2015;8(1):193–202. doi: 10.1161/CIRCEP.114.002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Martin T.P., Hortigon-Vinagre M.P., Findlay J.E., Elliott C., Currie S., Baillie G.S. Targeted disruption of the heat shock protein 20-phosphodiesterase 4D (PDE4D) interaction protects against pathological cardiac remodelling in a mouse model of hypertrophy. FEBS Open Bio. 2014;4:923–927. doi: 10.1016/j.fob.2014.10.011. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document