ABSTRACT
We have developed a phagebiotic composition using 8 virulent bacteriophages (2 strains of each species) which are able to lyse Acinetobacter baumannii, Klebsiella pneumoniae, Pseudomonas aeruginosa and Staphylococcus aureus. The unique character of the developed composition is ensured by particular properties of each bacteriophage comprising the preparation, including their range of lytic activity toward specific bacterial pathogens, morphology of their plaques, cycle of their development, restriction profile of their DNAs, specificity of their genomes (based on complete genome sequencing), and other properties. The preparation did not produce any signs of acute or chronic intoxication in the experimental animals. Therapeutic and prophylactic efficiency of the phagebiotic composition was demonstrated in the prevention and treatment of the experimental acute K. pneumoniae infection in mice. The investigations have shown that the preparation possesses a high therapeutic efficiency and is highly competitive with ciprofloxacin which is very effective against the infective strain K. pneumoniae. Our small-scale clinical trial was aimed to evaluate therapeutic effectiveness of the phagebiotic composition in an epidemiological emergency situation in an intensive care unit, caused by multi-resistant strains of Acinetobacter baumannii, Klebsiella pneumoniae and Pseudomonas aeruginosa. Seventy nine per cent of the initial samples from 14 patients’ endotracheal aspirate, blood and urine were contaminated. Twenty-four hours after the 3-day phage therapy (20 ml of cocktail at a titer for each phage 108 pfu/ml were introduced intragastrically through a tube once a day) contamination level dropped to 21%. Hence the obtained results enabled us to create a new phagebiotic composition that may be used as an alternative to antibiotics to treat these healthcare-associated infections.
KEYWORDS: Acinetobacter baumannii, healthcare-associated infections, Klebsiella pneumoniae, MRSA, phagebiotics, Pseudomonas aeruginosa
Introduction
According to the latest data from European Center for Disease Control (ECDC) and US Centers for Disease Control and Prevention (CDC), 10 ESCAPE bacteria are responsible for 2-thirds of all healthcare-associated infections (HAIs). The ESCAPE pathogens are multi-drug resistant strains of Enterococcus faecium, Staphylococcus aureus, Clostridium difficile, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacteriaceae including Klebsiella pneumoniae, Escherichia coli, Enterobacter species and Proteus species.1 According to the data from The Antimicrobial Chemotherapy Research Institute in Smolensk, the most frequent HAIs in Russia are caused by gram-negative pathogens: Pseudomonas aeruginosa (35%), Acinetobacter baumannii (15%) and representatives of genus Enterobacteriaceae (45%), including Klebsiella pneumoniae (14%) and Escherichia coli (13%).2
Recent advances in molecular biology, ecological interrelations between bacteriophages and their hosts, along with the growing concern about antibiotic-resistant strains of microorganisms necessitate a more thorough research of the possibilities of using phages in clinical practice. In this context, phages have been used to prevent and/or treat infectious diseases of bacterial etiology (including HAIs) for almost a century. Abedon T. S. et al. and Mai V. et al. in accordance with the definition of probiotics given by the World Health Organization, where they are described as having a healing effect on human organism when administered in adequate quantities, consider that it is possible to classify bacteriophage-based preparations as a new class of probiotics – phagebiotics.3,4 Given the ability of lytic phages to kill bacteria that cannot be killed by commonly available antibiotics, it seems prudent that this approach is rigorously evaluated in carefully designed animal and human trials.
Materials and methods
Using the clinical material obtained from ambulant cases a collection of 500 strains of Staphylococcus aureus, Acinetobacter baumannii, Pseudomonas aeruginosa and Klebsiella pneumoniae was created. Bacteriophages effective against these pathogens (2 strains of each species) were isolated and selected, focusing on their host range activity, titer, virulent nature and originality as well as the ability to retain their characteristics in storage, and sensitivity to adverse factors.5,6 Several tests were performed to characterize the bacteriophages’ stability: resistance to increased and decreased temperature, pH variations and chloroform. The resistance of phages to increased and decreased temperature was observed in the range of (−20°C) − 65°C. Plaque quantification from heated and chilled samples was carried out using Gratia double agar overlay method.7 We used chloroform treatment (and further centrifugation) as the cheapest method for eliminating bacterial cells and debris from phage-lysates to pre-sterilize them before microfiltration. Phage lysates were incubated for one hour at room temperature with buffer solutions calibrated to specific pH values to determine the bacteriophages’ resistance to acidic and alkaline pH values. We estimated the titer of the phages in the phage lysates after incubation using the Gratia method. A 80,000 × microscope was used to identify the morphology of phage particles. We carried out molecular genetics analysis, including PCR-confirmation of absence of known pathogenic loci. Virulent nature of our phages was proved by the absence of known integrases and using the on-line software PHACTS (www.phantome.org/PHACTS). Originality of the phages was confirmed by comparison of the genome of our phages with the earlier known DNAs of affine phages. The latter was based on a full genome sequencing (Ion Torrent semiconductor sequencing) and bioinformatics analysis carried out in the Ubuntu 12.04 operating system based on the Linux core. De-novo automatic genome assembly was performed using NEWBLER (Roche Diagnostics) software which worked well in this operating system. The obtained data was visualized using the BRIG software.8
The phagebiotic was used in liquid form and additionally contained 0.9% NaCl. The titer of each phage in the ready-to-use product was 108 pfu/ml.
The basic requirements for the preparation were safety and efficacy in prevention and treatment of infections. The safety of the bacteriophage cocktail was confirmed by the results of the toxicity tests (acute and chronic) performed on white outbred mice in compliance with Russian State Pharmacopoeia.9 We carried out all experiments using initially healthy animals. The mice had been under observation in quarantine during 2 weeks to check their health. All animal experiments were approved by the Institute’s Ethical Committee. Throughout the experiments, mice were given food and water ad libitum. Acute toxicity of the phagebiotic was evaluated via a single abdominal injection of 0.5 ml of the studied preparation in 10 outbred female mice (22.2 ± 1.2 g). Various parameters such as body mass dynamics, behavioral response, skin reaction and hematologic factors were subjected to close examination in the experimental animals within a period of 7 d and compared to those of the mice in the control group of identical age, mass and gender, which were given a single abdominal injection of saline solution. The same parameters were evaluated within a longer period of 20 d, where 10 white outbred mice were given daily abdominal injections of 0.5 ml of phagebiotic in comparison with the control group which were given saline solution following the same pattern. Additionally, on the 27th d of the experiment (7 d after the last intake of the preparation) samples of brain, heart, lungs, tongue, stomach, small and large intestine, kidneys, mesenteric lymph nodes and spleen of the animals from both groups were subjected to histological examination.
Pharmacokinetics of the phagebiotic were evaluated by the dynamics in bacteriophage concentration in organs and tissues of intact outbred white mice 1, 3, 6, 9, 12 and 24 h after the abdominal injection of a ∼108 pfu dose of preparation. Dissemination of phage particles was studied in the samples of blood, spleen, lungs, liver and lymphatic nodes.
Mean values and standard deviations were calculated for hematological parameters, amount of phage particles in the blood and parenchymal organs of mice. A t-test that was necessary to determine significant differences of hematological parameters in control and study groups of mice was carried out.
Therapeutic and prophylactic effects of the phage cocktail were proved experimentally on acute lethal Klebsiella infection in mice. We also used ciprofloxacin to compare effectiveness of phagebiotic and antibiotic in K. pneumoniae B2580 infected mice.
Human trials were aimed to evaluate therapeutic effectiveness of the cocktail in case of bacterial emergency in an intensive care unit (ICU), which was caused by multi-resistant strains of Acinetobacter baumannii, Klebsiella pneumoniae, Pseudomonas aeruginosa and Staphylococcus aureus. All of the human trials were carried out in accordance with the Declaration of Helsinki and with Russian national legislation. Within a 6-month period the experiment involved 55 patients in controlled ventilation after brain surgery. The patients were administered 20 ml of the preparation once a day within a period of 3–5 d via an intragastric tube.
Results
The composition of the phagebiotic includes 2 phage strains for each of the bacterial species of: A. baumannii, K. pneumoniae, P. aeruginosa and S. aureus, with an aggregate (for individual bacterial species) lytic spectrum on the strains of agents of nosocomial infections ranging from 56% to 95% (Table 1). The titer of the bacteriophages on non-lysogenic host bacteria reached 1010 pfu/ml. The results of the bacteriophages' stability testing showed a possibility of oral administration of phagebiotic taking into account gastric acidity.
Table 1.
Characteristics of the bacteriophage cocktail.
| Phage |
SCH1 |
SCH111 |
KPV15 |
KPV811 |
PA5 |
PA10 |
AP22 |
AM24 |
|---|---|---|---|---|---|---|---|---|
| Host species | S. aureus | S. aureus | K. pneumoniae | K. pneumoniae | P. aeruginosa | P. aeruginosa | A. baumannii | A. baumannii |
| Initial source and place of isolation | Clinical material, Chelyabinsk | Clinical material, Moscow | Waste water, Moscow region | Waste water, Moscow region | Waste water, Moscow region | Waste water, Moscow region | Clinical material, Moscow | Clinical material, Moscow |
| Host range activity (%/bacterial cultures) | 95% of S. aureus strains (included MRSA-strains) | 92.5% of K. pneumoniae strains (included pandrug-resistant) | 76% of P. aeruginosa strains (included antibiotic-resistant) | 56.25% of A. baumannii strains (included antibiotic-resistant) | ||||
| DNAs’ length, | 18023 | 18018 | 167034 | 42641 | 66182 | 91212 | 46387 | 97139 |
| bp / shape | linear | linear | linear | linear | linear | linear | circular | linear |
| Taxonomy of phages | Family Podoviridae Subfamily Picovirinae Genus P68virus | Family Podoviridae Subfamily Picovirinae Genus P68virus | Family Myoviridae | Family Podoviridae Subfamily Autographivirinae Genus Kp34virus | Family Myoviridae Genus Pbunavirus | Family Myoviridae Genus Pakpunavirus | Family Myoviridae Genus Ap22virus | Family Myoviridae |
The phage genomes were represented by double stranded DNA with the length from 18 kbp (staphylophages) to 167 kbp (one of Klebsiella pneumonia phages) (Table 1). According to bioinformatics analysis of the phages’ DNAs, virulent nature of all 8 strains was proved by the absence of known integrases, transcriptional repressors and other genes which determine temperate development of phage. Online application PHACTS also confirmed their virulent nature. The DNAs of phages* , contained in the phagebiotic, were compared with already known representatives of Podoviridae and Myoviridae families. The genetic maps allow us to demonstrate originality of our phages as they differ from already known phage maps published in NCBI GenBank more than 5%.
The safety of the phagebiotic composition was confirmed by the results of the investigation of its toxicity (acute and chronic) performed on white outbred mice. The preparation did not produce any signs of acute or chronic intoxication in the experimental animals. Phage injection did not cause the animals’ death and had no influence on their physiological condition, weight and morphology of their organs (liver, lungs, heart, thymus, spleen, base of tongue, lymph nodes, stomach, small intestine and colon, mesenterium and brain). Full blood counts in the animals of both study and control groups after abdominal administration of phagebiotic (study group) or saline solution (control group) in evaluation of acute and chronic toxicity did not show significant differences (P > 0.05).
Table 2 shows the dynamics in concentration of phage particles in blood, spleen, lungs, liver and lymph nodes of outbred white mice after an intraperitoneal injection of the preparation in a dose of ∼108 pfu in the case of KPV15.
Table 2.
The amount of phage particles in the blood and parenchymal organs of mice after a single intraperitoneal injection of the preparation exemplified by KPV15.
| Mean content of phage particles in 1 g(ml)* |
|||||
|---|---|---|---|---|---|
| Time after injection, hrs | Liver | Spleen | Lung | L/nodes | Blood |
| 1 | 102034 ± 26032 | 43431 ± 8652 | 80126 ± 17981 | 157604 ± 36218 | 20500 ± 5923 |
| 3 | 2797 ± 925 | 11326 ± 2785 | 6524 ± 2321 | 6916 ± 2564 | <10 |
| 6 | 4590 ± 1873 | 4620 ± 1986 | 5171 ± 2158 | 12987 ± 6857 | <10 |
| 9 | <10 | 46 ± 13 | <10 | <10 | <10 |
| 12 | 24 ± 9 | 98 ± 29 | 38 ± 11 | 96 ± 27 | <10 |
| 24 | <10 | 164 ± 47 | 96 ± 25 | 124 ± 42 | <10 |
Mean values and standard deviations are shown.
A single intraperitoneal injection of bacteriophage cocktail against hospital infection agents 24 or 12 hours before infecting the mice with K. pneumoniae B2580 strain culture resulted in a 100% survival. A single injection 6 h or 1 h before infection led to a 90% survival (Fig. 1).
Figure 1.
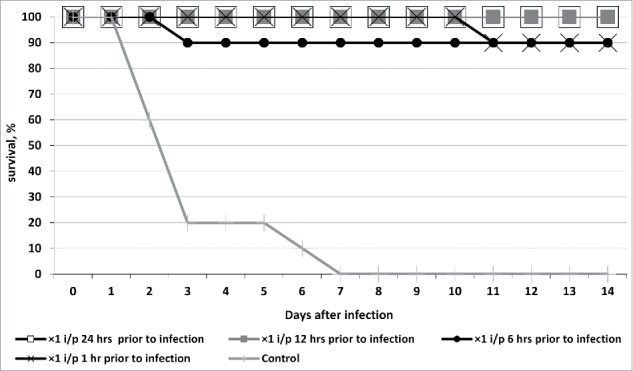
Survival of outbred white mice after parenteral (intraperitoneal - i/p) treatment with phagebiotic 1, 6, 12, and 24 h prior to a hypodermic infection with K. pneumoniae strain B2580. Animals from control group were not treated with phagebiotic.
The cocktail we have designed also offers high medicinal performance, on par with its antibiotic counterpart effective against K. pneumoniae (i.e., ciprofloxacin). Mice treated with the cocktail 1 and 6 h after the infection were completely sanitized from a highly virulent K. pneumoniae strain. Within 14 d after infection the animals showed no symptoms of acute Klebsiella infection. All of the animals not treated with the bacteriophage (control group) died between the 2nd and 5th d of the infection (Fig. 2).
Figure 2.
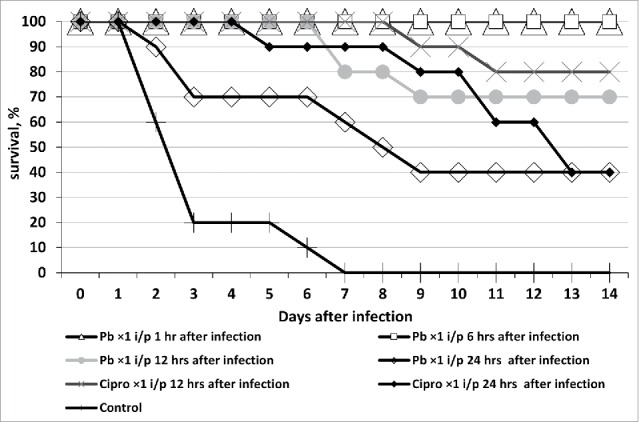
Survival of outbred white mice after parenteral (intraperitoneal - i/p) treatment with phagebiotic 1, 6, 12, and 24 h after a hypodermic infection with K. pneumoniae strain B2580. Animals from control group were not treated with phagebiotic (Pb) or ciprofloxacin (Cipro).
A small-scale clinical trial was aimed to evaluate therapeutic effectiveness of the phagebiotic composition in an epidemiological emergency situation in an intensive care unit, which was caused by multi-resistant strains of Acinetobacter baumannii, Klebsiella pneumoniae, Pseudomonas aeruginosa. When an epidemiological emergency situation caused by a multi-resistant K. pneumoniae strain was registered in the intensive care unit of a surgical clinic in Moscow, 13 patients in critical condition on mechanical ventilation were subjected to a triple sanitation of mouth and endotracheal tube with our phage cocktail. A single treatment included 5 ml of our cocktail with a titer of each phage of at least 108 pfu/ml. High percentage of successful eradication of K. pneumoniae shown in gray in the table confirms that further clinical trials of this product may prove to be useful (Table 3).
Table 3.
Preliminary data of first limited clinical trials of the bacteriophage cocktail* against nosocomial pathogens (“+”– presence of Klebsiella pneumoniae; “-”– absence of Klebsiella pneumoniae; ETA – endotracheal aspirate).
| No. of patient | Date of the initial survey | Sample | Klebsiella pneumoniae | Date of the secondary survey | Sample | Klebsiella pneumoniae |
|---|---|---|---|---|---|---|
| 1 | 07.07.2014 | ETA | + | 21.07.2014 | ETA | + |
| 2 | 06.05.2014 | ETA | + | 15.07.2014 | ETA | − |
| 3 | 07.07.2014 | ETA | + | 15.07.2014 | ETA | − |
| 4 | 30.06.2014 | ETA | + | 15.07.2014 | ETA | − |
| 5 | 02.07.2014 | ETA | + | 15.07.2014 | ETA | − |
| 6 | 09.07.2014 | ETA | + | 24.07.2014 | ETA | − |
| 7 | 08.07.2014 | ETA | + | 15.07.2014 | ETA | − |
| 8 | 06.05.2014 | Urine | + | 24.07.2014 | Urine | + |
| 9 | 03.07.2014 | Urine | + | 28.07.2014 | Urine | − |
| 10 | 25.06.2014 | Urine | + | 15.07.2014 | Urine | + |
| 11 | 07.04.2014 | ETA | + | 15.07.2014 | ETA | + |
| 12 | 17.06.2014 | ETA | + | 21.07.2014 | ETA | + |
| 13 | 07.07.2014 | ETA | + | 15.07.2014 | ETA | − |
Date of the bacteriophage application: 14.07.2014.
Another clinical trial was aimed to evaluate therapeutic effectiveness of the cocktail in yet another bacterial emergency in an intensive care unit, which was caused by multi-resistant strains of Acinetobacter baumannii, Klebsiella pneumoniae, Pseudomonas aeruginosa and Staphylococcus aureus. Seventy nine per cent of the initial samples from 14 patients’ endotracheal aspirate, blood and urine were contaminated. Twenty-four hours after the 3-day phage therapy (20 ml of cocktail at a titer for each phage 108 pfu/ml were introduced intragastrically through a tube once a day) contamination level dropped to 21%.
In the following trial we also managed to prove that when the cocktail is administered orally it is introduced to the blood system through the wall of the large intestine, and 24 h after the last intake individual bacteriophage strains can be isolated in patients’ urine, endotracheal aspirate and feces in a titer sufficient for its identification by both spot-test identification on a host strain and Gratia method (Table 4).
Table 4.
Bacteriophage isolation from the clinical material (after 4 days of phage therapy, on 5th day).
| No. of patient | PA 5 | PA 10 | KPV 15 | KPV 811 | AP 22 | AM 24 | SCH1/SCH111 | Sample |
|---|---|---|---|---|---|---|---|---|
| 1 | 20(*) | 25 | − | − | +++ | + | − | Feces |
| 2 | +++(**) | +++ | + | − | +++ | + | − | Feces |
| 3 | 4 × 104 | 3 × 105 | − | − | − | − | − | ETA |
| − | 10 | − | − | − | − | − | Feces | |
| 4 | + | 30 | + | − | ++ | − | ++++ | Feces |
| 5 | − | − | − | + | − | − | − | Urine |
| − | − | − | − | ++ | + | − | Feces | |
| 6 | +++ | +++ | ++ | + | +++ | ++ | − | Feces |
| 7 | 2 × 102 | ++++ | 2 × 102 | +++ | +++ | − | − | Urine |
| 5 × 104 | 4 × 105 | − | − | − | − | − | ETA | |
| 18 | 10 | − | − | ++ | + | − | Feces | |
| 8 | − | +++ | + | 104 | − | − | − | Urine |
| − | − | − | − | ++ | − | − | Feces | |
| 9 | − | − | − | − | 3 × 108 | − | − | Urine |
| 17 | 25 | ++ | − | +++ | ++ | − | Feces | |
| 10 | +++ | +++ | ++ | − | +++ | +++ | − | Feces |
numbers represent titer of bacteriophage by the Gratia method in PFU/ml.
«++++» zone of pure lysis on a sensitive host strain by spot-test, where fewer pluses represent an impure lysis, as a result of an insufficient bacteriophage concentration in the given sample. The «minus» sign signifies the absence of bacteriophage in the given sample.
PCR with specific primers was used to identify one of the bacteriophages included in the cocktail in the blood of the patients and healthy volunteers 24 h after the end of phage therapy. In the samples of 2 healthy volunteers and 3 patients bends corresponding to DNA of phage PA5 were found (Fig. 3).
Figure 3.
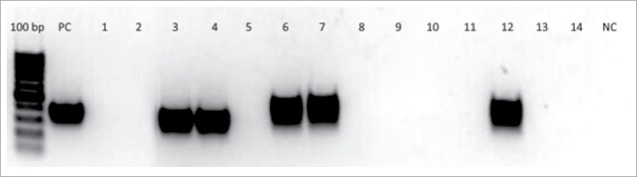
Results of double-nested PCR for identification of DNA of bacteriophage PA5 in the blood of 10 patients and 2 healthy volunteers. Gel electrophoresis lines of the 3rd round PCR products (amplicons): 100 bp – DNA ladder, PC – positive control (DNA of Pseudomonas aeruginosa PA5 phage), 1–2 - blood samples from healthy volunteers before PA5 phage administration, 3–4 – blood samples from healthy volunteers on 3rd day of PA5 phage administration, 5–14 – blood samples from 10 patients 24 h after end of intragastric administration of a bacteriophage cocktail including PA5 phage, NC – negative control (reaction mix). In the samples of 2 healthy volunteers (lines 3 and 4) and 3 patients (lines 6, 7, 12) bends corresponding to DNA of phage PA5 were found.
Conclusions
The results obtained from our clinical tests have confirmed our assumption that in order to develop prophylactic and therapeutic bacteriophage-based products against HAIs it is undesirable to use the classic approach of a fixed composition of bacteriophage strains in the cocktail. Under the influence of antibiotics and disinfectants, pathogenic microorganisms in a closed ecological system of a medical facility faster evolve into new strains, including multi-drug resistant forms. The way out of this situation may be a combination of a fixed composition of bacteriophage species in the cocktail with specifically selected bacteriophage strains active against actually persistent in a given hospital at a given moment bacterial strains from an existing collection of pheno- and genotypically characterized bacteriophages (Fig. 4).
Figure 4.
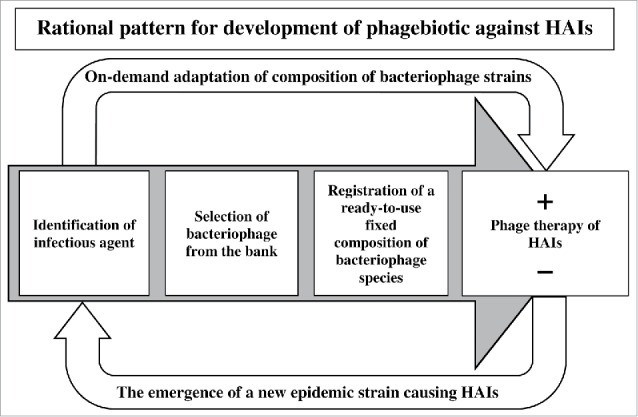
HAIs phage therapy model for clinical practice.
Footnotes
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Peterson LR. Bad Bugs, No Drugs: No ESCAPE Revisited. Clin Infect Dis 2009; 49(6):992-3; http://dx.doi.org/ 10.1086/605539 [DOI] [PubMed] [Google Scholar]
- [2].Reshedko GK, Ryabkova EL, Kretchikova OI, Sukhorukova MV, Shevchenko OV, Edelstain MV, Kozlov RS, on behalf of ROSNET study group Antimicrobial resistance patterns of gram-negative nosocomial pathogens in Russian ICUs. Clinical Microbiology and Antimicrobial Chemotherapy 2008; 10(2):96-112. (In Russian). [Google Scholar]
- [3].Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM. Phage treatment of human infections. Bacteriophage 2011; 1(2):66-85; PMID:22334863; http://dx.doi.org/ 10.4161/bact.1.2.15845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mai V, Ukhanova M, Visone L, Abuladze T, Sulakvelidze A. Bacteriophage administration reduces the concentration of Listeria monocytogenes in the gastrointestinal tract and its translocation to spleen and liver in experimentally infected mice. Int J Microbiol 2010; 2010:624234; PMID: 20652074; http://dx.doi.org/ 10.1155/2010/624234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Aleshkin A, Ershova O, Volozhantsev N, Popova A, Svetoch E, Rubalsky E. Phagebiotics in prevention and treatment of healthcare-associated infections Abstract Book “Bacteriophage 2016: 19th-21st January 2016 - Life Science Events”; P. 14 http://lifescienceevents.com/wp-content/uploads/PhageAbstracts2016-6.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Aleshkin A, Volozhantsev N, Popova A, Svetoch E, Rubal’sky E, Kiseleva I, Bochkareva S, Afanas'ev S. Phage-based cocktail for control of hospital-acquired pathogens. Phages 2015 Bacteriophage in Medicine, Food and Biotechnology: Conference handbook, 01–02 September 2015, St Hilda’s College, Oxford, UK; P. 17 http://lpmhealthcare.com/wp-content/uploads/2015/08/PHG15-Book.pdf. [Google Scholar]
- [7].Gratia A. Des relations numeriques entre bacteries lysogene set particules de bacteriophage. Annales de l'Institut Pasteur 1936; 57:652-76. [Google Scholar]
- [8].Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 2011; 12:402; PMID: 21824423; http://dx.doi.org/ 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Khabriev RU. Guidelines for experimental (pre-clinical) studies of new pharmacological agents. 2nd ed. Moscow: Meditsina Publishers ltd, 2005. (In Russian). [Google Scholar]


