Abstract
The HVEM (TNFRSF14) receptor gene is among the most frequently mutated genes in germinal center lymphomas. We report that loss of HVEM leads to cell autonomous activation of B cell proliferation and drives the development of GC lymphomas in vivo. HVEM deficient lymphoma B cells also induce a tumor supportive microenvironment marked by exacerbated lymphoid stroma activation and increased recruitment of T follicular helper (TFH) cells. These changes result from the disruption of inhibitory cell-cell interactions between the HVEM and BTLA (B and T Lymphocyte Attenuator) receptors. Accordingly, administration of the HVEM ectodomain protein (solHVEM(P37-V202)) binds BTLA and restores tumor suppression. To deliver solHVEM to lymphomas in vivo we engineered CD19-targeted chimeric antigen receptor (CAR) T cells that produce solHVEM locally and continuously. These modified CAR-T cells show enhanced therapeutic activity against xenografted lymphomas. Hence, the HVEM-BTLA axis opposes lymphoma development and our study illustrates the use of CAR-T cells as ‘micro-pharmacies’ able to deliver an anti-cancer protein.
Introduction
Most human lymphomas arise from germinal center (GC) B cells. These include diffuse large B cell lymphomas (DLBCL) and follicular lymphomas (FL) which continue to pose a significant health challenge. Recent genomic studies have yielded important new insight into lymphoma pathogenesis and have catalogued recurrent genomic lesions (Challa-Malladi et al., 2011; Cheung et al., 2010; Lohr et al., 2012; Morin et al., 2011; Okosun et al., 2014; Oricchio et al., 2011; Pasqualucci et al., 2014). In addition, the germinal center (GC) microenvironment has been discussed as a key factor in lymphoma development and as a predictor of clinical outcomes (Ame-Thomas et al., 2007; Amin et al., 2015; Dave et al., 2004; Lenz et al., 2008; Mourcin et al., 2012; Pangault et al., 2010). However, precise mechanisms linking the GC microenvironment to the pathogenesis of GC lymphomas are largely unknown.
The GC microenvironment is critical for most aspects of B cell function and likely contributes to lymphomagenesis. GCs are dynamic structures that are composed of multiple hematopoietic and stromal cell types (Chang and Turley, 2015; De Silva and Klein, 2015). For example, the main lymphoid stromal cell subtypes, fibroblastic reticular cells (FRCs) and follicular dendritic cells (FDCs), contribute to B cell recruitment, survival, and differentiation (Aguzzi et al., 2014; Fletcher et al., 2015). In turn, activated B cells produce the TNF family cytokines TNFα and LTα1β2 that stimulate FRCs and FDCs (Roozendaal and Mebius, 2011). CXCL13 derived from these stromal cells is the major attractant for follicular T helper (TFH) cells that in turn support B cells through CD40L and secretion of cytokines IL-4 and IL-21 (Crotty, 2014). Especially, FL B cells retain a strong dependence on the GC microenvironment, which is thought to form a permissive niche and engage in crosstalk with malignant B cells (Ame-Thomas and Tarte, 2014; Mourcin et al., 2012; Rehm et al., 2011).
Cancer specific gene alterations can shed light on tumor biology. For example, somatic mutations in the HVEM (Herpes Virus Entry Mediator; TNFRSF14) receptor gene are among the most frequent genetic lesions in GC lymphomas and have been variably associated with prognosis (Cheung et al., 2010; Launay et al., 2012; Lohr et al., 2012). Exactly how HVEM mutations contribute to the biology of GC lymphomas is not known.
Studies of the HVEM receptor in T lymphocytes inform our current knowledge of this receptor's function. In T lymphocytes HVEM engages in stimulating cell-cell interactions by binding to LIGHT or CD160 receptors, whereas HVEM binding to the BTLA receptor (B and T Lymphocyte Attenuator) results in an inhibitory signal (Bjordahl et al., 2013; Cai and Freeman, 2009; Costello et al., 2003; Pasero et al., 2012; Steinberg et al., 2011). Expression of HVEM and its partner receptors is lineage restricted. For example, normal B cells variably express HVEM and BTLA depending on their differentiation and activation stage but they lack LIGHT and CD160, whereas TFH cells are characterized by their high BTLA expression (M'Hidi et al., 2009; Murphy et al., 2006).
Our study examines the function of HVEM in GC lymphomagenesis using a genetically and pathologically accurate mouse model. We further explore strategies to restore HVEM function by delivering the HVEM ectodomain (solHVEM(Pro37-Val202)) to lymphomas in vivo.
Results
The interaction between the HVEM and BTLA receptors is lost in most human FLs
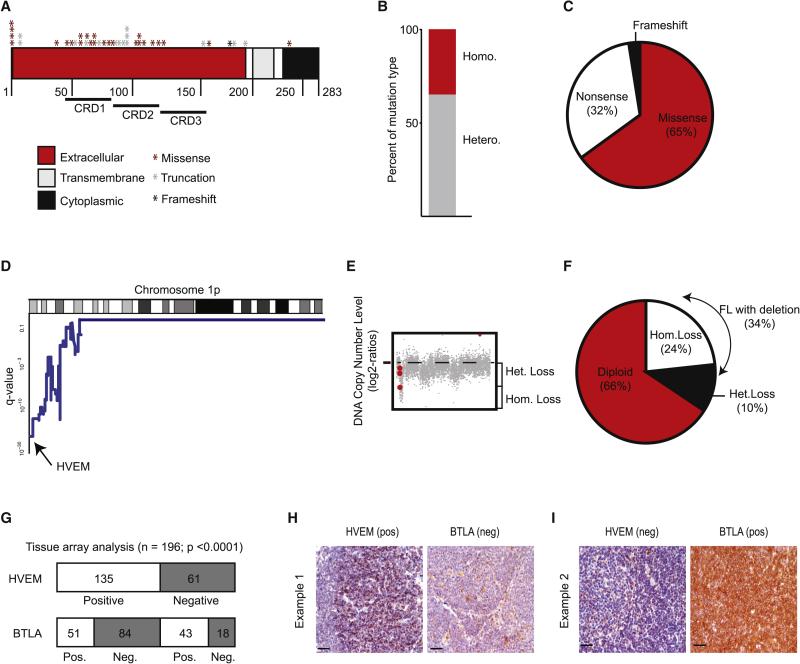
In a large collection (n = 141) of human FLs we find HVEM mutations in 28% (n = 40), and one third (35%) of these are homozygous mutations (Figure 1A-C)(Cheung et al., 2010; Launay et al., 2012; Lohr et al., 2012; Ross et al., 2007). HVEM mutations target the receptor's ectodomain and include missense (65%), nonsense (32.5%), and frame shift mutations (2.5%). Moreover, HVEM localizes to the minimal common region of the chromosome 1p36 deletion – a region that is commonly lost across B cell malignancies (Cheung et al., 2010; Fitzgibbon et al., 2007; Martin-Guerrero et al., 2013). Our meta-analysis of two separate series of array CGH data MSKCC: n = 64 (Oricchio et al., 2011); UNMC cohort: n = 198 (Bouska et al., 2014) shows that loss of the HVEM locus occurs in 34% of indolent FL samples (n = 262), and 37% of transformed FLs (n = 67) (Figure 1D-F, Suppl. Fig. 1A and B). GISTIC analysis indicates that 22-24% of these lesions are homozygous losses in both indolent and transformed samples (Figure 1E and Suppl. Fig. 1A). Hence, the genomic evidence indicates a powerful selection against the HVEM receptor gene during FL development.
Figure 1. The HVEM – BTLA interaction is disrupted in the majority of human FLs.
A, Summary of HVEM mutations in 141 FL samples; B, Distribution of copy number (CN) status in the 41 patients harboring a HVEM CN alteration, red: homozygous deletions, grey: heterozygous deletions; C, Percentage of each type of mutation found in FL patients; D, Chr. 1p36 deletions affect the HVEM locus (MSKCC cohort, n=64); E, GISTIC analysis indicates frequent homozygous HVEM deletions; F, Frequency of deletions by zygosity in indolent FL; G, Quantification of positive and negative cases represented on TMAs stained for HVEM and BTLA; H and I, immunohistochemical staining. In the first panel (H) strong staining with an anti-HVEM antibody was observed in the malignant cell population whereas BTLA remained largely negative. The second panel (I) is negative for HVEM but shows strong positivity for BTLA in all tumor cells. Original magnification x400, scale bars equal 50 μm. (See also Figure S1, and Table S1-S5).
We also examined HVEM protein expression in human FLs. We evaluated tissue microarrays comprising 198 FL samples for HVEM protein expression by immunohistochemistry. We scored a sample as HVEM positive when at least 20% of tumor cells showed specific staining. Using this cut-off, 61 samples (31.3%) were HVEM negative and 135 samples (68.7%) classified as HVEM positive (Figure 1G, Suppl. Fig. 1C). This proportion is consistent with the genomic data and we confirm reduced or absent protein expression in HVEM mutated or deleted samples for samples (n = 14) with available genomic and protein data (Suppl. Fig. 1D).
BTLA is a known HVEM binding partner and the only HVEM receptor expressed on normal and FL B cells (Murphy et al., 2006). Therefore, we evaluated BTLA expression across the lymphoma tissue arrays. For a positive BTLA score we demanded that tumor cells showed a stronger stain than reactive GC B cells, which are weakly positive and were used as on-slide controls. Using this cut-off for BTLA, 102 samples were negative (51.2%) and 94 samples (48.2%) scored as positive (Figure 1G, Suppl. Fig. 1E). Together, 145 of 198 samples (73%) were negative for either HVEM or BTLA and we wondered if these events might be linked. We tested their association using the chi-squared and found a significant negative (mutual exclusive) association such that HVEM positive tumors were more likely to lose BTLA than expected by chance (OR = 0.254; 95%CI 0.126 - 0.511; p < 0.0001) (Figure 1G-I, Suppl. Fig. 1F and 1G). We did not observe mutations or deletions of BTLA and it is most likely silenced transcriptionally. In this regard, we notice that BTLA expression is controlled by the KMT2D (MLL2) methyltransferase in FL (Ortega-Molina et al., 2015). Hence, the interaction between the HVEM and BTLA receptors is disrupted in the majority of human FLs indicating a potentially important tumor suppressor axis.
HVEM acts as a tumor suppressor in a mouse model of follicular lymphoma
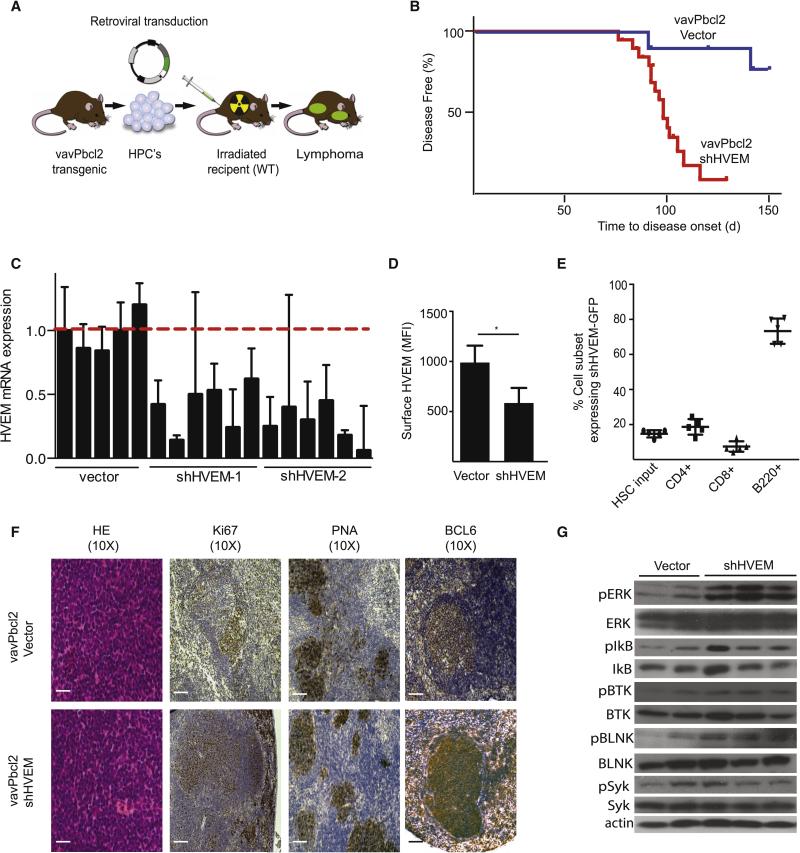
To elucidate the role of HVEM inactivation in FL development we took advantage of the vavPBcl2 model that recapitulates key aspects of the genetics and pathology of human BCL2-positive FLs (Egle et al., 2004; Oricchio et al., 2011). Briefly, we transduced vavPBcl2 hematopoietic progenitor cells (HPCs) isolated from fetal livers with retroviruses expressing short hairpin RNAs (shRNAs) against HVEM or empty vector controls. We then transplanted these cells into lethally irradiated mice, and monitored the recipients for lymphoma development (Figure 2A). Knockdown of HVEM (red, n = 19) caused a significant acceleration and increased penetrance of lymphoma development compared to controls (blue, n = 11). Ninety percent of control animals remained tumor free for > 100 days compared to only 10% of animals receiving the shHVEM (p < 0.01) (Figure 2B). We confirmed these results with a second shRNA against HVEM (Suppl. Fig. 2A). We also confirmed knockdown of the HVEM mRNA and HVEM surface expression by FACS (Figure 2C, 2D and Suppl. Fig. 2B). To test whether the HVEM knockdown in the B cell compartment was responsible for lymphoma development, we tracked the expression of the shHVEM co-expressed with the GFP reporter from the initial HPC infection into the derived hematopoietic compartments and did the same with HPCs expressing empty vector and GFP. The initial transduction efficiencies of the HPCs was ~15% and we found enrichment only in the FACS sorted shHVEM expressing lymphoma B cells (approx. 80%), by contrast we saw no enrichment with empty vector and GFP transduced HPCs (Figure 2E, Suppl. Fig. 2C). We noticed that the shHVEM and GFP were also expressed but not enriched in a fraction of whole CD8+ and CD4+ T cells (~15%) and specialized TFH cells (~2%). This may contribute to the observed phenotype although we could detect no significant change in HVEM mRNA levels in the total T cell population or in sorted TFH in association with the small proportion of GFPpos cells (Suppl. Fig. 2D and 2E and FACS not shown). We conclude that loss of HVEM drives specific B cell expansion leading to lymphoma development in vivo.
Figure 2. HVEM functions as a tumor suppressor in a mouse model of FL.
A, Schematic representation of vavPBcl2 mosaic mouse model; B, Kaplan-Meier analysis of disease free survival (blue: Vector, n=11; red: shRNA against HVEM, n=19); C, qRT-PCR analysis for relative HVEM expression on murine lymphomas; D, FACS quantification of HVEM surface expression in mouse tumors (n = 5, mean ± SD, *p < 0.01); E, Tracking expression of the shHVEM/GFP construct in indicated mouse cell populations, HSCs indicates initial infection efficiency before injection into mouse, (n=5); F, Representative pathology and immunohistochemistry for control (vavPBcl2/vector) and HVEM deficient (vavPBcl2/shHVEM) lymphomas, scale bars = 100 μm; G, Immunoblots for indicated signaling molecules on control (vector) and HVEM deficient (shHVEM) lymphomas. (See also Figure S2, and Table S6).
Pathological and molecular analyses of murine HVEM deficient lymphomas showed hallmarks of GC derived FLs and activation of key signaling molecules. For example, we found follicular architecture, expression of GC markers PNA, BCL6, and GL7 by immunohistochemistry and FACS analysis (Figure 2F, Suppl. Fig. 2F). Immunohistochemistry further showed increased Ki67 staining in HVEM deficient lymphomas (Suppl. Fig. 2G). FACS analysis showed that all lymphomas were largely composed of small B220+ and CD19+ B cells and showed no significant change in infiltrating CD3+ T cells (p = 0.14) (Suppl. Fig. 2H and I). A survey of signaling molecules further indicated activation and phosphorylation of signaling molecules related to the B cell receptor (BCR) pathway such as SYK, BTK, and BLNK, and their downstream targets ERK and IkB in HVEM deficient lymphomas compared to control lymphomas (Figure 2G). Deep sequencing-based BCR analysis revealed an oligoclonal disease and associated repertoire bias, with somatic hypermutation (SHM) yielding intraclonal diversity consistent with ongoing clonal evolution of a GC-driven disease (Suppl. Fig. 3).
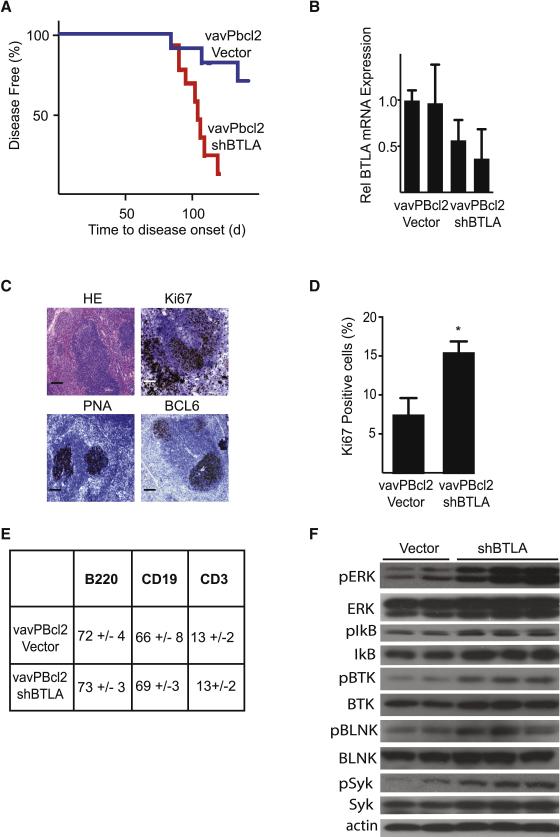
In human FL samples we had noticed a mutual exclusive pattern of HVEM and BTLA expression (Figure 1G-I). Studies in T cells have indicated that HVEM and BTLA can directly interact on the same cell - in cis (Cheung et al., 2009). These findings raise the possibility that loss of BTLA may similarly promote lymphoma development. We directly tested the effect of BTLA knockdown in the same vavPBcl2 mouse lymphoma model. Briefly, BTLA knockdown caused a significant acceleration of lymphoma development (n = 11 vector, n = 16 for BTLA, p < 0.01) (Figure 3A and 3B). Tumor pathology closely resembled the HVEM deficient tumors with follicular structures, higher Ki67 than controls, expression of the GC markers PNA and BCL6, and FACS analysis revealed cellular composition of predominant B220+ and CD19+ B cells, with similar CD3+ levels (Figure 3C-E, Suppl. Fig. 3D, compare to Figure 2). Similar to HVEM deficient lymphomas we observed increased phosphorylation of signaling molecules including BTK, BLNK, SYK, ERK, and IkB by immunoblots, whereas AKT was not consistently affected (Figure 3F, Suppl. Fig. 3E, and not shown). Hence, loss of either HVEM or BTLA can cooperate with Bcl2 and promote lymphoma development in vivo.
Figure 3. BTLA deficiency recapitulates the effect of HVEM loss on lymphoma development in vivo.
A, Kaplan-Meier analysis of disease free survival (blue: vector, n=11; red: shRNA against BTLA, n=16, p<0.01); B, qRT-PCR analysis of BTLA mRNA expression in control (vector) and BTLA (shBTLA) lymphomas; C, Pathological analysis of shBTLA tumors stained for representative sections including H&E, Ki67, PNA and BCL6, scale bars = 100 μm (control tumors are shown in Figure 2F); D, Quantification of Ki67 staining in shBTLA tumors (n=6, mean ± SD, *p<0.01); E, Surface analysis of vavPBcl2-vector and vavPBcl2-shBTLA tumors; F, Immunoblots for indicated signaling molecules on control (vector) and BTLA deficient (shBTLA) lymphomas. (See also Figure S3 and Table S6).
HVEM controls mitogenic signals in a cell autonomous and BTLA dependent manner
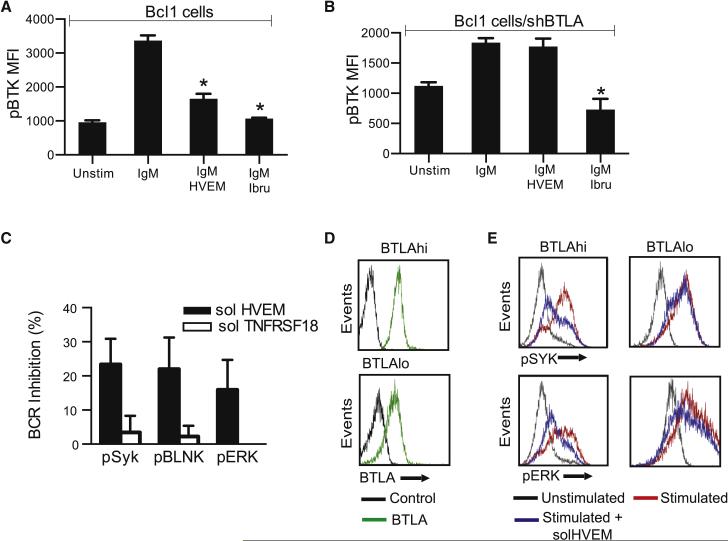
We have seen that loss of HVEM and BTLA lead to BCR activation in murine lymphomas B cells (Figures 2G and 3F). Activation of the BCR signal could be a direct effect related to BTLA's ability to bind CD79 or alternatively it could be secondary to changes in local cytokine levels (Vendel et al., 2009). In order to test whether HVEM has a direct, cell autonomous, and BTLA dependent effect on signaling we treated isolated lymphoma B cells with a purified soluble HVEM ectodomain protein fragment (solHVEM: Pro37 to Val202) that retains HVEM's binding properties (Cheung et al., 2005; del Rio et al., 2010). Briefly, we stimulated the BCR signaling pathway in Bcl1mouse lymphoma cells with anti-IgM in the presence or absence of solHVEM (10μg/ml) or the pharmacological BTK inhibitor ibrutinib (10nM) and measured BTK phosphorylation as an indicator of BCR pathway activation by flow cytometry. The addition of solHVEM blocked BTK phosphorylation and activation similar to the pharmacological inhibitor (Figure 4A). The ability of solHVEM to block the BCR signal transduction required BTLA and knockdown of BTLA prevented BTK inhibition in Bcl1 cells (Figure 4B). We made similar observations in primary human FL B cells. First, we stimulated purified human FL B cells with anti-IgG in the presence or absence of solHVEM (TNFRSF14; 10μg/ml) or the unrelated ectodomain of TNFRSF18 (10μg/ml) and confirmed HVEM specific inhibition of pSYK, pBLNK and pERK (Fig. 4C). Next, we analyzed BTLA expression across ten samples of purified human FL B cells by FACS and divided them into BTLAhi and BTLAlo groups (Figure 4D). We treated them as above and observed inhibition of SYK and ERK in BTLAhi cells whereas solHVEM had little effect in the BTLAlo cells (Figure 4E). Cumulative analysis of all ten primary human FL B cells confirmed a significant relationship between the ability of solHVEM to block SYK phosphorylation and BTLA surface expression (r = 0.697, p = 0.03) (Supp. Fig. 4A). We further confirmed solHVEM's (10μg/ml) inhibitory effect on BTK, SYK and ERK phosphorylation in DoHH2 lymphoma cells that express BTLA and carry a homozygous HVEM deletion (Suppl. Fig. 4B-F).
Figure 4. HVEM controls BCR signaling in lymphoma B cells.
A and B, Quantification of FACS analysis of phosphorylated BTK (pBTK) expression in Bcl1 cells after stimulation with anti-IgM in the presence of solHVEM (10μg/ml) or ibrutinib (10nM) in parental Bcl1 cells (A) or in BTLA deficient (Bcl1 shBTLA) cells (B); C, Percent BCR inhibition of indicated signaling molecules upon treatment with the solHVEM or the solTNFRSF18 proteins (10μg/ml) (mean ± SD, *p<0.05); D, FACS analysis of BTLA expression in purified primary human FL B cells identifies samples with high (BTLAhi) and low (BTLAlo) surface BTLA expression; E, FACS analysis for the indicated signaling molecules in human primary FL B cells that were BTLAhi or BTLAlo and stimulated with anti-human IgG (3min; 10 μg/ml and H202 1mM) in the presence or absence of solHVEM (10 μg/ml). (See also Figure S4).
HVEM deficient lymphomas have an excessive activation of the tumor stroma
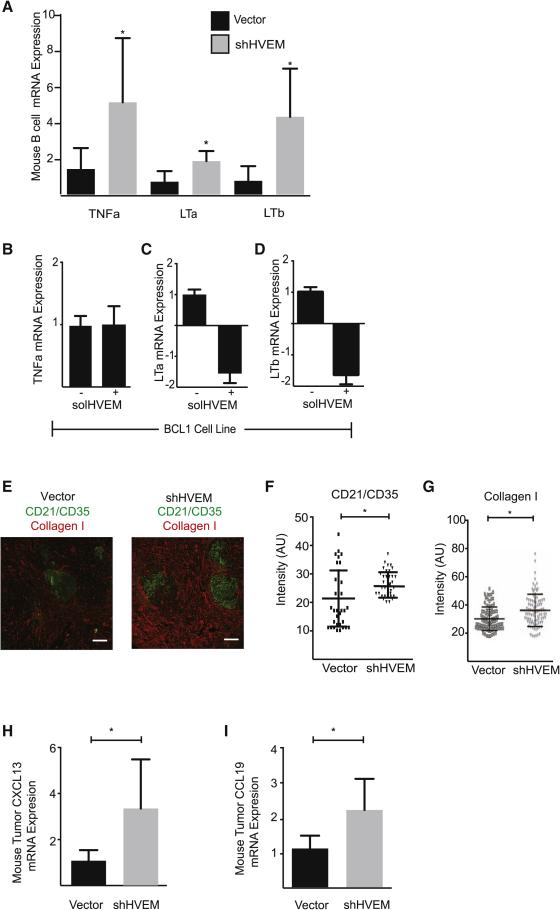
In human FLs the malignant B cells are admixed with an activated lymphoid stroma that provides support to the malignant B cells (Mourcin et al., 2012) (Suppl. Fig. 5A). The TNF family cytokines TNFα, LTα and LTβ are essential and non-redundant activators of the lymphoid stroma cells including the CD21Lpos follicular dendritic cells (FDCs) and the transglutaminasepos fibroblast reticular cells (FRCs) (Roozendaal and Mebius, 2011). An analysis of cytokine production in purified B220+ B cells from HVEM deficient lymphomas compared to control lymphomas revealed significantly increased production of all three stroma activating factors (n = 5, p < 0.05) (Figure 5A). Moreover, treatment of two different mouse lymphoma lines (Bcl1 and Myc/Bcl2) with the HVEM ectodomain (solHVEM; 10μg/ml) significantly decreased the expression of LTα and LTβ but did not change TNFα (p < 0.05); and differential regulation of these cytokines has been attributed to STAT5 activation (Lauenborg et al., 2015) (Figure 5B-D, Suppl. Fig. 5B-D).
Figure 5. Lymphoid stroma activation in HVEM deficient lymphomas.
A, Expression levels of stroma activating cytokines (LTα, LTβ, and TNFα) in B cells isolated from the spleens of vector and shHVEM mice (n=3, mean ± SD) by qRT-PCR; B-D, qRT-PCR expression analysis of TNFα (B), LTα (C), and LTβ (D) in Bcl1 cells 24h after treatment with solHVEM (10μg/ml); E, Immunohistofluorescence staining for the FDC marker CD21/35 and the FRC marker Collagen 1 on control lymphomas (vector) and HVEM knockdown lymphomas (shHVEM) (n=3, scale bars = 100 μm); F and G, Image quantification of CD21/35 (F) and collagen I (G) staining in control (Vector) and HVEM deficient (shHVEM) lymphomas based on 12 areas in the T-cell zone and 30 areas in the B cell zone per mice (cumulative number for 3 mice, * indicates p < 0.01 by parametric t-test; H and I, CXCL13 (H) and CCL19 (I) expression by qRT-PCR on control (vector) and HVEM knockdown (shHVEM) lymphomas (n=4, mean ± SD, * p< 0.05). (See also Figure S5).
The increased production of stroma activating TNF family cytokines was reflected in the abnormal lymphoid stroma activation in HVEM deficient tumors (Figure 5E). Quantitative analysis of microscopic images showed a significant increase of the CD21/CD35pos FDC network within follicles in HVEM deficient tumors compared to control tumors in the absence of immunization (n = 3; p < 0.01) (Figure 5F). Similarly, type I collagen density in perifollicular areas was significantly (p < 0.001) increased in HVEM deficient lymphomas reflecting activation of FRCs (Figure 5G). However, quantification of FRC branching did not reveal a quantitative increase in FRC cells and their network (Suppl. Fig. 5E and 5F). Further, consistent with these microscopic indicators of lymphoid stroma activation we observed significantly elevated expression of FDC and FRC derived cytokines - CXCL13 and CCL19 - in the HVEM deficient tumors compared to controls (n = 5; p < 0.05) (Mueller and Germain, 2009) (Figure 5H and 5I). Hence, HVEM deficiency contributes to the aberrant activation of lymphoid stroma cells.
Increased Follicular T helper (TFH) cells in HVEM deficient lymphomas
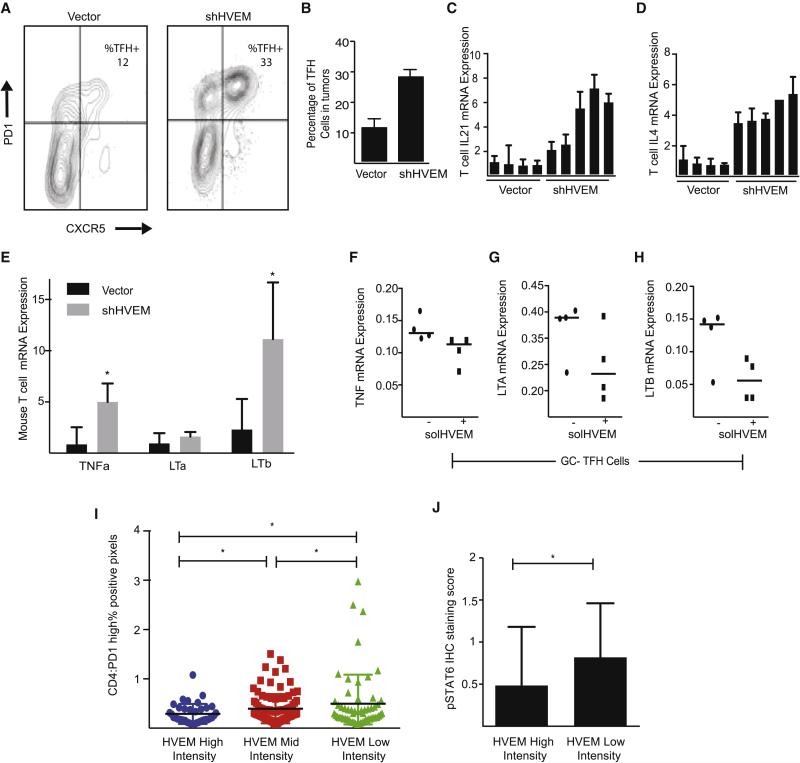
The stroma-derived cytokine CXCL13 is the main chemo-attractant for CXCR5pos follicular T helper cells (TFH) (Crotty, 2014). Consistent with the increased CXCL13 production in HVEM deficient lymphomas (Figure 5H) we observed a significant increase in the abundance of TFH cells in the HVEM deficient compared to control tumors (n = 3; p < 0.01) (Figure 6A and 6B). This increase in TFH cell numbers is associated with an elevated expression of TFH derived cytokines. Specifically, we find increased expression of IL-21, IL-4, and the stroma activating cytokines TNFα, LTα, and LTβ in FACS purified CD3+ T cells from HVEM deficient versus control lymphomas (n for each genotype = 5, p < 0.01) (Figure 6C - 6E). These TFH changes were not caused by aberrant shHVEM expression or loss of HVEM in TFH cells (Suppl. Fig. 2E). Intriguingly, we observed that the increased production of IL-21 and IL-4 by TFH cells was matched with an elevated expression of the IL-21 and IL-4 receptors on FACS purified lymphoma B cells from HVEM deficient lymphomas (p < 0.01) (Suppl. Fig. 6A and 6B). Human TFH cells are characterized by high-level expression of the BTLA receptor (Suppl. Figure 6C) and we wondered if HVEM directly affected these tumor infiltrating T cells. In order to test the direct effect of HVEM on TFH cells we treated purified human TFH cells with solHVEM as before in the presence or absence of stimulation with anti-CD3 and anti-CD28. SolHVEM did not affect TFH cell numbers or viability, and we observed reductions in the expression of LTα and LTβ, but not of TNFα, IL-21, or CXCL13 (Figure 6F - H, Suppl. Fig. 6D - F). Hence, genetically engineered murine HVEM deficient lymphomas show an increased ability to recruit IL-4 and IL-21 producing TFH cells.
Figure 6. Increased TFH cell recruitment in HVEM deficient lymphomas.
A and B, Representative FACS measurement (A) and quantification (B) of intratumoral TFH cells in control (vector) and HVEM deficient (shHVEM) murine lymphomas (n=3, mean ± SD); C and D, qRT-PCR measurement of IL-21 (C), and IL-4 (D) in sorted intra-tumoral T cells (vector: n=4, shHVEM: n=5); E, qRT-PCR measurement of the LTα, LTβ, and TNFα mRNA expression in T cells isolated from the spleens of vector and shHVEM mice (mean ± SD, * p < 0.05); F-H, qRT-PCR measurement of TNFα (F), LTα (G), and LTβ (H) in sorted TFH cell cultures (n = 4) and cultured with anti-CD3/anti-CD28 antibodies in presence or absence of solHVEM (10 μg/ml); I, Quantification of TFH cells (PD1hi fraction of CD4+ cells) in human FL samples grouped by percentage of HVEM positive tumor cells: high (> 80% HVEM positive cells; n = 24), medium (20% - 80% positive cells; n = 105), and low (< 20% positive cells; n = 59), *indicates p < 0.05); J, Quantification of immunohistochemical stains for phosphorylated STAT6 in human FL TMA samples comparing HVEM high (n=24) and HVEM low/negative (n=59) tumors, * indicates p < 0.05. (See also Figure S6).
We wondered if these changes might also be detectable in our collection of human FL samples. First, we measured TFH cell infiltration in relation to HVEM protein expression. We grouped all 198 tumors by high, medium, and low HVEM expression and quantified the percentage of PD1hi TFH cells as a fraction of all CD4 T cells. This revealed a significant increase in TFH infiltration (p = 0.032; comparing high to medium to low HVEM by one-way ANOVA) (Figure 6I). The TFH-derived cytokine IL-4 activates STAT6 (Pangault et al., 2010), therefore we examined STAT6 phosphorylation in tumors with > 90% of HVEM positive B cells (n = 24) and those that were essentially HVEM negative (i.e. < 10% HVEM positive B cells; n = 59). Consistent with increased numbers of IL-4 producing TFH cells we observed a significant increase in STAT6 phosphorylation in the HVEM negative FLs (p= 0.047) (Figure 6J). Hence, despite the genetic complexity of human FLs we can detect significant differences in TFH recruitment and B cell activation in HVEM deficient FLs.
The HVEM ectodomain protein has tumor suppressive effects
In order to test the potential anti-tumor effect of the solHVEM protein (HVEMPro37-Val202) we first characterized the expression of HVEM and BTLA across a panel of human and mouse lymphoma (mostly DLBCL) cell lines (Suppl. Fig. 7A). SolHVEM caused a significant growth inhibition in all BTLAhi lymphoma cells, whereas vehicle or the unrelated TNFRSF18 protein had no effects. Consistent with our results in human FL samples and primary cells (Figures 1 and 4), we found that BTLAlo cell lines generally expressed higher levels of HVEM and were insensitive to the solHVEM protein (Suppl. Fig. 7B).
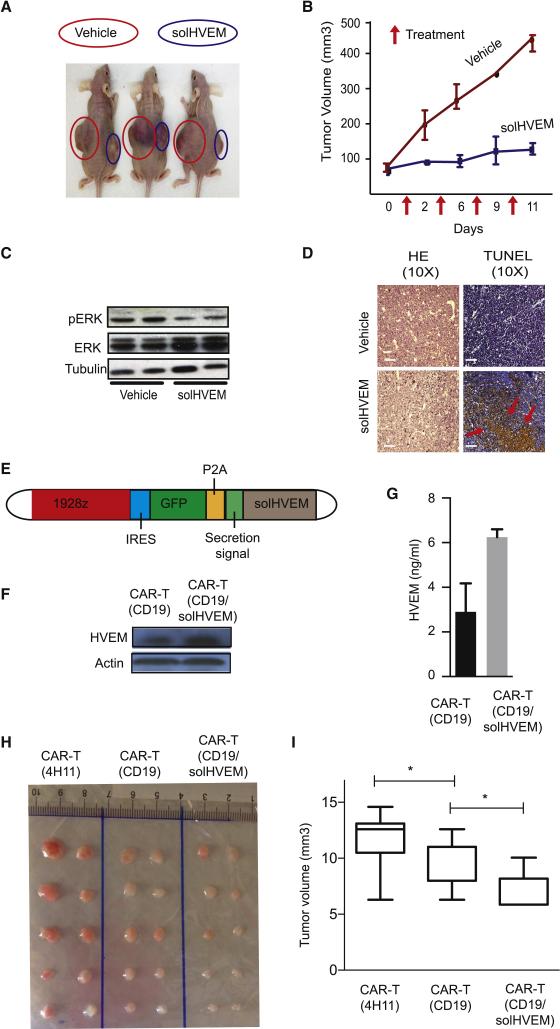
Next, we examined the activity of solHVEM against established lymphomas in vivo. We transplanted aggressive Myc/Bcl2 double positive murine lymphoma cells that express BTLA and lack HVEM (BTLAhi/HVEMlo) into the flanks of J:Nu nude mice and initiated treatment with intra-tumoral injection of solHVEM or vehicle (PBS) every three days for a total of four times once the engrafted tumors reached a volume of ~75mm3. Treatment with the solHVEM protein (20μg) prevented tumor growth, whereas the vehicle treated tumors expanded rapidly (n = 4; p < 0.01) (Figure 7A and 7B, Suppl. Fig. 7C and 7D). The effect of solHVEM was not merely cytostatic and TUNEL stains showed abundant apoptosis and immunoblots indicated ERK inhibition in vivo (Figure 7C and 7D). We also tested systemic injection of solHVEM (100μg, iv.) in Myc/Bcl2 murine lymphomas and observed only a modest effect (n =5; p = 0.12) (Suppl. Fig. 7E). These results indicate the powerful biological activity of solHVEM and they also highlight the need to achieve an adequate local exposure.
Figure 7. Restoring HVEM function for lymphoma therapy.
A, Representative picture of in vivo treatment of engrafted Myc/Bcl2 murine lymphomas by intratumoral injection of solHVEM (10μg) or vehicle; B, quantification of tumor volume response data from treatment study as in (A), injection times indicated by arrows; C, Immunoblot on lysates from solHVEM or vehicle treated lymphomas probed; D, Microscopic pathology on solHVEM or vehicle treated lymphomas stained as indicated, scale bars = 100 μm; E, Schematic of the CAR-T (CD19/solHVEM) retroviral construct; F, SolHVEM expression by immunoblot on lysates from CD19-directed CAR-T cells (CAR-T/CD19) and solHVEM producing CAR-Ts (CAR-T/CD19/solHVEM); G, SolHVEM secretion into supernatant measured by ELISA from indicated CAR-T cell types; H and I, In vivo CAR-T treatment studies of xenografted DoHH2 lymphomas treated with a single intravenous injection of 105 CAR-T cells; CAR-T(4H11) target an unrelated prostate antigen, CART/CD19 are CD-19 directed CAR-T cells, CAR-T/CD19/solHVEM are the solHVEM producing CAR-T cells; (H) image of tumors collected 1 month after CAR-T treatment; (I) quantification of tumor volume responses at end of study; *indicated p < 0.05. (See also Figure S7).
CAR-T cells can produce and secrete the solHVEM tumor suppressor
We engineered chimeric antigen receptor (CAR) T cells that seek out CD19 expressing B cells and that locally and continuously produce the solHVEM protein. Briefly, building on the reported CD19 CAR construct (Brentjens et al., 2013) we introduced the solHVEM coding sequence under separate transcriptional control and preceded by a secretion signal along with the GFP reporter for ease of tracking (Figure 7E). We confirmed that our CAR-T/solHVEM cells expressed and secreted the solHVEM protein by western blot on CAR-T lysates and by ELISA on CAR-T supernatants, respectively (Figure 7F and 7G). Initial tests showed that the solHVEM did not decrease T cell viability or impair CD3 and CD28 mediated T cell activation (Suppl. Fig. 7G and 7H). Next, we compared the ability of CAR-T and CAR-T/solHVEM cells to kill DoHH2 lymphoma cells in vitro and observed a significant enhancement in cell killing with CAR-T/solHVEM after 24h co-culture (Suppl. Fig. 7I). We tested the in vivo efficacy of CAR-T cells against xenografted DoHH2 lymphomas and treated lymphoma-bearing animals (tumor > 25mm3) with a single intravenous injection of 1×105 CAR-T cells. We compared CAR-T cells directed against an unrelated prostate antigen (4H11), regular CD19 directed CAR-T cells (CD19), and the HVEM producing and CD19 targeted CAR-T cells (CD19/solHVEM). We observed no frank toxicity or mortality with a single injection of CAR-T cells in either group (not shown). Compared to the control CAR-T (4H11) cells both the CD19-CAR-T cells and the solHVEM producing CAR-T cells produced significant therapeutic responses (n = 15; p < 0.05 for both comparisons). Moreover, the CAR-T/solHVEM cells were significantly more effective than the current CD-19 directed CAR-T cells (p = 0.016) (Figure 7H and 7I).
Discussion
Disruption of the HVEM-BTLA axis in germinal center lymphomas
The GC is the origin of most human B cell lymphomas and our study provides new insight into their pathogenesis. We find that the HVEM – BTLA interaction is disrupted in 75% of FLs indicating that it is a critical barrier to lymphoma development. The HVEM receptor gene is among the most frequent genetic targets in lymphomas and somatic mutations and chromosomal deletions typically result in loss of the receptor and some mutations may affect specific functions. BTLA is the only HVEM interacting receptor expressed in normal and GC lymphoma B cells and lymphomas that retain wild type HVEM are likely to silence expression of the BTLA receptor gene. BTLA is not a target of mutations or deletions, instead we notice that BTLA is a target of the KMT2D (MLL2) histone methyltransferase and KMT2D inactivation in lymphomas may contribute to silencing BTLA expression (Ortega-Molina et al., 2015). The high prevalence of these changes and the fact that they do not increase in transformed FLs suggests that the HVEM-BTLA axis is a failsafe mechanism that is disrupted during the early stages of GC lymphomagenesis.
Cell autonomous and microenvironmental consequences of HVEM loss
Our findings indicate that HVEM inactivation has dual effects on the lymphoma B cells and also shapes a tumor supportive niche. First, loss of HVEM stimulates BCR signaling and B cell growth in a cell autonomous and strictly BTLA dependent manner. The inhibitory BTLA receptor has two ITIM domains that can interact with BCR signaling molecules (CD79, SHP1/2) (Gavrieli et al., 2003; Vendel et al., 2009; Watanabe et al., 2003). Stimulation of BTLA by cell surface HVEM or soluble HVEM leads to inhibition of BCR signaling molecules and blocks lymphoma cell proliferation. In T cells this interaction can occur in cis on the same cell (Cheung et al., 2009). In B cell lymphomas this interaction may occur in cis leading to cell autonomous growth inhibition and also in trans, which would resemble a form of contact inhibition. Either way, disruption of the HVEM-BTLA interaction provides a mechanism of activating BCR related mitogenic signals in lymphoma B cells.
HVEM loss results in micro-environmental changes that support lymphoma growth. HVEM deficient B lymphocytes produce increased amounts of TNF family cytokines (TNFα, LTα, LTβ that are the key activators of lymphoid stroma cells such as FDCs and FRCs (Ame-Thomas et al., 2007; Guilloton et al., 2012; Roozendaal and Mebius, 2011). The activated lymphoid stroma in HVEM deficient mouse lymphomas closely resembles the abnormal stroma activation seen in human FLs (Mourcin et al., 2012). Human FL B cells show a unique dependence on stromal support that is mediated, at least in part, through increased CCL19 and CXCL13 mediated recruitment of TFH cells that in turn support B cells by producing IL-4, IL-21, and CD40L (Ame-Thomas et al., 2015{Pangault, 2010 #1807; Ame-Thomas et al., 2012; Pangault et al., 2010). These changes are readily evident in the genetically engineered mouse lymphomas, and we find activation of this supportive niche with increased TFH cells and IL-4 signaling also in human HVEM deficient FLs.
Direct and knock-on effects of HVEM loss
HVEM produces direct and BTLA dependent effects on B and TFH cells, in addition we observe secondary changes due to altered cytokine production. For example, lymphoid stromal cells do not express BTLA (not shown) and effects on the lymphoid stroma are mostly secondary to increased production of TNF family cytokines. On the other hand, BTLA is present at very high levels on TFH cells. Accordingly, we have seen that TFH cells are subject to both increased CXCL13 mediated recruitment and also direct effects of HVEM on TFH cells. Similarly, HVEM directly engages BTLA on lymphoma B cells and in addition TFH derived cytokines such as IL-4 and IL-21 provide further support B cell growth. Likely, HVEM has additional consequences and our study highlights how loss of HVEM disrupts a critical node that controls B cell growth in the GC environment.
Restoring the HVEM – BTLA interaction for therapy
HVEM is one of the most frequently mutated genes in FL and DLBCL and a therapeutic strategy tailored to HVEM deficient lymphomas is desirable. Notably, the interactions between the tumor suppressive HVEM and BTLA receptors occur at the cell surface and are, in principle, accessible for therapeutic attack. A soluble HVEM ectodomain protein (solHVEM) activates the inhibitory BTLA receptor resulting in impaired BCR signaling, partial normalization of cytokine production, apoptosis and tumor growth delay in vivo. However, this approach has two limitations. First, the anti-tumor effects of solHVEM depend on BTLA expression and an alternate strategy is needed to treat BTLA deficient lymphomas. Secondly, delivery of solHVEM to the lymphomas is challenging and may be achieved with bi-specific antibodies as we showed in (Oricchio et al., 2011), or alternatively by engineering HVEM producing cells as a vehicle.
CAR-T cells as “micro-pharmacies” to dispense solHVEM
There is much excitement about the use of genetically engineered CAR-T cells that can, in principle, attack any tumor antigen (Batlevi et al., 2016; Brentjens et al., 2003). Indeed this approach has shown impressive results especially against CD19+ B cell leukemias, however they appear to be less effective against lymphomas (Sadelain et al., 2015). To enhance the anti-lymphoma activity of CAR-T cells, we have engineered CD19 directed CAR-T cells to produce and secrete the solHVEM tumor suppressor protein. Given HVEM's biological activity and the high frequency of HVEM inactivation in FL and DLBCL we reasoned that local and continuous production would be superior to other forms of systemic delivery and may reduce unwanted off-target effects. Our findings indicate enhanced anti-lymphoma activity of solHVEM producing CAR-T cells. They provide proof-of-concept for the use of engineered immune cells as targeted delivery vehicles or “micro-pharmacies”. Additional studies are warranted to enhance the specific delivery of BTLA activating factors to HVEM deficient lymphomas and to explore cell-engineering technology as a platform for therapeutic delivery.
Methods and Resources
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to, and will be fulfilled by the Lead Contact Hans-Guido Wendel (wendelh@mskcc.org)
Experimental Model and Subject Details
Generation of Mice
All mice were housed in the specific-pathogen-free animal facility at the Memorial Sloan Kettering Cancer center and all animal studies were approved by the MSKCC IACUC committee (protocol-07-01-002). Unless indicated all mice studies used female mice on the C57Bl/6J background that were between 6 and 10 weeks of age. Wildtype mice were obtained from Jackson Laboratory (Bar Harbor, ME).
The vavPBcl2 mouse model is described (Egle et al., 2004) and adapted to adoptive transfer to retrovirally transduced HPCs (Wendel et al., 2004). In summary, we isolated vavPBcl2 transgenic fetal liver cells from vavPBcl2 heterozygous animals at embryonic day 14.5 (E14.5). The HPCs were grown in vitro for 4 days in a specially adapted growth medium containing IL3, stem cell factor, and IL6 and are retrovirally transduced with MSCV vectors directing the expression of shRNAs of interest. We transplant the genetically modified HPCs into lethally irradiated, syngeneic wild-type recipients and monitor disease onset weekly by palpation.
Human Samples
Subjects recruited after written informed consent recovery according to the Principles of the Declaration of Helsinki and the French National Cancer Institute (INCa) ethic committee recommendations. Samples comprised lymph nodes (LN) obtained from patients with follicular lymphoma (FL) and tonsils collected from children undergoing routine tonsillectomy. Tissues were cut into pieces and flushed using syringes and needles. Tonsil TFH were sorted using a FACSAria (Becton Dickinson) as CD3posCD4posCXCR5hiICOShiCD25neg cells with a purity greater than 98% as described (Mourcin et al., 2012; Pangault et al., 2010). Primary FL B cells were purified using the B cell isolation kit II (Miltenyi Biotech). Antibodies used in staining: Miltenyi CD3 (clone BW264/56), Beckman Coulter CD4 (clone 13B8.2), eBiosciences PD-1 (clone J105), and BD Biosciences CD25 (clone M-A251), CXCR5 (RF8B2), and BTLA (clone J168-540).
Method Details
Exon Sequencing of HVEM
Primers to amplify and sequence all coding exons and adjacent intronic sequences of HVEM were designed using the primer 3 program (http://primer3.ut.ee/) and sequence information generated using direct sequencing as described. Mutations were confirmed to be somatically acquired using unamplified lymphoma cell-derived DNA and paired CD3 cell-derived DNA from sorted cells as templates. (Li et al., 2014; Yildiz et al., 2015).
Deep Coverage Massively Parallel Re-Sequencing of HVEM
A customized multiplexed primer panel (Qiagen Gene Read Panel) was used to amplify all coding exons of HVEM. PCR products were pooled and sequencing libraries prepared using barcoded adapters. Sequencing was done on a HiSeq2000 sequencer. Bioinformatics nomination of sequence variants was performed using a custom algorithm developed by the University of Michigan bioinformatics core. Fastq files were uploaded to the Qiagen GeneRead data portal (http://ngsdataanalysis.sabiosciences.com) to trim primer regions from the reads and to align to the human genome (build hg19) using bowtie26. The aligned bam files were individually downloaded from the Qiagen portal and submitted to VarScan (2.3.6) for variant calling with default parameters. SnpEff (3.4B) was used to annotate the variants with gene names and predicted impact on amino acid sequence; dbNSFP (2.1) was used to annotate predicted functional impact based on standard tools (Sift, Polyphen, MutationTaster). Variants found in 1000 Genomes phase 2 were excluded9. Jacquard, a custom tool developed by the UM Bioinformatics Core, was used to combine all sample VCFs into a single matrix of variants by samples. All sequence variants with VAF > 15% were validated in stock T and paired N DNA using Sanger sequencing.
Array-CGH and Gistic Analysis
DNA from fresh frozen tissue or OCT-embedded was isolated and processed as previously described (Bouska et al., 2014; Oricchio et al., 2011). In summary, labeling and hybridization was done according to protocols performed by Agilent Technologies. Before submission to the Genomics core for labeling and hybridization to the 244K oligonucleotide array (Agilent Technologies), digestion efficiency of each DNA was checked by incubating 1 kb DNA ladder. Human male DNA served as the reference DNA. The slides were scanned at 5-um resolution using the G2565 Microarray Scanner System (Agilent) and Feature Extraction software (v9.1; Agilent Technologies) was used to generate the data. Data are available on GEO under accession no. GSE40989. Copy Number Data from the second dataset that consisted of 197 follicular lymphoma patients (UNMC dataset) has been generated using GeneChip Human Mapping 250K Nsp SNP array (Affymetrix) as described in (Bouska et al., 2014). To identify significantly amplified and deleted regions we used the Gistic 2.0 algorithm through the GenePattern web service (http://genepattern.broadinstitute.org) (Oricchio et al., 2014). GISTIC 2.0 (Beroukhim et al., 2010) has been run on segmented copy number data generated for each dataset using the DNAcopy package from Bioconductor (Olshen et al., 2004).
Immunohistochemical and TMA Methods
Immunohistochemistry (IHC) was applied to a tissue microarray (TMA) encompassing 1.5 mm duplicate cores of 199 formalin-fixed, paraffin-embedded (FFPE) tissue specimens from 186 patients diagnosed with FL (Kridel et al., 2015) 4μm sections were cut and IHC was performed on a Ventana BenchMark XT platform (Ventana, AZ) using a mouse monoclonal antibody against HVEM (dilution 1:50; clone 2G6-2C7; Abnova, Walnut, CA) and a rabbit polyclonal antibody against CD272/BTLA (dilution 1:100; Epitomics, cat. # S2379; Toronto, ON). Slides were evaluated by two hematopathologists (AM and RDG) for the percentage of positive tumor cells (in 10% increments) and staining intensity (0=negative, 1=weak, 2=moderate, 3=strong). Representative images were acquired with a Nikon DS-Fi1 camera connected to a Nikon Eclipse E600 microscope. The following antibodies were used to examine composition and functional properties of the microenvironment in human FL specimens: CD4(neat, clone SP35, Roche Ventana), PD-1 (dilution 1:50, clone MRQ-22, ESBE Cell Marque), pSTAT-6 (dilution 1:100, clone 18/P-Stat6, BD Biosciences). Immunohistochemical stained slides for CD4 and PD-1 were scanned with an Aperio ScanScope XT at 20x magnification. Analysis was performed utilizing the Aperio ImageScope viewer (Version 12.1.0; Aperio Technologies) and the Positive Pixel Count algorithm with optimized color saturation thresholds. Any staining was considered positive and the number of positive pixels was divided by the total pixel count. Phospho-STAT-6 expression was assessed semiquantitatively in intrafollicular T cells.
Immunohistofluorescence on Stromal Cells
Mouse spleens and human lymph nodes were snap frozen in OCT (Tissue-Tek OCT Compound). Twenty-micrometer sections were fixed in 4% PFA for 15min at room temperature. Sections were incubated for 1 hour with a blocking solution (PBS, 10% BSA, 10% Donkey serum, 0.1% Saponin) then incubated in a humidified chamber overnight at 4°C with the follow ing primary antibodies: CD21/CD35 (Rat IgG2b, dilution 1/50, BD Biosciences) and collagen I (Rabbit polyclonal, dilution 1/100, Abcam) for mouse spleens; and CD21L (Mouse IgM, dilution 1/100, Dako), Transglutaminase-2 (Mouse IgG1, dilution 1/50, Abcam), and CD20 (Polyclonal Rabbit, dilution 1/50, Abacam) from human lymph nodes. After washes, slides were incubated with the corresponding secondary antibodies (Jackson ImmunoResearch) and were finally mounted in Mowiol antifade reagent containing SytoxBlue (dilution 1/500, Invitrogen) and analyzed by confocal microscopy on a SP8 (Leica Microsystems). ImageJ software was used for image analysis.
TFH Stimulation
Purified TFH were cultured in IMDM 10% FCS with or without anti-CD3 (0.6μg/mL) and anti-CD28 (0.6μg/mL, Pelicluster Sanquin) MAbs in the presence or not of solHVEM (10μg/mL). After 3 days of culture, the number of viable TFH was evaluated by flow cytometry using count beads (Flow Count, Beckman Coulter) and Topro-3 staining (Invitrogen). CXCL13 and IL-21 were quantified in culture supernatants by ELISA (R&D Systems) according to manufacturer's instructions.
Real-Time Quantitative PCR
B and T cells were isolated from the spleens of mice using bead cell separation. Whole cell lysates were subject to either Pan T Cell Isolation Kit or the B Cell Isolation Kit (Miltenyl Biotec) and isolated subject to manufacturers instruction.
B and TFH samples were sorted before qRT-PCR by cell sorting using the Biomark system (Fluidigm). Total RNA was extracted from tumors, sorted T cells, and sorted B cells using Qiagen RNA extraction kit. Reverse transcription was performed on 1 μg of total RNA using the M-MulV reverse transcriptase (New England BioLabs). qRT-PCR analysis was performed by the ΔΔCt method using TaqMan Universal master mix on an ABI Prism 7000 Sequence Detection System (Applied Biosystems) (Mavrakis et al., 2008). Taqman Gene Expression assays were all received from Applied Biosystems: Gusb, IL-21, IL-4, IL-21ra, IL-4ra, HVEM, BTLA, LTα, TNFα, LTβ, CCL19, CXCL13.
Flow Cytometry
Single cell suspensions of each tumor genotype were obtained from mechanical dissociation and stained with representative surface maker antibody. After mechanical dissociation cells were collected in PBS-1%BSA and passed through a cell strainer to eliminate clumps and debris. Cells were spun down, suspended, and counted before primary antibody staining. Cells were stained with primary antibodies for at least 30 mins on ice in the dark. After staining cells were washed twice with PBS-1%BSA and analyzed on a BD LSRFortessa (Becton Dickinson, Franklin Lakes, NJ) Antibodies used in staining from BD Biosciences: B220 (clone RA3-6B2), CD19 (clone 1D3), FAS (clone Jo2), T and B cell activation antigen (GL7), IgG (A85-1), IgM (R6-60.2), CD3 (clone 17A2), or Biolegend: HVEM (clone HMHV-1B18), BTLA (clone 6A6).
Phospho-Flow Cytometry
Purified IgGpos FL B cells were stimulated using FITC-conjugated goat anti-human IgG (Invitrogen, 10 μ g/mL) in the presence of H2O2 (1 mM) with or without solHVEM (10 μ g/mL) or solTNFRSF18 (10 μ g/mL). The reaction was stopped by adding PFA at 4% final concentration for 15 min at room temperature. Fixed cells were permeabilized with methanol 80% for 20 min at −20°C in dark before washing and rehydratation with PBS-1% BSA. Phosphoprotein activation was quantified using Alexa 647-conjugated anti-pSyk (clone 17A/p-ZAP70), anti-pBLNK (clone j117-1278), or anti-pERK1/2 (clone 20A, BD Biosciences) and analyzed on B cells expressing clonal heavy and light chain gated using the anti-IgG FITC Ab and a PE-conjugated anti-kappa Ab (Southern Biotech). For phospho-BTK, phospho-Syk staining, DoHH2 or Bcl1 cells were pretreated for 60 min with either 10μg/mL of sHVEM (R&D Systems) or 10ng/mL Ibrutinib (ChemieTek PCI-32765) at 37°C in the presence of sh BTLA or without. Cells were fixed by adding equal volume of formaldehyde directly to the cells. Cells were incubated for 10 minutes at room temperature, washed 2x in PBS-1%BSA and the residual cells were permeabilized in 1mL of ice cold methanol (100%) for 30 min on ice. Cells were then washed twice and stained with the phospho-BTK (BD Biosciences clone N35-86) and phospho-Syk (BD Biosciences clone 17 A/P-Zap70) and analyzed on BD LSRFortessa.
Immunoblots
Immunoblots were performed from whole cell lysates or supernatant as previously described (Wendel et al., 2004). In brief, protein was extracted in 1X SDS buffer. Equal volumes of lysate were loaded per well of 10-12% SDS-PAGE gels and transferred to activated PVDF membranes (Millipore). Membranes were blocked in 5%BSA in TBST for 1 hour at room temperature followed by incubation with primary antibody in 5% BSA-TBST at 4°C overnight. All antibodies were used at dilutions of 1:1000 except for actin which was used at 1:10,000. Following incubation the membranes were washed 3X in TBST before being incubated with secondary antibody for 1hr at RT. Finally, the membranes were washed 3X in TBST before analysis. Proteins were visualized using enhanced chemiluminescence was used for detection (ECL; GE Healthcare). Antibodies were against, pSyk (Cell Signaling Technologies #2710), Syk (Cell Signaling Technologies #2712), pBTK (Cell Signaling Technologies # 5082), BTK (Cell Signaling Technologies # 3533) pERK (Cell Signaling Technologies #4370), ERK (Cell Signaling Technologies #9102), pBLNK (Abcam #174837), BLNK (Cell Signaling # 12168), IKb (Cell Signalling # 9242), pIKb (Cell Signalilng #2859), and actin (Sigma-Aldrich).
Sequencing of VDJ regions
RNA was prepared from potentially tumoral lymphoid tissues and from a normal mouse spleen as control. Concentrations of RNA were confirmed using a nanodrop and ~ 200ng RNA was used for sequencing. Transcripts were amplified using RACE-PCR starting with a reverse primer hybridizing within the μ CH1 exon. Expressed VDJ regions from μ heavy chain transcripts were sequenced through a next generation method. This strategy (Li et al., 2013) combined 5’ RACE PCR, pyrosequencing and precise repertoire analysis with quantification of the most frequent clonotypes using IMGT/High-V-QuestmRNA and associated tools available on IMGT (the International ImMunoGeneTics information website (www.imgt.org).
Cell Culture and Proliferation Assays
Lymphoma cell lines DoHH2, Ly-10, Granta, Su-DHL-6, Su-DHL-4, Su-DHL-10, Su-DHL-8, Raji were obtained from ATCC, authenticated, and maintained in RPMI- 1640 with 10% fetal bovine serum, 1% L-Glutamine and 1% penicillin/streptomycin (Oricchio et al., 2014). Mouse lymphoma cell line Myc/Bcl2 was maintained in IMDM-DMEM (50:50) with 10% fetal bovine serum, 1% L-Glutamine, and 1% penicillin/streptomycin. Bcl1 cell line was maintained in RPMI-1640 with 15% fetal bovine serum, 1% L-Glutamine and 1% penicillin/streptomycin, 0.05mM ME. Cell lines were seeded at 5 × 105 /mL and were treated with 10 μg/ml of sHVEM. After 24 hours cell number was counted using a hemocytometer for a total of 72 hours after treatment.
Treatment Studies
Transplant and treatment studies were generated as previously described (Schatz et al., 2011). In summary, subcutaneous injection of one million Myc/Bcl2 mouse lymphoma cells combined with Matrigel (BD) in the right and left flanks of mice J:Nu Nude (Foxn1 nu/ Foxn1 nu, Jackson Laboratories). Once tumors reached 75-mm3 mice were treated every three days by intra tumor injection with 20μg of sHVEM diluted in PBS (right flank) or with vehicle control (left flank). Tumor sizes were measured and recorded every three days. Tumors were weighed after the animals were sacrificed and tumors excised. In vivo efficacy of CAR-T treatment, xenografts were generated by s.c. injections of 10Mio DoHH2 human lymphoma cells mixed with Matrigel (BD) into flanks of NOD/SCID (NOD.CB17- Prkdcscid/J, Jackson Laboratory) mice. Upon visible tumor formation (25mm3), mice were given a single dose of 105 CAR-T. T cells containing prostate-specific membrane antigen (PSMA) scFv (Pegram et al., 2012) was used as a control CAR. Tumors were measured after the animals were sacrificed and tumors excised 4 weeks after the CAR-T cell injection.
CAR-T Cell Generation
SFG-1928z vector was modified to contain GFP and the human extracellular HVEM sequence by addition of P2A site followed by IgG Kappa secretion signal that precedes HVEM (P37-V202) sequence. Green fluorescent protein (GFP) sequence was included as a leader peptide following 1828z sequence by addition of internal ribosomal entry site (IRES). Human T cells were isolated from human PBMCs by density centrifugation, and activated and expanded by culturing with CD3/CD28 Dynobeads (Invitrogen) in presence of IL-2 (Peprotech) and phytohemagglutinin (Sigma). Transduction of T cells was performed on rectronectin (Takara) covered plates. Upon T cell transduction, GFP+ cells were sorted and further expanded using CD3/CD28 beads. The cytolytic capacity of transduced T cells was determined by co-culturing target and effector cells at particular cell ratios (Pegram et al., 2012). After 24 hours of co-culture, cells were harvested and stained for DAPI and Annexin V and assayed by flow cytometry to detect residual GFP-negative viable cells. HVEM expression was assessed via western blot analysis of T cells containing 1928-GFP construct alone in comparison to T cells containing 1928-GFP-HVEM construct. HVEM secretion was confirmed by ELISA assay of cell culture supernatant using the Origene Human HVEM ELISA kit.
Quantification and Statistical analysis
Statistical Methods
Sample sizes for comparisons between cell types or between mouse genotypes followed Mead's recommendations (Festing and Altman, 2002). Samples were allocated to their experimental groups according to their pre-determined type (i.e mouse genotype) and therefore there was no randomization. Investigators were not blinded to the experimental groups. Quantitative PCR data were obtained from independent biological replicates (n values indicated in the corresponding figure legends). Normal distribution and equal variance was confirmed in the large majority of data and, therefore, we assumed normality and equal variance for all samples. Based on this, we used the Student's t-test (two-tailed, unpaired) or ANOVA where appropriate to estimate statistical significance. Survival in mouse experiments was represented with Kaplan-Meier curves, and significance was estimated with the log-rank test. For association analysis between HVEM and BTLA expression in human FL tissue biopsies we performed a Chi-square test. Statistical parameters including the exact value of n and statistical significance are reported in the Results, Figures and Figure Legends. Statistical analysis was performed using Graph Pad PRISM 6.
Data and Software availability
Raw data files have been deposited in the NCBI Gene Expression Omnibus under accession number GSE40989.
Supplementary Material
Acknowledgements
We thank Irina Linkov (MSKCC), Shenqiu Wang (MSKCC), Ana Ortega-Molina (MSKCC), and Sylvain Prigent (Rennes University) for advice and reagents. Thanks to all the members of MSK Antitumor assessment core for technical assistance with mice; the MSK Laboratory of Comparative Pathology, MSK Flow Cytometry, MSK Molecular Cytology, and Microscopy Rennes Imaging Center (MRic-ALMF; UMS 6480 cores, and to the Centre de Ressources Biologiques (CRB)-Santé (BB-0033-00056) of Rennes hospital for its support in the processing of biological samples. This research was supported by funding from the American Cancer Society grant RSG-13-048-01-LIB (HGW), the Lymphoma Research Foundation (HGW), Cycle for Survival (HGW), the Steven A. Greenberg Foundation, Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and The Center for Experimental Therapeutics at Memorial Sloan Kettering Cancer Center (HGW), NIH grants RO1CA183876-03, 1RO1CA207217-01, 1R01CA19038-01 (HGW), Core Grant P30 CA008748 (HGW), NIH Spore P50 CA192937-01A1, LLS Score GC227931 (HGW), LLS 1318-15 (HGW); the Ligue Nationale contre le Cancer (KT), the Foundation ARC pour la recherche sur le cancer (AO 2011), the French Institut National du Cancer (INCA AAP PLBIO-13-085; KT), Follicular lymphoma grant from Lymphoma Research Foundation (WCC), NIH grant 1R01CA190384-01 (SNM), NIH Grants R01CA138738-05, PO1CA059350, PO1CA190174-01 (RJB); AM is supported by the Dr. Mildred-Scheel Cancer Foundation (Deutsche Krebshilfe), The Michael Smith Foundation for Health Research, and Lymphoma Canda; SNM is a Scholar of the Leukemia and Lymphoma Society; HGW is a Scholar of the Leukemia and Lymphoma Society. R.A. is supported by the FEDER (Fonds Européens de Développement Régional), the CPER (Contrat de plan Etat région Bretagne, axe biothérapie), and the ADHO (Association pour le Développement de l'Hémato-Oncologie).
Footnotes
Author contributions
M.B. initiated study and performed tumorigenesis experiments; D.S. performed signaling, immunotherapy and CAR-T studies; F.M. and R.A. performed microenvironment and human FL cell studies; R.J.B. provided CAR-T construct; V.S. cloned modified CAR-T construct; G.C., N.D.L., M.C., W.C.C, S.N.M. analyzed and provided human and murine genomic data; W.T., J.T.F., A.M., D.E., R.D.G. pathological analysis, IHC stains, TMA analyses; M.J. provided technical assistance; E.O. mouse lymphoma model; E.S. provided mouse assistance; K.T. supervised microenvironment and human FL cell studies; H.G.W designed the study wrote the manuscript and acts as lead contact.
Author information The authors declare no competing financial interests.
Supplementary information.
Supplemental information includes 7 Suppl. Figures and 6 Suppl. tables.
References
- Aguzzi A, Kranich J, Krautler NJ. Follicular dendritic cells: origin, phenotype, and function in health and disease. Trends Immunol. 2014;35:105–113. doi: 10.1016/j.it.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Ame-Thomas P, Hoeller S, Artchounin C, Misiak J, Braza MS, Jean R, Le Priol J, Monvoisin C, Martin N, Gaulard P, et al. CD10 delineates a subset of human IL-4 producing follicular helper T cells involved in the survival of follicular lymphoma B cells. Blood. 2015;125:2381–2385. doi: 10.1182/blood-2015-02-625152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ame-Thomas P, Le Priol J, Yssel H, Caron G, Pangault C, Jean R, Martin N, Marafioti T, Gaulard P, Lamy T, et al. Characterization of intratumoral follicular helper T cells in follicular lymphoma: role in the survival of malignant B cells. Leukemia. 2012;26:1053–1063. doi: 10.1038/leu.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ame-Thomas P, Maby-El Hajjami H, Monvoisin C, Jean R, Monnier D, Caulet-Maugendre S, Guillaudeux T, Lamy T, Fest T, Tarte K. Human mesenchymal stem cells isolated from bone marrow and lymphoid organs support tumor B-cell growth: role of stromal cells in follicular lymphoma pathogenesis. Blood. 2007;109:693–702. doi: 10.1182/blood-2006-05-020800. [DOI] [PubMed] [Google Scholar]
- Ame-Thomas P, Tarte K. The yin and the yang of follicular lymphoma cell niches: role of microenvironment heterogeneity and plasticity. Semin Cancer Biol. 2014;24:23–32. doi: 10.1016/j.semcancer.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Amin R, Mourcin F, Uhel F, Pangault C, Ruminy P, Dupre L, Guirriec M, Marchand T, Fest T, Lamy T, et al. DC-SIGN-expressing macrophages trigger activation of mannosylated IgM B-cell receptor in follicular lymphoma. Blood. 2015;126:1911–1920. doi: 10.1182/blood-2015-04-640912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlevi CL, Matsuki E, Brentjens RJ, Younes A. Novel immunotherapies in lymphoid malignancies. Nat Rev Clin Oncol. 2016;13:25–40. doi: 10.1038/nrclinonc.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjordahl RL, Steidl C, Gascoyne RD, Ware CF. Lymphotoxin network pathways shape the tumor microenvironment. Current opinion in immunology. 2013;25:222–229. doi: 10.1016/j.coi.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouska A, McKeithan TW, Deffenbacher KE, Lachel C, Wright GW, Iqbal J, Smith LM, Zhang W, Kucuk C, Rinaldi A, et al. Genome-wide copy-number analyses reveal genomic abnormalities involved in transformation of follicular lymphoma. Blood. 2014;123:1681–1690. doi: 10.1182/blood-2013-05-500595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, Bartido S, Stefanski J, Taylor C, Olszewska M, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5:177ra138. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentjens RJ, Latouche JB, Santos E, Marti F, Gong MC, Lyddane C, King PD, Larson S, Weiss M, Riviere I, et al. Eradication of systemic B-cell tumors by genetically targeted human T lymphocytes co-stimulated by CD80 and interleukin-15. Nat Med. 2003;9:279–286. doi: 10.1038/nm827. [DOI] [PubMed] [Google Scholar]
- Cai G, Freeman GJ. The CD160, BTLA, LIGHT/HVEM pathway: a bidirectional switch regulating T-cell activation. Immunological reviews. 2009;229:244–258. doi: 10.1111/j.1600-065X.2009.00783.x. [DOI] [PubMed] [Google Scholar]
- Challa-Malladi M, Lieu YK, Califano O, Holmes AB, Bhagat G, Murty VV, Dominguez-Sola D, Pasqualucci L, Dalla-Favera R. Combined Genetic Inactivation of β2-Microglobulin and CD58 Reveals Frequent Escape from Immune Recognition in Diffuse Large B Cell Lymphoma. Cancer Cell. 2011;20:728–740. doi: 10.1016/j.ccr.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JE, Turley SJ. Stromal infrastructure of the lymph node and coordination of immunity. Trends Immunol. 2015;36:30–39. doi: 10.1016/j.it.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Cheung KJ, Johnson NA, Affleck JG, Severson T, Steidl C, Ben-Neriah S, Schein J, Morin RD, Moore R, Shah SP, et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010;70:9166–9174. doi: 10.1158/0008-5472.CAN-10-2460. [DOI] [PubMed] [Google Scholar]
- Cheung TC, Humphreys IR, Potter KG, Norris PS, Shumway HM, Tran BR, Patterson G, Jean-Jacques R, Yoon M, Spear PG, et al. Evolutionarily divergent herpesviruses modulate T cell activation by targeting the herpesvirus entry mediator cosignaling pathway. Proc Natl Acad Sci U S A. 2005;102:13218–13223. doi: 10.1073/pnas.0506172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TC, Oborne LM, Steinberg MW, Macauley MG, Fukuyama S, Sanjo H, D'Souza C, Norris PS, Pfeffer K, Murphy KM, et al. T cell intrinsic heterodimeric complexes between HVEM and BTLA determine receptivity to the surrounding microenvironment. J Immunol. 2009;183:7286–7296. doi: 10.4049/jimmunol.0902490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello RT, Mallet F, Barbarat B, Schiano De Colella JM, Sainty D, Sweet RW, Truneh A, Olive D. Stimulation of non-Hodgkin's lymphoma via HVEM: an alternate and safe way to increase Fas-induced apoptosis and improve tumor immunogenicity. Leukemia. 2003;17:2500–2507. doi: 10.1038/sj.leu.2403175. [DOI] [PubMed] [Google Scholar]
- Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, Fisher RI, Braziel RM, Rimsza LM, Grogan TM, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nature reviews Immunology. 2015;15:137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio ML, Lucas CL, Buhler L, Rayat G, Rodriguez-Barbosa JI. HVEM/LIGHT/BTLA/CD160 cosignaling pathways as targets for immune regulation. J Leukoc Biol. 2010;87:223–235. doi: 10.1189/jlb.0809590. [DOI] [PubMed] [Google Scholar]
- Egle A, Harris AW, Bath ML, O'Reilly L, Cory S. VavP-Bcl2 transgenic mice develop follicular lymphoma preceded by germinal center hyperplasia. Blood. 2004;103:2276–2283. doi: 10.1182/blood-2003-07-2469. [DOI] [PubMed] [Google Scholar]
- Festing MF, Altman DG. Guidelines for the design and statistical analysis of experiments using laboratory animals. ILAR J. 2002;43:244–258. doi: 10.1093/ilar.43.4.244. [DOI] [PubMed] [Google Scholar]
- Fitzgibbon J, Iqbal S, Davies A, O'Shea D, Carlotti E, Chaplin T, Matthews J, Raghavan M, Norton A, Lister TA, et al. Genome-wide detection of recurring sites of uniparental disomy in follicular and transformed follicular lymphoma. Leukemia. 2007;21:1514–1520. doi: 10.1038/sj.leu.2404696. [DOI] [PubMed] [Google Scholar]
- Fletcher AL, Acton SE, Knoblich K. Lymph node fibroblastic reticular cells in health and disease. Nature reviews Immunology. 2015;15:350–361. doi: 10.1038/nri3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrieli M, Watanabe N, Loftin SK, Murphy TL, Murphy KM. Characterization of phosphotyrosine binding motifs in the cytoplasmic domain of B and T lymphocyte attenuator required for association with protein tyrosine phosphatases SHP-1 and SHP-2. Biochemical and biophysical research communications. 2003;312:1236–1243. doi: 10.1016/j.bbrc.2003.11.070. [DOI] [PubMed] [Google Scholar]
- Guilloton F, Caron G, Menard C, Pangault C, Ame-Thomas P, Dulong J, De Vos J, Rossille D, Henry C, Lamy T, et al. Mesenchymal stromal cells orchestrate follicular lymphoma cell niche through the CCL2-dependent recruitment and polarization of monocytes. Blood. 2012;119:2556–2567. doi: 10.1182/blood-2011-08-370908. [DOI] [PubMed] [Google Scholar]
- Kridel R, Mottok A, Farinha P, Ben-Neriah S, Ennishi D, Zheng Y, Chavez EA, Shulha HP, Tan K, Chan FC, et al. Cell of origin of transformed follicular lymphoma. Blood. 2015;126:2118–2127. doi: 10.1182/blood-2015-06-649905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauenborg B, Christensen L, Ralfkiaer U, Kopp KL, Jonson L, Dabelsteen S, Bonefeld CM, Geisler C, Gjerdrum LM, Zhang Q, et al. Malignant T cells express lymphotoxin alpha and drive endothelial activation in cutaneous T cell lymphoma. Oncotarget. 2015;6:15235–15249. doi: 10.18632/oncotarget.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launay E, Pangault C, Bertrand P, Jardin F, Lamy T, Tilly H, Tarte K, Bastard C, Fest T. High rate of TNFRSF14 gene alterations related to 1p36 region in de novo follicular lymphoma and impact on prognosis. Leukemia. 2012;26:559–562. doi: 10.1038/leu.2011.266. [DOI] [PubMed] [Google Scholar]
- Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, Xu W, Tan B, Goldschmidt N, Iqbal J, et al. Stromal gene signatures in large-B-cell lymphomas. The New England journal of medicine. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Kaminski MS, Li Y, Yildiz M, Ouillette P, Jones S, Fox H, Jacobi K, Saiya-Cork K, Bixby D, et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123:1487–1498. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lefranc MP, Miles JJ, Alamyar E, Giudicelli V, Duroux P, Freeman JD, Corbin VD, Scheerlinck JP, Frohman MA, et al. IMGT/HighV QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nat Commun. 2013;4:2333. doi: 10.1038/ncomms3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M'Hidi H, Thibult ML, Chetaille B, Rey F, Bouadallah R, Nicollas R, Olive D, Xerri L. High expression of the inhibitory receptor BTLA in T-follicular helper cells and in B-cell small lymphocytic lymphoma/chronic lymphocytic leukemia. American journal of clinical pathology. 2009;132:589–596. doi: 10.1309/AJCPPHKGYYGGL39C. [DOI] [PubMed] [Google Scholar]
- Martin-Guerrero I, Salaverria I, Burkhardt B, Szczepanowski M, Baudis M, Bens S, de Leval L, Garcia-Orad A, Horn H, Lisfeld J, et al. Recurrent loss of heterozygosity in 1p36 associated with TNFRSF14 mutations in IRF4 translocation negative pediatric follicular lymphomas. Haematologica. 2013;98:1237–1241. doi: 10.3324/haematol.2012.073916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis KJ, Zhu H, Silva RL, Mills JR, Teruya-Feldstein J, Lowe SW, Tam W, Pelletier J, Wendel HG. Tumorigenic activity and therapeutic inhibition of Rheb GTPase. Genes Dev. 2008;22:2178–2188. doi: 10.1101/gad.1690808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourcin F, Pangault C, Amin-Ali R, Ame-Thomas P, Tarte K. Stromal cell contribution to human follicular lymphoma pathogenesis. Front Immunol. 2012;3:280. doi: 10.3389/fimmu.2012.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SN, Germain RN. Stromal cell contributions to the homeostasis and functionality of the immune system. Nature reviews Immunology. 2009;9:618–629. doi: 10.1038/nri2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KM, Nelson CA, Sedy JR. Balancing co-stimulation and inhibition with BTLA and HVEM. Nature reviews Immunology. 2006;6:671–681. doi: 10.1038/nri1917. [DOI] [PubMed] [Google Scholar]
- Okosun J, Bodor C, Wang J, Araf S, Yang CY, Pan C, Boller S, Cittaro D, Bozek M, Iqbal S, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46:176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Oricchio E, Ciriello G, Jiang M, Boice MH, Schatz JH, Heguy A, Viale A, de Stanchina E, Teruya-Feldstein J, Bouska A, et al. Frequent disruption of the RB pathway in indolent follicular lymphoma suggests a new combination therapy. J Exp Med. 2014;211:1379–1391. doi: 10.1084/jem.20132120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oricchio E, Nanjangud G, Wolfe AL, Schatz JH, Mavrakis KJ, Jiang M, Liu X, Bruno J, Heguy A, Olshen AB, et al. The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma. Cell. 2011;147:554–564. doi: 10.1016/j.cell.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Molina A, Boss IW, Canela A, Pan H, Jiang Y, Zhao C, Jiang M, Hu D, Agirre X, Niesvizky I, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21:1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangault C, Ame-Thomas P, Ruminy P, Rossille D, Caron G, Baia M, De Vos J, Roussel M, Monvoisin C, Lamy T, et al. Follicular lymphoma cell niche: identification of a preeminent IL-4-dependent T(FH)-B cell axis. Leukemia. 2010;24:2080–2089. doi: 10.1038/leu.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero C, Speiser DE, Derre L, Olive D. The HVEM network: new directions in targeting novel costimulatory/co-inhibitory molecules for cancer therapy. Current opinion in pharmacology. 2012;12:478–485. doi: 10.1016/j.coph.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Khiabanian H, Fangazio M, Vasishtha M, Messina M, Holmes AB, Ouillette P, Trifonov V, Rossi D, Tabbo F, et al. Genetics of follicular lymphoma transformation. Cell reports. 2014;6:130–140. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegram HJ, Lee JC, Hayman EG, Imperato GH, Tedder TF, Sadelain M, Brentjens RJ. Tumor-targeted T cells modified to secrete IL-12 eradicate systemic tumors without need for prior conditioning. Blood. 2012;119:4133–4141. doi: 10.1182/blood-2011-12-400044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm A, Mensen A, Schradi K, Gerlach K, Wittstock S, Winter S, Buchner G, Dorken B, Lipp M, Hopken UE. Cooperative function of CCR7 and lymphotoxin in the formation of a lymphoma-permissive niche within murine secondary lymphoid organs. Blood. 2011;118:1020–1033. doi: 10.1182/blood-2010-11-321265. [DOI] [PubMed] [Google Scholar]
- Roozendaal R, Mebius RE. Stromal cell-immune cell interactions. Annual review of immunology. 2011;29:23–43. doi: 10.1146/annurev-immunol-031210-101357. [DOI] [PubMed] [Google Scholar]
- Ross CW, Ouillette PD, Saddler CM, Shedden KA, Malek SN. Comprehensive analysis of copy number and allele status identifies multiple chromosome defects underlying follicular lymphoma pathogenesis. Clin Cancer Res. 2007;13:4777–4785. doi: 10.1158/1078-0432.CCR-07-0456. [DOI] [PubMed] [Google Scholar]
- Sadelain M, Brentjens R, Riviere I, Park J. CD19 CAR Therapy for Acute Lymphoblastic Leukemia. Am Soc Clin Oncol Educ Book. 2015:e360–363. doi: 10.14694/EdBook_AM.2015.35.e360. [DOI] [PubMed] [Google Scholar]
- Schatz JH, Oricchio E, Wolfe AL, Jiang M, Linkov I, Maragulia J, Shi W, Zhang Z, Rajasekhar VK, Pagano NC, et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med. 2011;208:1799–1807. doi: 10.1084/jem.20110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg MW, Cheung TC, Ware CF. The signaling networks of the herpesvirus entry mediator (TNFRSF14) in immune regulation. Immunological reviews. 2011;244:169–187. doi: 10.1111/j.1600-065X.2011.01064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendel AC, Calemine-Fenaux J, Izrael-Tomasevic A, Chauhan V, Arnott D, Eaton DL. B and T lymphocyte attenuator regulates B cell receptor signaling by targeting Syk and BLNK. J Immunol. 2009;182:1509–1517. doi: 10.4049/jimmunol.182.3.1509. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Gavrieli M, Sedy JR, Yang J, Fallarino F, Loftin SK, Hurchla MA, Zimmerman N, Sim J, Zang X, et al. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nature immunology. 2003;4:670–679. doi: 10.1038/ni944. [DOI] [PubMed] [Google Scholar]
- Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- Yildiz M, Li H, Bernard D, Amin NA, Ouillette P, Jones S, Saiya-Cork K, Parkin B, Jacobi K, Shedden K, et al. Activating STAT6 mutations in follicular lymphoma. Blood. 2015;125:668–679. doi: 10.1182/blood-2014-06-582650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.