Abstract
PomA is a membrane protein that is one of the essential components of the sodium-driven flagellar motor in Vibrio species. The cytoplasmic charged residues of Escherichia coli MotA, which is a PomA homolog, are believed to be required for the interaction of MotA with the C-terminal region of FliG. It was previously shown that a PomA variant with neutral substitutions in the conserved charged residues (R88A, K89A, E96Q, E97Q, and E99Q; AAQQQ) was functional. In the present study, five other conserved charged residues were replaced with neutral amino acids in the AAQQQ PomA protein. These additional substitutions did not affect the function of PomA. However, strains expressing the AAQQQ PomA variant with either an L131F or a T132M substitution, neither of which affected motor function alone, exhibited a temperature-sensitive (TS) motility phenotype. The double substitutions R88A or E96Q together with L131F were sufficient for the TS phenotype. The motility of the PomA TS mutants immediately ceased upon a temperature shift from 20 to 42°C and was restored to the original level approximately 10 min after the temperature was returned to 20°C. It is believed that PomA forms a channel complex with PomB. The complex formation of TS PomA and PomB did not seem to be affected by temperature. Suppressor mutations of the TS phenotype were mapped in the cytoplasmic boundaries of the transmembrane segments of PomA. We suggest that the cytoplasmic surface of PomA is changed by the amino acid substitutions and that the interaction of this surface with the FliG C-terminal region is temperature sensitive.
Bacteria can swim by rotating helical flagellar filaments like a screw. The molecular motor is embedded in the membrane at the base of the flagellar filament. It can rotate by using the electrochemical potential of specific ions across the cytoplasmic membrane. Bacterial flagella are categorized based on the coupling ion. The flagellar motor of Escherichia coli is a proton-driven motor (30), and the polar flagella of Vibrio species are sodium driven (6, 18, 20, 26).
Based on studies of E. coli and Salmonella enterica serovar Typhimurium, approximately 50 genes are known to be required for flagella assembly and function (9, 29). In the proton-driven flagellar motor of E. coli, MotA, MotB, and FliG are components required for flagellar rotation (11). MotA has four transmembrane segments and a relatively large cytoplasmic domain between the second and third transmembrane segments (54). MotB has a single transmembrane segment and a peptidoglycan-binding motif in the C-terminal domain (15, 16). MotA and MotB form a complex and function as proton channels (12, 42). This complex is proposed to be the stator of the flagellar motor and surrounds the rotor structure (21). Genetic studies demonstrated that FliG forms a switch complex with FliN and FliM (47). FliG binds to both the C-ring proteins (FliM and FliN) and the MS-ring protein (FliF) (37, 44). However, the precise location of FliG is not known (43).
Partial structures of FliG from the thermophile Thermotoga maritima, including the middle and C-terminal domains, were determined at atomic resolution (14, 28). The structures showed that the functionally important charged residues are clustered along the ridges toward the outside of the FliG domain. On the other hand, the crystal structures of the stator components are not yet available. The membrane topologies have only been modeled as shown in Fig. 1. A model of the stator was generated based on the assumption that the force-generating unit is a 1:1 complex of MotA and MotB and the five transmembrane helices comprise an ion channel. The model was revised based on recent results showing the stoichiometry of the force-generating unit as a 4:2 complex of MotA and MotB (11, 13, 24) and other previous results (39, 40, 49).
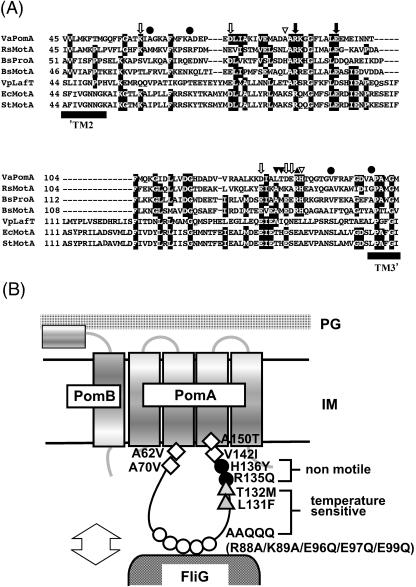
FIG. 1.
(A) Alignments of the PomA cytoplasmic loop with homologous proteins of various species. VaPomA, V. alginolyticus PomA; Rs MotA, R. sphaeroides MotA; BsProA, B. subtilis hypothetical protein A; BsMotA, B. subtilis MotA; VpLafT, V. parahaemolyticus LafT; EcMotA, E. coli MotA; StMotA, S. enterica serovar Typhimurium MotA. The residues are shown by white letters in black boxes when more than four identical residues are present at the aligned position. Black arrows indicate the two conserved charged residues. White arrows indicate the charged residues which were changed to neutral residues in this study. Black circles indicate the suppressor mutations of the TS PomA mutant. The paired black triangles and black or white inverted triangles show the sites of the previously isolated double mutations. (B) Mutations mapped in a schematic diagram of motor. ○, substitutions of AAQQQ (R88A, K89A, E96Q, E97Q, and E99Q); •, nonmotile mutations; ▵, substitutions required for the TS phenotype in addition to the AAQQQ substitutions; □, suppressor mutations of the TS mutant.
In the sodium-driven motor of Vibrio species, such as Vibrio alginolyticus, Vibrio cholerae, and Vibrio parahaemolyticus, PomA,PomB, MotX, and MotY have been identified as essential components of the stator (2, 17, 33, 48). PomA and PomB, which are homologs of E. coli MotA and MotB, are thought to form a complex as a sodium ion channel (39, 40, 51). MotX and MotY, which are localized in the outer membrane (35), have no homologous components in the proton-driven motor (31, 32, 34, 36), and their function is currently unclear.
It is likely that the motors of the proton-driven and sodium-driven flagella work essentially by the same mechanism. MotA of Rhodobacter sphaeroides, whose motor is proton driven (38), can replace PomA and function with PomB, MotX, and MotY in V. alginolyticus as the sodium motor component (1). MomB is a chimeric protein between the N terminus of MotB from R. sphaeroides and the C terminus of PomB. It functions as a Na+-driven motor component with MotX and MotY in V. alginolyticus (3). PotB is a chimeric protein that has the transmembrane domain of the N terminus of PomB and the periplasmic domain of the C terminus of E. coli MotB. Surprisingly, PotB functions with PomA as a sodium motor component not only in V. alginolyticus but also in E. coli (4). MotA and MotB of E. coli can function in Vibrio cells as the proton motor components without MotX and/or MotY. These results indicate that the Vibrio sodium motor is converted into a proton motor and also that the E. coli proton motor can be changed into a sodium motor (4, 17).
Site-directed mutagenesis of the proton-driven motor of E. coli suggested that the charged residues R90 and E98 in the cytoplasmic domain of MotA have electrostatic interactions with five charged residues in the FliG C-terminal domain: K264, R281, D288, D289, and R297. These interactions are thought to be critical for flagellar rotation (27, 53, 55). The two charged residues in MotA are conserved in the cytoplasmic domain of PomA (R88 and E96). Several charged residues (K89, E97, and E99) are located near R88 and E96 of PomA. It was recently reported that a PomA mutant with neutral substitutions at all of these charged residues (R88A, K89A, E96Q, E97Q, E99Q; AAQQQ) is functional (52). The five charged residues of E. coli FliG, K264, R281, D288, D289, and R297, are conserved in V. alginolyticus FliG (K284, R301, D308, D309, and R317, respectively). We have introduced single or multiple substitutions for the charged residues. Most proteins with substitutions at these five conserved positions in FliG still conferred significant motility. All of the triple-substitution proteins constructed in that study (which correspond to those that resulted in the loss of motor function in E. coli) also allowed motility (50). These lines of evidence suggest that other charged residues are required for flagellar rotation in the sodium-driven motor.
In the present study, we first tried to identify other important charged residues of the cytoplasmic domain in PomA. Next, we examined the cumulative effects of the conserved charged residues and other mutations in the cytoplasmic domain. We found that the combination of AAQQQ and L131F or T132M substitutions (AAQQQ+L131F and AAQQQ+T132M PomA mutants) conferred a temperature-sensitive phenotype (TS phenotype) for motility. As far as we know, this is the first study to report TS mutations in any of the mot genes. Our results suggest that the cytoplasmic interface of PomA and the cytoplasmic boundaries of the second and third transmembrane segments of PomA might act synergistically in force generation.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and media.
The strains and plasmids used are listed in Table 1. V. alginolyticus was cultured at 30°C in VC medium (0.5% [wt/vol] polypeptone, 0.5% [wt/vol] yeast extract, 0.4% [wt/vol] K2HPO4, 3% [wt/vol] NaCl, 0.2% [wt/vol] glucose) or VPG500 medium (1% [wt/vol] polypeptone, 0.4% [wt/vol] K2HPO4, 500 mM NaCl, 0.5% [wt/vol] glycerol). E. coli was cultured in Luria-Bertani broth (1% [wt/vol] tryptone peptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl). Kanamycin was added to a final concentration of 100 μg/ml for V. alginolyticus and 50 μg/ml for E. coli. For the swarming assay, overnight cultures were spotted on 0.25% agar-VPG500 plates that contained 100 μg of kanamycin/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| V. alginolyticus strain | ||
| VIO5 | Rifr, Pof+, Laf− | 36 |
| NMB190 | VIO5 ΔpomA (211-bp deletion) | 1 |
| NMB191 | VIO5 ΔpomAB | 51 |
| E. coli strain | ||
| JM109 | 50 | |
| Plasmid | ||
| pSU41 | kan (Kmr), Plac lacZα | 7 |
| pYA301 | pSU41 0.8 kb BamHI-BamHI (pomA+) | 22 |
| pYA303 | pSU41 1.9 kb BamHI-SacI (pomAB+) | 22 |
Kmr, kanamycin resistant; Rifr, rifampicin resistant; Plac, lac promoter.
Site-directed mutagenesis.
We used a PCR method as previously described (22) to introduce the substitutions into PomA. A pair of sense and antisense primers that contained mismatches at the mutation site was synthesized, and the full-length plasmid that contained the pomA gene was amplified by using the primers. The presence of the mutations was confirmed by DNA sequencing.
Measurement of swimming speed.
Overnight cultures in VC medium were inoculated into VPG500 medium at 100-fold dilution and cultivated for 3 h at 30°C. Cells were harvested by centrifugation and suspended in TMN500 buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 5 mM glucose, 500 mM NaCl). The cell suspensions were incubated for 10 min at the temperature setting for measurement. Suspensions were diluted 100-fold into the same buffer containing 20 mM serine, which was used to suppress the directional change of swimming, and incubated at the same temperature. Cell motility was observed with a dark-field microscope on a microwarm plate, which was used to control the temperature, and recorded on videotape. Swimming speed was determined as previously described (5). The average swimming speed was obtained by measuring at least 20 swimming tracks from the two independent experiments.
Detection of PomA proteins in Vibrio cells.
Vibrio cells that harbored vector only or plasmids carrying wild-type or mutant pomA genes were cultured in VPG500 medium at various temperatures. The cells were harvested at an optical density at 660 nm of 1.0 and suspended in distilled water. The same volume of sodium dodecyl sulfate (SDS) loading buffer was added to cell suspensions, which were boiled at 100°C for 5 min. These samples were separated by SDS-polyacrylamide gel (12%) electrophoresis (PAGE), and immunoblotting was performed using anti-PomA antibody (PomA91).
Detection of PomA-PomB interaction.
Vibrio cells that harbored plasmids carrying His-tagged wild-type or mutant pomA genes were cultured in VC medium at 30°C for 3 h. The cells were harvested by centrifugation, washed with V buffer (25 mM Tris-Cl [pH 7.5], 300 mM NaCl, 10 mM MgCl2), and resuspended in 20TMPD (20 mM Tris-Cl [pH 8.0], 5 mM MgSO4, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT]) containing 5 μg of DNase I/ml. The cell suspension was passed through a French press twice at 500 kg/cm2. After unbroken cells were removed by low-speed centrifugation (10,000 × g for 5 min at 4°C), the membrane was recovered by ultracentrifugation (150,000 × g for 1 h at 4°C). The membrane pellet was homogenized with 20TPD (20 mM Tris-Cl [pH 8.0], 0.5 mM PMSF, 1 mM DTT) containing 20% (wt/vol) glycerol and stored at −80°C. Total cell membrane (10 mg/ml of protein) was solubilized in 20TNPD (20 mM Tris-Cl [pH 8.0], 0.15 M NaCl, 0.5 mM PMSF, 1 mM DTT) containing 10% (wt/vol) glycerol, 5 mM imidazole, and 2.5% (wt/vol) CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} and incubated for 10 min on ice. Solubilized samples were centrifuged at 150,000 × g for 30 min at 4°C. Ni-nitrilotriacetic acid (Ni-NTA) agarose was added to the solubilized fraction, and the sample solution was incubated for 1 h at 4°C. The supernatant was removed by centrifugation, and the Ni-NTA pellet was washed three times with 20TNPD containing 10% (wt/vol) glycerol, 5 mM imidazole, and 1% (wt/vol) CHAPS at 20°C or 42°C. PomB and His-tagged PomA proteins were eluted by 20TNPD that contained 10% (wt/vol) glycerol, 200 mM imidazole, and 1% (wt/vol) CHAPS at 20°C or 42°C. The same volume of SDS loading buffer was added to the eluted solution, and the samples were boiled at 100°C for 5 min. These samples were separated by SDS-PAGE as described above. PomA and PomB proteins were detected by immunoblotting with the anti-PomA (PomA91) and anti-PomB (PomB93) antibodies.
Isolation of motile suppressors from the TS mutant.
Single colonies of the TS PomA mutant were inoculated on VPG500 swarm plates by toothpick and incubated at 37°C for 2 days. The revertant cells, which recovered swarming ability, were picked from the edge of the swarm ring, and single colonies were isolated. After the swarming ability of the single colonies was checked, plasmids were extracted from the revertant cells. The plasmids were reintroduced into NMB190, the pomA mutant, to confirm the mutations.
RESULTS
Charged residues of V. alginolyticus PomA.
Based on an alignment with E. coli MotA, two charged residues (R90 and E98 of MotA) are well-conserved not only in V. alginolyticus PomA but also in other bacterial species (Fig. 1). In the cytoplasmic domain between the second and third transmembrane segments in V. alginolyticus PomA, there are 14 positively charged residues and 17 negatively charged residues. Some of these residues were conserved in E. coli MotA, as well as in proteins from other species (Fig. 1A). To identify the charged residues that are required for flagellar rotation in the cytoplasmic domain of PomA, K60, D75, D128, D133, or E134 were individually substituted with neutral amino acid residues as the sixth mutation (K60A, D75N, D128N, D133N, and E134Q) in the AAQQQ PomA protein. These PomA variants were expressed from plasmids in ΔpomA strain NMB190. Based on the swarming assay, motility was not affected by the additional substitutions tested in this study. The swimming speeds of these mutants were as fast as those of the wild type or the AAQQQ mutant (data not shown).
Mutations in the cytoplasmic domain of PomA.
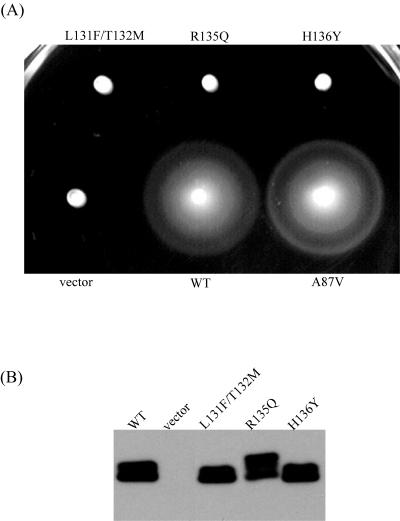
We could not find functionally critical residues for flagellar rotation in PomA in a homology search with E. coli MotA or other homologous proteins. To get clues for studying the cytoplasmic domain of PomA, we explored the mutations in this region. It was previously reported that double substitutions in the cytoplasmic region, A87V and H136Y, L131F and T132M, and R135Q and T169I (transmembrane segment 3), abolished the motility of V. alginolyticus (25). Each substitution was introduced individually into wild-type PomA to determine which substitution(s) affected flagellar rotation. Vibrio cells expressing each of the mutant proteins, PomA with an A87V substitution (PomA-A87V), PomA-L131F, or PomA-T132M, swarmed as well as cells with wild-type PomA (Fig. 2A and 3A). On the other hand, Vibrio cells expressing PomA-R135Q or PomA-H136Y did not swarm at all (Fig. 2A). In addition, we confirmed under the microscope that strains carrying these two PomA variants were unable to swim (data not shown). These PomA variants were detected at levels similar to that of wild-type PomA, and the PomA bands were doublets as previously published (52) (Fig. 2B). Furthermore, PomA-H136A or PomA-H136D conferred the ability to swim, but PomA-R135A, PomA-R135K, PomA-R135D, and PomA-R135E did not (data not shown). These results suggest that the charged residue R135, which is not conserved in E. coli, is critical for the motor rotation of V. alginolyticus (Fig. 1).
FIG. 2.
Nonfunctional PomA variants. (A) Swarms of NMB190 cells that contain vector, wild-type PomA (WT), or single or double amino acid-substituted PomA. The cells were grown at 30°C for 5.5 h. (B) Immunoblot of PomA proteins in whole-cell extracts. NMB190 cells harboring the vector only, plasmid containing wild-type PomA, or mutant PomA were harvested and suspended in distilled water. This suspension was subjected to SDS-PAGE and detected with anti-PomA antibody.
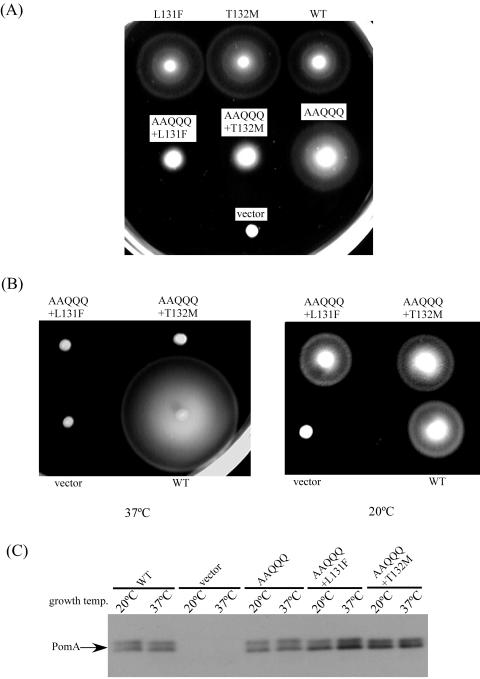
FIG. 3.
Temperature-sensitive mutants of the motor protein. (A) Swarms of NMB190 cells expressing wild-type (WT) PomA or PomA-L131F, PomA-T132M, AAQQQ PomA, AAQQQ+L131F PomA, or AAQQQ+T132M PomA. The cells were grown at 30°C for 5.5 h. (B) Swarms of NMB190 cells expressing AAQQQ+L131F and AAQQQ+T132M PomA. The cells were grown at 37°C for 5.5 h or at 20°C for 10 h. (C) Immunoblot of PomA proteins in whole-cell extracts. The cells were grown to an optical density at 660 nm of 1.0 at 20°C or 37°C in VPG500 medium. NMB190 cells harboring vector only or the plasmid containing wild-type PomA or cells containing mutant PomA were harvested and suspended in distilled water. This suspension was subjected to SDS-PAGE, and PomA was detected with anti-PomA antibody.
Mutations in the motor protein causing temperature sensitivity.
As mentioned above, cells with L131F and T132M substitutions in PomA have swarming abilities similar to those of cells with wild-type PomA. When we introduced these substitutions into AAQQQ PomA as the sixth substitution (AAQQQ+L131F and AAQQQ+T132M), the additional change significantly reduced swarming ability at 30°C (Fig. 3A). The swarming abilities of strains carrying the AAQQQ+L131F and AAQQQ+T132M PomA proteins were examined at various temperatures. We detected a clear difference in swarming abilities at 20°C and 37°C. The mutant proteins did not allow swarming at 37°C, but the strain with the wild-type PomA swarmed well at this temperature. At 20°C, the strains with mutant and wild-type PomA proteins demonstrated similar swarming abilities (Fig. 3B). The swarming abilities of strains with a single PomA substitution (L131F or T132M) were similar to those of strains with wild-type PomA at 37°C (data not shown). When cells were grown at 20°C and 37°C, there was almost no difference in the amount of mutant or wild-type PomA protein (Fig. 3C).
Next, we measured the swimming speeds at various temperatures to investigate the motor function of AAQQQ+L131F PomA. The swimming speed of the wild-type strain was 87 μm/s at 42°C (Table 2). AAQQQ+L131F PomA did not confer swimming at this temperature. At 30°C, cells with AAQQQ+L131F PomA could swim; however, the swimming speed (34 μm/s) was slower than that of the wild type (62 μm/s). At 20°C, the wild type and the mutant carrying AAQQQ+L131F PomA swam at similar speeds (Table 2). These results indicate that the motor function of this form of PomA is comparable to that of wild-type PomA at 20°C.
TABLE 2.
Swimming speeds of cells expressing TS PomA
| Temp (°C) | Swimming speed (μm/s) ± SD of NMB190 expressing:
|
|
|---|---|---|
| WT PomA | AAQQQ + L131F PomA | |
| 42 | 87 ± 17 | NAa |
| 30 | 62 ± 10 | 34 ± 9 |
| 20 | 46 ± 9 | 42 ± 9 |
NA, not applicable because motile cells were not observed.
When PomB was coexpressed with AAQQQ+L131F PomA from the same plasmid in the ΔpomAB strain (NMB191), the fraction of swimming cells was higher than that in the strain expressing AAQQQ+L131F PomA alone at various temperatures (Fig. 4A). In the strain that expressed TS PomA alone, 20% of the cells could swim at 30°C. On the other hand, at 30°C the fraction of swimming cells was similar to that of the wild-type in the strain coexpressing PomB and TS PomA. The TS phenotype was alleviated by coexpression of PomB; however, cells were completely nonmotile at 42°C. This result shows that the TS phenotype is alleviated by an increase in the amount of PomB.
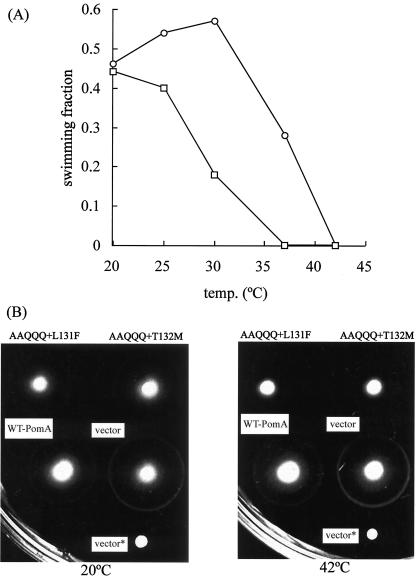
FIG. 4.
(A) Swimming ability of cells expressing TS PomA. The fractions of swimming cells expressing AAQQQ+L131F PomA alone in NMB190 (squares) or with PomB in NMB191 (circles) from the plasmid are shown. (B) Effect on swarm formation of the TS PomA mutation in VIO5 cells. Shown are swarms of VIO5 cells that harbor the vector only, a plasmid encoding wild-type PomA, and plasmids encoding AAQQQ+L131F or AAQQQ+T132M PomA. The cells were grown at 20°C for 10 h or 42°C for 2.5 h. vector*, NMB190 cells harboring vector (nonmotile control).
We investigated the dominant effect of TS PomA against wild-type PomA. Strain VIO5 has wild-type polar flagella. VIO5 cells expressing the TS PomA protein swarmed as well as cells expressing wild-type PomA at 20°C. On the other hand, at the restrictive temperature (42°C), VIO5 cells expressing TS PomA had reduced swarming ability (Fig. 4B).
Recovery of swimming ability of TS PomA mutant cells.
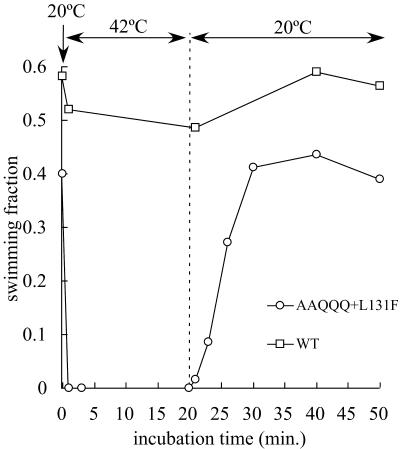
To investigate whether or not the TS phenotype was reversible, the recovery of swimming was measured at low temperature after treatment at high temperature. In these experiments, chloramphenicol (25 μg/ml) was added to the buffer to prevent de novo synthesis of proteins. This concentration corresponds to 20 times the MIC of chloramphenicol for V. alginolyticus (data not shown). Approximately the same fractions of cells expressing wild-type PomA were capable of swimming at 20 and 42°C. On the other hand, the swimming fraction of cells expressing AAQQQ+L131F PomA immediately dropped to zero when the temperature was increased from 20 to 42°C. When the temperature was returned to 20°C, the fraction of swimming cells gradually increased and was restored to its original value in approximately 10 min (Fig. 5).
FIG. 5.
Recovery of swimming ability of the TS PomA mutants. The fractions of swimming NMB190 cells expressing AAQQQ+L131F PomA (circles) and wild-type PomA (squares) are shown. The cell suspension was prepared in a manner similar to that for the measurement of swimming speed, except that it contained 25 μg of chloramphenicol/ml in TMN500 buffer. This cell suspension was incubated at 20°C for 10 min, then placed at 42°C, and returned to 20°C. The number of swimming cells was counted at the incubation temperature on a microwarm plate. The value was the average of results from two independent experiments.
Interaction of TS PomA with PomB.
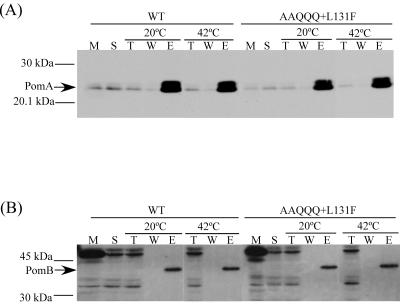
It has been shown that PomA interacts with PomB to form a complex (39, 40, 46, 49, 51). We speculated that the mutations causing temperature sensitivity might affect the PomA-PomB interaction at high temperatures. A His tag was fused to the N terminus of TS PomA, and the fusion protein was expressed in NMB190. This His-tagged TS PomA protein conferred the same TS phenotype as the original TS PomA. After the membrane fraction was isolated and solubilized with CHAPS, which can solubilize the membrane with minimal dissociation of the PomA/PomB complex (T. Atsumi, personal communication), the His-tagged TS PomA proteins were bound to Ni-NTA agarose. After elution, the His-tagged TS PomA and PomB were detected by immunoblotting. We detected similar amounts of TS PomA and wild-type PomA at each elution temperature. In addition, no differences were detected in the amount of coeluted PomB with TS PomA or wild-type PomA (Fig. 6). Based on this result, it appears that temperature does not affect the interaction of TS PomA with PomB.
FIG. 6.
Interaction of TS PomA with PomB. The His-tagged PomA/PomB complex was isolated as described in Materials and Methods. PomA and PomB proteins were detected with anti-PomA (A) and anti-PomB (B) antibodies. M, total membrane; S, solubilized fraction; T, flowthrough; W, washed fraction; E, eluted fraction.
Suppressors of the TS PomA mutation.
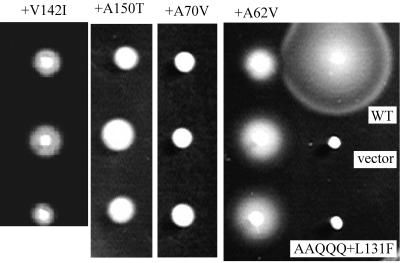
Motile suppressors of TS PomA were obtained by long incubation (1 to 2 days) on swarm plates at 37°C. Eleven motile suppressors were obtained, including five revertants and six pseudorevertants. In all five revertants, the L131F substitution was returned to leucine. Four of the five revertants had the original codon (CTT), and in one revertant, TTT was changed to TTG. In the six pseudorevertants, the following additional substitutions were found in four positions: A62V (two isolates), A70V (single isolate), V142I (two isolates), and A150T (single isolate) (Fig. 7). The newly substituted residues were larger than the original residues and were clustered around the boundaries of the cytoplasmic domain and the second or third transmembrane segment (Fig. 1).
FIG. 7.
Motile suppressors of the TS PomA mutants. Shown are swarms of NMB190 cells expressing no PomA (vector), wild-type PomA (WT), AAQQQ+L131F PomA, or the suppressor mutants (TS PomA-V142I, TS PomA-A150T, TS PomA-A70V, or TS PomA-A62V). The plasmids were extracted from the revertant cells and reintroduced into the pomA mutant (NMB190), and single colonies were inoculated into a semisolid agar plate. The cells were grown at 37°C for 2.5 h.
Minimum requirements for amino acid substitutions conferring the temperature-sensitive phenotype.
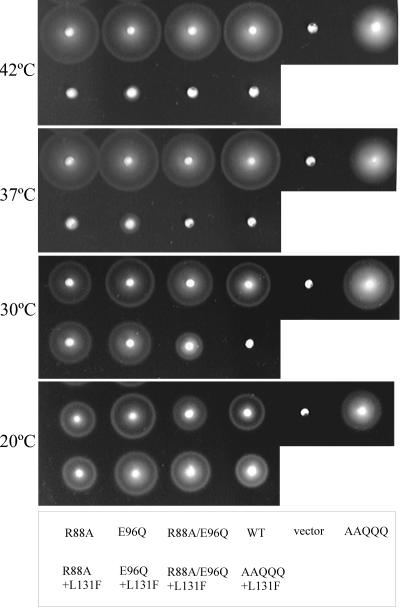
To identify which of the five substitutions in AAQQQ PomA affects the TS phenotype, three PomA variants, PomA-R88A+L131F, PomA-E96Q+L131F, and PomA-R88A/E96Q+L131F (PomA with the triple substitution of R88A and E96Q [the conserved charged residues] with L131F), were constructed (Fig. 8). PomA-R88A+L131F and PomA-E96Q+L131F conferred motilities at 20°C and 30°C similar to that of wild-type PomA. However, the swarming ability of cells expressing these two PomA variants was remarkably reduced at temperatures over 37°C. PomA-R88A/E96Q+L131F and wild-type PomA conferred similar motilities at 20°C, but the swarming ability of the mutant cells was reduced at 30°C. AAQQQ+L131F PomA and PomA-R88A/E96Q+L131F did not confer the ability to swarm at temperatures over 37°C. When each of the AAQQQ substitutions in TS PomA (AAQQQ+L131F PomA) was returned to the original charged residue, only those proteins with R88 or E96 resulted in a strongly reduced TS phenotype, but the other substitutions had no effect (data not shown). These results may suggest that R88A and E96Q are important mainly for the TS phenotype with L131F and that the other substitutions may also have a slight effect on the TS phenotype. Without the L131F substitution, the R88A and/or E96Q substitutions did not affect swarming ability at any temperature. It is worth noting that the swarming ability of the strain expressing the AAQQQ PomA mutant was slightly reduced at 37 and 42°C.
FIG. 8.
Minimum amino acid substitutions required to confer the TS phenotype. NMB190 cells expressing no PomA (vector), wild-type PomA (WT), and the PomA mutants (PomA-R88A, PomA-E96Q, PomA-R88A/E96Q, AAQQQ PomA, PomA-R88A+L131F, PomA-E96Q+L131F, PomA-R88A/E96Q+L131F, or AAQQQ+L131F PomA) were grown at 20°C (for 10 h), 30°C (for 5 h), 37°C (for 3.5 h), and 42°C (for 3.5 h).
DISCUSSION
In bacterial flagellar movement, the stator and rotor cooperate to generate torque from the ion influx, which results in motor rotation. In the proton-driven flagellar motor of E. coli, the charged residues R90 and E98 in the cytoplasmic domain of the stator protein MotA are critical for flagellar rotation. However, these conserved charged residues in PomA are apparently not essential for flagellar rotation in Vibrio (52). In the present study, the secondary conserved charged residues (Fig. 1A) were replaced with neutral residues in the AAQQQ PomA mutant to identify additional charged residues in the cytoplasmic domain of PomA that may be required for motor rotation. However, all of the sextet mutants were functional. The substitution of D128 in PomA, which corresponds to E150 of MotA (a moderately critical residue in E. coli) (11, 53), did not affect function. This result may suggest that the conserved charged residues in PomA are a subset of residues essential for sodium-driven rotation. Additional residues other than the conserved or the secondary conserved charged residues may be involved in electrostatic interactions with FliG. Alternatively, common residues required for specific electrostatic interactions may not be present in the stator components of the H+-driven and Na+-driven motors. The R135 residue may be a candidate for an important charged residue required for the rotation of the Na+-driven motor of V. alginolyticus. This Arg residue is not conserved in E. coli and S. enterica serovar Ttyphimurium; however, it is conserved in Vibrio species, R. sphaeroides, and Bacillus subtilis (Fig. 1).
We herein reported that the AAQQQ+L131F and AAQQQ+T132M PomA proteins exhibited a temperature-sensitive phenotype for motor function (Fig. 3). The strain expressing the AAQQQ+L131F PomA protein was nonmotile at temperatures over 37°C, and the swimming ability of this strain was restored at 20°C even though protein synthesis was inhibited (Fig. 5). Thus, the de novo synthesis of PomA is not required for restoration of motility. The same levels of PomB were associated with TS PomA and wild-type PomA under both permissive and restrictive temperatures (Fig. 6). This result suggests that PomB and TS PomA can interact with each other to maintain the proper complexes even at restrictive temperatures. The TS phenotype caused by the AAQQQ+L131F mutations was suppressed by mutations that were clustered at the border of the cytoplasmic domain and in the transmembrane segment. These results suggest that the cytoplasmic boundaries of the second and third transmembrane segments are critical for the function of PomA to sustain the structure of the cytoplasmic domain. The cytoplasmic boundaries of PomA and its cytoplasmic interface to FliG might synergistically act in force generation. Taking this evidence together, we speculated that PomA mutations in the cytoplasmic domain might affect the structure of PomA or the assembly of the stator complex to the rotor and could result in the prevention of interactions between PomA and FliG to cooperatively generate torque. At this time, however, we do not have evidence to indicate whether or not the regions around L131F or AAQQQ interact directly with FliG in wild-type PomA.
Strains carrying PomA variants with the double substitutions R88A or E96Q and L131F exhibited a TS phenotype above 37°C. Furthermore, the triple substitution of R88A and E96Q (the conserved charged residues) with L131F appeared to be sufficient for conferring the TS phenotype (Fig. 8). These results may suggest that the region near these two conserved charged residues has an important role in flagellar rotation. The other substitutions seem to affect the TS phenotype slightly. This is not surprising because it is easy to imagine that the region around the conserved residues is affected by other nearby mutations.
The TS phenotype was alleviated by coexpressing PomB from the plasmid (Fig. 4). This effect may be explained by an increase in the number of TS PomA/PomB complexes. Probably both functional and nonfunctional TS PomA/PomB complexes exist in the cell at 30°C. If the total number of PomA/PomB complexes is increased by the overproduction of both PomA and PomB from the plasmid, the number of the functional complexes should be increased. Thus, the possibility of the assembly of a functional TS PomA/PomB complex with the rotor is higher when PomB is provided from a plasmid than when it is provided from the chromosome.
As far as we know, other TS mot mutants have not been isolated; however, temperature-sensitive Mot− fliG and Mot− fliN mutants were reported for S. enterica serovar Typhimurium (45). The mutants are capable of assembling a complete flagellum but are nonmotile at restrictive temperatures. Mutations in the switch protein genes fliG, fliM, and fliN gave three phenotypes: Mot−, Che−, and Fla−. The amino acid substitutions of these TS FliG mutants were mapped to F236L, D244Y, and L259R in the C-terminal domain of S. enterica serovar Typhimurium FliG (19). The other Mot mutants that mapped to the fliG gene are clustered in this region. The crystal structure of the middle and C-terminal regions of T. maritima FliG has been reported (14). The middle and C-terminal regions of the T. maritima FliG molecule consist of two compact globular domains linked by an α-helix and an extended segment that contains a well-conserved Gly-Gly motif. The C-terminal region has a ridge containing the charged residues that are believed to interact with the charged residues of the MotA cytoplasmic domain (11, 14, 28). The Mot− region is located in the middle of the C-terminal region that is between the ridge surface region and the well-conserved Gly-Gly motif. Mutations in the Mot− region may affect the ridge region that is the interface to the cytoplasmic domain of MotA.
The interactions between the cytoplasmic region of PomA or MotA and the C-terminal region of FliG are thought to be essential for force generation. In PomA, the region from L131 to H136 may contribute to torque generation together with the region from R88 to E99, which is the conserved charged region. In the proton-driven motor, it seems that the protonation and deprotonation of an Asp residue in the MotB transmembrane segment induces a conformational change of the MotA cytoplasmic region to generate the rotational force (23). The force-generating unit was inferred to be a 4:2 complex of PomA (MotA) and PomB (MotB) (11, 13, 24, 39, 40, 49). Recent data suggest that the motor complex may have a larger structure than previously assumed or that the minimal force-generating unit is not a 4:2 complex (46). Presently, however, we do not know the mechanism of generation of rotational force, although many models have been presented (8, 10, 41). We anticipate that our present data will assist in the understanding of the mechanism once high-resolution data on the stator complex are available.
Acknowledgments
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan; the Japan Science and Technology Corporation (to M.H. and T.Y.); and the Soft Nano-Machine Project of Japan Science and Technology Agency (to T.Y. and M.H.).
REFERENCES
- 1.Asai, Y., I. Kawagishi, E. Sockett, and M. Homma. 1999. Hybrid motor with the H+-and Na+-driven components can rotate Vibrio polar flagella by using sodium ions. J. Bacteriol. 181:6322-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai, Y., S. Kojima, H. Kato, N. Nishioka, I. Kawagishi, and M. Homma. 1997. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J. Bacteriol. 179:5104-5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai, Y., R. E. Sockett, I. Kawagishi, and M. Homma. 2000. Coupling ion specificity of chimeras between H+- and Na+-driven motor proteins, MotB and PomB, in Vibrio polar flagella. EMBO J. 19:3639-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asai, Y., T. Yakushi, I. Kawagishi, and M. Homma. 2003. Ion-coupling determinants of Na+-driven and H+-driven flagellar motors. J. Mol. Biol. 327:453-463. [DOI] [PubMed] [Google Scholar]
- 5.Atsumi, T., Y. Maekawa, T. Yamada, I. Kawagishi, Y. Imae, and M. Homma. 1996. Effect of viscosity on swimming by the lateral and polar flagella of Vibrio alginolyticus. J. Bacteriol. 178:5024-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atsumi, T., L. McCarter, and Y. Imae. 1992. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355:182-184. [DOI] [PubMed] [Google Scholar]
- 7.Bartolome, B., Y. Jubete, E. Martínez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 8.Berg, H. C. 2000. Constraints on models for the flagellar rotary motor. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg, H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19-54. [DOI] [PubMed] [Google Scholar]
- 10.Berry, R. M., and J. P. Armitage. 1999. The bacterial flagella motor. Adv. Microb. Physiol. 41:291-337. [DOI] [PubMed] [Google Scholar]
- 11.Blair, D. F. 2003. Flagellar movement driven by proton translocation. FEBS Lett. 545:86-95. [DOI] [PubMed] [Google Scholar]
- 12.Blair, D. F., and H. C. Berg. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439-449. [DOI] [PubMed] [Google Scholar]
- 13.Braun, T. F., L. Q. Al-Mawsawi, S. Kojima, and D. F. Blair. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 43:35-45. [DOI] [PubMed] [Google Scholar]
- 14.Brown, P. N., C. P. Hill, and D. F. Blair. 2002. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21:3225-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chun, S. Y., and J. S. Parkinson. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239:276-278. [DOI] [PubMed] [Google Scholar]
- 16.De Mot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 17.Gosink, K. K., and C. C. Häse. 2000. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J. Bacteriol. 182:4234-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Häse, C. C., and J. J. Mekalanos. 1999. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 96:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irikura, V. M., M. Kihara, S. Yamaguchi, H. Sockett, and R. M. Macnab. 1993. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawagishi, I., Y. Maekawa, T. Atsumi, M. Homma, and Y. Imae. 1995. Isolation of the polar and lateral flagellum-defective mutants in Vibrio alginolyticus and identification of their flagellar driving energy sources. J. Bacteriol. 177:5158-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan, S., M. Dapice, and T. S. Reese. 1988. Effects of mot gene expression on the structure of the flagellar motor. J. Mol. Biol. 202:575-584. [DOI] [PubMed] [Google Scholar]
- 22.Kojima, S., Y. Asai, T. Atsumi, I. Kawagishi, and M. Homma. 1999. Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by mutations of putative channel components. J. Mol. Biol. 285:1537-1547. [DOI] [PubMed] [Google Scholar]
- 23.Kojima, S., and D. F. Blair. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041-13050. [DOI] [PubMed] [Google Scholar]
- 24.Kojima, S., and D. F. Blair. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26-34. [DOI] [PubMed] [Google Scholar]
- 25.Kojima, S., M. Kuroda, I. Kawagishi, and M. Homma. 1999. Random mutagenesis of the pomA gene encoding the putative channel component of the Na+-driven polar flagellar motor of Vibrio alginolyticus. Microbiology 145:1759-1767. [DOI] [PubMed] [Google Scholar]
- 26.Kojima, S., K. Yamamoto, I. Kawagishi, and M. Homma. 1999. The polar flagella motor of Vibrio cholerae is driven by an Na+ motive force. J. Bacteriol. 181:1927-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lloyd, S. A., and D. F. Blair. 1997. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J. Mol. Biol. 266:733-744. [DOI] [PubMed] [Google Scholar]
- 28.Lloyd, S. A., F. G. Whitby, D. F. Blair, and C. P. Hill. 1999. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 400:472-475. [DOI] [PubMed] [Google Scholar]
- 29.Macnab, R. M. 1999. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J. Bacteriol. 181:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manson, M. D., P. Tedesco, H. C. Berg, F. M. Harold, and C. van der Drift. 1977. A proton motive force drives bacterial flagella. Proc. Natl. Acad. Sci. USA 74:3060-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarter, L. L. 1994. MotX, the channel component of the sodium-type flagellar motor. J. Bacteriol. 176:5988-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarter, L. L. 1994. MotY, a component of the sodium-type flagellar motor. J. Bacteriol. 176:4219-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okabe, M., T. Yakushi, Y. Asai, and M. Homma. 2001. Cloning and characterization of motX, a Vibrio alginolyticus sodium-driven flagellar motor gene. J. Biochem. 130:879-884. [DOI] [PubMed] [Google Scholar]
- 35.Okabe, M., T. Yakushi, M. Kojima, and M. Homma. 2002. MotX and MotY, specific components of the sodium-driven flagellar motor, colocalize to the outer membrane in Vibrio alginolyticus. Mol. Microbiol. 46:125-134. [DOI] [PubMed] [Google Scholar]
- 36.Okunishi, I., I. Kawagishi, and M. Homma. 1996. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 178:2409-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oosawa, K., T. Ueno, and S.-I. Aizawa. 1994. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J. Bacteriol. 176:3683-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Packer, H. L., D. M. Harrison, R. M. Dixon, and J. P. Armitage. 1994. The effect of pH on the growth and motility of Rhodobacter sphaeroides WS8 and the nature of the driving force of the flagellar motor. Biochim. Biophys. Acta 1188:101-107. [DOI] [PubMed] [Google Scholar]
- 39.Sato, K., and M. Homma. 2000. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:5718-5722. [DOI] [PubMed] [Google Scholar]
- 40.Sato, K., and M. Homma. 2000. Multimeric structure of PomA, the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:20223-20228. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt, R. 2003. Helix rotation model of the flagellar rotary motor. Biophys. J. 85:843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stolz, B., and H. C. Berg. 1991. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol. 173:7033-7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki, H., K. Yonekura, and K. Namba. 2004. Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J. Mol. Biol. 337:105-113. [DOI] [PubMed] [Google Scholar]
- 44.Tang, H., T. F. Braun, and D. F. Blair. 1996. Motility protein complexes in the bacterial flagellar motor. J. Mol. Biol. 261:209-221. [DOI] [PubMed] [Google Scholar]
- 45.Vogler, A. P., M. Homma, V. M. Irikura, and R. M. Macnab. 1991. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J. Bacteriol. 173:3564-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yakushi, T., M. Kojima, and M. Homma. 2004. Isolation of Vibrio Na+-driven flagellar motor complex composed of PomA and PomB solubilized by sucrose monocaprate. Microbiology 150:911-920. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi, S., H. Fujita, A. Ishihara, S.-I. Aizawa, and R. M. Macnab. 1986. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J. Bacteriol. 166:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yorimitsu, T., and M. Homma. 2001. Na+-driven flagellar motor of Vibrio. Biochim. Biophys. Acta 1505:82-93. [DOI] [PubMed] [Google Scholar]
- 49.Yorimitsu, T., M. Kojima, T. Yakushi, and M. Homma. 2004. Multimeric structure of the PomA/PomB channel complex in the Na+-driven flagellar motor of Vibrio alginolyticus. J. Biochem. 135:43-51. [DOI] [PubMed] [Google Scholar]
- 50.Yorimitsu, T., A. Mimaki, T. Yakushi, and M. Homma. 2003. The conserved charged residues of the C-terminal region of FliG, a rotor component of Na+-driven flagellar motor. J. Mol. Biol. 334:567-583. [DOI] [PubMed] [Google Scholar]
- 51.Yorimitsu, T., K. Sato, Y. Asai, I. Kawagishi, and M. Homma. 1999. Functional interaction between PomA and PomB, the Na+-driven flagellar motor components of Vibrio alginolyticus. J. Bacteriol. 181:5103-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yorimitsu, T., Y. Sowa, A. Ishijima, T. Yakushi, and M. Homma. 2002. The systematic substitutions around the conserved charged residues of the cytoplasmic loop of Na+-driven flagellar motor component PomA. J. Mol. Biol. 320:403-413. [DOI] [PubMed] [Google Scholar]
- 53.Zhou, J. D., and D. F. Blair. 1997. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol. 273:428-439. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, J. D., R. T. Fazzio, and D. F. Blair. 1995. Membrane topology of the MotA protein of Escherichia coli. J. Mol. Biol. 251:237-242. [DOI] [PubMed] [Google Scholar]
- 55.Zhou, J. D., S. A. Lloyd, and D. F. Blair. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 95:6436-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]