Abstract
Objectives
The radiology report is the most important source of clinical imaging information. It documents critical information about the patient’s health and the radiologist’s interpretation of medical findings. It also communicates information to the referring physicians and records that information for future clinical and research use. Although efforts to structure some radiology report information through predefined templates are beginning to bear fruit, a large portion of radiology report information is entered in free text. The free text format is a major obstacle for rapid extraction and subsequent use of information by clinicians, researchers, and healthcare information systems. This difficulty is due to the ambiguity and subtlety of natural language, complexity of described images, and variations among different radiologists and healthcare organizations. As a result, radiology reports are used only once by the clinician who ordered the study and rarely are used again for research and data mining. In this work, machine learning techniques and a large multi-institutional radiology report repository are used to extract the semantics of the radiology report and overcome the barriers to the re-use of radiology report information in clinical research and other healthcare applications.
Material and methods
We describe a machine learning system to annotate radiology reports and extract report contents according to an information model. This information model covers the majority of clinically significant contents in radiology reports and is applicable to a wide variety of radiology study types. Our automated approach uses discriminative sequence classifiers for named-entity recognition to extract and organize clinically significant terms and phrases consistent with the information model. We evaluated our information extraction system on 150 radiology reports from three major healthcare organizations and compared its results to a commonly used non-machine learning information extraction method. We also evaluated the generalizability of our approach across different organizations by training and testing our system on data from different organizations.
Results
Our results show the efficacy of our machine learning approach in extracting the information model’s elements (10-fold cross-validation average performance: precision: 87%, recall: 84%, F1 score: 85%) and its superiority and generalizability compared to the common non-machine learning approach (p-value < 0.05).
Conclusions
Our machine learning information extraction approach provides an effective automatic method to annotate and extract clinically significant information from a large collection of free text radiology reports. This information extraction system can help clinicians better understand the radiology reports and prioritize their review process. In addition, the extracted information can be used by researchers to link radiology reports to information from other data sources such as electronic health records and the patient’s genome. Extracted information also can facilitate disease surveillance, real-time clinical decision support for the radiologist, and content-based image retrieval.
Keywords: Natural language processing, Information extraction, Discriminative sequence classifier, Radiology report narrative
1. Introduction
Radiology report narrative encompasses critical information about many body parts and health conditions and is a major component of the evidence for clinical diagnosis and disease treatment. In addition, radiology reports provide a rich source of information for disease surveillance, information retrieval, and clinical decision support. However, the free text format of radiology reports and the complexity of natural language make it difficult to extract and re-use report information for clinical care and biomedical research.
Despite this complexity, radiology report narrative mostly follows a common information model consisting of specific semantic elements, such as uncertainty, anatomy, observations, and their modifiers [1]. These common elements capture the essence of report semantics and summarize report information content. Using this information model as a framework for information extraction provides structured details for clinical and research applications and could be generalizable to a wide variety of radiology studies and healthcare organizations. However, identifying and extracting these information model elements is a challenging task due to the ambiguity and subtlety of natural language, the complexity of the described images, and the stylistic variations among radiologists and healthcare organizations.
In this paper, we first present an imaging report information model from an earlier radiology reporting system [1] that defines and summarizes the information content of a radiology report. This information model covers the majority of clinically significant information in radiology reports and is applicable to a wide variety of diagnostic radiology study types. Then we propose an automatic natural language processing (NLP) system to extract clinically significant concepts from the radiology report according to this information model. This system uses a named-entity recognition sequence classifier to identify the information model elements and extract them from the reports. Our approach is applied and evaluated on de-identified radiology reports from three major healthcare organizations: Mayo Clinic (Mayo), MD Anderson Cancer Center (MDA), and Medical College of Wisconsin (MCW).
The main contribution of our work is using existing machine learning techniques to build an information extraction system that can accurately identify significant terms and phrases in radiology reports according to a radiology-specific information model. Our information extraction system yields structured data from the radiology report to link with other clinical and genomic data sources for translational research, information retrieval, disease surveillance, and clinical decision support. The structured data extracted from the radiology report can also improve search of imaging reports for healthcare monitoring and help clinicians and researchers review and understand the reports.
2. Related work
Radiology reports previously have been analyzed using NLP techniques to extract clinically important findings and recommendations [2–4]. The Lexicon Mediated Entropy Reduction (LEXIMER) system extracts and classifies phrases with important findings and recommendations from radiology reports through lexicon-based hierarchical decision trees [3]. In another approach [4], sentences in radiology reports that include clinically important recommendation information were identified though a maximum entropy classifier. Both of these systems provide binary classification (containing or not containing important findings or recommendations) rather than extracting most key clinical concepts using a detailed information model. Our approach enables the re-use of the extracted information for numerous clinical and research purposes, rather than just for the two purposes for which LEXIMER was tailored.
More general NLP techniques previously have been used to classify and extract information from radiology report narrative [5–17]. In earlier work, Medical Language Extraction and Encoding System (MEDLEE) extracted information from Columbia-Presbyterian Medical Center’s chest radiology report repository [5]. MEDLEE uses a controlled vocabulary and grammatical rules to translate text to a structured database format. MEDLEE’s results were evaluated for 24 clinical conditions based on 150 manually labeled radiology reports [6]. However, in separate studies the authors reported decreases in MEDLEE performance when it was applied to multiple organizations’ chest radiology reports [7] and when it was applied to more complex narrative reports from CT and MR head images [8].
In other related work, the Radiology Analysis tool (RADA) was developed to extract key medical concepts and their attributes from radiology reports and to convert them to a structured database format through a specialized glossary of domain concepts, attributes, and predefined grammar rules [9]. Mayo Clinic’s Clinical Text Analysis and Knowledge Extraction System (cTAKES) provides a dictionary-based named-entity recognizer to highlight the Unified Medical Language System (UMLS) Metathesaurus terms in text, in addition to other NLP functionalities, such as tokenizing, part of speech tagging, and parsing [10]. As two other widely used UMLS dictionary-based approaches, Health Information Text Extraction (HITEx) from Brigham and Women’s Hospital and Harvard Medical School finds UMLS matches to tag principal diagnoses [11] and MetaMap from National Library of Medicine finds UMLS concepts in biomedical literature [12]. A drawback of MEDLEE and other dictionary-based and rule-based annotation and information extraction systems is their limited coverage and generalizability [13]. Building an exhaustive list of terms and rules to model language and extract domain concepts is extremely time consuming. As a result, these dictionary-based and rule-based methods usually suffer from lower recall compared to their precision. In addition, even in the presence of extensive dictionaries and rule bases, the results may be still suboptimal due to the interactions between rules and natural language variations and ambiguity [13].
In related statistical NLP work [14], a statistical dependency parser is combined with controlled vocabulary to capture the relationships between concepts and formalize findings and their properties in a structured format. In another statistical approach, SymText and MPLUS NLP systems combine a controlled terminology, a syntactic context-free grammar parser and Bayesian network-based semantic grammar to code findings in radiology reports [15–17]. Recent related work explored the use of different machine learning methods such as support vector machines and Bayesian networks to classify chest CT scans for invasive fungal and mold diseases at report and patient levels [18,19]. These methods were specialized and evaluated in a limited domains and were not built to extract and summarize the free text information content in multi-institutional radiology reports according to an explicit information model. Our approach improves on the above approaches because a new corpus of annotated reports is not needed to create systems for each new purpose. Also, because our information model is not specific to one type of radiology exam or organization, and it is more generalizable in the domain of radiology.
3. Material and methods
First, we describe the information model that we use to summarize radiology reports, which provides a framework to build and evaluate our information extraction system. Then, we present our radiology report repository and the set of features extracted from the reports in our NLP approach. Our information extraction system is built around a core named-entity recognition method. We propose three different named-entity recognition methods for our information extraction task: (1) dictionary-based method, (2) conditional Markov model (CMM) and (3) conditional random field model (CRF). The first method is commonly used in lexicon-based annotation and information extraction systems and serves as a baseline for two other machine learning methods. Finally, we explain the evaluation mechanism for our system.
3.1. Information model
Our information model provides a framework to summarize radiology reports and to build and evaluate our information extraction system. Our information model’s level of detail is optimized to extract and organize the NLP system’s results, so they are beneficial to clinical research and decision support and surveillance systems. However, the model is also simple enough to enable rapid manual annotation of a training set for our NLP system. This information model has 5 classes of concepts: anatomy, anatomy modifier, observation, observation modifier, and uncertainty. These 5 classes represent the vast majority of clinically significant information contained in radiology reports. For example, the existing information model elements in the report statement “a 1 cm calcified mass probably is present in the anterior right upper lobe” are shown in Table 1.
Table 1.
Information model classes and their examples.
| Information model class | Example |
|---|---|
| Anatomy | “Right upper lobe” |
| Anatomy modifier | “Anterior” |
| Observation | “Mass” |
| Observation modifier | “Calcified”, “1 cm” |
| Uncertainty | “Probably is present” |
Our information model was originally developed to support an earlier radiology reporting system [1], and served as the underpinnings for a widely used terminology to represent information in radiology reports [20]. The model was refined for our information extraction task by one of the authors (CL). Our information model is compatible with the body of literature on representative radiology report information models [21–26]. For comparison, in the information model that was used as a basis for MEDLEE information extraction system [22], radiology findings are defined by observation concept class and various qualifier concept classes for observations, including body location, location qualifier, certainty, degree, temporal, quantity, and property. This information model has a hierarchical structure, which can be used for logical inferences. MEDLEE information model can be considered as a detailed version of our information model in this work. The observation, body location, location qualifier, and certainty classes in MEDLEE information model are identical to observation, anatomy, anatomy modifier, and uncertainty classes in our information model. However, in our information model, as a simplification, we combined degree, temporal, quantity, and property observation qualifier classes as the observation modifier class. This simplification facilitates the manual annotation effort for building training data set and still captures the observation qualifiers’ information that is needed for our information extraction system’s applications.
3.2. Radiology report data set
The source of the radiology report narrative in this work is the RadCore database. RadCore is a multi-institutional database of radiology reports aggregated in 2007 from three major healthcare organizations: Mayo Clinic, MD Anderson Cancer Center, and Medical College of Wisconsin. RadCore radiology reports were collected under institutional review board approval from those three organizations. There were no major differences in the formatting of chest CT radiology reports in these different organizations. The reports were de-identified by their source organization before submission to RadCore database. This project is approved by the Stanford institutional review board. Table 2 shows the number of radiology reports from each organization in RadCore data set.
Table 2.
Data source organizations and their radiology report counts in RadCore database.
| Data source organization | Number of radiology reports |
|---|---|
| Mayo Clinic | 812 |
| MD Anderson Cancer Center | 5000 |
| Medical College of Wisconsin | 1893,819 |
3.3. Training set construction
Given the large amount of data in RadCore radiology report repository and our limited resources, we restricted our focus to chest CT radiology reports to keep the manual annotation requirements tractable. We extracted a representative subset of radiology reports with the same exam type, chest CT, to create our manually annotated multi-institutional corpus. These annotated data provide a key information source for training and evaluation of our information extraction system. Chest CT reports often contain complex observations and findings about a number of vital organs and pathologic conditions. This level of complexity provides a challenging test for our information extraction system and can show the generalizability of our approach for other radiology studies.
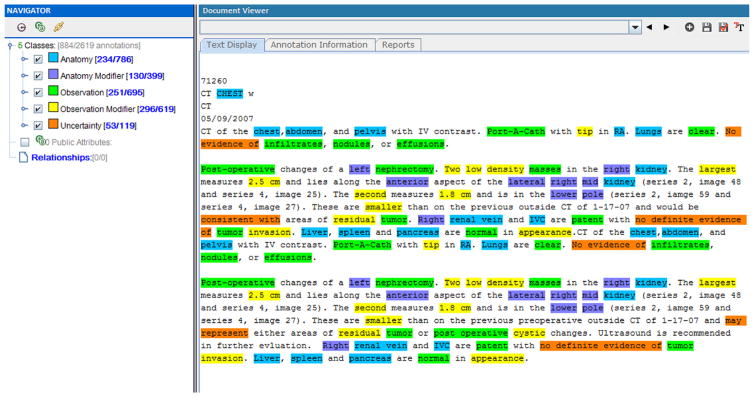
The training set was built through random selection of chest CT reports in the RadCore database. In total, 150 reports, 50 reports from each organization, were automatically selected from the repository. In the manual labeling process one of the authors (CL), highlighted terms and phrases that belong to our information model concept classes. We used the Extensible Human Oracle Suite of Tools (eHOST) [27], an open source annotation tool, for manual annotation. Fig. 1 shows a screenshot of a manually labeled radiology report in eHOST. Manual annotations were exported in XML format by eHost and parsed so they could be used to train and evaluate our information extraction system.
Fig. 1.
A sample manually annotated radiology report in eHOST.
To evaluate the quality of the manual annotations and to estimate the complexity of the annotation task, we calculated inter-annotator agreement for a subset of our data set. We randomly selected 5 radiology reports from each organization in our data set. These 15 reports constituted 10% of our annotated data set. We asked an independent radiologist (GB, see acknowledgments) to annotate these reports according to our information model. We calculated the total percentage of agreements between two annotators. To remove the effect of agreements by chance, we also calculated Cohen’s Kappa coefficient [28], a widely accepted agreement metric in NLP, for these two sets of annotations.
3.4. Feature extraction
We used a combination of semantic and syntactic features in training our machine learning models. The list of these features is as follows:
Part of speech tags
Part of speech tags are each word’s grammatical category, such as verb, adjective, and adverb. The widely-used Stanford Part of Speech Tagger [29] was employed to extract these tags.
Word stems
Word stems are canonical representations of words after removing their morphological variations. We used the Porter stemmer [30], the de facto standard English stemming algorithm, to extract word stems. For example, verbs “performed” and “performs” are both mapped to the canonical shape “perform”.
Word n-grams
A word’s n-grams are all the word’s substrings of length n or less. To keep the computation tractable, we only extracted prefix and suffix substrings with less than 6 characters as word n-gram features.
Word shape
Word shapes are orthographic signatures that encode words’ capitalization, inclusion of numbers and other non-alphabetic character information. We used Stanford CoreNLP toolkit [31] to extract word shapes.
Negation
We used NegEx [32], a widely used clinical text mining tool, to determine negations. NegEx first identifies negation triggers in text based on its dictionary, then uses a set of rules to determine which terms fall within the scope of those triggering terms [32].
RadLex lexicon
RadLex® [20] is a controlled lexicon for radiology terminology. RadLex lexicon is organized in a hierarchal structure and available in Web Ontology Language (OWL) format. We used RadLex to identify each term’s memberships in semantic classes. The current version of RadLex (3.11) contains 58,065 terms in 34,446 classes. RadLex is freely available from the Radiological Society of North America (RSNA). Considering our information model and the hierarchical structure of RadLex lexicon, we chose relevant roots for each information model class and flattened the sub-trees under those roots for use as dictionaries. Because there is no clear distinction between anatomy and observation modifiers in RadLex and the common use of location, density, and orientation modifiers to describe both anatomical structures and observations, we extracted a single dictionary for both anatomy modifier and observation modifier concept classes. Table 3 shows the dictionaries used, their corresponding RadLex roots, and the number of entries in each.
Table 3.
Concept dictionaries with their RadLex roots and entity counts.
| Dictionary | RadLex roots | Number of entries |
|---|---|---|
| Anatomy | Anatomical structure Immaterial anatomical entity Anatomical set |
37,907 |
| Modifier | RadLex descriptor | 1217 |
| Observation | Pathophysiologic finding Benign finding Portion of body substance Object Imaging observation |
3573 |
| Uncertainty | Certainty descriptor | 23 |
3.5. Named-entity recognition
To identify and extract the terms and phrases in radiology reports that belong to our information model’s concept classes, we built a named-entity recognition module. Given the module’s key role in our annotation and information extraction system, we developed and evaluated three different named-entity recognition methods: dictionary-based, CMM, and CRF methods to find the most effective approach. The dictionary-based method, which is not a machine learning approach, is commonly used in biomedical applications and serves as a baseline for the two latter machine learning approaches. In this project, we used CMM and CRF training infrastructure in Stanford Named-Entity Recognizer toolkit [33] to build our named-entity recognition annotation models. These machine learning approaches rely on our labeled training data and their extracted features to recognize the common patterns for identifying information model elements.
3.5.1. Dictionary-based method
As a baseline, we leveraged the widely used cTAKES dictionary-based named-entity recognition methodology in this work. Because no radiology terminology is included in the current version of cTAKES’s dictionary, we used cTAKES system description [10] and RadLex to implement the dictionary-based baseline method for radiology reports. As a result, our dictionary-based method is a representative of the cTAKES annotation method and provides a comparison between our system and a commonly used annotation system for biomedical applications.
In our dictionary-based method we used RadLex to build a dictionary for each concept class in the information model with the exception of the modifier dictionary, which had two corresponding concept classes: anatomy modifier and observation modifier (Table 3). As mentioned before, this is due to the shared set of modifiers used for both anatomical structures and observations. Therefore, in this method we merged and treated anatomy modifier and observation modifier concept classes as a single concept class.
In the dictionary-based method, the RadLex terms in each dictionary were converted to their canonical forms through the Porter stemming algorithm and stored in a table. The report terms were also converted to their canonical forms through the Porter stemming algorithm and looked up in the dictionary tables. The terms and phrases that existed in a dictionary table were annotated by the dictionary’s information model class. In our method, if a term or phrase was matched with more than one dictionary entry through multiple text spans, we considered the longest text span as the matched entry for annotation. For example, in phrase “deep vein thrombosis” although individual terms “vein” and “thrombosis” exist in our dictionaries, only the longest match, “deep vein thrombosis” is annotated and used for information extraction.
3.5.2. Conditional Markov model
Conditional Markov models or CMMs, also known as maximum-entropy Markov models (MEMMs), are sequence classifiers [34]. CMM classifiers are commonly used for sequence labeling tasks such as part of speech tagging and named-entity recognition. In NLP applications, for each word in the sequence of input words, CMMs make a single annotation decision at a time, conditioned on the features from the word and its surroundings words, as well as previous decisions. As a result, sequence classifiers such as CMMs include surrounding context in decision making to annotate each word in text [34].
CMMs combine maximum-entropy classifiers with Markov chains from hidden Markov models (HMMs). Similar to HMMs, CMMs use the Markov assumption and the Viterbi algorithm to search over label space. However, they use a maximum entropy framework for features and normalization [34]. CMMs are discriminative models. Discriminative models are based on conditional probability distributions and are considered to be more effective for NLP tasks than generative models such as HMMs, which are based on joint probability distributions [35]. CMMs do not consider features as independent. Rather than learning a joint probability distribution for input features and output labels as generative models do, they find parameters that maximize the conditional probability of output labels given the input features [35]. We refer the reader to [34] for more details about CMM classifiers.
In this work, we used the CMM training infrastructure in Stan-ford Named-Entity Recognizer toolkit [33] to build a named-entity recognition model to annotate and extract information from radiology reports.
3.5.3. Conditional random field model
Conditional random fields (CRFs) are another form of sequence classifier, which are used in the state-of-the-art part of speech tagging and named-entity recognition systems [36]. CRFs, similar to CMMs, are discriminative models. A CRF model includes an estimation of the conditional distribution of output labels given the input features with an associated graphical structure. We used a linear chain graphical structure in this work, which predicts sequences of annotation labels for the sequences of input words from radiology reports. The CRF model considers previously assigned labels, surrounding terms, and their features as context for annotation of a single word. The major difference between CRFs and CMMs is their method for probability normalization. Probabilities in CRFs are normalized globally for a sequence. In contrast, probabilities in CMMs are normalized locally for each state in the sequence. The global normalization in CRFs improves model’s general accuracy, but increases its computational complexity. We refer the readers to [37,38] for a detailed discussion of CRFs.
Because discriminative models such as CMMs and CRFs are conditional, dependencies among the input features do not need to be explicitly represented in their graphical structure. These discriminative models do not impose any assumptions on the dependencies and probability distributions of input features. As a result, in the model training process, discriminative models divide the feature weights for correlated and overlapping features instead of considering the features as additional pieces of evidence [37]. Therefore, in contrast to generative models, possible repeated and correlated features do not affect the performance or cause over fitting in discriminative models such as CMMs and CRFs. Often, rich arrays of features with potential overlaps are used to train CRF and CMM models for NLP applications without side effects from feature correlations [37].
We used the CRF training functionality provided in Stanford Named-Entity Recognizer toolkit [33] for training a named-entity recognition model for information extraction from radiology reports in this work.
3.6. Evaluation
To evaluate our information extraction system with CMM and CRF named-entity recognition methods and to compare them to the dictionary-based baseline method, we performed a 10-fold cross-validation for each method on our data set of 150 manually annotated radiology reports. We computed standard metrics of precision, recall, and F1 score for each model and aggregated the results through micro-averaging. The cross-validation removes selection bias and evaluates the methods on the entire labeled data set. In this evaluation, we measured precision, recall, and F1 score for the non-machine learning dictionary-based method on all ten cross-validation test partitions as well. We compared the performance of these three methods for each information model concept class in 10-fold cross-validation using Student’s t-test [39]. Throughout our evaluation we used 0.05 as the significance level for our statistical comparisons.
To evaluate the generalizability of our machine learning approach, we built 3 additional models for CMM and CRF methods. Each model was trained on data from two organizations and evaluated by measuring precision, recall, and F1 score on data from the third organization. The precision, recall, and F1 score of the dictionary-based method was also measured on each test set. The performance of all three methods was compared using Student’s t-test.
4. Results
In our inter-annotator agreement evaluation, two annotators agreed in 85.5% of their annotations. Kappa coefficient for these two sets of annotations is 0.75. For comparison, Table 4 shows the distribution of concept classes in the manual annotations and the distribution of concept classes in the annotation results of our three information extraction methods in cross-validation. The results show the number of annotations in the dictionary-based method is considerably lower than the number of manual and machine learning annotations. Table 5 shows the results of our 10-fold cross-validation on 150 manually annotated radiology reports for all 5 information model classes. Precision, recall, F1 scores, the number of true positives (TP), false positives (FP), and false negatives (FN) are listed for three variations of our information extraction system with dictionary-based, CMM, and CRF named-entity recognition methods.
Table 4.
The number and percentage of annotated concept classes in manual annotations and annotation results of different information extraction methods in cross-validation.
| Manual annotation
|
Dictionary-based
|
CMM
|
CRF
|
|||||
|---|---|---|---|---|---|---|---|---|
| Count | (%) | Count | (%) | Count | (%) | Count | (%) | |
| Anatomy | 4411 | 31 | 3017 | 31 | 4513 | 33 | 4510 | 33 |
| Anatomy modifier | 1837 | 13 | – | – | 1699 | 12 | 1690 | 12 |
| Observation | 3466 | 24 | 2336 | 24 | 3291 | 24 | 3259 | 24 |
| Observation modifier | 3199 | 22 | – | – | 3124 | 23 | 3129 | 23 |
| Uncertainty | 1455 | 10 | 628 | 6 | 1253 | 9 | 1219 | 9 |
| Modifier | – | – | 3712 | 38 | – | – | – | – |
| Total | 14,368 | 100 | 9693 | 100 | 13,880 | 100 | 13,807 | 100 |
Table 5.
Results of 10-fold cross-validation for different named-entity recognition methods.
| Method | Concept | Precision (%) | Recall (%) | F1 score (%) | TP | FP | TN |
|---|---|---|---|---|---|---|---|
| Dictionary-based | Anatomy | 77.7 | 53.2 | 63.1 | 2345 | 672 | 2066 |
| Modifier | 63.3 | 46.6 | 53.7 | 2349 | 1363 | 2687 | |
| Observation | 72.7 | 49.0 | 58.5 | 1697 | 639 | 1769 | |
| Uncertainty | 90.1 | 38.9 | 54.3 | 566 | 62 | 889 | |
| Total | 71.8 | 48.4 | 57.8 | 6957 | 2736 | 7411 | |
| CMM | Anatomy | 90.5 | 92.6 | 91.5 | 4084 | 429 | 327 |
| Anatomy modifier | 80.2 | 74.2 | 77.1 | 1363 | 336 | 474 | |
| Observation | 89.4 | 84.9 | 87.1 | 2941 | 350 | 525 | |
| Observation modifier | 81.4 | 79.5 | 80.4 | 2543 | 581 | 656 | |
| Uncertainty | 91.1 | 78.5 | 84.3 | 1142 | 111 | 313 | |
| Total | 87.0 | 84.0 | 85.5 | 12,073 | 1807 | 2295 | |
| CRF | Anatomy | 90.6 | 92.6 | 91.6 | 4085 | 425 | 326 |
| Anatomy modifier | 80.8 | 74.4 | 77.5 | 1366 | 324 | 471 | |
| Observation | 89.4 | 84.1 | 86.7 | 2915 | 344 | 551 | |
| Observation modifier | 81.2 | 79.5 | 80.3 | 2542 | 587 | 657 | |
| Uncertainty | 90.7 | 76.0 | 82.7 | 1105 | 114 | 350 | |
| Total | 87.0 | 83.6 | 85.3 | 12,013 | 1794 | 2355 |
Our Student’s t-test comparison on the performance of these three methods shows that both CMM and CRF methods outperform the dictionary-based method with statistical significance (p-value < 0.05) for all information model classes in 10-fold cross-validation. Table 6 presents the p-values for Student’s t-test comparison between F1 scores of these three methods. This Student’s t-test comparison also shows that there is no statistically significant difference between CMM and CRF methods’ performances for all information model classes in cross-validation (Table 6).
Table 6.
p -Values for Student’s t-test comparison between F1 scores of dictionary-based, CMM and CRF methods.
| Concept | Dictionary-based/CMM (p-value) | Dictionary-based/CRF (p-value) | CMM/CRF (p-value) |
|---|---|---|---|
| Anatomy | 2.34E – 18 | 2.52E – 18 | 0.96 |
| Anatomy modifier | – | – | 0.76 |
| Observation | 2.25E – 14 | 1.83E – 14 | 0.63 |
| Observation modifier | – | – | 0.90 |
| Uncertainty | 8.55E – 11 | 1.42E – 10 | 0.39 |
| Total | 4.44E – 18 | 3.64E – 18 | 0.77 |
Table 7 shows precision, recall, and F1 scores of different named-entity recognition methods in our information extraction system when they are trained and tested on different organization’s data to evaluate the generalizability of our approach. The Student’s t-test comparison for F1 scores of these three methods, summarized in Table 8, shows both CMM and CRF methods outperform the dictionary-based method with a statistical significance (p-value < 0.05). There is no statistically significant difference between the performances of the CMM and CRF methods.
Table 7.
Generalizability evaluation results across different organizations.
| Method | Training organizations | Test organization | Precision (%) | Recall (%) | F1 score (%) | TP | FP | TN |
|---|---|---|---|---|---|---|---|---|
| Dictionary-based | N/A | MC | 73.4 | 47.9 | 58.0 | 1476 | 534 | 1605 |
| N/A | MCW | 70.7 | 49.2 | 58.0 | 3581 | 1487 | 3693 | |
| N/A | MDA | 72.7 | 47.4 | 57.3 | 1900 | 715 | 2113 | |
| CMM | MCW, MDA | MC | 83.0 | 79.2 | 81.0 | 2439 | 499 | 642 |
| MC, MDA | MCW | 78.9 | 67.7 | 72.9 | 4924 | 1319 | 2350 | |
| MC, MCW | MDA | 85.7 | 78.5 | 81.9 | 3150 | 527 | 863 | |
| CRF | MCW, MDA | MC | 83.3 | 78.9 | 81.0 | 2430 | 486 | 651 |
| MC, MDA | MCW | 78.4 | 68.2 | 72.9 | 4961 | 1369 | 2313 | |
| MC, MCW | MDA | 85.4 | 77.7 | 81.3 | 3117 | 535 | 896 |
Table 8.
p -Values for Student’s t-test comparison between different methods’ F1 scores when they are trained and tested on data from different organizations.
| Training/test organizations | Dictionary-based/CMM (p-value) | Dictionary-based/CRF (p-value) | CMM/CRF (p-value) |
|---|---|---|---|
| (Mayo, MCW)/MDA | 7.07E – 05 | 8.86E – 05 | 0.83 |
| (Mayo, MDA)/MCW | 4.56E – 02 | 3.81E – 02 | 0.98 |
| (MCW, MDA)/Mayo | 3.16E – 04 | 2.51E – 04 | 1.00 |
5. Discussion
5.1. Methods and results review
The main contribution of this work is the use of existing machine learning frameworks and NLP syntactic and semantic features to build an information extraction system that can accurately identify terms and phrases in radiology reports according to a radiology-specific information model. Our syntactic features are derived from grammatical structures, word forms, and morphology, such as part of speech tags, word stems, word n-grams, and word shapes. Our semantic features relate to the meaning and interpretation of words and phrases, such as negation and memberships in RadLex ontological classes.
To build an effective information extraction system, we investigated three different methods, dictionary-based, CMM, and CRF, for named-entity recognition as the core component of this system. CMM and CRF, which are both machine learning methods, had strong performances, with precision of 87%, recall of 84%, and F1 scores of 85% on average in our 10-fold cross-validation. We observed 15% gain in precision, 35% gain in recall, and 28% gain in F1 score for CMM and CRF methods on average compared to the commonly used dictionary-based method in our cross-validation evaluation. Our evaluation showed that the performance of CMM and CRF methods was equally strong with no statistically significant difference.
The major drawback of the dictionary-based method is its lack of coverage and generalizability. In general, having access to dictionaries with exhaustive lists of concepts in the domain of interest is challenging. Even in the presence of these dictionaries, the variability and ambiguity of natural language do not match the simplicity of the dictionary lookup methods [13]. Many anatomy and observation modifiers describing location, density, and orientation cannot be distinguished without analyzing their context. For example, “3rd” could apply to an anatomic structure such as a rib, or to a nodule, which is an observation. As a result, the dictionary-based method cannot differentiate between anatomy and observation modifiers in our information model. In our dictionary-based method we modified the information model to consolidate anatomy modifier and observation modifier concept classes as a single modifier class. This modification made the task of information extraction easier for the dictionary-based method compared to two other machine learning methods which need to differentiate between anatomy and observation modifiers. This simplification in dictionary-based method provided a strong baseline against which to measure our machine learning methods’ performances. Our comparison to this dictionary-based baseline suggests that dictionaries such as RadLex are more instrumental for radiology report information extraction in combination with semantic and syntactic features.
5.2. Inter-annotator agreement
The reference standard annotations in this study were generated by one radiologist (CL) who also contributed to creating the information model. This might have introduced some biases in the annotations. To explore these potential biases, we asked a second independent radiologist (GB) to annotate 10% of our data set without specific training. The inter-annotator agreement measure for this subset showed reasonably high agreement between the two annotators (Kappa coefficient = 0.75). This level of inter-annotator agreement shows the integrity of our reference standard annotations. The existing disagreements between the annotators demonstrate the challenges of the manual annotation process, caused by the complexity of radiology report language. We expect providing a comprehensive set of annotation guidelines with expressive examples will improve the quality of the reference standard annotations and the inter-annotator agreement.
5.3. Error analysis
We investigated the most common incorrect annotation classes for each method in our error analysis. Table 9 shows the top pairs of correct and incorrect annotation classes in cross-validation on our entire data set. As shown in this table, the number of errors in the dictionary-based method is almost as three times larger than the number of errors in the machine learning methods. For further insight into the causes of errors, we randomly selected 5 radiology reports from each organization to manually review the annotation errors by each method. These 15 radiology reports cover 10% of our data set and 1287 errors (803 dictionary-based errors, 231 CMM errors and 253 CRF errors).
Table 9.
Top pairs of correct and incorrect annotation classes by each method in cross-validation.
| Dictionary-based
|
CMM
|
CRF
|
||||||
|---|---|---|---|---|---|---|---|---|
| True label | Assigned label | (%) | True label | Assigned label | (%) | True label | Assigned label | (%) |
| Modifier | None | 26 | Observation modifier | None | 13 | Observation modifier | None | 13 |
| Anatomy | None | 16 | Uncertainty | None | 9 | Uncertainty | None | 10 |
| Observation | None | 14 | Anatomy modifier | Anatomy | 9 | Anatomy modifier | Anatomy | 9 |
| Uncertainty | None | 11 | Observation | Observation modifier | 8 | Observation | Observation modifier | 8 |
| None | Modifier | 6 | None | Observation modifier | 7 | Observation | None | 8 |
| Total number of errors | 8107 | Total number of errors | 2842 | Total number of errors | 2888 | |||
According to our manual review, 22% of the dictionary-based method’s errors are caused by the low coverage of radiology report terms in RadLex. As a result, various terms from the information model classes are not annotated by the dictionary-based method. For example, the term “diverticulosis” does not exist in RadLex, and therefore is missed in the dictionary-based method’s annotations. We expect extending the concept dictionaries will improve the dictionary-based method’s performance. In addition, because the dictionary terms are used as features in our machine learning methods as well, we expect this extension will address some common missed annotations in all three methods such as “side” and “surface”.
The remaining 78% of the annotation errors in the dictionary-based method are because of the context-free nature of this method. For example, the term “normal” is classified as a modifier in RadLex class hierarchy and is part of the modifier dictionary. However, in sentence “the heart appears normal”, normal is an observation rather than a modifier. Of note, both CMM and CRF methods identify “normal” correctly as an observation in this sample sentence.
For CMM and CRF methods, 28% of the errors are caused by annotating anatomy and observation terms as anatomy and observation modifiers and vice versa. In these cases the machine learning models and their NLP features fail to disambiguate between the concepts terms and their modifiers. For example, in phrase “right upper lobe”, “right” and “upper” are parts of an anatomical structure’s name and our machine learning methods correctly identify “right” and “upper” belong to the anatomy class. However, in phrase “right upper lung”, despite of the similar language pattern, “right” and “upper” are modifiers for an anatomy (lung) rather than part of the anatomy’s name. Our machine learning methods incorrectly annotate “right” and “upper” as anatomy instead of anatomy modifier in the latter example. We plan to expand our annotated training data and enrich our NLP features to address these errors in our machine learning methods. Expanding the training data will inform and refine the machine learning models for such error cases and adding new features that capture terms’ distributional semantics and co-occurrences patterns [40] address the annotation errors even without observing similar cases in the training set.
In addition, 24% of errors in CMM and CRF methods are due to manual annotation errors and inconsistent annotations in the training set. For example terms “except” and “otherwise” were not consistently annotated by uncertainty class in the training set. We expect increasing the number of annotators and performing quality control on reference standard annotations will address these errors. The remaining 48% of errors in CMM and CRF methods includes a wide range of radiology report terms such as “blood”, “cell”, “sensitive”, and “posteriorly”. We associate these errors with the lack of indicative features in our machine learning methods. We expect adding syntactic and semantic features to our machine learning methods, such as grammatical dependencies and distributional semantics, improves their performances for these cases.
We also compared the results of these three methods to each other. Table 10 shows the agreements between different methods on their cross-validation results. Our comparison shows the results of CMM and CRF methods are different from the dictionary-based method in nearly 27% of all cases in cross-validation. However, CMM and CRF methods disagree in less than 2% of the cases.
Table 10.
Agreement between three methods on cross-validation results.
| Method | Dictionary-based (%) | CMM (%) | CRF (%) |
|---|---|---|---|
| Dictionary-based | 100.0 | 72.9 | 73.0 |
| CMM | 72.9 | 100.0 | 98.2 |
| CRF | 73.0 | 98.2 | 100.0 |
We examined the validity of the annotations when the dictionary-based method agreed and disagreed with the machine learning methods. When CMM and CRF methods agreed with the dictionary-based method, the annotations were correct in 95% of cases. In disagreements between the machine learning methods and the dictionary-based method, the CRF and CMM annotations were correct in 43% of cases, while the dictionary-based method’s annotations were correct in 29% of cases. Therefore, we expect a high level of validity for annotations on which the dictionary-based method and the machine learning methods agree.
For further investigation, we divided the cross-validation results of the machine learning methods into two groups. The first group contains the terms and phrases that can be found in RadLex and the second group contains the terms and phrases that cannot be found in RadLex. We calculated the precisions, recalls and F1 scores in these two groups for both CMM and CRF methods, which are show in Table 11.
Table 11.
The performances of the machine learning methods on data with RadLex terms versus the data without RadLex terms.
| Method | Test data | Precision (%) | Recall (%) | F1 score (%) | TP | FP | TN |
|---|---|---|---|---|---|---|---|
| CMM | With RadLex terms | 90.4 | 91.0 | 90.7 | 8190 | 870 | 807 |
| Without RadLex terms | 80.6 | 72.3 | 76.2 | 3883 | 937 | 1488 | |
| CRF | With RadLex terms | 90.4 | 90.8 | 90.6 | 8170 | 865 | 827 |
| Without RadLex terms | 80.5 | 71.6 | 75.8 | 3843 | 929 | 1528 |
We observed that both machine learning methods achieved reasonable performance in cases without RadLex terms (average F1 score of 76%). However, their performance metrics in these cases are lower than the performance metrics in cases containing RadLex terms (average F1 score decrease by 15%). Table 11 shows that the performance of the machine learning methods is not determined entirely by RadLex. However, using RadLex dictionaries as input features does influence the quality of the results.
Unlike the dictionary-based method, our machine learning methods may consider terms outside the dictionaries. In the machine learning named-entity recognition methods, the dictionaries provide additional features to many other semantic and syntactic features for training. And their influence on named-entity recognition results is determined by their weights in trained CMM and CRF models.
We investigated cases in which CMM and CRF methods disagreed in our manual annotation review. Among 3641 terms in these reviewed radiology reports, CMM and CRF methods disagreed on annotations for only 71 terms. These disagreements include mostly uncommon patterns and language in the data set. These rare cases are barely covered in the training data set and slight feature weight differences between CMM and CRF models resulted in disagreements in these uncommon cases. For example in the sentence “no destructive bony lesions are present”, “bony” is referring to anatomy, while “no” is uncertainty, “destructive” is observation modifier and “lesions” is observation. In this sentence, while CRF method annotates the sentence correctly, the CMM method does not assign any annotation to the term “bony”, because it is very uncommon in our training data set for observations and observation modifiers (“lesions” and “destructive” in this sentence) to be separated by a term from another class. As another example, CMM correctly annotates “appearance” as an observation modifier in the sentence of “the liver is normal in appearance without focal hepatic lesions”. However, because of this uncommon language in the training data set and its proximity to “without” from the uncertainty class, CRF incorrectly annotates “appearance” as an uncertainty. We expect adding more annotated data for these uncommon patterns and language and using them for further model training will decrease the small number of disagreements between the machine learning methods.
Finally, we examined annotation errors in our cross-organization study through manual review. We observed the error types in cross-organization analysis are similar to the error types in cross-validation. This is due to the similarities in the pattern of information model concepts in radiology reports across different organizations, which are also reflected in comparable cross-validation and cross-organization results. Of note, we did not observe any spelling errors in the review of the radiology reports. All reports in our data set are dictated by radiologists using speech recognition systems [41]. These speech recognition systems have built-in dictionaries, perform spell check, and therefore almost always include correctly spelled terms and phrases in radiology reports [41].
5.4. Features analysis
We extracted the feature weights in both CMM and CRF models to identify the features with the most influence on the results. Tables 12 and 13 show the list of top 5 influential features for each information model class in CMM and CRF models. As shown in these tables, dictionary terms, part of speech (POS) tags, word n-grams, and surrounding words have strong effect on the results of both machine learning methods.
Table 12.
Top five features in our CMM name entity recognition model for each information model class.
| Anatomy | Anatomy modifier | Observation | Anatomy modifier | Uncertainty |
|---|---|---|---|---|
|
|
|
|
|
Table 13.
Top five features in our CRF name entity recognition model for each information model class.
| Anatomy | Anatomy modifier | Observation | Observation modifier | Uncertainty |
|---|---|---|---|---|
| 1. Anatomy dictionary | 1. Prefix “bi” | 1. Observation dictionary | 1. Modifier dictionary | 1. Uncertainty dictionary |
| 2. Suffix “ary” | 2. Prefix “mid” | 2. Prefix “nod” | 2. Adjective-adjective POS tags | 2. Combination of adjective-proposition POS tags |
| 3. “Lobe” and following word shapes | 3. Prefix “bas” | 3. “Changes” as the following word | 3. “Normal” and the following words | 3. Prefix “poss” |
| 4. “Lobe” and following words | 4. “Hepatic” and the pervious words | 4. Proper noun POS tag | 4. Adjective-noun-noun POS tags | 4. “Possibl” word stem |
| 5. “Chest” stem word | 5. Noun-preposition-determiner POS tags | 5. Suffix “tomy” | 5. Cardinal number POS tag | 5. “evidence” as the previous word |
As Tables 12 and 13 suggest, in addition to considering the words and their features, CMM and CRF methods make annotation decisions based on the surrounding context and previous annotations. In contrast, the dictionary-based method statically maps a word to an annotation in the dictionary lookup table without considering the word’s context. Also, because CMM and CRF are discriminative models, correlated and overlapping features are not as susceptible to over fitting.
5.5. Generalizability
We evaluated the generalizability of our machine learning information extraction methods across different healthcare organizations by training and testing them on data from different sources. Our results showed the strong performance of CMM and CRF methods on novel data from different organizations (average precision: 82%, average recall: 75%, average F1 score: 79%). We observed the average gain of 10% in precision, 27% in recall, and 21% in F1 score when our machine learning approach is applied to a different organization’s data compared to the dictionary-based method. The dictionary-based method, which uses RadLex lexicon, is not dependent on a particular organization, and as is shown in Table 7, its results are consistent across different organizations. We did not observe a statistically significant difference between the performances of CMM and CRF methods in terms of generalizability. The cross-organizational evaluation shown in Table 7 demonstrates that the syntactic and semantic features learned from radiology reports in CMM and CRF methods are effective in identifying the information model concepts in a new organization’s radiology reports.
5.6. Impact
Given our cross-validation and generalizability results, our information extraction system can provide an infrastructure to develop and improve various biomedical information systems. For example, our information extraction system can improve the performance of a radiology report information retrieval system’s matching algorithm through annotating queries and reports according to the information model. Our information extraction system can be combined with other text analysis systems such as constituency and dependency parsers and text classifiers to provide summaries of radiology reports. The resulting annotations can be joined with other information sources, such as electronic health records, for case prioritization or disease surveillance. Therefore, we expect the results of our information extraction system being used for improving other biomedical applications rather than being directly provided to radiologists and other healthcare providers.
5.7. Limitations and future work
Due to limited resources for manual annotation, we restricted the focus of our information extraction task to chest CT reports. Despite this restriction, chest CT report narrative covers observations and findings from many vital organs and conditions and is representative of the complexity of radiology report narrative for other imaging modalities and body regions. Even with a relatively small set of training data, our results show the robustness and generalizability of our approach across different organizations. In fact, none of the NLP techniques described in this work is specific to an information model, narrative, or organization. The developed techniques are applicable to other types of narrative with different information models and data sources as well.
As mentioned in the analysis of inter-annotator agreement, the reference standard annotations in this work are generated by one domain expert. To address any potential biases in these annotations we plan to leverage multiple annotators instructed with annotation guidelines and examples for manual annotation in the future extension of our work. We will use the majority vote among these overlapping annotations to remove potential disagreements, biases, and noise in annotations. We expect this will increase the reliability of our annotated training set and improve our machine learning information extraction system. In addition, as suggested by our error analysis, we plan to expand the size and richness of our training data by including features such as grammatical dependencies and distributional semantics to improve the reliability of our machine learning information extraction system.
As future work, we also plan to extend the domain of our system beyond chest CT reports to other types of radiology reports and clinical notes. We also plan to expand the richness of our information model. This includes adding more concept classes to the information model and splitting the existing classes to more detailed subclasses. In addition, to demonstrate the application of this work in other healthcare information systems, we plan to use the presented information extraction system to summarize clinically significant information in radiology reports and prioritize the urgency of each report as a part of a real-time clinical decision support system for radiologists.
6. Conclusions
We described a machine learning information extraction system to extract clinically significant concepts from radiology reports according to a published information model. This information model covers the majority of clinically significant information contained in radiology reports and is applicable across organizations. We investigated two machine learning methods, CMM and CRF, and a commonly used dictionary-based method as baseline for named-entity recognition in our system. We evaluated our methods using a data set containing 150 manually annotated radiology reports from three major healthcare organizations. The machine learning information extraction system performed equally effectively with both CMM and CRF named-entity recognition methods (average F1 score: 85%). Our results showed the strength and generalizability of our machine learning approach compared to the dictionary-based approach.
The extracted information from radiology reports can be used to link radiology report information to the patient’s electronic health record or to genomic data. The same results can enable clinicians to prioritize the report review process and to rapidly identify reports that need further follow up. The extracted report information can facilitate automated identification of patients for clinical trials based on imaging features, accelerate disease surveillance, and enable real-time clinical decision support systems for radiologists. In addition, by attaching the extracted concepts to the images themselves, content-based image retrieval becomes possible. Therefore, the described information extraction system provides substantial utility to support biomedical research and clinical practice.
Acknowledgments
The authors would like to thanks Graham Bay (GB) for his help with manual annotation quality control. The authors would like to thank Daniel Rubin, Chuck Kahn, Kevin McEnery and Brad Erickson for their help compiling the RadCore database and Daniel Rubin for providing access to the database.
References
- 1.Langlotz Curtis P, Meininger Lee. Enhancing the expressiveness and usability of structured image reporting systems. In: Marc Overhage J, editor. Proceedings of the AMIA symposium. Los Angeles, CA: American Medical Informatics Association; 2000. p. 467. [PMC free article] [PubMed] [Google Scholar]
- 2.Dreyer Keith J, Kalra Mannudeep K, Maher Michael M, Hurier Autumn M, Asfaw Benjamin A, Thomas Schultz, et al. Application of recently developed computer algorithm for automatic classification of unstructured radiology reports: validation study 1. Radiology. 2005;234(2):323–9. doi: 10.1148/radiol.2341040049. [DOI] [PubMed] [Google Scholar]
- 3.Dreyer Keith J. Information theory entropy reduction program. 8,756,234. US Patent. issued June 17, 2014.
- 4.Meliha Yetisgen-Yildiz, Gunn Martin L, Fei Xia, Payne Thomas H. A text processing pipeline to extract recommendations from radiology reports. J Biomed Inf. 2013;46(2):354–62. doi: 10.1016/j.jbi.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Carol Friedman, George Hripcsak, William DuMouchel, Johnson Stephen B, Clayton Paul D. Natural language processing in an operational clinical information system. Nat Lang Eng. 1995;1(01):83–108. [Google Scholar]
- 6.George Hripcsak, Austin John HM, Alderson Philip O, Carol Friedman. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports 1. Radiology. 2002;224(1):157–63. doi: 10.1148/radiol.2241011118. [DOI] [PubMed] [Google Scholar]
- 7.George Hripcsak, Kuperman Gilad J, Carol Friedman. Extracting findings from narrative reports: software transferability and sources of physician disagreement. Methods Inf Med. 1998;37(1):1–7. [PubMed] [Google Scholar]
- 8.Elkins Jacob S, Carol Friedman, Bernadette Boden-Albala, Sacco Ralph L, George Hripcsak. Coding neuroradiology reports for the Northern Manhattan Stroke Study: a comparison of natural language processing and manual review. Comput Biomed Res. 2000;33(1):1–10. doi: 10.1006/cbmr.1999.1535. [DOI] [PubMed] [Google Scholar]
- 9.Johnson David B, Taira Ricky K, Cardenas Alfonso F, Aberle Denise R. Extracting information from free text radiology reports. Int J Digit Libr. 1997;1(3):297–308. [Google Scholar]
- 10.Savova Guergana K, Masanz James J, Ogren Philip V, Jiaping Zheng, Sunghwan Sohn, Kipper-Schuler Karin C, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inf Assoc. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sergey Goryachev, Margarita Sordo, Zeng Qing T. A suite of natural language processing tools developed for the I2B2 project. In: Bates David W, editor. Proceedings of the AMIA symposium. Vol. 2. Washington DC: American Medical Informatics Association; 2006. p. 931. [PMC free article] [PubMed] [Google Scholar]
- 12.Aronson Alan R. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. In: Suzanne Bakken., editor. Proceedings of the AMIA symposium. Washington DC: American Medical Informatics Association; 2001. p. 17. [PMC free article] [PubMed] [Google Scholar]
- 13.Taira Ricky K, Soderland Stephen G. A statistical natural language processor for medical reports. In: Lorenzi Nancy M, editor. Proceedings of the AMIA symposium. Washington, DC: American Medical Informatics Association; 1999. p. 970. [PMC free article] [PubMed] [Google Scholar]
- 14.Taira Ricky K, Soderland Stephen G, Jakobovits Rex M. Automatic structuring of radiology free-text reports 1. Radiographics. 2001;21(1):237–45. doi: 10.1148/radiographics.21.1.g01ja18237. [DOI] [PubMed] [Google Scholar]
- 15.Peter Haug, Spence Koehler, Min Lau Lee, Ping Wang, Roberto Rocha, Huff Stan. A natural language understanding system combining syntactic and semantic techniques. In: Ozbolt Judy G, editor. Proceedings of the annual symposium on computer application in medical care. Washington DC: American Medical Informatics Association; 1994. p. 247. [PMC free article] [PubMed] [Google Scholar]
- 16.Haug Peter J, Spence Koehler, Min Lau Lee, Ping Wang, Roberto Rocha, Huff Stanley M. Experience with a mixed semantic/syntactic parser. In: Gardner Reed M, editor. Proceedings of the annual symposium on computer application in medical care. New Orleans, LA: American Medical Informatics Association; 1995. p. 284. [Google Scholar]
- 17.Christensen Lee M, Haug Peter J, Marcelo Fiszman. MPLUS: a probabilistic medical language understanding system. In: Stephen Johnson., editor. Proceedings of the ACL-02 workshop on natural language processing in the biomedical domain. Vol. 3. Stroudsburg, PA: Association for Computational Linguistics; 2002. pp. 29–36. [Google Scholar]
- 18.David Martinez, Ananda-Rajah Michelle R, Hanna Suominen, Slavin Monica A, Thursky Karin A, Lawrence Cavedon. Automatic detection of patients with invasive fungal disease from free-text computed tomography (CT) scans. J Biomed Inf. 2015;53:251–60. doi: 10.1016/j.jbi.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Ananda-Rajah Michelle R, David Martinez, Slavin Monica A, Lawrence Cavedon, Michael Dooley, Allen Cheng, et al. Facilitating surveillance of pulmonary invasive mold diseases in patients with haematological malignancies by screening computed tomography reports using natural language processing. PLoS ONE. 2014;9(9):e107797. doi: 10.1371/journal.pone.0107797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langlotz Curtis P. RadLex: a new method for indexing online educational materials. Radiographics. 2006;26(6):1595–7. doi: 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- 21.Friedman C, Cimino JJ, Johnson SB. A schema for representing medical language applied to clinical radiology. J Am Med Inf Assoc. 1994;1(3):233–48. doi: 10.1136/jamia.1994.95236155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carol Friedman, Huff Stanley M, Hersh William R, Edward Pattison-Gordon, Cimino James J. The Canon Group’s effort: working toward a merged model. J Am Med Inf Assoc. 1995;2(1):4–18. doi: 10.1136/jamia.1995.95202547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell Douglas S, Edward Pattison-Gordon, Greenes Rober A. Experiments in concept modeling for radiographic image reports. J Am Med Inf Assoc. 1994;1(3):249–62. doi: 10.1136/jamia.1994.95236156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rocha RA, Huff SM, Haug PJ, Evans DA, Bray BE. Evaluation of a semantic data model for chest radiology: application of a new methodology. Methods Inf Med. 1998;37(4–5):477–90. [PubMed] [Google Scholar]
- 25.Bidgood W., Dean Jr The SNOMED DICOM microglossary: controlled terminology resource for data interchange in biomedical imaging. Methods Inf Med. 1998;37(4–5):404–14. [PubMed] [Google Scholar]
- 26.Dean Bidgood W, Bruce Bray, Nicolas Brown, Rossi Mori Angelo, Spackman Kent A, Alan Golichowski, et al. Image acquisition context procedure description attributes for clinically relevant indexing and selective retrieval of biomedical images. J Am Med Inf Assoc. 1999;6(1):61–75. doi: 10.1136/jamia.1999.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. [accessed April 2015];eHost: A tool for semantic annotation and lexical curation. 〈 https://code.google.com/p/ehost/〉.
- 28.Jean Carletta. Assessing agreement on classification tasks: the kappa statistic. Comput Linguist. 1996;22(2):249–54. [Google Scholar]
- 29.Kristina Toutanova, Dan Klein, Manning Christopher D, Yoram Singer. In: Marti Hearst, Mari Ostendorf., editors. Feature-rich part-of-speech tagging with a cyclic dependency network; Proceedings of the 2003 Conference of the North American chapter of the association for computational linguistics on human language technology; Edmonton, AB: Association for Computational Linguistics; 2003. pp. 173–80. [Google Scholar]
- 30.Van Rijsbergen Cornelis J, Edward Robertson Stephen, Porter Martin F. New models in probabilistic information retrieval. Cambridge: Computer Laboratory, University of Cambridge; 1980. [Google Scholar]
- 31.Manning Christopher D, Mihai Surdeanu, John Bauer, Jenny Finkel, Bethard Steven J, David McClosky. In: Philip Resnik, Rebecca Resnik, Margaret Mitchell., editors. The Stanford CoreNLP natural language processing toolkit; Proceedings of 52nd annual meeting of the association for computational linguistics: system demonstrations; Baltimore, MD: Association for Computational Linguistics; 2014. pp. 55–60. [Google Scholar]
- 32.Chapman Wendy W, Will Bridewell, Paul Hanbury, Cooper Gregory F, Buchanan Bruce G. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inf. 2001;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 33.Rose Finkel Jenny, Trond Grenager, Christopher Manning. In: Kareem Darwish, Mona Diab, Nizar Habash., editors. Incorporating nonlocal information into information extraction systems by Gibbs sampling; Proceedings of the 43rd annual meeting on association for computational linguistics; Ann Arbor, MI: Association for Computational Linguistics; 2005. pp. 363–70. [Google Scholar]
- 34.Adwait Ratnaparkhi. A maximum entropy model for part-of-speech tagging. In: Eric Brill, Kenneth Church., editors. Proceedings of the conference on empirical methods in natural language processing. Vol. 1. Philadelphia, PA: Association for Computational Linguistics; 1996. pp. 133–42. [Google Scholar]
- 35.Kristina Toutanova, Manning Christopher D. In: Vijay-Shanker K, Chang-Ning Huang, editors. Enriching the knowledge sources used in a maximum entropy part-of-speech tagger; Proceedings of the 2000 Joint SIGDAT conference on Empirical methods in natural language processing and very large corpora: held in conjunction with the 38th annual meeting of the association for computational linguistics; Hong Kong, China: Association for Computational Linguistics; 2000. pp. 63–70. [Google Scholar]
- 36.John Lafferty, Andrew McCallum, Pereira Fernando CN. In: Brodley CE, Danyluk AP, editors. Conditional random fields: Probabilistic models for segmenting and labeling sequence data; Proceedings of the 18th international conference on machine learning; Williamstown, MA: Morgan Kaufmann Publishers; 2001. [Google Scholar]
- 37.Charles Sutton, Andrew McCallum. An introduction to conditional random fields for relational learning. In: Getoor L, Taskar B, editors. Book chapter in Introduction to statistical relational learning. MIT Press; 2006. pp. 93–128. [Google Scholar]
- 38.Charles Sutton, Andrew McCallum. An introduction to conditional random fields. 2010. (arXiv preprint arXiv:1011.4088) [Google Scholar]
- 39.Student. The probable error of a mean. Biometrika. 1908;6(1):1–25. [Google Scholar]
- 40.Tomas Mikolov, Ilya Sutskever, Kai Chen, Corrado Greg S, Dean Jeff. In: Burges Christopher JC, Leon Bottou, Zoubin Ghahramani, Weinberger Kilian Q, editors. Distributed representations of words and phrases and their compositionality; Proceedings of 27th Annual Conference on Neural Information Processing Systems (NIPS); Lake Tahoe, NV: Advances in Neural Information Processing Systems; 2013. pp. 3111–9. [Google Scholar]
- 41.Pezzullo John A, Tung Glenn A, Rogg Jeffrey M, Davis Lawrence M, Brody Jeffrey M, Mayo-Smith William W. Voice recognition dictation: radiologist as transcriptionist. J Digit Imaging. 2008;21(4):384–9. doi: 10.1007/s10278-007-9039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]