Abstract
The four gene products of the accessory gene regulator (agr) P2 operon of Staphylococcus aureus assemble a quorum-sensing system: AgrA and AgrC resemble a two-component signal transduction system, and AgrB and AgrD are required to produce an autoinducing peptide. Upon activation, this quorum-sensing system positively regulates the transcription of the P2 operon as well as the P3 operon, whose transcript, RNAIII, regulates the expression of virulence genes. Four groups of S. aureus have been identified based on the agr sequences and the group-specific interaction between the autoinducing peptide and AgrC. AgrB is a transmembrane protein involved in the processing of AgrD propeptide, and its interaction with AgrD is also group specific. In this study, a series of chimeric AgrBs were constructed by swapping between group I and group II AgrBs, and these mutants were used to analyze the group-specific segment(s) in AgrB that was responsible for AgrD processing. Our results revealed that the first transmembrane α-helix and the extracellular loop 1 of group I AgrB were decisive in the specific processing of group I AgrD. In contrast, two hydrophilic segments of group II AgrB played a crucial role in the group-specific processing of group II AgrD. We also found that several chimeric AgrBs were capable of processing AgrD from both groups, suggesting that all AgrB homologues may utilize the same or a similar mechanism in the processing of AgrDs.
The coordinate expression of Staphylococcus aureus virulence factors is primarily controlled by an accessory gene regulator (agr) (4, 10). This locus consists of two transcripts: RNAII and RNAIII, which are transcribed divergently from P2 and P3 operons, respectively. RNAIII is the actual effector of the Agr response, i.e., it positively regulates the expression of secreted proteins and negatively regulates cell surface proteins. The four proteins, AgrA, -B, -C, and -D, encoded by the P2 operon assemble a quorum-sensing system regulating the transcription of RNAIII as well as the expression of Agr proteins (10, 12). Among the four Agr proteins, AgrC is a sensor kinase of a bacterial two-component signal transduction pathway with an N-terminal transmembrane domain and a C-terminal histidine kinase domain that is phosphorylated upon ligand binding (5). AgrA resembles a response regulator, although its dynamic phosphorylation and DNA binding activity have yet to be confirmed (1, 13). AgrD is the precursor for an autoinducing peptide (AIP) that is secreted into the culture supernatant and functions as a ligand for AgrC (4-6). AgrD is a membrane protein, with its N-terminal portion anchored in the membrane via an amphipathic helix that is required for its function and its stability (19). AgrB is required for the production of the mature AIP (3, 4). This transmembrane protein contains six transmembrane segments, four of which are hydrophobic while the other two are hydrophilic with several highly positively charged amino acid residues, and is involved in the proteolytic cleavage of AgrD propeptide (18). However, the detailed mechanisms of how AgrD propeptide is processed to form the mature AIP and how the mature AIP is secreted remain largely obscure.
The interaction between the mature AIP and AgrC is group specific (3, 6). Based on the agr sequences and the specific recognition between the AIP and AgrC, four groups of S. aureus have been identified so far (2, 3, 9). The mature AIP activates the agr responses in strains that belong to the same group, while it inhibits those in strains from heterologous groups (2, 3, 7-9). However, conflicting results have been reported on the activities of group IV AIP. Lyon et al. (7) showed that synthetic group I AIP could weakly activate the agr response in group IV strain and group IV AIP could strongly activate that in group I strains. In contrast, McDowell et al. (9) reported that synthetic group IV AIP activated the agr response in group IV strains but inhibited that in group I strains. The AIPs of S. aureus consist of seven to nine amino acid residues, and five of the residues form a thiolactone macrocycle by a thioester bond between a conserved cysteine and the carboxyl group of the C-terminal residue (2-4, 7-9).
Similarly, the interaction between AgrB and AgrD is group specific. Studies of AIP production from S. aureus strains containing either agrB or agrD or both genes from different groups show that group I AgrB (AgrB1) processes group I AgrD (AgrD1) and group III AgrD (AgrD3) but not group II AgrD (AgrD2) and vice versa (3). In this study, we made a series of chimeric proteins by swapping between AgrB1 and group II AgrB (AgrB2), and we used these mutant proteins to identify the AgrB segment(s) responsible for group-specific processing of AgrD in S. aureus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. aureus strains and plasmids used in this study are listed in Table 1. S. aureus cells grown overnight at 37°C on GL plates (11) were used to inoculate liquid cultures. CY-GP broth (11) was used for routine culture of S. aureus, supplemented with chloramphenicol (5 μg/ml), erythromycin (5 μg/ml), or both when necessary. Cell growth was monitored with a Klett-Summerson colorimeter with a green (540-nm) filter (Klett, Long Island City, N.Y.) except in the AIP activity assay, where cells were cultured in a VERSAmax microplate reader (Molecular Devices) and monitored at an optical density at 650 nm.
TABLE 1.
S. aureus plasmids and strains used in this study
| Plasmid or strain | Genotype and description | Reference |
|---|---|---|
| Plasmids | ||
| pRN5548 | Vector carrying a staphylococcal inducible PblaZ promoter | 14 |
| pRN6441 | Cloning vector | 14 |
| pRN6683 | agr P3-blaZ fusion | 4 |
| pRN6958 | agrD2 in pRN5548 | 3 |
| pGJ2001 | agrB1 in pRN5548ΔPstI | 18 |
| pGJ2002 | agrB2 in pRN5548ΔPstI | This study |
| pGJ4002 | agrD1 in pRN6441 | 18 |
| pLZ2004 | agrB1-6xhis in pRN5548 | 18 |
| pLZ2005 | agrB2-6xhis in pRN5548 | This study |
| pLZ2006 | Chimeric agrB (1P2) in pRN5548 | This study |
| pLZ2007 | Chimeric agrB (1H2P1) in pRN5548 | This study |
| pLZ2008 | Chimeric agrB (1H2) in pRN5548 | This study |
| pLZ2009 | Chimeric agrB (2P1) in pRN5548 | This study |
| pLZ2010 | Chimeric agrB (2H1P2) in pRN5548 | This study |
| pLZ2011 | Chimeric agrB (2H1) in pRN5548 | This study |
| pLZ2012 | Chimeric agrB (1C2) in pRN5548 | This study |
| pLZ2013 | Chimeric agrB (1D2) in pRN5548 | This study |
| pLZ2014 | Chimeric agrB (1E2) in pRN5548 | This study |
| pLZ2015 | Chimeric agrB (1F2) in pRN5548 | This study |
| pLZ2016 | Chimeric agrB (1G2) in pRN5548 | This study |
| pLZ2017 | Chimeric agrB (2C1) in pRN5548 | This study |
| pLZ2018 | Chimeric agrB (2F1) in pRN5548 | This study |
| pLZ2019 | Chimeric agrB (2C1F2) in pRN5548 | This study |
| pLZ2020 | Chimeric agrB (1C2F1) in pRN5548 | This study |
| pLZ2021 | Chimeric agrB (1Q2) in pRN5548 | This study |
| pLZ2022 | Chimeric agrB (2Q1) in pRN5548 | This study |
| pLZ2023 | Chimeric agrB (2H1P2Q1) in pRN5548 | This study |
| pLZ2024 | Chimeric agrB (2F1Q2) in pRN5548 | This study |
| pLZ2025 | Chimeric agrB (2F1H2) in pRN5548 | This study |
| pLZ2026 | Chimeric agrB (2F1Q2) in pRN5548 | This study |
| pLZ2027 | Chimeric agrB (2F1H2P1Q2) in pRN5548 | This study |
| pLZ2028 | Chimeric agrB (1F2H1) in pRN5548 | This study |
| pLZ2029 | Chimeric agrB (1P2Q1) in pRN5548 | This study |
| pLZ2030 | Chimeric agrB (1F2H1P2Q1) in pRN5548 | This study |
| pLZ4001 | agrD2 in pRN6441 | This study |
| Strains | ||
| RN6390B | Group I agr+ strain | 14 |
| SA502A | Group II agr+ strain, ATCC27217 | 3 |
| RN6911 | RN6390B agr 1057-4546::tetM | 14 |
| GJ2035 | RN6911(pI524) | 18 |
| GJ2001 | GJ2035(pGJ2001) | 18 |
| LZ4001 | GJ2035(pLZ4001) | This study |
Construction of plasmids.
All S. aureus plasmids used in this study were based on either pRN5548 (expression vector carrying a staphylococcal inducible PblaZ promoter) or pRN6441 (cloning vector) (13) (Table 1). Plasmid pGJ2002 was constructed by digesting pRN6956 (3) with SphI and SalI, blunting with DNA polymerase I large fragment (Klenow), and ligating. To construct the C-terminal six-histidine-tagged (His6-tagged) AgrB2-expressing plasmid pLZ2005, a PCR product with oligonucleotides GJ#77 (5′-TTAAGTGTATTTTTGTTTACCT-3′ in agrB2) and LZ#3 (5′-ATTAATGATGATGATGATGATGATCCTCCTTAGGGAA-3′ [His6 codons and a stop codon are underlined, and the agrB2 sequence is italicized]) as primers and pGJ2002 as template, was generated. The PCR product was digested with HgiAI and cloned into the HgiAI and BspHI (blunted with Klenow) sites of pGJ2002. pLZ4001 carrying agrD2 was constructed by cloning a ClaI fragment of pRN6958 and pRN6963 (3) into the ClaI site of pRN6441.
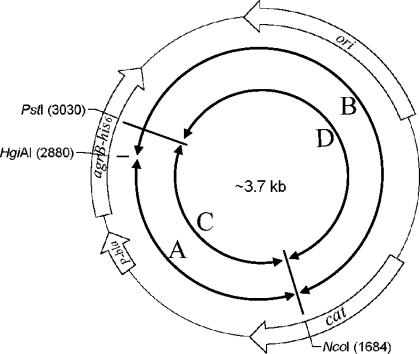
Two methods were used to construct plasmids carrying chimeric agrB-his6 genes. (i) Restriction endonuclease digestion method: plasmid carrying the agrB1-his6 or agrB2-his6 gene or a derived plasmid containing a chimeric agrB-his6 gene was digested with NcoI (in the chloramphenicol acetyltransferase gene of pRN5548) and HgiAI or PstI (both sites are in the same locations of both agrB1 and agrB2 genes). The DNA fragments were purified from agarose gels. Appropriate combinations of DNA fragments were then ligated (Fig. 1). Expression plasmids pLZ2006 to pLZ2011 and pLZ2024 to pLZ2030 were made by this method (Table 2). (ii) PCR-based method: PCR products were generated with Pfu Turbo DNA polymerase (Stratagene) using oligonucleotide GJ#44 or GJ#45 (containing the NcoI site in the chloramphenicol acetyltransferase gene of pRN5548) and oligonucleotide with sequence within either the agrB1 or agrB2 gene beginning from the expected switch point as primers (oligonucleotide primers used in this study are listed in Table 3) and plasmid carrying either the agrB1-his6 or agrB2-his6 gene or a derived plasmid containing a chimeric agrB-his6 gene as templates. Oligonucleotide primers were 5′ phosphorylated with T4 polynucleotide kinase before use in these PCRs. The DNA fragments were digested with NcoI and purified from agarose gels. Appropriate combinations of DNA fragments were then ligated to generate plasmids pLZ2012 to pLZ2023 (Table 2). All the constructs made were transformed into S. aureus GJ2035 (18) by the protoplast method (11). The sequences of chimeric agrB-his6 genes were confirmed by DNA sequencing.
FIG. 1.
Schematic presentation of plasmid carrying WT or chimeric agrB-his6. DNA fragments (A, B, C, and D) generated by restriction enzyme digestion were used to construct new chimeric agrB genes.
TABLE 2.
Construction of chimeric AgrB expression plasmidsa
| Method and plasmid | Chimeric agrB | Fragment 1
|
Fragment 2
|
||
|---|---|---|---|---|---|
| Template | Primersb | Template | Primersb | ||
| Enzymatic digestion-based subcloning | |||||
| pLZ2006 | 1P2 | pLZ2004-C | pLZ2005-D | ||
| pLZ2007 | 1H2P1 | pLZ2008-C | pLZ2004-D | ||
| pLZ2008 | 1H2 | pLZ2004-A | pLZ2005-B | ||
| pLZ2009 | 2P1 | pLZ2005-C | pLZ2004-D | ||
| pLZ2010 | 2H1P2 | pLZ2005-A | pLZ2006-B | ||
| pLZ2011 | 2H1 | pLZ2005-A | pLZ2004-B | ||
| pLZ2024 | 2F1Q2 | pLZ2018-C | pLZ2021-D | ||
| pLZ2025 | 2F1H2 | pLZ2018-A | pLZ2005-B | ||
| pLZ2026 | 2F1Q2 | pLZ2005-C | pLZ2021-D | ||
| pLZ2027 | 2F1H2P1Q2 | pLZ2018-A | pLZ2026-B | ||
| pLZ2028 | 1F2H1 | pLZ2015-A | pLZ2004-B | ||
| pLZ2029 | 1P2Q1 | pLZ2004-C | pLZ2022-D | ||
| pLZ2030 | 1F2H1P2Q1 | pLZ2015-A | pLZ2023-B | ||
| PCR-based subcloning | |||||
| pLZ2012 | 1C2 | pLZ2004 | GJ#76/GJ#44 | pLZ2005 | GJ#78/GJ#45 |
| pLZ2013 | 1D2 | pLZ2004 | GJ#75/GJ#44 | pLZ2005 | GJ#77/GJ#45 |
| pLZ2014 | 1E2 | pLZ2004 | GJ#29/GJ#44 | pLZ2005 | GJ#49/GJ#45 |
| pLZ2015 | 1F2 | pLZ2004 | LZ#4/GJ#44 | pLZ2005 | LZ#5/GJ#45 |
| pLZ2016 | 1G2 | pLZ2004 | LZ#6/GJ#44 | pLZ2005 | LZ#7/GJ#45 |
| pLZ2017 | 2C1 | pLZ2005 | LZ#36/GJ#44 | pLZ2004 | LZ#35/GJ#45 |
| pLZ2018 | 2F1 | pLZ2005 | LZ#34/GJ#44 | pLZ2004 | LZ#33/GJ#45 |
| pLZ2019 | 2C1F2 | pLZ2005 | LZ#36/GJ#44 | pLZ2015 | LZ#35/GJ#45 |
| pLZ2020 | 1C2F1 | pLZ2012 | LZ#34/GJ#44 | pLZ2004 | LZ#33/GJ#45 |
| pLZ1021 | 1Q2 | pLZ2004 | LZ#27/GJ#44 | pLZ2005 | LZ#26/GJ#45 |
| pLZ2022 | 2Q1 | pLZ2005 | LZ#38/GJ#44 | pLZ2004 | LZ#39/GJ#45 |
| pLZ2023 | 2H1P2Q1 | pLZ2010 | LZ#38/GJ#44 | pLZ2004 | LZ#39/GJ#45 |
TABLE 3.
Oligonucleotides used in this study
| Primer | Nucleotide sequence (5′→3′) | Location |
|---|---|---|
| GJ#29 | CGTAATTAACGTAAACAG | agrB1 (1944-1961)a |
| GJ#44 | CTATTATTCCATGGACTTCATTTAC | Around the NcoI site of pRN5548 |
| GJ#45 | GTAAATGAAGTCCATGGAATAATAG | Complementary to GJ#44 |
| GJ#49 | CACTTATCCTATATGTTAATACG | agrB2 (187-209)b |
| GJ#75 | AATATAGGCAATAGTATACAT | agrB1 (1911-1931)a |
| GJ#76 | TTTACCTATATTTTTAGCTAAG | agrB1 (1880-1901)a |
| GJ#78 | ATACTTGTAACATATAGTATTTC | agrB2 (127-149)b |
| LZ#4 | AAATGTTAAATTCGTAATTAAC | agrB1 (1952-1973)a |
| LZ#5 | ATGTTAATACGCTATAATG | agrB2 (199-223)b |
| LZ#6 | TCTTCTTATTAAATAAAATGTT | agrB1 (1869-1888)a |
| LZ#7 | AATGCACATGGTGCT | agrB2 (214-228)b |
| LZ#26 | AAGTATTTATCTATAATTATGTAT | agrB2 (424-447)b |
| LZ#27 | TTTTCGTTTAATAAGTCGCA | agrB1 (2179-2198)a |
| LZ#33 | TATTTAATAAGAAGACATGCAC | agrB1 (1974-1995)a |
| LZ#34 | ATAGGATAAGTGTGTAACT | agrB2 (180-198)b |
| LZ#35 | TTAATTGTTATGTATACTATTGCCT | agrB1 (1902-1926)a |
| LZ#36 | TTTAAAAAAATTACCAACTATAATTTGC | agrB2 (99-126)b |
| LZ#38 | TTTACGTTTTACTAATTTTATTGGT | agrB2 (399-423)b |
| LZ#39 | AAATATTATGCGATTATTGTTAGT | agrB1 (2199-2222)a |
Expression of chimeric AgrBs and WT AgrDs in S. aureus.
Plasmid carrying either the agrB1-his6 or agrB2-his6 gene or a chimeric agrB-his6 gene was transformed into S. aureus strain GJ4002 or LZ4001 by electroporation (17). These strains were grown to 75 Klett units in CY-GP broth (11) and induced with 0.5 μg of methicillin/ml for 5 h to express His6-tagged wild-type (WT) or chimeric AgrB and WT AgrD.
AIP activity assays.
AIPs produced by induced S. aureus strains were assayed by measuring the β-lactamase activity of the reporter strains [RN6390B(pRN6683) for group I and SA502A(pRN6683) for group II] according to the method described previously (3).
Whole-cell lysate preparation and Western blot hybridization.
S. aureus cells were suspended in 1× sucrose-sodium maleate-MgCl2 (11) containing 10 μg of lysostaphin/ml and incubated for 30 min at 37°C. The protoplasts were then pelleted and lysed by addition of phosphate-buffered saline (16) supplemented with 1 mM phenylmethanesulfonyl fluoride and protease inhibitor (protease inhibitor cocktail set II; Calbiochem). The cell lysates were briefly sonicated, and the unlysed cells and cell debris were removed by centrifugation at 7,000 × g for 10 min at 4°C. Total protein concentration of the supernatant was measured with the Bio-Rad protein assay dye reagent and the optical density at 595 nm (Bio-Rad Laboratories). Whole-cell lysates in sodium dodecyl sulfate (SDS) sample buffer (16) were incubated at 70°C for 10 min before loading onto SDS-polyacrylamide gel electrophoresis gels (16). The separated proteins were electrophoretically transferred to Hybond-P polyvinylidene difluoride membranes (Amersham Pharmacia Biotech). After blocking overnight at 4°C in Tris-buffered saline (16) plus 0.05% Tween 20 and 3% bovine albumin (fraction V; Sigma), the membranes were incubated in the blocking buffer with the primary antibody (1:2,000 dilution of mouse anti-tetraHis monoclonal antibody; QIAGEN) for 1 h at room temperature. The blots were washed extensively and probed with horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Amersham Pharmacia Biotech). The immunoblots were detected with an ECL Plus Western blotting detection system (Amersham Pharmacia Biotech) according to the manufacturer's instructions, followed by exposure to a HyperfilmECL film (Amersham Pharmacia Biotech).
RESULTS
Construction and expression of chimeric AgrBs in S. aureus.
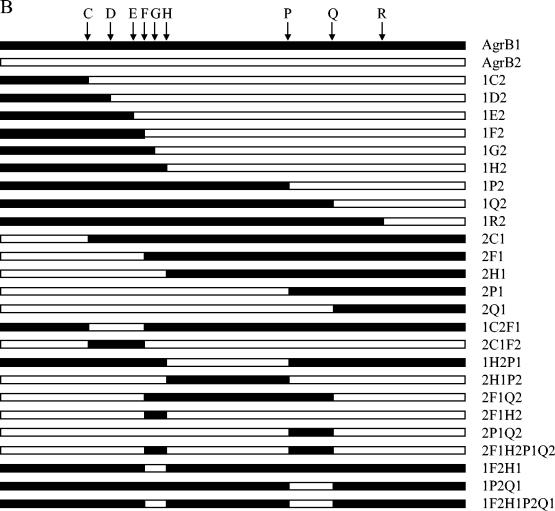
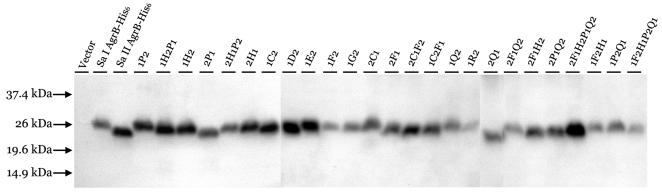
Since the interaction between AgrB and AgrD is group specific, we reasoned that there would be a segment(s) within AgrB that was responsible for the group specificity. To identify this segment(s), we made a series of chimeric agrB genes by swapping between agrB1 and agrB2 using either enzymatic digestion-based subcloning or PCR-based subcloning methods. A total of 25 chimeric agrB genes were constructed. The expected chimeric AgrBs are shown in Fig. 2. Plasmid encoding the His6-tagged WT AgrB1 or AgrB2 or the chimeric AgrB was transformed into agr-null S. aureus strain GJ2035 (18). We noted that the His6-tagged either AgrB1 or AgrB2 processed its cognate AgrD to produce amounts of mature AIPs comparable to those of WT AgrBs (data not shown), suggesting that the addition of His6 residues at the C terminus of AgrB had no effects on its function. Western blot hybridization analysis with an anti-tetraHis monoclonal antibody as a probe revealed bands with estimated molecular masses between 22 and 26 kDa from whole-cell lysates of S. aureus cells carrying those agrB constructs (Fig. 3). No bands were observed in lysates from cells containing the cloning vector (Fig. 3). The sizes of the proteins detected were consistent with those of the predicted molecular masses of the His6-tagged AgrBs, and the amount of chimeric AgrB protein in each lane was comparable to that of the WT AgrB-His6 protein, except that 1F2, 1R2, 2F1Q2, and 1F2H1P2Q1 were expressed at lower amounts, suggesting that these four chimeric AgrBs were less stable than the WT and other chimeric AgrBs.
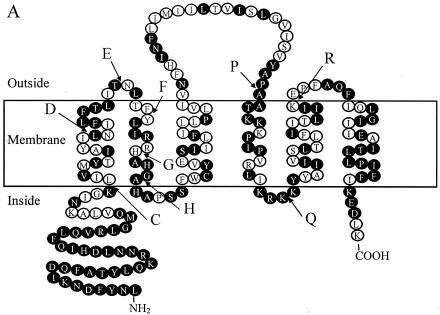
FIG. 2.
The chimeric AgrBs. (A) Transmembrane topological presentation of AgrB1 (NCBI accession no. CAA36781). Identical amino acid residues between AgrB1 and AgrB2 (NCBI accession no. AAB63264) are highlighted. The switch points between AgrB1 and AgrB2 are indicated. (B) Schematic presentations of WT and chimeric AgrBs. 1, AgrB1; 2, AgrB2. Swapping points are indicated by letters. For example, 1H2P1 means that the region between the N terminus and point H is from AgrB1, the region between point H and P is from AgrB2, and the region between point P and the C terminus is from AgrB1.
FIG. 3.
Western blot analysis of whole-cell lysates from induced cells expressing WT or chimeric AgrB-His6 proteins. The same amount of total proteins was loaded in each lane. The positions of the molecular mass markers are indicated to the left by arrows. The names of AgrBs expressed are indicated at the top of each lane.
The first transmembrane α-helix and the extracellular loop 1 of AgrB1 are decisive in the specific processing of AgrD1.
To identify the region(s) in AgrB1 responsible for its specific processing of AgrD1, we took advantage of the existence of two restriction endonuclease cleavage sites (HgiAI and PstI) that are in the same locations in agrB1 and agrB2 genes (Fig. 2A, points H and P in AgrB). Various DNA fragments were switched between these two agrB genes, and the resulting plasmids carrying chimeric agrB genes (1H2, 1P2, 2H1, 2P1, 1H2P1, or 2H1P2) were transformed into S. aureus strain GJ4002, which contains plasmid pGJ4002 carrying the WT agrD1 gene under the control of a staphylococcal Pbla promoter (18). The ability of these chimeric AgrBs to process the AgrD1 propeptide to generate mature AIP1 was then tested by measuring the AIP1 activities of the culture supernatants from induced cells using RN6390B(pRN6683) as a reporter strain (4). These results are shown in Fig. 4. As expected, no AIP1 was produced from cells expressing AgrD1 alone or with AgrB2. Cells expressing chimeric AgrBs (1H2, 1P2, and 1H2P1) containing the AgrB1 sequence from the N terminus to point H (amino acid residues 1 to 75) processed AgrD1 and produced amounts of mature AIP1 comparable to that of the positive control, whereas cells expressing chimeric AgrBs containing the same region from AgrB2 (2H1, 2P1, and 2H1P2) did not, a finding that indicated that the N-terminal 75 amino acid residues in AgrB1 determined the group specificity.
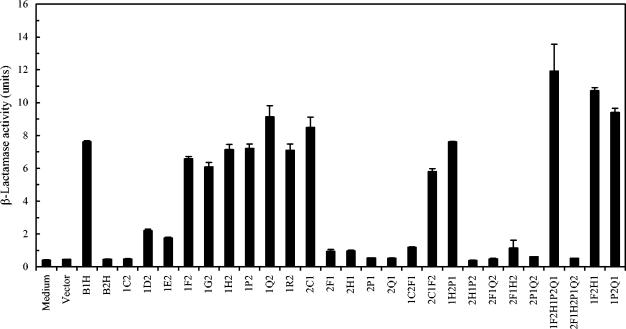
FIG. 4.
AIP1 activities produced from S. aureus strains coexpressing AgrD1 and the WT and chimeric AgrBs. Culture supernatants were prepared from S. aureus cells expressing WT or chimeric AgrBs, and AgrD1 and AIP1 activities were assayed by measuring the β-lactamase activity, using RN6390B(pRN6683) as the reporter strain as described previously (4). Values are means from three independent experiments with standard errors as indicated.
To further narrow down the segment(s) in AgrB1 that was responsible for its group specificity, we then made several new chimeric agrB genes using PCR-based methods. These new constructs (1C2, 1D2, 1E2, 1F2, 1G2, 2C1, 2F1, 1C2F1, and 2C1F2) were tested for their abilities to process AgrD1 and to produce mature AIP1. Cells expressing 1C2 did not generate any AIP1 activities (Fig. 4). Of note, the first 34 amino acid residues from the N terminus were conserved among all S. aureus strains (Fig. 2A), and the first switch point (point C) was designed at the beginning of the first transmembrane α-helix (leucine 43 according to our proposed AgrB topology model [18]). When more of the N-terminal 75 amino acid residues from AgrB1 were added, the chimeric AgrBs 1D2 and 1E2 showed low but detectable activities in producing AIP1. Cells expressing chimeric AgrB containing the region from the N terminus to point F of AgrB1 (1F2) restored activity to approximately 85% of WT AgrB1 activity, and the addition of more AgrB1 sequence (1G2) did not further increase activity. In contrast, replacing the region between the N terminus and point F of AgrB1 with the same region of AgrB2 (2F1) eliminated its activity, whereas the presence of AgrB2 sequence down to lysine 42 in AgrB1 had no effects on its activity, as seen in cells expressing 2C1 (Fig. 4). These results suggested that the region between point C and F in AgrB1 was required for its specific processing of AgrD1, which was further confirmed by our results showing that cells expressing 2C1F2 that embedded AgrB1 sequence only between C and F in AgrB2 produced AIP1 (approximately 80% of the WT AgrB), and cells expressing mutant 1C2F1 that embedded AgrB2 sequence between C and F in AgrB1 did not. Taking together, these results suggested that the region from lysine 42 to threonine 66 of AgrB1 that formed the first transmembrane α-helix and the extracellular loop 1 (18) is essential for the group-specific interaction between AgrB1 and AgrD1.
Other chimeric AgrBs constructed in this study were also tested for their abilities to process AgrD1 to generate mature AIP1. As shown in Fig. 4, only those that contained the AgrB1 C-to-F region (1F2H1P2Q1, 1F2H1, and 1P2Q1) showed AIP1 activities, whereas those that did not contain the C-to-F region (2F1Q2, 2F1H2, 2P1Q2, and 2F1H2P1Q2) did not, further confirming that the region from lysine 42 to threonine 66 of AgrB1 is required for its specific interaction with AgrD1.
The two hydrophilic segments in AgrB2 are essential for the specific processing of AgrD2.
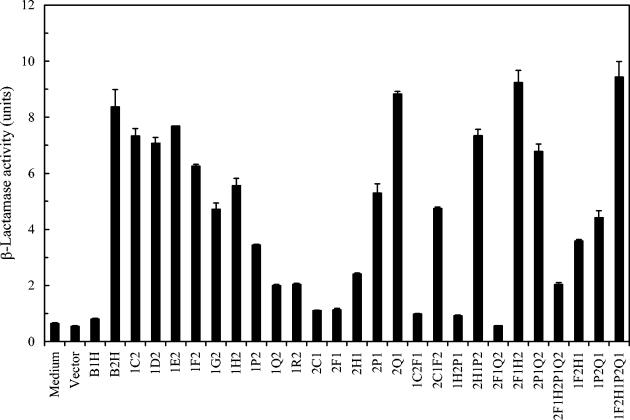
The same set of chimeric AgrBs used for the determination of AgrB1 on its specific interaction with AgrD1 were also coexpressed with WT AgrD2. The abilities of these chimeric AgrBs to generate mature AIP2 were assayed in an attempt to determine the region(s) in AgrB2 that is involved in the specific interaction with AgrD2. As shown in Fig. 5, cells with His6-tagged AgrB1 expressed together with AgrD2 did not produce AIP2 activity, and cells expressing His6-tagged AgrB2 were normally functional to produce AIP2 from AgrD2. Replacing the region between the N terminus and point C of AgrB2 by the same region of AgrB1 (1C2) had no significant effects on AgrB2 activity. Progressively adding AgrB1 sequences to AgrB2 (1D2, 1E2, 1F2, 1G2, 1H2, and 1P2) gradually reduced AgrB2 activities. When two newly constructed chimeric AgrBs (1Q2 and 1R2) were tested, the AgrB2 activities were further reduced to less than 20% of the WT AgrB2. Similarly, replacing the region between the N terminus and point C (2C1) or between the N terminus and point H (2F1) of AgrB1 with the same region of AgrB2 did not restore any AgrB2 activity. However, when more AgrB2 sequence was added to AgrB1 (2H1), low but detectable AgrB2 activity was observed. When the AgB1 regions were between the N terminus and point P and between the N terminus and point Q, these two chimeric AgrBs (2P1 and newly constructed 2Q1) had approximately 60 and 100% AgrB2 activity, respectively. These results suggested that the region between point C and point Q in AgrB2 was responsible for AgrB2 group specificity.
FIG. 5.
AIP2 activities produced from S. aureus strains coexpressing AgrD2 and the WT and chimeric AgrBs. AIP2 activities were assayed by measuring the β-lactamase activity with SA502A(pRN6683) as the reporter strain as described previously (3). Values are means from three independent experiments with standard errors as indicated.
Cells coexpressing AgrD2 and 1C2F1 or 1H2P1 produced no detectable AIP2 activity. In contrast, 2C1F2 and 2H1P2 retained approximately 55 and 90% of WT AgrB2 activity, respectively, suggesting that the region between point C and F but not between H and P in AgrB2 had a certain effect on AgrB2 group specificity. When seven more chimeric AgrBs were constructed and analyzed, interesting results were obtained. First, cells expressing 2F1Q2 produced no AIP2 activity, indicating that the two regions between point F and H and between P and Q were required for AgrB2 to process AgrD2 and to generate mature AIP2. Second, chimeric AgrBs, 2F1H2 and 2P1Q2 (replacement of the F-to-H region and P-to-Q region of AgrB2 by AgrB1, respectively) had no significant effects on AgrB2 activities. However, when both regions in AgrB2 were replaced by AgrB1 sequences, the resulting chimeric AgrB (2F1H2P1Q2) totally lost its AgrB2 activity. Third, replacing the F-to-H or P-to-Q region alone in AgrB1 with the same region of AgrB2 (1F2H1 or 1P2Q1) retained approximately 40 to 50% of WT AgrB2 activity, and the replacement of both regions in AgrB1 (1F2H1P2Q1) totally restored its AgrB2 activity. Taking together, these results indicated that the joint action of the two hydrophilic regions (F to H and P to Q) were essential for AgrB2 to process AgrD2 to generate mature AIP2.
Chimeric AgrBs that can process both AgrD1 and AgrD2 or none.
On comparison of the activities of the chimeric AgrBs we constructed and analyzed, we observed that eight chimeric AgrBs (1F2, 1G2, 1H2, 1P2, 2C1F2, 1F2H1P2Q1, 1F2H1, and 1P2Q1) (Fig. 4 and 5) were able to process both AgrD1 and AgrD2 to generate their corresponding AIPs. Among them, four chimeric AgrBs (1Q2, 1F2H1P2Q1, 1F2H1, and 1P2Q1) had higher AgrB1 activities in the processing of AgrD1 to generate mature AIP1 (Fig. 4). We also observed that four chimeric AgrBs (2F1, 2H1, 2C1F2, and 2F1Q2) had either no detectable or significant activities in the processing of both AgrD1 and AgrD2 (Fig. 4 and 5).
DISCUSSION
Four groups of S. aureus strains have been classified according to their Agr sequences and the specific interactions between the AgrC proteins and AIPs. Studies on AgrB and AgrD show that the interaction between them is also group specific. In this study, we constructed chimeric AgrBs with sequences from AgrB1 and AgrB2 swapped at various switch points to identify the AgrB segment(s) that is responsible for the group-specific interaction. Computer analysis has shown that both AgrB1 and AgrB2 have similar hydrophobicity profiles and are likely to have similar membrane topologies (18). Because the overall sequences were not changed in the chimeric AgrBs, the secondary structure of AgrB would not be expected to have significant changes. Our results clearly showed that the region between lysine 42 and threonine 66 (the first transmembrane α-helix and the extracellular loop 1) of AgrB1 was the most crucial segment in determining the specific processing of AgrD1. However, the same segment in AgrB2 was not important in its group-specific interaction with AgrD2. Instead, the two hydrophilic segments (methionine 67 to glycine 75 and alanine 126 to lysine 141) in AgrB2 were required for its specific processing of AgrD2.
The AgrD1 and AgrD2 sequences and their corresponding AIPs are different. (An alignment of these AgrDs is shown below [AgrD1, NCBI accession no. CAA36782; AgrD2, NCBI accession no. AAB63265]. The AIP sequences are shown in bold.)
AgrD1, MNTLFNLFFDFITGILKNIGNIAAYSTCDFIMDEVEVPKELTQLHE
AgrD2, MNTLVNMFFDFIIKLAKAIGIVGGVNACSSLFDEPKVPAELTNLYDK
The processing of AgrD to generate mature AIP is a complicated process involving proteolytic cleavage at two sites, the formation of a thioester bond between the sulfhydryl group of the cysteine residues and the carboxyl group of the C-terminal amino acid residue of the AIP sequence and the secretion of the mature AIP. AgrB is a membrane protein with six transmembrane segments (18). It has been proposed that the four α-helices form a channel and the two hydrophilic segments are located inside (18). So, it is possible that the interactions between AgrB and AgrD occur in the intramembranous and/or juxtamembranous compartment. Although the segments responsible for the group-specific interaction in AgrB1 and AgrB2 were found to be different, the folding of AgrB may bring together the transmembrane segments; thus, the first transmembrane α-helix and the two hydrophilic segments may form a group-specific interactive domain.
AgrB has been proposed both as an endopeptidase that is involved in proteolytic cleavages of AgrD and as an oligopeptide transporter that secretes the mature AIP. The proteolytic reactions in AgrBs are likely to be conserved, whereas processes such as substrate binding and mature AIP secretion could be group specific. Recently, our laboratory has identified two amino acid residues (histidine 77 and cysteine 83) in both S. aureus AgrB1 (L. Zhang, R. Qiu, and G. Ji, unpublished data) and Staphylococcus intermedius AgrB (W. Pei and G. Ji, unpublished data) that are required for its activity. Most possibly these two amino acid residues form a catalytic center of a putative endopeptidase. These two amino acids are at the same positions and conserved among all AgrBs identified so far, and they are located juxtapositionally on the inner surface of the cytoplasmic membrane according to the AgrB1 topological model (Fig. 2A) (18). It is likely the catalytic activities are conserved among these AgrBs. The true nature of this putative endopeptidase is unclear. It is possible AgrB is a cysteine endopeptidase, since cysteine and histidine are the two most common conserved residues that form the catalytic centers of cysteine peptidases (15), though AgrBs have no sequence homologies with other cysteine peptidase and mutations in other conserved residues among AgrBs that have catalytic potential found in other types of proteases did not totally eliminate its activity (W. Pei, R. Qiu, L. Zhang, and G. Ji, unpublished data). Furthermore, when all the chimeric AgrB mutants were tested, eight were able to process AgrD propeptide from both groups to produce significant levels of AIP activities. Of note, the two hydrophilic segments were relatively conserved in both AgrBs, with three and four amino acid differences, respectively. It is interesting that chimeric AgrB 1F2H1P2Q1, which consisted of AgrB1 sequence with only seven amino acids from AgrB2, was able to process both AgrDs to produce comparable or even higher AIP activities than the WT AgrB. These results suggested that while the interaction between AgrB and AgrD is group specific, the mechanisms of the AgrD processing to generate AIP by the AgrB protein are the same or similar.
Acknowledgments
We thank Saibal Dey for helpful comments on the manuscript.
This work was supported by National Institutes of Health grant RO1AI46445 and a Uniformed Services University of Health Sciences grant to G.J.
REFERENCES
- 1.Janzon, L., S. Lofdahl, and S. Arvidson. 1989. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol. Gen. Genet. 219:480-485. [DOI] [PubMed] [Google Scholar]
- 2.Jarraud, S., G. J. Lyon, A. M. Figueiredo, L. Gerard, F. Vandenesch, J. Etienne, T. W. Muir, and R. P. Novick. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 4.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lina, G., S. Jarraud, G. Ji, T. Greenland, A. Pedraza, J. Etienne, R. P. Novick, and F. Vandenesch. 1998. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 6.Lyon, G. J., J. S. Wright, A. Christopoulos, R. P. Novick, and T. W. Muir. 2002. Reversible and specific extracellular antagonism of receptor-histidine kinase signaling. J. Biol. Chem. 277:6247-6253. [DOI] [PubMed] [Google Scholar]
- 7.Lyon, G. J., J. S. Wright, T. W. Muir, and R. P. Novick. 2002. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry 41:10095-10104. [DOI] [PubMed] [Google Scholar]
- 8.Mayville, P., G. Ji, R. Beavis, H. Yang, M. Goger, R. P. Novick, and T. W. Muir. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 96:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDowell, P., Z. Affas, C. Reynolds, M. T. Holden, S. J. Wood, S. Saint, A. Cockayne, P. J. Hill, C. E. Dodd, B. W. Bycroft, W. C. Chan, and P. Williams. 2001. Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol. Microbiol. 41:503-512. [DOI] [PubMed] [Google Scholar]
- 10.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 11.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 12.Novick, R. P. 1999. Regulation of pathogenicity in Staphylococcus aureus by a peptide-based density-sensing system, p. 129-145. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 13.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, F. Vandenesch, and S. Moghazeh. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446-458. [DOI] [PubMed] [Google Scholar]
- 14.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawlings, N. D., and A. J. Barrett. 1994. Families of cysteine peptidases. Methods Enzymol. 244:461-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, L., L. Gray, R. P. Novick, and G. Ji. 2002. Transmembrane topology of AgrB, the protein involved in the post-translational modification of AgrD in Staphylococcus aureus. J. Biol. Chem. 277:34736-34742. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, L., J. Lin, and G. Ji. 2004. Membrane anchoring of the AgrD N-terminal amphipathic region is required for its processing to produce a quorum sensing pheromone in Staphylococcus aureus. J. Biol. Chem. 279:19448-19456. [DOI] [PubMed] [Google Scholar]