Abstract
Mutants with defects in components of the glutathione-glutaredoxin (GSH/Grx) system of Rhodobacter capsulatus were constructed to study its role in defense against oxidative stress and the redox-dependent formation of photosynthetic complexes. The lack of the glutaredoxin 3 gene (grxC) or the glutathione synthetase B gene (gshB) resulted in lower growth rates under aerobic conditions and higher sensitivity to oxidative stress, confirming the role of the GSH/Grx system in oxidative stress defense. Both mutants are highly sensitive to disulfide stress, indicating a major contribution of the GSH/Grx system to the thiol-disulfide redox buffer in the cytoplasm. Like mutations in the thioredoxin system, mutations in the GSH/Grx system affected the formation of photosynthetic complexes, which is redox dependent in R. capsulatus. Expression of the genes grxC, gshB, grxA for glutaredoxin 1, and gorA for glutathione reductase, all encoding components of the GSH/Grx system, was not induced by oxidative stress. Other genes, for which a role in oxidative stress was established in Escherichia coli, acnA, fpr, fur, and katG, were strongly induced by oxidative stress in R. capsulatus. Mutations in the grxC, and/or gshB, and/or trxC (thioredoxin 2) genes affected expression of these genes, indicating an interplay of the different defense systems against oxidative stress. The OxyR and the SoxRS regulons control the expression of many genes involved in oxidative stress defense in E. coli in response to H2O2 and superoxide, respectively. Our data and the available genome sequence of R. capsulatus suggest that a SoxRS system is lacking but an alternative superoxide specific regulator exists in R. capsulatus. While the expression of gorA and grxA is regulated by H2O2 in E. coli this is not the case in R. capsulatus, indicating that the OxyR regulons of these two species are significantly different.
Two major redox systems are involved in maintaining a reduced environment in the Escherichia coli cytosol: the glutathione-glutaredoxin (GSH/Grx) system and the thioredoxin system (reviewed in references 8 and 15). The tripeptide glutathione (l-γ-glutamyl-l-cysteinylglycine) and the small protein thioredoxin (12 kDa) are reductants in many cellular reactions (14, 17, 29, 31, 34). GSH functions to reduce cellular disulfide bonds, often in conjunction with glutaredoxin proteins (8). The E. coli redox buffer in the cytoplasm is mostly composed of GSH. The intracellular concentration is approximately 5 mM, and it is kept almost completely reduced (1, 19). GSH reductase reduces glutathione disulfide, which is formed upon oxidation, at the expense of NADPH. NADPH is recycled by glucose-6-phosphate dehydrogenase. GSH is not essential for the survival of E. coli (10). E. coli contains three glutaredoxins, glutaredoxins 1 to 3 (encoded by the genes grxA,- B, and -C), and two thioredoxins, 1 and 2 (encoded by trxA and trxC). Most glutaredoxins, all thioredoxins, and thioredoxin reductases (encoded by trxB) contain the conserved motif CX1X2C in their active sites and share the thioredoxin fold composed of three helices and a four-stranded beta sheet.
The electron transfer through disulfide bond exchange reactions in the cytoplasm recycles enzymes, such as the essential ribonucleotide reductase (14), phosphoadenosine-phosphosulfate reductase (31), methionine sulfoxide reductase (6), and arsenate reductase (22). Changes in the thiol-disulfide redox status of proteins are also important for the control of protein function. Oxidation of critical cysteine residues can activate or inactivate a given protein. Thiol-redox reactions are involved in the regulation of photosynthetic enzymes by light (7) or in regulation of transcription factors such as NF-κB (32).
The increased production of reactive oxygen species during oxidative stress leads to the oxidation of protein thiols. Some proteins, like the transcriptional activator OxyR, are transiently activated by the formation of disulfide bonds under oxidative stress conditions (42). The expression of many of the H2O2-inducible genes is regulated by OxyR in E. coli (43). Members of the OxyR regulon with established functions in the oxidative stress response are katG (hydroperoxidase I), ahpCF (alkylhydroperoxide reductase), gorA (glutathione reductase), grxA (glutaredoxin 1), trxC (thioredoxin 2), fur (repressor of iron uptake), dps (unspecific DNA binding protein), oxyS (regulatory RNA), agn43 (protein of the outer membrane), and fhuF (ferric reductase). Several E. coli genes are regulated by the SoxRS system in response to treatment with superoxide-generating agents. These genes include sodA (Mn superoxide dismutase), nfo (endonuclease IV for DNA repair), zwf (glucose-6-phosphate dehydrogenase), tolC (outer membrane protein), fur, micF (regulatory RNA affecting ompF expression), acrAB (multidrug efflux pump), fumC (fumarase C), acnA (aconitase), nfsA (nitroreductase A), fpr (ferredoxin/flavodoxin reductase), fldA, fldB (flavodoxin), and ribA (GTP cyclohydrolase).
A number of E. coli mutant strains lacking components of the thioredoxin and GSH/Grx systems have been created and analyzed. Thioredoxin 1 (TrxA), thioredoxin 2 (TrxC), and thioredoxin reductase (TrxB) are not essential for survival. While E. coli trxA and trxB mutants show increased sensitivity to H2O2, this phenotype is not observed in trxC mutants (30, 37). E. coli strains with mutations in components of the GSH system are also viable. gshA, gshB (encoding the two enzymes for glutathione synthesis), and gorA (encoding glutathione reductase) mutants exhibit higher sensitivity to diamide, which causes direct oxidation of GSH (19). Under normal growth conditions these mutants show the wild-type phenotype, as also observed for grxA and grxC mutants (2). A study using E. coli strains with mutations in components of both the thioredoxin and the GSH/Grx system revealed that E. coli requires either of the two systems to grow well aerobically (29). It was demonstrated in the same study that mutants lacking components of the thioredoxin or the GSH/Grx system are defective in reducing cytoplasmic disulfide bonds.
Rhodobacter capsulatus is a facultatively photosynthetic bacterium. It can convert energy by aerobic respiration and forms only very low amounts of photosynthetic complexes at high oxygen tension. At low oxygen tension, a photosynthetic apparatus is synthesized which is used for ATP generation by cyclic photophosphorylation under anaerobic conditions. The ancestors of the modern family Rhodospirillaceae developed anoxygenic photosynthesis under the reducing atmosphere of early Earth. With the increase of oxygen in the atmosphere generated by the cyanobacterial photosynthetic complexes, they had to develop defense mechanisms for protection against harmful reactive oxygen species. The study of the defense systems against oxidative stress of Rhodospirillaceae will provide valuable information on the evolution of these systems.
R. capsulatus, like E. coli, harbors at least two thioredoxins, encoded by the trxA and trxC genes. The almost completed genome sequence (http://www.integratedgenomics.com/genomereleases.html#list1) also identifies genes forglutaredoxin 1 (grxA), glutaredoxin 3 (grxC), GSH synthetase (gshA and gshB), and glutathione reductase (gorA). In order to get first insights into the role of the GSH/Grx system of R. capsulatus, we inactivated the grxC and/or gshB gene or one of these genes in combination with trxC. The gshB mutation interrupts a basic reaction of the GSH/Grx system. The two glutaredoxins may have partially overlapping functions. Therefore, the grxC mutation was expected to affect the function of the GSH/Grx system less severely than the gshB mutation. Since we have previously shown a role of thioredoxin 2 in the oxidative stress response of R. capsulatus (20), we were interested in seeing how R. capsulatus can cope with defects in both the GSH/Grx and the thioredoxin systems. We characterized the mutants in regard to growth under different oxygen tension and the resistance to substances which generate oxidative stress. Furthermore, we analyzed how the lack of components of the GSH/Grx system affects the regulation of other genes with putative functions in the defense against oxidative stress.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. E. coli cultures were grown in Luria-Bertani broth at 37°C; R. capsulatus cultures were grown at 32°C in a malate minimal salt medium (9). The cells were grown aerobically by incubating 100 ml of culture in 500-ml baffled flasks under vigorous shaking. The dissolved oxygen concentration was determined to be 200 μM by using a Pt/Ag oxygen electrode (Bachofer). For semiaerobic growth conditions, 40 ml of cell culture was incubated in 50-ml flasks (dissolved oxygen concentration, 3 to 10 μM). Cells were grown in the light in screw-cap flasks filled to the top for anaerobic conditions. For oxygen shift experiments, cells were grown semiaerobically to an optical density at 660 nm (OD660) of 0.3 to 0.4 and then transferred to the 500-ml flasks for aerobic growth. When required, antibiotics were added to the growth medium at the following concentrations: kanamycin, 25 μg ml−1; and spectinomycin, 10 μg ml−1.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi (lac-proAB) F′[traD36 proAB+lac1qlacZΔM15] | Stratagene |
| SM10 | recA thi thr leu; chromosomal RP4-2 (Tc::Mu) | 33 |
| S17-1 | recA hsdR pro; chromosomal RP4-2 (Tc::Mu Km::Tn7) | 33 |
| R. capsulatus | ||
| SB1003 | Wild type | 41 |
| SB1003 trxC | SB1003 ΔtrxC::Kmr | 20 |
| SB grxC::Km | SB1003 ΔgrxC::Kmr | This study |
| SB grxC | SB1003 ΔgrxC::ΩSpr | This study |
| SB trxC grxC | SB1003 ΔtrxC::Kmr ΔgrxC::ΩSpr | This study |
| SB gshB::Km | SB1003 ΔgshB::Kmr | This study |
| SB gshB | SB1003 ΔgshB::ΩSpr | This study |
| SB trxC gshB | SB1003 ΔtrxC::Kmr ΔgshB::ΩSpr | This study |
| SB grxC gshB | SB1003 ΔgrxC::Kmr ΔgshB::ΩSpr | This study |
| SB grxC(pRKgrxC) | Plasmid containing cloned grxC gene in SB1003 ΔgrxC::ΩSpr | This study |
| SB gshB(pRKgshB) | Plasmid containing cloned gshB gene in SB1003 ΔgshB::ΩSpr | This study |
| Plasmids | ||
| pRK415 | A broad-host-range plasmid | 18 |
| pUTC51 | 1.9-kb Pst1 fragment containing the trxA gene of R. sphaeroides under the control of the ptrc promoter and the lacIq | 3 |
| pUC4KSAC | Apr, source of the Kmr cassette | 4 |
| pPHU281 | TcrlacZ′ mob(RP4) | 16 |
| pHP45Ω | pBR322 plasmid with a 2.0-kb DNA segment consisting of an antibiotic resistance gene (spectinomycin/streptomycin) flanked by short inverted repeats carrying transcription and translation termination signals | 27 |
| pPHΔRcgrxC | 810-bp fragment containing the upstream and downstream sequences of the R. capsulatus grxC stretch cloned into pPHU281 | This study |
| pPHΔRcgrxC::Km | 1.3-kb kanamycin cartridge cloned into pPHΔRcgrxC | This study |
| pPHΔRcgrxC::ΩSp | 2.2-kb ΩSp cartridge cloned into pPHΔRcgrxC | This study |
| pPHΔRcgshB | 546-bp fragment containing the upstream and downstream sequences of the R. capsulatus gshB stretch cloned into pPHU281 | This study |
| pPHΔRcgshB::Km | 1.3-kb kanamycin cartridge cloned into pPHΔRcgshB | This study |
| pPHΔRcgshB::ΩSp | 2.2-kb ΩSp cartridge cloned into pPHΔRcgshB | This study |
| pRKgrxC | 643-bp fragment containing the grxC gene and its putative promoter region in plasmid pRK415 | This study |
| pRKgshB | 1,417-bp fragment containing the gshB gene and its putative promoter region in plasmid pRK415 | This study |
Mutant construction.
In order to generate mutants, DNA fragments, one containing upstream sequences and part of the genes encoding the N-terminal amino acids (using primers P1 and P2 for grxC, P5 and P6 for gshB) and one containing part of the genes encoding the C-terminal amino acids and downstream sequences of the target genes (using primers P3 and P4 for grxC, P7 and P8 for gshB) were amplified by PCR with primers introducing suitable restriction sites for cloning. The sequences of the primers used for PCR are listed in Table 2. The PCR products were cloned into plasmid pPHU281 (16) to generate pPHUΔgrxC and pPHUΔgshB. The kanamycin or the Ω spectinomycin cartridge (4, 27) was inserted between the two fragments to get plasmids pPHUΔgrxC::Km, pPHUΔgrxC::ΩSp, pPHUΔgshB::Km, and pPHUΔgshB::ΩSp, respectively. These plasmids were transformed into E. coli strain SM10 or S17-1 (33) and diparentally conjugated into SB1003 (41) or SB trxC (20). The resulting colonies were screened for a double-crossover recombination event based on the antibiotic resistances. The correct insertions of the resistance cassettes in the isolated grxC and gshB single mutants, SB grxC and SB gshB, and double mutants SB trxC grxC, SB trxC gshB, and SB grxC gshB (Table 1) were confirmed by Southern analysis as described previously (20). For complementation, DNA fragments containing the putative promoter region and the whole gene length were amplified with primers P1 and P4 for grxC and primers P9 and P10 for gshB and cloned into pRK415 (18) to generate pRKgrxC and pRKgshB. The two plasmids were conjugated into the single mutant SB grxC or SB gshB to get SB grxC(pRKgrxC) and SB gshB(pRKgshB).
TABLE 2.
Primers used in this study
| Primer | Sequencea |
|---|---|
| For cloning | |
| P1 | 5′-GGAAGAATTCGTTGCGGCG-3′ |
| P2 | 5′-CCAAGGGCGGATCCTGGC-3′ |
| P3 | 5′-CGTCAAGGATCCGTTCTGGG-3′ |
| P4 | 5′-GCCCGCTGCAGGCACGG-3′ |
| P5 | 5′-ATGAGGATCCGCGGCACGGTAT-3′ |
| P6 | 5′-TGATCGACGACCTGCAGACCTC-3′ |
| P7 | 5′-GGAGAGCGTCGAATTCTCGT-3′ |
| P8 | 5′-TGATCGGATCCTGGCAGAGT-3′ |
| P9 | 5′-CTGCAGTTGCAGCCGAACATCAGGTG-3′ |
| P10 | 5′-GAATTCAGGTCTGCCGGATCGGTCAT-3′ |
| For RT-PCR | |
| RcacnA-RT>A | 5′-GCCGCGCTCAAGGTGGTGCT-3′ |
| RcacnA-RT>B | 5′-GGTCAACGACCGCCGGAACG-3′ |
| Rcfpr-RT>A | 5′-TGGTCGAGGCGCTGCAAGAG-3′ |
| Rcfpr-RT>B | 5′-GAGGCGAGGTTGTCGGTGAT-3′ |
| Rcfur-RT>A | 5′-ACGTGGACTGCGCTGGCTTG-3′ |
| Rcfur-RT>B | 5′-ACGGTGGCGAGCGAGACCTT-3′ |
| Rcgor-RT>A | 5′-ATGTGACCGACCGGATCAAC-3′ |
| Rcgor-RT>B | 5′-AATGTCAACCGCTTCGCTCA-3′ |
| RcgrxA-RT>A | 5′-ATGACCGCACTCGACCAGAT-3′ |
| RcgrxA-RT>B | 5′-CTTGTCGTCGAAGAGCTTGT-3′ |
| RcgshB-RT>A | 5′-CGTCGTCAACGATCCGTTCT-3′ |
| RcgshB-RT>B | 5′-AGGATGACATCGCCATGCTC-3′ |
| RckatG-RT>A | 5′-GCCTCGGTCGCCGATGTGAT-3′ |
| RckatG-RT>B | 5′-CACCGGCTCCAGCACGTCAA-3′ |
Bold letters within a primer indicate a restriction site.
RT-PCR.
Total RNA was isolated from R. capsulatus cultures as described previously (26). For reverse transcription-PCR (RT-PCR) analysis the reverse transcription reaction was carried out in a final volume of 25 μl containing 30 pmol of reverse primer, 2 μg of total RNA, a 1 mM concentration of each of four deoxyribonucleoside triphosphates, and 1.5 U of reverse transcriptase (Promega) at 42°C for 1 h and 99°C for 2 min to inactivate the reverse transcriptase. PCR was carried out in a final volume of 50 μl containing 5 μl of RT solution, 1 U of Taq polymerase (QIAGEN), and 2 pmol each of the oligonucleotide primers (Table 2). Amplification was carried out by an initial denaturation step at 96°C for 3 min followed by 20 to 28 cycles of 96°C for 1 min, 56 to 62°C for 40 s, and 72°C for 30 s. A sample lacking reverse transcriptase was included for each reaction as a control for DNA contamination. Reaction products were subjected to electrophoresis on a 10% polyacrylamide gel.
Determination of survival rates.
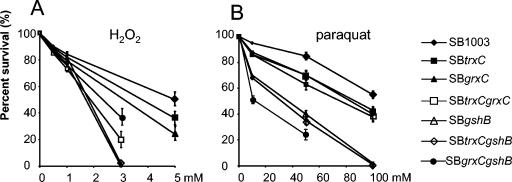
Cultures were grown under semiaerobic conditions until the OD660 reached 0.3 to 0.4; then agents which generate oxidative stress were added at the concentrations indicated in Fig. 1 for 1 h and dilutions were plated. Survival of 100% corresponds to the viable cell number determined immediately before the addition of the oxidative agents. The percentage of colonies grown from the treated cultures is shown as percent survival in Fig. 1. The mean values plus the standard deviations are given from three independent experiments. Hydrogen peroxide and methyl viologen (paraquat) were purchased from Sigma.
FIG. 1.
Survival rates of wild-type and mutant strains after treatment with different concentrations of H2O2 (A) or paraquat (B). The bars indicate the standard errors of the measurements from three independent experiments. Determination of survival rates is described in Materials and Methods. At concentrations where no colonies were detected, no value for the survival rate is given.
Spectral analysis.
In vivo absorption spectra of crude intracytoplasmic membranes were obtained by sonicating cells suspended in 10 mM Tris-HCl-1.0 mM EDTA-15 mM NaN3. Following clarification by centrifugation at 10 000 × g for 5 min, the soluble fraction was scanned from 500 to 900 nm with a Perkin-Elmer Lambda 12 spectrophotometer.
Zone of inhibition assays.
For zone of inhibition assays, cultures were grown semiaerobically overnight at 32°C and then diluted to an OD660 of 0.2. Cultures were then grown to an OD660 of 0.3 to 0.4 (midlogarithmic growth phase). Inhibition assays were carried out on plates as described previously (20). The diameter of the zone of growth inhibition indicated the sensitivity. All assays were performed three to four times in duplicate.
RESULTS AND DISCUSSION
To investigate the role of the GSH/Grx system in the defense against oxidative stress, we constructed mutants lacking GrxC and/or GshB or one of these components in combination with TrxC. In these constructs, the kanamycin or Ω spectinomycin resistance cartridge was used, respectively, to generate the single mutants SB grxC and SB gshB and the double mutants, SB trxC grxC, SB trxC gshB, and SB grxC gshB (Table 1).
While the kanamycin cartridge allows the transcription of downstream genes, the Ω spectinomycin cartridge harbors transcriptional terminators which block transcription of downstream genes. Both gshB and grxC genes are followed on the chromosome by genes which are transcribed in the same direction. The operonal organization of these genes is not known. Therefore, we had to consider polar effects on downstream genes by insertion of the Ω spectinomycin cartridge. The phenotypes of grxC and gshB mutants carrying either the kanamycin or the Ω spectinomycin cartridge did not differ significantly. Thus, the differences in phenotypes and expression patterns of the mutants compared to the parental strains can be attributed to the lack of expression of grxC or gshB, and we show results only for the mutants containing the Ω spectinomycin cartridge. Furthermore, introduction of the intact grxC or gshB gene on a plasmid into the mutants restored the wild-type phenotype, as shown below. The Ω spectinomycin cartridge was used to generate the double mutants, which have the trxC or grxC gene inactivated by insertion of the kanamycin cartridge.
Effect of mutations in the GSH/Grx system on the growth rate under different oxygen conditions.
To see how mutations in the GSH/Grx system affect the growth of R. capsulatus, we incubated cultures of the different strains either under high oxygen tension (200 μM dissolved oxygen) or anaerobically in the light. The doubling times are given in Table 3. All strains with mutations in the gshB and/or grxC gene showed reduced growth rates at high oxygen tension but no markedly different growth rates under anaerobic conditions when compared to the wild-type strain. Even lower growth rates than those under aerobic conditions were detected in the mutant strains when the cultures were shifted from low oxygen (3 to 10 μM dissolved oxygen) to high oxygen (200 μM dissolved oxygen) tension, while this transition had no significant effect on the growth of the wild type. These data strongly suggest that the GSH/Grx system is involved in defense against oxidative stress. The growth rate of the gshB mutant strain under aerobic conditions was significantly lower than that of the grxC mutant strain. Only one set of gshA gshB genes is present in R. capsulatus. Consequently the gshB mutant completely fails to synthesize GSH. Besides glutaredoxin 3 (encoded by grxC), R. capsulatus harbors glutaredoxin 1 (encoded by grxA). It is conceivable that the glutaredoxins of R. capsulatus have partially overlapping functions, as known for E. coli (2). As shown below, the gshB mutation also has stronger effects on the formation of photosynthetic complexes and on the sensitivity to oxidative stress compared to the grxC mutation.
TABLE 3.
Doubling times of growth for the wild type and grxC, gshB, trxC grxC, and trxC gshB mutants under aerobic and anaerobic conditions and with a shift from semiaerobic to aerobic conditions
| Straina | Doubling time (h) under conditionb
|
||
|---|---|---|---|
| Aerobic | Anaerobic | Shift from semiaerobic to aerobic | |
| SB1003 | 2.1 ± 0.15 | 12.2 ± 0.66 | 2.5 ± 0.16 |
| SB trxC | 2.2 ± 0.20 | 12.0 ± 0.70 | 2.6 ± 0.21 |
| SB grxC | 3.6 ± 0.3 | 12.0 ± 0.55 | 5.0 ± 0.33 |
| SB trxC grxC | 4.0 ± 0.35 | 12.8 ± 0.36 | 7.2 ± 0.25 |
| SB gshB | 6.0 ± 0.33 | 12.0 ± 0.86 | 10.0 ± 0.16 |
| SB trxC gshB | 6.8 ± 0.25 | 13.5 ± 0.55 | 11.0 ± 0.25 |
| SB grxC gshB | 6.2 ± 0.22 | 12.3 ± 0.54 | 10.5 ± 0.33 |
For strain characteristics, see Table 1.
Mean values ± standard deviations from three independent experiments.
The strain with the trxC gshB double mutation exhibited the lowest growth rates among all strains, and the trxC grxC, grxC gshB, and trxC gshB double mutants showed slightly lower growth rates than the grxC and gshB single mutants under aerobic conditions and after a shift from semiaerobic to aerobic conditions (Table 3). The trxC mutation alone does not significantly affect the growth of R. capsulatus. These results imply a coordinate action of the GSH/Grx and thioredoxin systems during aerobic growth or adaptation to an increase in oxygen tension. Some essential enzymes may strongly depend on the presence of the reductive pathway, the GSH/Grx system. One candidate for such an enzyme is aerobic ribonucleotide reductase, which requires the reduction of a disulfide bond to complete its catalytic cycle and which, in vitro, can use thioredoxin or glutaredoxins for this purpose (2). Since the anaerobic ribonucleotide reductase does not use the thioredoxin or GSH/Grx system (24), the ability of these mutants to grow well under anaerobic conditions is understandable.
Effect of mutations in the GSH/Grx system on oxygen-regulated formation of photosynthetic complexes.
The formation of photosynthetic complexes in Rhodobacter is under redox control. The two-component system RegB/RegA is a central regulator of this redox control pathway. However, other proteins are also involved in the redox-regulated expression of photosynthesis genes (reviewed in references 11 and 12). Among these proteins are the single thioredoxin of Rhodobacter sphaeroides, TrxA, and the TrxA and TrxC proteins of R. capsulatus. TrxA and TrxC affect the redox-dependent transcription of the puf and puc genes in opposite ways (20, 26). The puf and puc operons encode pigment binding proteins and other proteins required for the formation of photosynthetic complexes. To see whether mutations in the GSH/Grx system affect the formation of photosynthetic complexes in R. capsulatus, cell extracts from wild-type and mutant strains, which were grown under low oxygen tension to the identical optical density, were prepared. A spectral analysis of these cell extracts revealed marked differences in the amounts of photosynthetic complexes for the different strains (Fig. 2). The reaction center specifically absorbs at 803 and 875 nm. The light-harvesting (LH) I complex absorbs at 870 nm. The LHII complex, which is the dominant pigment protein complex under low oxygen, absorbs at 800 and 850 nm, covers the reaction center-specific absorbance, and overlaps with the LHI specific absorbance at 870 nm.
FIG. 2.
Absorption spectra of cell extracts of the wild-type strain, strains lacking thioredoxin 2 (trxC), glutaredoxin 3 (grxC), or glutathione synthetase B (gshB), and grxC- and gshB- complemented strains. Cultures were grown semiaerobically in malate medium supplemented with appropriate antibiotics overnight. Samples were prepared as described in Materials and Methods. Cell extracts were scanned from 500 to 900 nm with a Perkin-Elmer Lambda 12 spectrophotometer. Strains are listed in Table 1.
The highest amount of photosynthetic complexes is found in wild-type cells and strain SB trxC. The mutation in grxC resulted in a moderate reduction of the specific absorbance, which was similar in the trxC grxC double mutant. The mutation of the gshB gene resulted in a stronger reduction of photosynthetic complexes, which was even more severe in the trxC gshB double mutant. Since the doubling times of the two single mutants under semiaerobic conditions were similar, the reduced formation of photosynthetic complexes in the gshB mutant cannot be considered a consequence of reduced growth rate. Although the trxC mutant accumulates levels of photosynthetic complexes similar to those of the wild-type strain, SB1003 (20 and Fig. 2), the trxC gshB double mutant accumulated almost no photosynthetic complexes, indicating a cooperative action of thioredoxin 2 and GSH. When we transferred the intact grxC or gshB gene into the single mutant, the complemented strains showed almost wild-type absorption. We conclude that indeed the lack of grxC or gshB leads to the reduced formation of photosynthetic complexes in the mutant strains. Since the formation of photosynthetic complexes is redox dependent, the reduced amounts of photosynthetic complexes may be the consequence of altered redox potentials in the mutants. The following observations favor this hypothesis and support the importance of both GSH and TrxC for the reduction of disulfide bonds in the cytoplasm of Rhodobacter: (i) TrxC contributes to defense against diamide-generating stress (20), (ii) mutants SB gshB and SB trxC gshB are hypersensitive to diamide (see below), (iii) the growth inhibition zones (diameter) caused by the reductant dithiothreitol (1,000 mM) were 14.3 ± 0.058 mm for wild-type SB1003 but 12.0 ± 0.076 mm for SB trxC. These observations suggest less reducing conditions in the gshB and trxC mutants when compared to the wild type. Most likely, nonnative disulfide bonds accumulate in a number of cytoplasmic proteins in the double mutant SB trxC gshB. Such “unwanted” disulfide bonds could affect the function and stability of cytoplasmic proteins, thus affecting cell physiology, e.g., expression of photosynthesis genes, formation of photosynthesis complexes, and bacteriochlorophyll biosynthesis.
Effect of mutations in the GSH/Grx system on resistance against oxidative stress.
It is well established that the GSH/Grx system plays an important role in keeping cytoplasmic proteins in a reduced state and is involved in the oxidative stress response (reviewed in references 8, 29, and 35). To test whether the GSH/Grx system of R. capsulatus has a similar role, we tested the resistance of the different strains to hydrogen peroxide and paraquat. Paraquat causes the production of superoxide radicals, which are converted to hydrogen peroxide by superoxide dismutase. As shown in Fig. 1, mutants SB trxC and SB grxC are slightly more sensitive to H2O2 and paraquat than SB1003. The same was true for strain SB trxC grxC after paraquat treatment. However, strain SB trxC grxC was highly sensitive to H2O2. Moreover, SB gshB, SB trxC gshB, and SB grxC gshB are significantly more sensitive to H2O2 and paraquat compared to wild-type SB1003 and the other mutants, especially at high concentrations of the oxidative agents. Since GSH is a major reductant in the cytoplasm (5 mM in the cells of E. coli and up to 10 mM in eukaryotic cells), a high sensitivity of gshB mutants to H2O2 and paraquat is understandable.
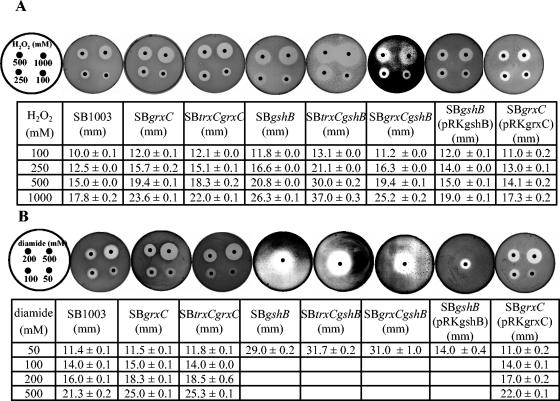
Next we qualitatively determined the sensitivity of the log-phase grxC or gshB single or double mutants to inhibiting concentrations of various oxidants by an inhibition zone method (20). No clear zones of inhibition formed when paraquat was utilized on the plates, so these data were not considered. To support our hypothesis that the cytoplasmic redox buffer in the mutants is changed, we monitored zone inhibition by the thiol-oxidizing agent diamide. To ensure reproducibility, only log-phase cells were used. The results are shown in Fig. 3. The inhibition zones for H2O2 and diamide were slightly larger for both SB grxC and SB trxC grxC mutants than that for the wild type. Both gshB single and double mutants showed higher sensitivity to these components than the wild type and the grxC mutants. The sensitivity order was: SB trxC gshB > SB grxC gshB ≃ SB gshB > SB trxC grxC ≃ SB grxC > SB1003. When the grxC and gshB mutants were complemented with the respective wild-type genes, they showed resistance to H2O2 and diamide similar to that of the wild type (Fig. 3). The hypersensitivity to diamide and increased sensitivity to H2O2 of gshB single and double mutants implies that GSH is a major factor constituting the thiol-disulfide redox buffer in the cytoplasm in R. capsulatus.
FIG. 3.
Resistance of R. capsulatus SB1003, SB grxC, SB trxC grxC, SB gshB, SB trxC gshB, SB grxC gshB, SB grxC(pRKgrxC), and SB gshB(pRKgshB) to hydrogen peroxide (A) and diamide (B) in the zone inhibition assay. A 0.4-ml portion of a culture in exponential growth phase (OD660 = 0.35) was mixed with prewarmed (42°C) top agar (0.7% agar) and layered onto malate minimal salt medium plates. After the top agar had hardened, 5.5-mm-diameter filter paper disks containing 5 μl of oxidative agents as indicated were placed on the plates. Mean values of the diameter (millimeters) of the inhibition zones plus the standard deviations from four different experiments are displayed.
Effect of mutations in the GSH/Grx system on expression of genes with putative functions in the oxidative stress response.
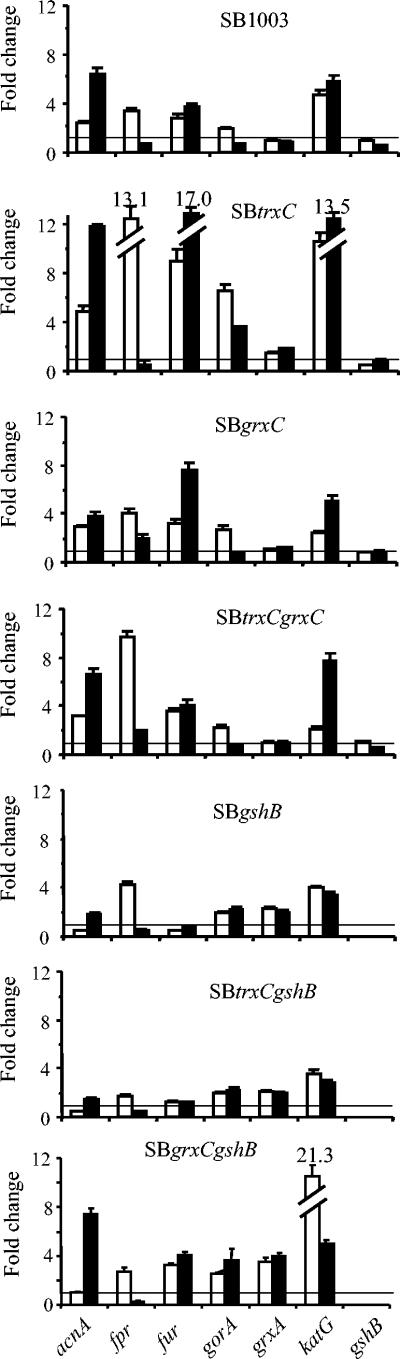
In E. coli many genes are activated in response to oxidative stress (reviewed in reference 36). Hydrogen peroxide induces a number of genes by oxidizing the OxyR transcription factor. The R. capsulatus genome encodes an OxyR homolog (RRC03461) which is involved in defense against oxidative stress (41a), while SoxRS homologs are missing. We tested R. capsulatus genes, whose counterparts are under control of OxyR or SoxRS in E. coli and are known to contribute to the defense against oxidative stress. These are the grxA, gorA, fur, acnA, and fpr genes. Furthermore, we analyzed how the mutations in the GSH/Grx system affect expression of these genes. We also tested the expression of the gshB gene in the strains SB trxC, SB grxC, and SB trxC grxC. Expression levels were quantified by semiquantitative RT-PCR and are displayed in Fig. 4. Since expression of the grxC gene was independent of oxidative stress and did not vary between the wild type and trxC or gshB mutants, these results are not included in Fig. 4.
FIG. 4.
Expression of various genes after treatment of cultures with hydrogen peroxide (black bars) or paraquat (white bars). The wild type and trxC, grxC, and gshB single or double mutants were treated for 15 min with 1 mM hydrogen peroxide (H2O2) or paraquat. The intensity of RT-PCR product bands in agarose gels following ethidium bromide staining was quantified and corrected for the intensity of the PCR product of the rpoZ gene (internal standard). Data represent the means of three RT- PCR amplifications. Values from treated samples are given as fold change compared to those from the corresponding control without treatment, which was considered as 1. Standard deviations are indicated by error bars.
In R. capsulatus wild-type strain SB1003, hydrogen peroxide stimulated the expression of acnA, fur, and katG by factors of 3.7 to 6.4, while fpr, gorA, and gshB expression was repressed. The addition of paraquat stimulated the expression of the genes acnA, fpr, fur, gorA, and katG by factors of 1.9 to 4.7 but not the expression of the gshB gene. The induction of acnA by superoxide in E. coli results in the synthesis of higher levels of aconitase A, which is resistant to superoxide and can therefore keep the tricarbonic acid cycle functional because of a factor that protects its activity from oxidative damage (39). The fur gene encodes a regulatory protein which represses genes required for iron uptake. Upon oxidative stress a stronger repression of iron uptake can prevent the formation of hydroxyl radicals by the Fenton reaction. Highly induced expression of acnA and fur in response to peroxide and superoxide stress implies a role for AcnA and Fur against oxidative stress in R. capsulatus. The fpr gene product, ferredoxin/flavodoxin reductase, is involved in the reduction of Fe-S clusters (5). Its induction by paraquat, but not by H2O2 suggests that the regulator of fpr is superoxide specific. Expression of grxA was oxidative stress independent in this study. In order not to miss the effect by selecting only one time point, we also monitored the expression of gorA and grxA at 0, 5, 15, and 30 min after addition of H2O2. Again, gorA and grxA expression was independent of oxidative stress generated by H2O2 (1 and 5 mM). As a positive control, the katG gene was analyzed, which was strongly upregulated by 1 mM and even 5 mM H2O2 in this time course. Thus, the expression pattern of some of these genes in wild-type R. capsulatus differed significantly from that reported for E. coli. The expression of the grxA and gorA genes is H2O2 dependently regulated by OxyR in E. coli (23, 28, 38). The expression of acnA and fpr is regulated by SoxRS (13, 21) and also induced by H2O2 in E. coli but showed a stronger response to H2O2 than to paraquat in R. capsulatus. In R. capsulatus, both katG and fur showed strong induction by H2O2 and paraquat, as observed in E. coli. The lack of induction of grxA, grxC, gorA, and gshB by H2O2 suggests that these genes do not belong to the OxyR-like regulon, while katG and fur are supposed to be members of such a regulon in R. capsulatus. In agreement with this is the finding that katG no longer responds to H2O2 in an oxyR mutant strain of R. capsulatus (41a).
The trxC mutant showed a much higher extent of induction by H2O2 of acnA, fur, gorA, and katG and stronger paraquat-induced expression of acnA, fpr, fur, gorA, and katG than the wild type, confirming the role of TrxC in the response to oxidative stress and demonstrating interplay of different defense systems (20). Expression of grxA, grxC, and gshB was similar in the trxC mutant and the wild type and independent of H2O2 in either strain, implying that expression of genes for the GSH/Grx system is independent of thioredoxin 2.
The oxidative stress-dependent expression pattern in the grxC mutants resembled that of the wild type with a few exceptions: hydrogen peroxide resulted in repression of fpr expression in the wild type but slight induction of fpr expression in grxC and trxC grxC mutants. As observed in the trxC single mutant, the trxC grxC double mutant showed stronger induction of fpr gene expression after paraquat treatment than the wild type. The grxC gene itself did not respond to H2O2 (data not shown), and both mutants SB grxC and SB trxC grxC showed constant grxA mRNA levels, like the wild type, suggesting that the expression of grxA is not dependent on the presence of GrxC.
The expression patterns of the single mutant SB gshB and double mutants SB trxC gshB and SB grxC gshB differed significantly from that detected for the parental wild-type strain. In contrast to the wild type, these mutant strains showed a two- to fourfold induction of the grxA and gorA genes after hydrogen peroxide or paraquat treatment. These data suggest that the expression of genes of the GSH/Grx system depends on the GSH level. All other genes, acnA, fur, and katG, showed a lower induction in SB gshB and SB trxC gshB than in SB1003. Both SB gshB and SB trxC gshB showed a similar pattern of gene expression in our studies, indicating that GSH is one of the major factors affecting expression of acnA, fur, and katG. In E. coli and Haemophilus influenzae, GSH was found to be involved in the expression of katG (25, 40).
Although the grxC mutant showed a response to oxidative stress similar to that of SB1003, the double mutant SB grxC gshB showed significantly changed expression patterns of some genes when compared to the single mutant SB gshB. Again, an effect of GSH on expression of genes of the GSH/Grx system was observed: grxA and gorA were upregulated by H2O2 and paraquat in strain SB grxC gshB. Surprisingly, the responses of acnA to H2O2, fur to H2O2 and paraquat, and katG to H2O2, were similar in SB grxC gshB and the wild type. Moreover, the highest induction (21.3 fold) of katG by paraquat in SB grxC gshB implies a second regulatory pathway of katG expression by another regulator besides OxyR. By checking the basal level of acnA, fur, and katG in the wild type and all mutants, it was established there was no difference observed among these strains except that the expression level of the katG gene in gshB single and double mutants was threefold higher than that in wild type. This is in agreement with the hypothesis that some OxyR protein molecules may be locked in an oxidized, activated state in the gshB mutant because the reductant of oxidized OxyR, GrxA, which needs to be reduced by GSH, cannot act timely and properly to reduce OxyR. It demonstrates that the expression of katG may be regulated dominantly by the OxyR regulator.
Conclusions.
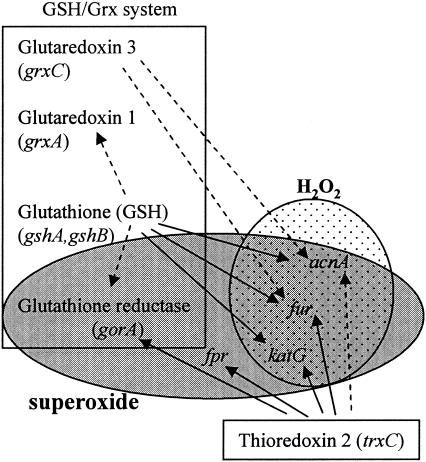
Taken together our data reveal that the response to oxidative stress in R. capsulatus is significantly different from the well-studied response of E. coli. The OxyR regulon may consist of different genes and does not include gorA and grxA as in E. coli. SoxR and SoxS are not encoded by the almost completed R. capsulatus genome sequence, and the gene is also lacking in the completed genome sequence of R. sphaeroides; however, an alternative regulator which mediates a superoxide-specific response can be postulated to control the expression of fpr and gorA in Rhodobacter. Figure 5 gives a schematic overview of the different regulatory units identified in the oxidative stress response of R. capsulatus and of the influence of the components of the thioredoxin and GSH/Grx systems on this response. The analysis of gene expression in the single and double mutants revealed that GSH has a major impact on the regulation of the oxidative stress response and underlines the interplay between different regulatory systems involved in the oxidative stress response. GSH strongly affects acnA and fur expression and the regulation of genes for components of the GSH/Grx system. Future studies should be aimed at the identification of cis regulatory elements in the promoter regions of the genes we analyzed and the identification of proteins interacting with these domains. In combination with expression studies of the different mutants, such studies will shed more light on the regulation of the oxidative stress response of R. capsulatus.
FIG. 5.
Schematic model for regulation of genes by the GSH/Grx system and TrxC in response to oxidative stress in R. capsulatus. Dotted oval, H2O2 stimulon; gray oval, superoxide stimulon; rectangles, GSH/Grx system and Trx 2. Solid arrows represent strong effects, and dashed arrows represent moderate effects, which may be direct or indirect through additional components.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft (Kl 563/16-1) and by Fonds der Chemischen Industrie.
REFERENCES
- 1.Aslund, F., and J. Beckwith. 1999. Bridge over troubled waters: sensing stress by disulfide bond formation. Cell 96:751-753. [DOI] [PubMed] [Google Scholar]
- 2.Aslund, F., B. Ehn, A. Miranda-Vizuete, C. Pueyo, and A. Holmgren. 1994. Two additional glutaredoxins exist in Escherichia coli: glutaredoxin 3 is a hydrogen donor for ribonucleotide reductase in a thioredoxin/glutaredoxin 1 double mutant. Proc. Natl. Acad. Sci. USA 91:9813-9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assemat, K., P. M. Alzari, and J. D. Clement-Metral. 1995. Conservative substitutions in the hydrophobic core of Rhodobacter sphaeroides thioredoxin produce distinct functional effects. Prot. Sci. 4:2510-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barany, F. 1985. Two-codon insertion mutagenesis of plasmid genes by using single-stranded hexameric oligonucleotides. Proc. Natl. Acad. Sci. USA 82:4202-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bittel, C., C. L. Tabares, M. Armesto, N. Carrillo, and N. Cortez. 2003. The oxidant-responsive diaphorase of Rhodobacter capsulatus is a ferredoxin (flavodoxin)-NADP(H) reductase. FEBS Lett. 553:408-412. [DOI] [PubMed] [Google Scholar]
- 6.Boschi-Muller, S., S. Azza, and G. Branlant. 2001. E. coli methionine sulfoxide reductase with a truncated N terminus or C terminus, or both, retains the ability to reduce methionine sulfoxide. Prot. Sci. 10:2272-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchanan, B. B. 1991. Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin/thioredoxin system. Perspective on its discovery, present status, and future development. Arch. Biochem. Biophys. 288:1-9. [DOI] [PubMed] [Google Scholar]
- 8.Carmel-Harel, O., and G. Storz. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54:439-461. [DOI] [PubMed] [Google Scholar]
- 9.Drews, G. 1983. Mikrobiologisches Praktikum. Springer-Verlag, Berlin, Germany.
- 10.Fuchs, J. A., and H. R. Warner. 1975. Isolation of an Escherichia coli mutant deficient in glutathione synthesis. J. Bacteriol. 124:140-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregor, J., and G. Klug. 2002. Oxygen-regulated expression of genes for pigment binding proteins in Rhodobacter capsulatus. J. Mol. Microbiol. Biotechnol. 4:249-253. [PubMed] [Google Scholar]
- 12.Gregor, J., and G. Klug. 1999. Regulation of bacterial photosynthesis genes by oxygen and light. FEMS Microbiol. Lett. 179:1-9. [DOI] [PubMed] [Google Scholar]
- 13.Gruer, M. J., and J. R. Guest. 1994. Two genetically-distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology 140:2531-2541. [DOI] [PubMed] [Google Scholar]
- 14.Holmgren, A. 1976. Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc. Natl. Acad. Sci. USA 73:2275-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmgren, A. 1989. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 264:13963-13966. [PubMed] [Google Scholar]
- 16.Hübner, P., B. Masepohl, W. Klipp, and T. A. Bickle. 1993. nif gene expression studies in Rhodobacter capsulatus: ntrC-independent repression by high ammonium concentrations. Mol. Microbiol. 10:123-132. [DOI] [PubMed] [Google Scholar]
- 17.Kalinina, E. V., A. N. Saprin, V. S. Solomka, N. P. Shcherbak, N. S. Chermnykh, and L. A. Piruzian. 2001. Role of the antioxidant system and redox-dependent regulation of transcription factors bcl-2 and p53 in forming resistance of human K562 erythroleukemia cells to doxorubicin. Vopr. Onkol. 47:595-600. [PubMed] [Google Scholar]
- 18.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 19.Kosower, N. S., and E. M. Kosower. 1978. The glutathione status of cells. Int. Rev. Cytol. 54:109-160. [DOI] [PubMed] [Google Scholar]
- 20.Li, K., E. Hartig, and G. Klug. 2003. Thioredoxin 2 is involved in oxidative stress defence and redox-dependent expression of photosynthesis genes in Rhodobacter capsulatus. Microbiology 149:419-430. [DOI] [PubMed] [Google Scholar]
- 21.Liochev, S. I., A. Hausladen, W. F. J. Beyer, and I. Fridovich. 1994. NADPH:ferredoxin oxidoreductase acts as a paraquat diaphorase and is a member of the soxRS regulon. Proc. Natl. Acad. Sci. USA 91:1328-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Messens, J., G. Hayburn, A. Desmyter, G. Laus, and L. Wyns. 1999. The essential catalytic redox couple in arsenate reductase from Staphylococcus aureus. Biochemistry 38:16857-16865. [DOI] [PubMed] [Google Scholar]
- 23.Michán, C., M. Manchado, G. Dorado, and C. Pueyo. 1999. In vivo transcription of the Escherichia coli oxyR regulon as a function of growth phase and in response to oxidative stress. J. Bacteriol. 181:2759-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulliez, E., S. Ollagnier, M. Fontecave, R. Eliasson, and P. Reichard. 1995. Formate is the hydrogen donor for the anaerobic ribonucleotide reductase from Escherichia coli. Proc. Natl. Acad. Sci. USA 92:8759-8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oktyabrsky, O. N., G. V. Smirnovam, and N. G. Muzyka. 2001. Role of glutathione in regulation of hydroperoxidase I in growing Escherichia coli. Free Radic. Biol. Med. 31:250-255. [DOI] [PubMed] [Google Scholar]
- 26.Pasternak, C., K. Haberzettl, and G. Klug. 1999. Thioredoxin is involved in oxygen-regulated formation of the photosynthetic apparatus of Rhodobacter sphaeroides. J. Bacteriol. 181:100-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 28.Prieto-Alamo, M.-J., J. Jurado, R. Gallardo-Madueno, F. Monje-Casas, A. Holmgren, and C. Pueyo. 2000. Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J. Biol. Chem. 275:13398-13405. [DOI] [PubMed] [Google Scholar]
- 29.Prinz, W. A., F. Aslund, A. Holmgren, and J. Beckwith. 1997. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J. Biol. Chem. 272:15661-15667. [DOI] [PubMed] [Google Scholar]
- 30.Ritz, D., H. Patel, B. Doan, M. Zheng, F. Aslund, G. Storz, and J. Beckwith. 2000. Thioredoxin 2 is involved in the oxidative stress response in Escherichia coli. J. Biol. Chem. 275:2505-2512. [DOI] [PubMed] [Google Scholar]
- 31.Russel, M., P. Model, and A. Holmgren. 1990. Thioredoxin or glutaredoxin in Escherichia coli is essential for sulfate reduction but not for deoxyribonucleotide synthesis. J. Bacteriol. 172:1923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulze-Osthoff, K., H. Schenk, and W. Droge. 1995. Effects of thioredoxin on activation of transcription factor NF-κB. Methods Enzymol. 252:253-264. [DOI] [PubMed] [Google Scholar]
- 33.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1:784-791. [Google Scholar]
- 34.Smirnova, G. V., N. G. Muzyka, M. N. Glukhovchenko, T. A. Krasnykh, and O. N. Oktyabrsky. 1999. Oxidative stress resistance of Escherichia coli strains deficient in glutathione biosynthesis. Biochemistry (Moscow) 64:1111-1116. [PubMed] [Google Scholar]
- 35.Song, J. J., J. G. Rhee, M. Suntharalingam, S. A. Walsh, D. R. Spitz, and Y. J. Lee. 2002. Role of glutaredoxin in metabolic oxidative stress. Glutaredoxin as a sensor of oxidative stress mediated by H2O2. J. Biol. Chem. 277:46566-46575. [DOI] [PubMed] [Google Scholar]
- 36.Storz, G., and M. Zheng. 2000. Oxidative stress, p. 47-59. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 37.Takemoto, T., Q. M. Zhang, and S. Yonei. 1998. Different mechanisms of thioredoxin in its reduced and oxidized forms in defense against hydrogen peroxide in Escherichia coli. Free Radic. Biol. Med. 24:556-562. [DOI] [PubMed] [Google Scholar]
- 38.Tao, K. 1997. OxyR-dependent induction of Escherichia coli grx gene expression by peroxide stress. J. Bacteriol. 179:5967-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varghese, S., Y. Tang, and J. A. Imlay. 2003. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J. Bacteriol. 185:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vergauwen, B., F. Pauwels, and J. J. Van Beeumen. 2003. Glutathione and catalase provide overlapping defenses for protection against respiration-generated hydrogen peroxide in Haemophilus influenzae. J. Bacteriol. 185:5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yen, H. C., and B. Marrs. 1976. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J. Bacteriol. 126:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Zeller, T., and G. Klug. Detoxification of hydrogen peroxide and expression of catalase genes in Rhodobacter. Microbiology, in press. [DOI] [PubMed]
- 42.Zheng, M., F. Aslund, and G. Storz. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718-1721. [DOI] [PubMed] [Google Scholar]
- 43.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]