Abstract
The binding of the essential cell division protein FtsN of Escherichia coli to the murein (peptidoglycan) sacculus was studied. Soluble truncated variants of FtsN, including the complete periplasmic part of the protein as well as a variant containing only the C-terminal 77 amino acids, did bind to purified murein sacculi isolated from wild-type cells. FtsN variants lacking this C-terminal region showed reduced or no binding to murein. Binding of FtsN was severely reduced when tested against sacculi isolated either from filamentous cells with blocked cell division or from chain-forming cells of a triple amidase mutant. Binding experiments with radioactively labeled murein digestion products revealed that the longer murein glycan strands (>25 disaccharide units) showed a specific affinity to FtsN, but neither muropeptides, peptides, nor short glycan fragments bound to FtsN. In vivo FtsN could be cross-linked to murein with the soluble disulfide bridge containing cross-linker DTSSP. Less FtsN, but similar amounts of OmpA, was cross-linked to murein of filamentous or of chain-forming cells compared to levels in wild-type cells. Expression of truncated FtsN variants in cells depleted in full-length FtsN revealed that the presence of the C-terminal murein-binding domain was not required for cell division under laboratory conditions. FtsN was present in 3,000 to 6,000 copies per cell in exponentially growing wild-type E. coli MC1061. We discuss the possibilities that the binding of FtsN to murein during cell division might either stabilize the septal region or might have a function unrelated to cell division.
Division of the rod-shaped bacterium Escherichia coli includes the formation of two new polar caps of the daughter cells. Division is facilitated by the so-called divisome, a ring structure at the tip of the inward-growing septum (12, 44). About twelve known essential cell division proteins (Fts proteins) were shown to localize at this site (47). Probably the best characterized Fts protein is FtsZ, a homolog of eukaryotic tubulin, which is the first known protein that localizes at the division site and which forms a ring-like polymeric structure (5, 16, 39, 40, 52). The localization of all other cell division proteins depends on the presence of FtsZ. It is assumed that FtsZ may provide not only the platform for the assembly of the other components of the divisome but also the force for constriction by its ability to utilize energy from GTP hydrolysis. The FtsZ ring is stabilized by and maybe connected to the membrane via ZipA (21, 29, 30, 38) and FtsA, which has an actin-like fold (2, 42, 43, 49, 55, 59). The assembly of the divisome then continues with the sequential localization of the predicted ABC transporter FtsEX (54), followed by the membrane proteins FtsK (3), FtsQ (7, 9), FtsL (15, 23, 27), YgbQ (now termed FtsB) (8), and FtsW (6, 46) at the FtsZ-FtsA-ZipA ring. After FtsW, the monofunctional murein transpeptidase penicillin-binding protein 3 (PBP3; also named FtsI) localizes at the site of division (48, 61, 62), followed by FtsN (1, 13, 14) and the periplasmic N-acetylmuramyl-l-alanine amidase AmiC (4, 34). A comprehensive summary of the localization studies with strains that carried fts alleles and at the same time produced green fluorescent protein (GFP) constructs of the different cell division proteins was recently presented (11).
The precise function of most of the cell division proteins and the interactions between them during the assembly of the divisome and during the progression of the inwards growing constriction remains elusive. In this publication we report our studies on the role of FtsN in cell division. Interestingly, FtsN was shown to be a multicopy suppressor of the ftsA12(Ts), ftsI23(Ts), ftsQ1(Ts), ftsK44(Ts), and ftsK3531(Ts) alleles (13, 20), and it suppressed the lethality of overproduced MalE-MinE (50). FtsN has a short cytoplasmic domain of 33 amino acids, followed by a membrane-spanning region from amino acids 34 to 53 (according to the output of PHD, derived from the predictprotein server at http://cubic.bioc.columbia.edu/predictprotein/). Most of the protein (amino acids 54 to 319) is located in the periplasm (14), and the information for the localization at the cell division site is supplied by the periplasmic domain (1). The localization of FtsN to the septum was found to depend on the activity of PBP3 (1, 57), and four amino acids in PBP3 that were modeled in close proximity to the cytoplasmic membrane were recently found to be important for the recruitment of FtsN to the cell division site (63). A GFP-AmiC fusion protein (but not GFP-AmiA) does localize at the site of cell division in an FtsN-dependent fashion (4, 35). The C-terminal region of FtsN has a weak sequence similarity to the noncatalytic domain of certain murein hydrolases, e.g., CwlC from Bacillus subtilis, that was suspected to represent a murein-binding site. The solution structure and domain architecture of the periplasmic part of FtsN was recently solved by nuclear magnetic resonance (64). Interestingly, there is a long glutamine-rich stretch (amino acids 48 to 247) that appears not to be structured, except for three short helices near the cytoplasmic membrane. The C terminus (amino acids 243 to 319) has a small globular domain with a βαββαβ-fold, the region similar in sequence to CwlC. Here we demonstrate that FtsN binds to isolated murein, possibly to the longer glycan chains in the sacculus, via the C-terminal domain. In addition, we present evidence that binding of FtsN to murein occurs during cell division. However, binding of FtsN to murein was found not to be essential for cell division under standard laboratory conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The E. coli strains used in this study were wild-type MC1061 (10), MC6RP1 (17), and the mutant MHD52 (MC1061 ΔamiA::Cm ΔamiB ΔamiC::Kan), which lacks three murein hydrolytic amidases (34). For the preparation of radiolabeled murein, the m-diaminopimelic acid (m-Dap) auxotrophic strain W7 (dap lys) (32) was used. Strain JOE565 is the MC4100ftsN::km araD+ strain harboring the plasmid pBAD33-ftsN (11). In addition, strain MC6RP41 (22) with the temperature-sensitive ftsI655 allele was used in this study. Unless otherwise stated, the cells were grown in Luria broth (LB) medium containing the appropriate antibiotics at 37°C in a shaking water bath.
Expression of FtsN variants from pWKS30.
FtsN variants were expressed under the control of the chromosomal ftsN promoter from the low-copy-number plasmid pWKS30 (58). For this purpose, DNA fragments starting 200 bp upstream of the start codon of the ftsN gene and containing the full-length gene or different truncated ftsN genes were amplified by PCR using chromosomal DNA as a template. The upstream primer 5′-CGATATGGATCCGGAAGCTATGCTGTTATTGC-3′ was combined with different downstream primers, and the PCR products were cloned into the low-copy plasmid pWKS30 (58) to yield plasmids pWKS30-ftsN, pWKS30-ftsN1-241, pWKS30-ftsN1-138, and pWKS30-ftsN1-52 (the numbers indicate the first and last amino acid of the produced FtsN form) by using standard protocols. The downstream primers were as follows: for pWKS30-ftsN, primer 5′-CACTACGAATTCTACGGTATTGCCCAACGTGG-3′; for pWKS30-ftsN1-241, primer 5′-TATTGAATTCTCACTCCGCCGTCGGTTTTGGC-3′; for pWKS30-ftsN1-138, primer 5′-CATTGAATTCTCATTCAACCAGCTGCGTTG-GC-3′; for pWKS30-ftsN1-52, primer 5′-GATGGAATTCTCAGTACAGA-CCACCGATAAAGG-3′. The resulting plasmids were transformed into JOE565 for FtsN depletion experiments and into MC6RP41 (ftsI655) cells to investigate suppression of the Ts phenotype. Cells harboring pWKS30 and its derivatives were grown with 50 μg of ampicillin/ml. The presence of the truncated forms of FtsN was verified by analyzing cell lysates by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot followed by detection with FtsN antiserum, with the exception of the shortest fragment, which was not recognized by the serum.
Preparation of cells for light microscopy.
Full-length FtsN was depleted in JOE565 as previously described (11). JOE565 strains containing different pWKS30 plasmids were grow overnight at 37°C in LB medium containing 50 μg of ampicillin/ml, 10 μg of chloramphenicol/ml, 40 μg of kanamycin/ml, and 0.2% arabinose. The cells were inoculated 1:250 in LB containing the antibiotics and 0.2% arabinose or 0.2% glucose and were grown at 37°C until an optical density (OD) of 0.2 to 0.3 was reached. Each culture was then diluted 1:100 into fresh medium of the same composition and was grown again until an OD of 0.2 to 0.3 was reached. After such serial culturing, the cells harboring pWKS30 grown in glucose-containing medium were filamentous. The cells were fixed as described previously (11), washed twice with 1 ml of phosphate-buffered saline (PBS), resuspended in PBS, and examined by light microscopy.
Suppression of the ftsI(Ts) phenotype.
The pWKS30-based plasmids containing different ftsN gene fragments were transformed into MC6RP41. Serial dilutions of cultures were plated twice, and the plates were incubated for 18 h at 30 and 42°C, respectively. The relative plating efficiency (high versus low temperature) was determined as CFU/milliliter at 42°C divided by CFU/milliliter at 30°C, as described previously (13).
Purification of FtsN variants.
Soluble forms of FtsN and truncated variants were purified as described elsewhere (64). The concentration of the FtsN variants was determined using both the bicinchoninic acid test (Pierce) and the Lowry test (Bio-Rad, Hercules, Calif.). Both methods gave the same protein concentrations.
Isolation of murein.
Murein was prepared according to the published standard procedure (24). Briefly, cells growing exponentially with aeration were chilled rapidly, harvested by centrifugation, resuspended in ice-cold water, and dropped into a boiling 4% SDS solution. After boiling for 30 min, the crude sacculi were collected by ultracentrifugation, washed free of SDS with water by repeated centrifugation-resuspension steps. High-molecular-weight glycogen was digested with 100 μg of α-amylase/ml in 10 mM Tris-HCl buffer (pH 7.0) for 2 h at 37°C. Bound lipoprotein and protein contaminations were digested with 200 μg of pronase/ml for 90 min at 60°C. After the addition of 1% SDS the sacculi were boiled for 15 min. The SDS was removed by repeated centrifugation-resuspension in water, and the purified sacculi were stored in water containing 0.05% sodium azide at 4°C at a concentration of about 1 mg/ml. In one preparation, the pronase treatment was omitted. Instead, the sacculi were washed three times with a solution of 20 mM Tris-HCl (pH 7.0), 0.1% Triton X-100 and were resuspended and stored in the same buffer. This results in murein that still contains covalently bound lipoprotein. In the case of MC6RP1 the cells were grown with and without a 10-μg/ml concentration of the cell division inhibitor furazlocillin (generous gift from Bayer, Wuppertal, Germany), and the sacculi were prepared by a method yielding highly purified murein sacculi for electron microscopy, as described in detail in a previous publication (17). The concentration of different murein samples was compared by digestion of the samples with cellosyl followed by separation by high-performance liquid chromatography (HPLC) (see below) and quantification of the UV absorption of the muropeptides. [3H]m-Dap-labeled murein (60,000 cpm/μg) was prepared from strain W7 according to a published procedure (25), whereas murein with the label in the glycan strands (7,000 cpm/μg) was prepared from MC1061 cells that grew in the presence of [3H]GlcNAc (36).
Enzymatic digestion of murein.
Murein, either unlabeled or radioactively labeled in either the peptide or the glycan part, was digested with different enzymes to generate disaccharide-peptide subunits (muropeptides), peptides, and murein glycan strands, respectively. A mixture of about 80 different muropeptides was produced by digestion with cellosyl from Streptomyces globisporus for 6 to 18 h at 37°C in a 10 mM sodium phosphate buffer (pH 4.8) (24). Alternatively, the murein was digested into glycan and the peptide part with human amidase in a 50 mM sodium phosphate buffer (pH 7.9) (33). The murein glycan strands were further processed either by cellosyl yielding mainly GlcNAcMurNAc disaccharide residues at the conditions described above or by the membrane-bound lytic transglycosylase MltA in a solution of 10 mM Tris-maleate, 10 mM MgCl2, 0.2% Triton X-100 (pH 5.2) at 30°C, yielding GlcNAc-1,6-anhydroMurNAc disaccharides. MltA was purified from E. coli as described previously (56). After all digestions, the pH of the sample was adjusted to 4 to 5 and the enzymes were removed by boiling for 8 min, followed by a short centrifugation.
HPLC analysis of murein fragments.
Muropeptides were analyzed after reduction with sodium borohydride by reversed-phase HPLC at 55°C in a 135-min linear gradient from 50 mM sodium phosphate (pH 4.31) to 75 mM sodium phosphate (pH 4.95), 15% methanol (24, 26). Murein glycan strands were separated according to their degree of oligomerization by reversed-phase HPLC at 50°C in a slightly convex gradient from 100 mM sodium phosphate (pH 2.0) to 100 mM sodium phosphate (pH 2.0), 11% acetonitril. This method allowed the separation of glycans from 1 to 30 disaccharide units; longer glycans were eluted together in an ensuing methanol step (33). The detection occurred for both methods at 205 nm and, if radioactively labeled murein was analyzed, with the help of a radioactivity flowthrough detector (Canberra Packard).
Protein methods.
Proteins were separated by SDS-PAGE as described in a published protocol (41). For immunodetection of FtsN or OmpA, the proteins were blotted on a nitrocellulose membrane (Western blot). The membrane was blocked by incubation for 30 min with a solution of 0.1 M Tris-HCl (pH 7.5), 0.9% NaCl, 0.2% Tween-20 (TTBS) containing 5% dehydrated skim milk. After four washes with TTBS, the blot membrane was incubated for 1.5 h with a 1:2,000 dilution of the polyclonal α-FtsN antiserum (this work) or α-OmpA antiserum (kindly provided by H. Schwarz, Max Planck Institute for Developmental Biology, Tübingen, Germany), respectively, in TTBS containing 0.5% dehydrated skim milk. After washing (four times for 10 min each wash), a 1:40,00 dilution of goat anti-rabbit horseradish peroxidase conjugate (Amersham Life Science) was added for 1.5 h. The membrane was then washed as described above and the FtsN or OmpA band was visualized by peroxidase reaction using chloronaphtol as a substrate. For this visualization, the blot membrane was developed with fresh chloronaphtol reagent (0.5 mg of chloronaphtol/ml in 0.1 M Tris-HCl [pH 7.4] plus 0.5 μl of 30% hydrogen peroxide/ml) until the bands appeared.
Preparation and purification of FtsN antisera.
Serum against FtsN was produced from rabbits at Eurogentec company (Herstal, Belgium), using purified FtsN58-319GSH6 protein for immunization. For immunoaffinity purification of the serum according to a standard procedure (31), 2 mg of FtsN58-319GSH6 was coupled to 500 μl of Affigel-10 beads (Bio-Rad) according to the recommendation of the manufacturer. Two milliliters of serum was diluted with 40 ml of 10 mM Tris-HCl (pH 7.5) and incubated with the FtsN-containing beads for 1 h at 6°C. The beads were then filled into a column and washed with 20 ml of 0.5 M NaCl. The elution of the FtsN-specific immunoglobulins occurred with 4 M MgCl2. The immunoglobulin-containing fractions were pooled, dialyzed against PBS, and concentrated. After the addition of 50% glycerol the purified serum was stored at −20°C.
Assay for binding of FtsN and control proteins to murein.
Murein (about 100 μg) from a 1-mg/ml suspension was collected by centrifugation in a Beckmann TLA-100 rotor at 90,000 rpm at 4°C for 30 min. Murein was resuspended in a 90-μl solution of 10 mM Tris-maleate, 10 mM MgCl2, 50 mM NaCl, 0.02% NaN3 (pH 6.8) (binding buffer I), and 3 μg of FtsN variants was added in a volume of 10 μl. A control sample contained FtsN but no murein. In other assays, a mix of molecular weight marker proteins or an extract with the soluble proteins from E. coli was included. After 30 min of incubation on ice, the samples were centrifuged (see above) and the murein pellet was resuspended in 200 μl of cold binding buffer I and recovered by centrifugation. The pellet was then resuspended in 2% SDS solution and stirred for 1 h. The supernatant of the binding step (S), the wash (W), and the resuspended pellet (P) were analyzed by discontinuous SDS-PAGE (12 or 16% acrylamide). FtsN was detected either by zinc staining (Bio-Rad) according to the protocol of the manufacturer or with the purified antiserum after blotting on a nitrocellulose membrane (see above, protein methods). Care was taken that the same amount of the sacculi preparations were used in the binding assays. This was ensured by digestion of murein by the muramidase cellosyl and quantification of the total UV signal of the released muropeptides separated by HPLC as described above (data not shown).
Assay for binding of murein fragments to FtsN.
The following different murein digestion products were prepared for binding studies. [3H]m-Dap-labeled murein was digested with cellosyl to yield labeled muropeptides or with human serum amidase to yield labeled peptides and nonlabeled glycan strands. If [3H]GlcNAc-labeled murein was digested with human serum amidase, labeled glycan strands and unlabeled peptides were obtained. These glycans could be further digested with either cellosyl or MltA (see above) to yield labeled disaccharides or 1,6-anhydrodisaccharides. For the binding experiments, 8,000 cpm of [3H]m-Dap-labeled or of [3H]GlcNAc-labeled murein fragments (0.1 to 1 μg) were incubated with 20 μg of FtsN58-319GSH6 in a total volume of 100 μl of 10 mM Tris-HCl, 10 mM MgCl2, 50 mM NaCl, 0.02% NaN3 (pH 8.0) (binding buffer II). Control samples did not contain FtsN. The samples were added to a suspension of Ni-nitrilotriacetic acid (NTA) beads (total of 300 μl) that had been washed three times with 1 ml of binding buffer II and were incubated for 2 h at 6°C with gentle agitation. After centrifugation, the beads were washed with 1 ml of binding buffer II before the bound protein, together with eventually bound cell wall fragments, was released from the beads by incubation for 30 min at room temperature with binding buffer II containing 1.6 M imidazole. Aliquots of the supernatant of the binding step, the wash, and the imidazole elution were taken for scintillation counting, and the percentage of bound radioactive murein fragments was determined. If the bound radioactive murein fragments were subjected to HPLC analysis, the method was changed as follows. First, the amount of murein fragments was increased to 2.8 μg (170,000 cpm) of muropeptides (cellosyl digestion of [3H]m-Dap-labeled murein) or to 7.2 μg (50,400 cpm) of peptide plus glycan strands (amidase digestion of [3H]GlcNAc-labeled murein). Second, the imidazole was omitted and the elution of FtsN together with bound murein fragments from the Ni-NTA beads was achieved by an incubation with 200 μl of 50 mM sodium phosphate (pH 3.7) for 30 min followed by boiling for 8 min. The sample was centrifuged briefly, and the supernatant containing the eluted murein fragments was collected.
In vivo cross-linking of proteins to murein.
For the cross-linking of proteins to murein, a published procedure was applied (37, 51) with modifications. We omitted the hydrophobic DSP cross-linker but used its water-soluble derivative DTSSP (Sigma). DTSSP can cross-link amino groups with a distance of less than 1.2 nm, and because of its internal disulfide bond it is cleavable by reducing agents such as β-mercaptoethanol. Briefly, cells of a 100-ml culture grown at 37°C with vigorous shaking were harvested at an OD of 0.4 to 0.5 and were resuspended in 2 ml of 50 mM sodium phosphate, 20% sucrose (pH 7.4). DTSSP was dissolved to 40 mg/ml in the same buffer, and 250 μl of this solution was added to the cell suspension. The samples were incubated for 1 h at 25°C, and then the cells were sedimented by centrifugation, washed once with 3 ml of PBS, and resuspended in 3 ml of PBS. This suspension was dropped into 3 ml of boiling 8% SDS solution and kept at 100°C for 3 h. After an incubation at 80°C for 18 h, the murein pellet was collected by ultracentrifugation at 30°C at 90,000 rpm for 30 min in a Beckmann TLA 100.3 rotor. The pellet was washed once with 3 ml of 1% SDS and finally was resuspended with 1% SDS to a final volume of 150 μl. Aliquots were subjected to SDS-PAGE after boiling in sample buffer that did or did not contain 10% β-mercaptoethanol. Proteins bound to murein via DTSSP were released in the presence of β-mercaptoethanol. After Western blot, FtsN and OmpA were detected with polyclonal antisera as described above.
Determination of the amount of FtsN per CFU.
Cells of wild-type MC1061 were grown at 37°C in LB medium until an OD of 0.2 to 0.4 was reached. The culture was placed on ice and the CFU was determined by the preparation of serial dilutions, plating on LB agar, and counting of the colonies after 24 h of incubation at 37°C. A 24-ml aliquot of the exponential-phase culture was centrifuged, and the cell pellet was resuspended in 150 μl of PBS. One-hundred fifty microliters of SDS-PAGE sample buffer was then added, and the sample was boiled for 10 min. Aliquots of 40 μl were either left unsupplemented or were supplemented with 15 to 120 ng of FtsN58-319GSH6. After SDS-12% PAGE and Western blotting, full-length FtsN and FtsN58-319GSH6 were detected by the purified antiserum (that was raised against FtsN58-319GSH6) and staining (see above). The amount of FtsN present in the cell lysate was estimated semiquantitatively by comparison of the band intensities of FtsN and the different amounts of added FtsN58-319GSH6.
RESULTS
Binding of different FtsN variants to purified murein sacculi.
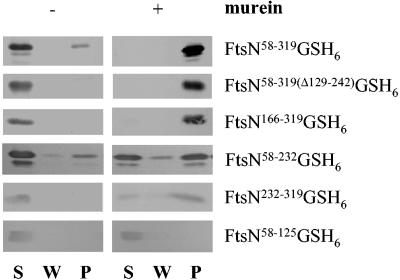
We studied a possible binding of soluble His-tagged forms of FtsN to murein in a simple pull-down experiment (see Materials and Methods). In addition to the full-length periplasmic part of FtsN (FtsN58-319GSH6), the following five truncated forms of FtsN were studied: FtsN58-319GSH6, with an internal deletion from amino acids 129 to 242; two His-tagged truncated forms, with the last C-terminal 154 or 77 amino acids; and two His-tagged forms from amino acids 58 to 125 and 58 to 232 (see Fig. 6). In control experiments, the FtsN variants were treated in the same way but without murein. Only small amounts of the FtsN variants pelleted without murein in this assay, whereas the bulk of the proteins remained in the supernatant (Fig. 1). Four variants of FtsN did bind to murein sacculi isolated from wild-type cells, all containing the C-terminal 77 amino acids. Although a fraction of the shortest C-terminal form of FtsN was found in the supernatant, this was probably because the binding capacity of murein was exceeded by the 0.3 nmol of protein in the reaction mixture. In fact, in a titration experiment the maximum binding capacity of murein for FtsN58-319GSH6 was estimated at 6 μg (0.2 nmol) of protein per 100 μg of murein (data not shown). Two forms without the C-terminal part showed a reduced binding (FtsN58-232GSH6) or no binding (FtsN58-125GSH6) to murein. We conclude that murein-binding is mainly mediated by the C-terminal globular domain of FtsN.
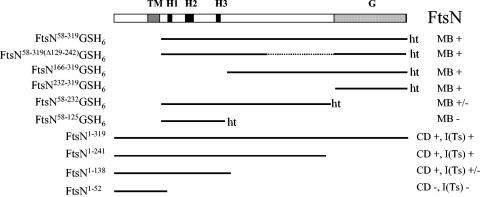
FIG. 6.
Regions in FtsN required for murein binding, cell division, and suppression of ftsI655(Ts). The purified FtsN proteins used in murein-binding studies and the FtsN variants that were produced from pWKS30 plasmids are shown on the left side. The following different regions in FtsN are indicated: TM, transmembrane helix (amino acids 34 to 47); periplasmic helices H1 (58 to 65), H2 (80 to 92), and H3 (117 to 123); and the C-terminal globular domain G (243 to 319). Proteins for murein binding studies contained a C-terminal His-tag (ht). MB+, binding to murein; MB−, no binding to murein; CD+, cell division in a FtsN depletion strain; CD−, no cell division in a FtsN depletion strain; I(Ts)+, suppression of ftsI655(Ts); I(Ts)−, no suppression of ftsI655(Ts).
FIG. 1.
Binding of different His-tagged FtsN variants to murein. Purified FtsN variants (3 μg each), as indicated to the right, were incubated without (control experiment, left side) or with about 100 μg of murein sacculi isolated from wild-type MC1061 (right side). Murein was pelleted by ultracentrifugation and was washed once with buffer. After another centrifugation step, the pellet was resuspended in buffer with 2% SDS. Aliquots of the supernatant after incubation with FtsN (S), the wash fraction (W), and the resuspended pellet (P) were separated on a SDS-16% PAGE, and after blotting on nitrocellulose FtsN was detected with a specific antiserum and chloronaphtol staining. All four FtsN variants containing the C-terminal domain bound to murein sacculi.
We tested if proteins generally bind to murein in two experiments. In the first experiment, the proteins of a molecular weight marker kit were tested. One protein, phosphorylase b from rabbit muscle, bound to murein, whereas the other five marker proteins did not (data not shown). In the second experiment, out of a total soluble protein extract from E. coli there were a few proteins binding to murein, some of which were those present at highest abundance. Most of the proteins present in the extract did not bind (data not shown).
We also tested for possible murein hydrolase activity of FtsN at various conditions, but we were not able to detect the release of soluble fragments from radioactively labeled murein by FtsN (data not shown).
Binding of FtsN58-319GSH6 to different murein preparations.
To look for the basic attributes of murein required for FtsN binding, murein purified from different sources or by alternative protocols was checked for the binding of the full-length periplasmic FtsN58-319GSH6 variant (Fig. 2). In contrast to sacculi of wild-type cells that were purified by treatment with a protease, FtsN58-319GSH6 did not bind to murein sacculi that still contained covalently bound Braun's lipoprotein, and less FtsN58-319GSH6 bound to sacculi isolated from a strain lacking thee amidases (AmiA, AmiB, and AmiC) that form chains of nonseparated cells. These sacculi were prepared by the standard method for analytical purposes and might still contain minor impurities. To exclude the possibility that FtsN58-319GSH6 would bind to murein-contaminating compounds, sacculi were prepared according to a method involving a more extensive SDS extraction procedure which yields murein of the highest quality for the use in electron microscopy (see Materials and Methods). The derivative FtsN58-319GSH6 did bind to sacculi isolated from the strain MC6RP1 by this method and from MC1061 purified by the standard method at similar levels (Fig. 2). On the other hand, binding of FtsN58-319GSH6 was significantly reduced when the assay was performed with murein from MC6RP1 in which the cell division was blocked by the addition of furazlocillin.
FIG. 2.
Binding of FtsN58-319GSH6 to different murein preparations. The soluble, His-tagged variant of FtsN (2 μg) was incubated with about 100 μg of purified murein sacculi from wild-type MC1061 (lanes 1), with sacculi from the wild type that contained bound lipoprotein (lanes 2), with sacculi from MHD51 cells that form chains due to the lack of murein hydrolase amidases (lanes 3), with sacculi from MC6RP1 cells (lanes 4), or with sacculi from MC6RP1 cells that were blocked in cell division (lanes 5). One control sample did not contain murein (lanes C). After ultracentrifugation, the murein pellet was washed once and, after another sedimentation, was resuspended in 2% SDS buffer. Aliquots of the supernatant after incubation with FtsN (S), the wash fraction (W), and the resuspended pellet (P) were separated on an SDS-12% PAGE, and the proteins were detected by zinc staining. The applied FtsN bound quantitatively to the murein of MC1061 and of MC6RP1, whereas the amount of FtsN binding to sacculi from chain-forming cells of MHD52 or from filamentous cells of MC6RP1 grown in the presence of furazlocillin was reduced. FtsN did not bind to murein that still contained covalently bound lipoprotein.
Murein digestion products as ligands for FtsN58-319GSH6.
To determine the structural subunits of the mureins that were recognized by FtsN58-319GSH6, different radioactively labeled murein digestion products were generated that could be detected in a binding assay with Ni-NTA to capture His-tagged FtsN58-319GSH6. First, murein with the radioactive label in the m-Dap residue at position 3 in the peptide part was digested with muramidase or amidase, and binding of the digestion products to FtsN58-319GSH6 was tested. Both muropeptides as well as peptides lacking the disaccharide moieties, but not the glycan strands, were radioactively labeled. As shown in Table 1, neither muropeptides nor peptides bound to FtsN58-319GSH6 in measurable amounts. The binding experiment was repeated, and the supernatant as well as the FtsN-bound muropeptide sample were analyzed by HPLC with a radioactivity flowthrough detector. As shown in Fig. 3A, no signal of a minor muropeptide binding specifically to FtsN58-319GSH6 could be detected.
TABLE 1.
Binding of muropeptides or peptides to FtsN
| Enzyme | [3H]M-Dap-murein digestion productsa | % Total radioactivity of product:
|
|
|---|---|---|---|
| Bound to FtsN | Bound to control beads | ||
| Muramidase | Muropeptides | 2.0 | 2.3 |
| Amidase | Peptides + glycans | 2.5 | 1.9 |
Products in boldface were radioactive.
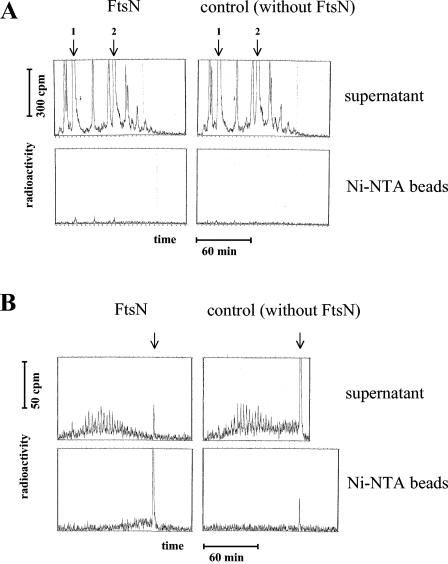
FIG. 3.
Binding of FtsN58-319GSH6 to muropeptides (A) or murein glycan strands (B). (A) FtsN was incubated with a digestion of 3H-labeled murein (label in the peptide part) with cellosyl. As a control, the radioactive muropeptides were treated without the addition of FtsN. Ni-NTA beads were then added to bind the His-tagged protein. The beads were washed three times and then were eluted at pH 3.7. The supernatant of the incubation with (or without) FtsN as well as the eluate from the Ni-NTA beads were analyzed by HPLC that was connected to a radioactivity flowthrough detector using the muropeptide method. The sensitivity was adjusted to high to detect minor compounds. The positions of the major monomer, GlcNAc-MurNAc(l-Ala-d-Glu-m-Dap-d-Ala) (arrow 1), and dimer, GlcNAc-MurNAc(l-Ala-d-Glu-m-Dap-[d-Ala]-d-Ala-m-Dap-d-Glu-l-Ala)MurNAc-GlcNAc (arrow 2), are indicated. No specific binding of any major or minor muropeptide to FtsN was detected. (B) FtsN was incubated with a digestion of 3H-labeled murein (label in the glycan part) with amidase. As a control, the radioactive murein glycan strands were treated without the addition of FtsN. Ni-NTA beads were then added to bind the His-tagged protein. The beads were washed three times and then were eluted at pH 3.7. The supernatant of the incubation with (or without) FtsN as well as the eluate from the Ni-NTA beads were analyzed by HPLC that was connected to a radioactivity flowthrough detector using the murein glycan strand method. The arrows indicate the position of the long glycans with more than 30 disaccharide units that elute together in a methanol step. The specific binding to FtsN was a property only of the glycans with a higher degree of oligomerization.
In a second approach, murein labeled in both N-acetylamino sugars (GlcNAc and MurNAc) was digested with purified human serum amidase, yielding radioactively labeled glycan strands and nonlabeled peptides. Incubation of FtsN58-319GSH6 with the amidase digestion products resulted in a significant proportion (27.3%) of the labeled glycan strands getting bound to FtsN58-319GSH6 (Table 2). However, when the glycan strands were processed to disaccharides, either to GlcNAcMurNAc with cellosyl or to GlcNAc-1,6-anhydroMurNAc with a lytic transglycosylase, no binding to FtsN58-319GSH6 occurred. There was no significant binding of glycan strand material to Ni-NTA beads in control experiments with seven other soluble His-tagged proteins (data not shown).
TABLE 2.
Binding of murein glycan strands to FtsN
| Enzyme(s) | [3H]GlcNAc- murein digestion productse | % Total radioactivity of product:
|
|
|---|---|---|---|
| Bound to FtsN | Bound to control beads | ||
| Amidase | Glycans + peptides | 27.3 | 1.0 |
| Amidase + muramidase | GMa + peptides | 1.0 | NDd |
| Amidase + LTb | GM(anh)c + peptides | 3.5 | ND |
GM, GlcNAc-MurNAc.
LT, lytic transglycosylase.
GM(anh), GlcNAc-1,6-anhydroMurNAc.
ND, not determined.
Products in boldface were radioactive.
To analyze the properties of the FtsN58-319GSH6-bound material, the binding experiment was repeated with a larger amount of both FtsN and amidase digestion products, and the retained glycan strands were eluted and subjected to HPLC analysis. As shown in Fig. 3B, only the glycans of the higher degree of oligomerization bound to FtsN58-319GSH6. In fact, analysis of glycan chain length distribution in the supernatant after the FtsN58-319GSH6 binding reaction showed an almost total deficit of long (more than about 25 disaccharide units) chains compared to that of the corresponding control sample (Fig. 3B, supernatant panels). The missing glycan chains, though, could be recovered from the FtsN58-319GSH6 fraction, which was essentially free of short glycan chains. In the control experiment, the long glycan material bound only marginally to Ni-NTA in the absence of FtsN58-319GSH6 (Fig. 3B, Ni-NTA bead panels). Thus, we conclude that FtsN binds to the murein sacculi by a specific interaction with the long, but not with the short, glycan strands. This is in accordance with the finding that digestion of the glycans to disaccharide units abolished their ability to bind to FtsN58-319GSH6 (Table 2).
In vivo cross-linking of FtsN to murein.
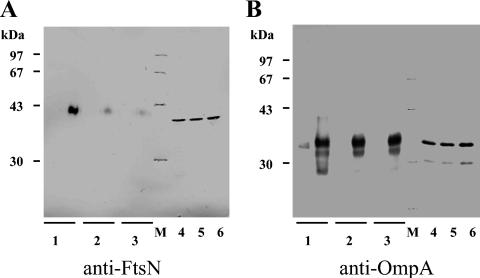
After showing that purified FtsN58-319GSH6 binds to isolated murein sacculi and to the long murein glycan strands, we tested if one can cross-link native FtsN to murein in vivo. Following a published procedure (37), we found that FtsN could indeed be cross-linked to murein with DSP, a hydrophobic cross-linker containing a disulfide bond (data not shown). Because a fraction of the DSP precipitated upon addition to the cell suspension, we were unable to quantitatively remove the excess of cross-linker by centrifugation. Therefore, we preferred to use a water-soluble form of DSP, the DTSSP cross-linker. Indeed, we found that FtsN was cross-linked to murein in wild-type cells and could be released from the purified murein in the presence of the reducing agent β-mercaptoethanol (Fig. 4). The electrophoretic mobility of the released FtsN was slightly changed, presumably because the covalently linked part(s) of cross-linker molecules changed the charge and molecular weight of the protein. No FtsN was released without β-mercaptoethanol, indicating that FtsN was bound to murein via cross-linker molecules. Cells with blocked cell division or cells of the chain-forming MHD52 mutant contained similar amounts of FtsN compared to those of wild-type cells. However, the amount of FtsN that was cross-linked to murein in filamentous wild-type cells after inhibition of cell division or in chaining cells of the triple amidase mutant MHD52 was smaller than the amount bound to sacculi from wild-type cells with intact cell division and separation (Fig. 4A). In contrast, the outer membrane protein OmpA that was shown to bind to murein and that has no role in cell division was cross-linked to murein in approximately the same amounts in cells of the wild type, the wild type with blocked cell division, and the multiple-amidase mutant with blocked cell separation (Fig. 4B).
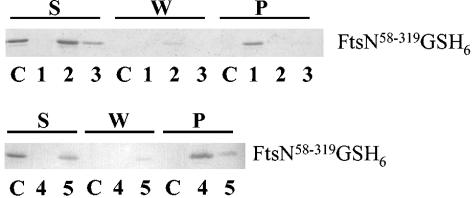
FIG. 4.
In vivo cross-linking of FtsN to murein. Exponentially growing cells of wild-type MC1061 without (sample 1) and with (sample 2) inhibition of cell division and cells of MHD52 lacking three amidases (sample 3) were incubated with DTSSP cross-linker, and murein was isolated as described in Materials and Methods. The applied samples correspond to 16 ml of cell culture (A) and 0.533 ml (B). As control samples, the resuspended cells (without cross-linker) of MC1061 (lane 4), MC1061 with blocked cell division (lane 5), and MHD52 (lane 6) corresponding to 0.4 ml (A) or 13 μl (B) of cell culture were analyzed. The aliquots were subjected to SDS-12% PAGE after they were boiled either without or with the addition of β-mercaptoethanol to the sample buffer. After blotting on nitrocellulose, FtsN (A) and, as a control, OmpA (B) proteins were detected with polyclonal antisera and visualized by chloronaphtol staining. FtsN as well as OmpA could be cross-linked to murein in vivo. A larger amount of FtsN was cross-linked to murein in wild-type cells compared to that in the wild type with blocked cell division or cells of the chain-forming amidase mutant. M, molecular size marker.
Amount of FtsN per cell.
We have determined the amount of FtsN per CFU in exponentially growing wild-type MC1061 cells with the help of the purified serum against FtsN58-319GSH6. In six independent experiments, amounts of 17 to 41 ng of FtsN per 108 CFU were estimated semiquantitatively (data not shown). From these experiments, the abundance of FtsN was calculated to be 4,650 ± 1,780 molecules per cell.
The murein-binding domain of FtsN is not essential for cell division or for suppression of ftsI655(Ts) phenotype.
To test which part of FtsN is required for cell division, we produced full-length or different truncated FtsN forms from the low-copy-number plasmid pWKS30 in the strain JOE565 that has a chromosomally inactivated ftsN gene and harbors the plasmid pBAD33-ftsN, in which the ftsN gene is under the control of the arabinose inducible promoter that is repressed with glucose. As expected, glucose-grown JOE565 with the empty pWKS30 plasmid grew as filaments, and cell division was enabled with pWKS30-ftsN that contained the full-length ftsN gene (Fig. 5). Also, JOE565 cells with plasmids that produced only truncated forms of FtsN lacking the C-terminal murein-binding domain, namely FtsN1-241 or FtsN1-138, could divide. However, division was not possible if the shortest form, FtsN1-52, was present in glucose-grown JOE565. These results indicate that the region between amino acids 53 and 137 is essential for division and that the murein-binding domain (amino acids 243 to 319) is not required for cell division under these conditions.
FIG. 5.
Production of truncated FtsN variants in an ftsN depletion strain. Strain JOE565 with the chromosomally inactivated ftsN gene and the pBAD33-ftsN plasmid was transformed with the low-copy plasmid pWKS30 or with pWKS30 derivatives that contained either the full-length ftsN gene or truncated forms. If grown with glucose, the expression of ftsN from pBAD-33-ftsN is repressed and only the ftsN forms from pWKS30 plasmids are expressed. Cells were fixed with 2% formaldehyde prior to light microscopy. Cell division was blocked without FtsN (empty pWKS30 plasmid) and with the production of FtsN1-52, whereas division was possible in the presence of FtsN1-138, FtsN1-241, and of full-length FtsN. Bar, 10 μm.
We found that the ftsI655(Ts) phenotype could be, like ftsI23 (13), suppressed by increased levels of FtsN, and that this suppression does not depend on the presence of the murein-binding site in the overproduced FtsN molecules. Table 3 shows the results of two independent experiments.
TABLE 3.
Suppression of ftsI655(Ts) phenotype in MC6RP41 by overproduction of various FtsN variants
| Plasmid | Relative efficiency of plating at 42°C:
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| pWKS30 | 2.9 × 10−5 | 7.5 × 10−6 |
| pWKS30-ftsN | 0.93 | 0.33 |
| pWKS30-ftsN1-241 | 0.88 | 0.46 |
| pWKS30-ftsN1-138 | 6.6 × 10−3 | 2.4 × 10−3 |
| pWKS30-ftsN1-52 | 1.1 × 10−6 | 9.7 × 10−8 |
Figure 6 summarizes our results on the participation of different regions in FtsN in murein binding, cell division, and suppression of the ftsI655(Ts) phenotype.
DISCUSSION
The precise function of most of the cell division proteins and the interplay between them during septation still has to be elucidated. In this study we have investigated the role of the essential cell division protein FtsN. We show (i) that FtsN binds to murein via its periplasmic C-terminal domain, (ii) that the binding is to the murein glycan strands and not to the peptide part of murein, (iii) that the murein-binding domain of FtsN is not essential for division under standard laboratory conditions, and (iv) that the periplasmic region close to the membrane anchor is required for cell division. In what follows we discuss these findings.
Binding studies with various purified FtsN variants indicate that the C-terminal 77 amino acids, which constitute the structured region of the protein (64), function as a murein-binding site, possibly also with some contribution of the flanking region. Murein is a polymeric compound isolated from whole cells by an SDS-extraction procedure followed by further purification steps, including amylase and protease treatments. To exclude the possibility that it might contain minor impurities of cellular origin, we also isolated murein sacculi according to a protocol for use in electron microscopy giving the highest possible purity. FtsN still binds to murein sacculi isolated in this way, making it very likely that FtsN indeed binds to murein and not to a contaminating substance. It has to be pointed out that murein is a closed bag-shaped molecule. In the binding experiments FtsN was added to the murein sacculi from outside, whereas in vivo FtsN would reach murein from the inside because of its amino terminus anchoring it to the cytoplasmic membrane. Such topological and sterical constrains might be the reason why FtsN did not bind to sacculi that still contained (on the outside) covalently bound lipoprotein.
Murein contains different functional groups and charged amino acids at neutral pH. Thus, various proteins functionally unrelated to murein metabolism might bind to murein merely by electrostatic forces. Indeed, we found that phosphorylase b from rabbit muscle as well as some highly abundant proteins from an E. coli extract bound to murein (data not shown). Nevertheless, our control experiments also showed that murein binding does not seem to be a property of the majority of randomly chosen proteins. The following facts indicate that the murein binding of FtsN might be related to a specific function: (i) the interaction with murein occurs via the C-terminal globular domain that is, like the murein sacculus, located in the periplasm; (ii) the murein-binding domain of FtsN is present in other proteins related to murein, e.g., amidase CwlC from Bacillus subtilis; and (iii) FtsN is placed in the recruitment hierarchy for the septal ring between PBP3 and AmiC, both of which are involved in murein metabolism. The periplasmic region of FtsN starts with the first part (amino acids 48 to 123 containing three short helices), followed by a nonstructured, presumably flexible region (amino acids 124 to 242) and the C-terminal globular domain (amino acids 243 to 319) (Fig. 6) (64). If fully stretched, the periplasmic portion of FtsN could span about 70 nm, which is about three to four times more than the thickness of the periplasm (45). Thus, it appears that the protein that is anchored to the cytoplasmic membrane at the cell division site bridges via the flexible part to the murein sacculus.
Our experiments on the binding of radioactively labeled murein fragments to the soluble FtsN58-319GSH6 revealed that neither muropeptides nor peptides were able to bind to the protein. However, long (more than ∼25 disaccharide units) murein glycan strands, devoid of the peptide moieties, specifically and efficiently bind to FtsN58-319GSH6, but no binding of strands shorter than ∼25 disaccharide units was observed. The structural basis for the recognition of only long but not short glycans by FtsN is unknown. Although one cannot unequivocally disprove such a possibility, it seems highly unlikely that the long glycans carry some substitution that is recognized by FtsN, because the long glycan material can be quantitatively processed to disaccharide subunits and there are no indications of the presence of covalently attached compounds (33). On the other hand, only the long murein glycan strands either might be able to adopt a special conformation that is prone for the binding to FtsN or might allow the binding of several FtsN molecules in a cooperative way. It would be extremely difficult to prepare long glycans of defined length from murein in amounts needed to solve the structure in complex with FtsN.
In accordance with the results of the binding experiments with purified FtsN58-319GSH6 and isolated murein or murein glycan strands, FtsN could be cross-linked to murein in vivo with the water-soluble cross-linker DTSSP, indicating that FtsN binds to murein in vivo. We have indications that the binding of FtsN to murein occurs during cell division. FtsN58-319GSH6 binding was significantly reduced when murein sacculi from cells blocked in cell division by the inhibition of PBP3 were used in the binding assay. Furthermore, the amount of FtsN that was cross-linked to murein in vivo was also reduced upon inhibition of cell division, whereas the amount of cross-linked OmpA, an outer membrane protein that was shown to bind to murein but has no role in cell division (53, 60), was not affected. Thus, the binding of FtsN to murein seems to occur mainly in dividing cells. It is possible that FtsN binds primarily to new (septal) murein. Considering the binding of FtsN to longer glycan strands, the following questions remain. Do long glycans have a specific localization at the division site in the sacculus? Does the inhibition of PBP3 affect the distribution of long glycans in the sacculus in such a way that there are fewer binding sites for FtsN? Unfortunately, it is not possible with the presently available methods to visualize the long glycan strands on isolated sacculi.
By production of truncated FtsN forms with simultaneous depletion of full-length FtsN, we were able to demonstrate that the murein-binding site of FtsN is not required for cell division. When repressed, the pBAD promoter on the pBAD33-ftsN plasmid is very tight (28). It is unlikely that cell division was enabled by a few copies of full-length FtsN that were still produced upon depletion and was active only in the presence (but not in the absence) of certain truncated FtsN variants. Murein binding of FtsN might stabilize the septal region, but this function seems not to be essential under standard laboratory conditions. A possible stabilizing function might be supported by the finding of the relatively high copy number of FtsN per cell of 3,000 to 6,000. Other cell division proteins (FtsQ, FtsL) are extremely scarce (ca. 20 to 40 molecules/cell), and so far only FtsZ was found to be present in a higher number of about 20,000 molecules per cell (5). Because FtsN is required for the septal recruitment of AmiC (4), an amidase involved in the splitting of the septum during division (34), it is possible that the murein-bound C-terminal domain of FtsN could be the receptor for AmiC at the division site. This function would not be essential, because in the absence of AmiC other murein hydrolases are active in splitting the septum in an FtsN-independent way (34). However, unlike a deletion mutant lacking AmiC, the cells producing only a truncated FtsN lacking the murein-binding domain did not show a mild chaining phenotype, indicating that AmiC might be functional in the absence of the murein-binding site of FtsN. Alternatively, the binding of FtsN to murein could have a function that is not directly linked to the septation process.
We found that the periplasmic region in FtsN between amino acids 53 and 138 is required for division. This part is located C terminal of the cytoplasmic transmembrane helix and contains three short helices. Our observation is in line with the finding that the part of FtsN required for its recruitment to the septum was found to be present in the periplasm (1). Four amino acids that are important for the localization of FtsN at the septum were identified in the noncatalytic domain and close to the membrane anchor of PBP3 (63). Thus, there might be a site of interaction between FtsN and PBP3 in close proximity to the membrane that is important for the septal recruitment and/or function of FtsN. Overproduction of FtsN was shown to suppress fts mutations in ftsA, ftsQ, ftsI, and ftsK (13, 20), and a recent study using two-hybrid assays identified possible interactions of FtsN with FtsN itself, FtsQ, FtsW, and PBP3 (18). Considering the more than 30-fold excess in copy number of FtsN over the approximately 130 copies per cell of PBP3 (19) and the fact that localization of FtsN depends on the presence and activity of PBP3 (1), one could also envision a more complex mechanism for FtsN localization in which initial or ongoing septal murein synthesis with participation of PBP3 is required for the septal recruitment of FtsN. However, it is still not known how FtsN, after being recruited to the septum, participates in the process of septation and what the basis of its essentiality for cell division is.
Acknowledgments
We are grateful to J. C. Chen from the J. Beckwith Laboratory, Harvard Medical School, Boston, Mass., for the strain JOE565, K. Hantke from the Universität Tübingen for plasmid pWKS30, H. Schwarz from the Max-Planck-Institut für Entwicklungsbiologie, Tübingen, Germany, for the antiserum against OmpA, and F. Götz from the Department of Microbial Genetics, Universität Tübingen, for his support.
This project was supported in part by the European Commission within the SANITAS network (QLK3-CT-2000-00079). M.A.P. was supported by grant no. BCM2001-2264 from the PGC and an institutional grant from Fundación Ramón Areces to the CBMSO.
REFERENCES
- 1.Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25:303-309. [DOI] [PubMed] [Google Scholar]
- 2.Addinall, S. G., and J. Lutkenhaus. 1996. FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol. 178:7167-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begg, K. J., S. J. Dewar, and W. D. Donachie. 1995. A new Escherichia coli cell division gene, ftsK. J. Bacteriol. 177:6211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhardt, T. G., and P. A. de Boer. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi, E., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 6.Boyle, D. S., M. M. Khattar, S. G. Addinall, J. Lutkenhaus, and W. D. Donachie. 1997. ftsW is an essential cell-division gene in Escherichia coli. Mol. Microbiol. 24:1263-1273. [DOI] [PubMed] [Google Scholar]
- 7.Buddelmeijer, N., M. E. Aarsman, A. H. Kolk, M. Vicente, and N. Nanninga. 1998. Localization of cell division protein FtsQ by immunofluorescence microscopy in dividing and nondividing cells of Escherichia coli. J. Bacteriol. 180:6107-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buddelmeijer, N., N. Judson, D. Boyd, J. J. Mekalanos, and J. Beckwith. 2002. YgbQ, a cell division protein in Escherichia coli and Vibrio cholerae, localizes in codependent fashion with FtsL to the division site. Proc. Natl. Acad. Sci. USA 99:6316-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carson, M. J., J. Barondess, and J. Beckwith. 1991. The FtsQ protein of Escherichia coli: membrane topology, abundance, and cell division phenotypes due to overproduction and insertion mutations. J. Bacteriol. 173:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casabadan, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 11.Chen, J. C., and J. Beckwith. 2001. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42:395-413. [DOI] [PubMed] [Google Scholar]
- 12.Cooper, S. 1991. Synthesis of the cell surface during the division cycle of rod-shaped, gram-negative bacteria. Microbiol. Rev. 55:649-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai, K., Y. Xu, and J. Lutkenhaus. 1993. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts). J. Bacteriol. 175:3790-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai, K., Y. Xu, and J. Lutkenhaus. 1996. Topological characterization of the essential Escherichia coli cell division protein FtsN. J. Bacteriol. 178:1328-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniel, R. A., E. J. Harry, V. L. Katis, R. G. Wake, and J. Errington. 1998. Characterization of the essential cell division gene ftsL(yIID) of Bacillus subtilis and its role in the assembly of the division apparatus. Mol. Microbiol. 29:593-604. [DOI] [PubMed] [Google Scholar]
- 16.de Boer, P., R. Crossley, and L. Rothfield. 1992. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359:254-256. [DOI] [PubMed] [Google Scholar]
- 17.de Pedro, M. A., J. C. Quintela, J. V. Holtje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Lallo, G., M. Fagioli, D. Barionovi, P. Ghelardini, and L. Paolozzi. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149:3353-3359. [DOI] [PubMed] [Google Scholar]
- 19.Dougherty, T. J., K. Kennedy, R. E. Kessler, and M. J. Pucci. 1996. Direct quantitation of the number of individual penicillin-binding proteins per cell in Escherichia coli. J. Bacteriol. 178:6110-6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Draper, G. C., N. McLennan, K. Begg, M. Masters, and W. D. Donachie. 1998. Only the N-terminal domain of FtsK functions in cell division. J. Bacteriol. 180:4621-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erickson, H. P. 2001. The FtsZ protofilament and attachment of ZipA—structural constraints on the FtsZ power stroke. Curr. Opin. Cell Biol. 13:55-60. [DOI] [PubMed] [Google Scholar]
- 22.Garcia del Portillo, F., and M. A. de Pedro. 1990. Differential effect of mutational impairment of penicillin-binding proteins 1A and 1B on Escherichia coli strains harboring thermosensitive mutations in the cell division genes ftsA, ftsQ, ftsZ, and pbpB. J. Bacteriol. 172:5863-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghigo, J. M., and J. Beckwith. 2000. Cell division in Escherichia coli: role of FtsL domains in septal localization, function, and oligomerization. J. Bacteriol. 182:116-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glauner, B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172:451-464. [DOI] [PubMed] [Google Scholar]
- 25.Glauner, B., and J. V. Höltje. 1990. Growth pattern of the murein sacculus of Escherichia coli. J. Biol. Chem. 265:18988-18996. [PubMed] [Google Scholar]
- 26.Glauner, B., J. V. Höltje, and U. Schwarz. 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 263:10088-10095. [PubMed] [Google Scholar]
- 27.Guzman, L. M., J. J. Barondess, and J. Beckwith. 1992. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J. Bacteriol. 174:7716-7728. [PMC free article] [PubMed] [Google Scholar]
- 28.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hale, C. A., and P. A. de Boer. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175-185. [DOI] [PubMed] [Google Scholar]
- 30.Hale, C. A., and P. A. de Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Hartmann, R., J. V. Höltje, and U. Schwarz. 1972. Targets of penicillin action in Escherichia coli. Nature 235:426-429. [DOI] [PubMed] [Google Scholar]
- 33.Harz, H., K. Burgdorf, and J. V. Höltje. 1990. Isolation und separation of the glycan strands from murein of Escherichia coli by reversed phase high-performance liquid chromatography. Anal. Biochem. 190:120-128. [DOI] [PubMed] [Google Scholar]
- 34.Heidrich, C., M. F. Templin, A. Ursinus, M. Merdanovic, J. Berger, H. Schwarz, M. A. de Pedro, and J. V. Höltje. 2001. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41:167-178. [DOI] [PubMed] [Google Scholar]
- 35.Ize, B., N. R. Stanley, G. Buchanan, and T. Palmer. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol. Microbiol. 48:1183-1193. [DOI] [PubMed] [Google Scholar]
- 36.Kraft, A. R., J. Prabhu, A. Ursinus, and J. V. Höltje. 1999. Interference with murein turnover has no effect on growth but reduces β-lactamase induction in Escherichia coli. J. Bacteriol. 181:7192-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leduc, M., K. Ishidate, N. Shakibai, and L. Rothfield. 1992. Interactions of Escherichia coli membrane lipoproteins with the murein sacculus. J. Bacteriol. 174:7982-7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, Z., A. Mukherjee, and J. Lutkenhaus. 1999. Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol. 31:1853-1861. [DOI] [PubMed] [Google Scholar]
- 39.Lowe, J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203-206. [DOI] [PubMed] [Google Scholar]
- 40.Lowe, J., and L. A. Amos. 1999. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 18:2364-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lutgenberg, B. J., J. Meijers, R. Peters, P. van der Hoek, and L. van Alphen. 1975. Electrophoretic resolution of the “major outer membrane protein” of Escherichia coli K12 into four bands. FEBS Lett. 58:254-258. [DOI] [PubMed] [Google Scholar]
- 42.Ma, X., D. W. Ehrhardt, and W. Margolin. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA 93:12998-13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma, X., Q. Sun, R. Wang, G. Singh, E. L. Jonietz, and W. Margolin. 1997. Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J. Bacteriol. 179:6788-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 45.Matias, V. R., A. Al-Amoudi, J. Dubochet, and T. J. Beveridge. 2003. Cryotransmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 185:6112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mercer, K. L., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nanninga, N. 2001. Cytokinesis in prokaryotes and eukaryotes: common principles and different solutions. Microbiol. Mol. Biol. Rev. 65:319-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen-Disteche, M., C. Fraipont, N. Buddelmeijer, and N. Nanninga. 1998. The structure and function of Escherichia coli penicillin-binding protein 3. Cell Mol. Life Sci. 54:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pichoff, S., B. Vollrath, and J. P. Bouche. 1997. MinCD-independent inhibition of cell division by a protein that fuses MalE to the topological specificity factor MinE. J. Bacteriol. 179:4616-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pucciarelli, M. G., and F. Garcia del Portillo. 2003. Protein-peptidoglycan interactions modulate the assembly of the needle complex in the Salmonella invasion-associated type III secretion system. Mol. Microbiol. 48:573-585. [DOI] [PubMed] [Google Scholar]
- 52.Raychaudhuri, D., and J. T. Park. 1992. Escherichia coli cell division gene ftsZ encodes a novel GTP-binding protein. Nature 359:251-254. [DOI] [PubMed] [Google Scholar]
- 53.Rosenbusch, J. P. 1974. Characterization of the major envelope protein from Escherichia coli. Regular arrangement on the peptidoglycan and unusual dodecyl sulfate binding. J. Biol. Chem. 249:8019-8029. [PubMed] [Google Scholar]
- 54.Schmidt, K. L., N. D. Peterson, R. J. Kustusch, M. C. Wissel, B. Graham, G. J. Phillips, and D. S. Weiss. 2004. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 186:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van den Ent, F., and J. Löwe. 2000. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 19:5300-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vollmer, W., M. von Rechenberg, and J. V. Höltje. 1999. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 274:6726-6734. [DOI] [PubMed] [Google Scholar]
- 57.Wang, L., M. K. Khattar, W. D. Donachie, and J. Lutkenhaus. 1998. FtsI and FtsW are localized to the septum in Escherichia coli. J. Bacteriol. 180:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 59.Wang, X., J. Huang, A. Mukherjee, C. Cao, and J. Lutkenhaus. 1997. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, Y. 2002. The function of OmpA in Escherichia coli. Biochem. Biophys. Res. Commun. 292:396-401. [DOI] [PubMed] [Google Scholar]
- 61.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weiss, D. S., K. Pogliano, M. Carson, L. M. Guzman, C. Fraipont, M. Nguyen-Disteche, R. Losick, and J. Beckwith. 1997. Localization of the Escherichia coli cell division protein Ftsl (PBP3) to the division site and cell pole. Mol. Microbiol. 25:671-681. [DOI] [PubMed] [Google Scholar]
- 63.Wissel, M. C., and D. S. Weiss. 2004. Genetic analysis of the cell division protein FtsI (PBP3): amino acid substitutions that impair septal localization of FtsI and recruitment of FtsN. J. Bacteriol. 186:490-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, J. C., F. van den Ent, D. Neuhaus, J. Brevier, and J. Löwe. 2004. Solution structure and domain architecture of the divisome protein FtsN. Mol. Microbiol. 52:651-660. [DOI] [PubMed] [Google Scholar]