Abstract
Background/Aims
Chronic liver disease is a major widespread cause of death, and whole liver transplantation is the only definitive treatment for patients with end-stage liver diseases. However, many problems, including donor shortage, surgical complications and cost, hinder their usage. Recently, tissue-engineering technology provided a potential breakthrough for solving these problems. Three-dimensional (3D) printing technology has been used to mimic tissues and organs suitable for transplantation, but applications for the liver have been rare.
Methods
A 3D bioprinting system was used to construct 3D printed hepatic structures using alginate. HepG2 cells were cultured on these 3D structures for 3 weeks and examined by fluorescence microscopy, histology and immunohistochemistry. The expression of liver-specific markers was quantified on days 1, 7, 14, and 21.
Results
The cells grew well on the alginate scaffold, and liver-specific gene expression increased. The cells grew more extensively in 3D culture than two-dimensional culture and exhibited better structural aspects of the liver, indicating that the 3D bioprinting method recapitulates the liver architecture.
Conclusions
The 3D bioprinting of hepatic structures appears feasible. This technology may become a major tool and provide a bridge between basic science and the clinical challenges for regenerative medicine of the liver.
Keywords: Hep G2 cell, Printing, three-dimensional
INTRODUCTION
For patients suffering from end-stage liver disease including chronic liver fibrosis or alcoholic cirrhosis, whole liver transplantation is the only definitive treatment. Although thousands of liver transplants have been performed over recent decades, most patients die while waiting their turn due to a shortage of livers for transplantation. To overcome this, many scientists have suggested the use of hepatocyte transplantation instead of liver organ transplantation. However, hepatocyte transplantation requires an unlimited source of hepatocytes. Furthermore, the utilization of primary hepatocytes has limitation by itself for the application due to their extremely slow proliferation, and is rarely optimal for long-term culture systems.
To obtain a number of hepatocytes, direct differentiation technology using pluripotent stem cells (PSCs) has been developed.1–4 Since PSCs can replicate themselves infinitely and differentiate into most of cell types including hepatocytes, they are considered as an alternative cell source for supplying unlimited number of hepatocytes for transplantation.5–8 However, although PSC-derived hepatocyte-like cells have representative features of primary hepatocytes, repopulation capacity in vivo, as well as restoration of the liver function,9–11 most researchers are aware of the inherent limitations of cells grown under two-dimensional (2D) conditions.12–14
In general, tissues and organs have characteristic surfaces of unique shape, and are composed of cells interacting physiologically and biochemically. In order to better understand disease mechanisms and obtain cells for disease modelling and drug screening, three-dimensional (3D) structures are desirable15,16 since the true 3D microenvironment of target tissues and organs can never be mimicked by 2D models.
3D tissue-engineering technology has been advanced and considered to solve the problems of 2D culture system and better capture the complexity of in vivo environments. As a general rule, in 3D modelling the formation of a scaffold is a prerequisite step for reconstructing the unique structures of target tissues and organs for determining cell behavior and controlling physical cell-to-cell connections.17–19 Recently tissue-engineering methods combined with 3D printing technology have emerged as the most useful and efficient tools for producing complicated scaffold structures.20–22 Therefore, 3D bioprinting technology may successfully mimic complicating complex organs with systemic 3D microarchitecture including liver suitable for transplantation, and eventually overcome a plenty of obstacles including donor shortage, surgical complications and exorbitant cost for liver transplantation.20–22
In the present study, we utilized 3D bioprinting technology for reconstructing liver tissues or organs. To achieve this goal, we used HepG2 cells, one of the liver cancer-derived cells to be applied to 3D printing technology. Newly fabricated, multiple layered-3D structures were efficiently modeled by mixing alginate with HepG2 cells. Finally, we suggest that mimicking the 3D hepatic structure by using 3D printing technology not only assists the HepG2 cells to stably repopulate, but also improves their gene expression profiles.
MATERIALS AND METHODS
1. Cell culture and maintenance
HepG2 cells, human hepatoblastoma cells derived from hepatocellular carcinoma were cultured in in 75T Flasks in DMEM (11965; Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (10378; Gibco) at 37°C in a CO2 incubator. The medium was changed every 2 to 3 days and the cells were used after about 14 passages.
2. Lentivirus production
Lentiviruses were produced as previously described. mCherry was packaged by cotransfection with psPAX2 lentiviral packaging plasmid and pCMV-VSV-G plasmid in human embryonic kidney 293T cells. Culture supernatants were harvested after 48 hours and 72 hours. The virus was resuspended in the culture medium, aliquoted and stored at −80°C. Lentiviral transduction of the mCherry was carried out in culture medium supplemented with 4 μg/mL polybrene.
3. Three-dimensional bioprinting of HepG2 cells with alginate
Three-dimensional printed strands of encapsulated cells were made with a 3D bioprinting system fabricated by the Korea Institute of Machinery and Materials (KIMM). Before 3D printing, liver cells were prepared in 3% alginate solution. The nozzle size of the printer was 400 μm, the strand pitch was 1,000 μm and the 3D printed hydrogel structures measured 25 mm by 25 mm. For cell printing, a continuous air pressure of 300 kPa was supplied to the dispenser, and cell-encapsulated alginate was extruded onto 10 cm2 culture dish. After printing, the 3D alginate hydrogel was soaked with 1% calcium chloride solution for solid crosslinking, washed with phosphate buffered saline (PBS) and placed in HepG2 culture medium.
4. Quantitative polymerase chain reaction analysis
Total RNAs were isolated using TRIZOL Reagent (Gibco) Samples of 2 μg RNA were reverse transcribed with a Transcriptor First Strand cDNA Synthesis Kit (Roche; Branford, CT, USA), and real-time PCR was performed using 10 μL of LightCycler 480 SYBR Green1 Master (Roche, Branford, CT, USA), 1 μL of cDNA and oligonucleotide primers, on an LC488 qPCR system (Roche). Reactions were carried out in triplicate; the primers used for human Albumin, ASGPR1, AFP, CK19, Cyp1A2, TAT and GAPDH are listed in Table 1. The PCR cycles consisted of 45 cycles of 95°C, 60°C, and 72°C for 15 seconds each. Melting curves and melting peak data were obtained to characterize the PCR products.
Table 1.
Oligonucleotide Primers for Real-Time PCR
| Gene | Primer | Primer Sequence (5′-3′) | PCR product (bp) |
|---|---|---|---|
| Albumin | Forward | GCACAGAATCCTTGGTGAACAG | 100 |
| Reverse | ATGGAAGGTGAATGTTTCAGCA | ||
| ASGR1 | Forward | CAGCAACTTCACAGCGACCA | 96 |
| Reverse | AGCTGGGACTCTAGCGACTT | ||
| AFP | Forward | AGACTGCTGCAGCCAAAGTGA | 80 |
| Reverse | GTGGGATCGATGCTGGAGTG | ||
| CK19 | Forward | TCCGAACCAAGTTTGAGACG | 186 |
| Reverse | CCCTCAGCGTACTGATTTCC | ||
| Cyp1A2 | Forward | CGGACAGCACTTCCCTGAGA | 124 |
| Reverse | AGGCAGGTAGCGAAGGATGG | ||
| TAT | Forward | TGGGGACCCTACTGTGTTTGG | 105 |
| Reverse | ATGGATGGGGCATAGCCATT | ||
| GAPDH | Forward | TGCACCACCAACTGCTTAGC | 87 |
| Reverse | GGCATGGACTGTGGTCATGAG |
PCR, polymerase chain reaction; ASGR1, asialoglycoprotein receptor 1; AFP, alpha-fetoprotein; CK19, cytokeratin 19; Cyp1A2, cytochrome P450 superfamily of enzymes 1A2; TAT, tyrosine amino-transferase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
5. Immunocytochemistry
For immunofluorescence staining, cultured HepG2 cells were fixed in 4% paraformaldehyde in PBS for 15 minutes at room temperature. After washing three times with PBS, the cells were incubated with PBS containing 0.3% Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA) for 10 minutes at room temperature followed by blocking of nonspecific binding sites with 5% goat serum (S-1000; Vector Laboratories, Inc., Burlingame, CA, USA) in PBS for 40 minutes. After washing, the samples in wells were incubated with primary antibodies as follows: rabbit antihuman albumin antibody (CL2513A; Cedarlane, Burlington, ON, Canada) and rabbit antihuman α-1-fetoprotein (A0008; DAKO, Glostrup, Denmark) in 350 μL of 2% goat serum at 4°C overnight. Next, they were incubated with goat antimouse or antirabbit secondary antibody for 1h. After washing, they were counterstained with 3 μM DAPI (D-1306; Invitrogen, Carlsbad, CA, USA) for 4 minutes with shaking 70 to 80 rpm. Cells in the culture dishes were mounted with mounting solution (0100-01; Southern Biotech, Birmingham, AL, USA), and examined under a fluorescence microscope or a Thermo confocal laser scanning microscope.
6. Hematoxylin & eosin staining and immunohistochemistry
Sections were deparaffinized by immersion in xylene for 10 minutes and ethanol three times for 7 minutes, and stained with hematoxylin (Sigma-Aldrich) for 5 minutes. They were then counterstained with eosin for 1 minute, after washing with running water for 5 minutes. Finally, they were washed with running water and dehydrated in 95% ethanol for 5 minutes.
The 3D printed mCherry-HepG2 alginate scaffolds were fixed in 4% buffered formalin and embedded in paraffin. Four-micrometer thick sections were cut and mounted on silane-coated glass slides. All sections were deparaffinized and rehydrated with graded alcohols. After washing endogenous peroxidase was blocked with 3% H2O2 diluted in methanol for 15 minutes at room temperature. Antigen retrieval was performed by treating the slides in pH 6.0 citric acid buffer in an autoclave. The slides were incubated in a moist chamber with rabbit antihuman albumin antibody (CL2513A, 1:200; CEDARLANE), rabbit anti-human α-fetoprotein (A0008, 1:200; DAKO), and Ki-67 (1:200), overnight at 4°C. After one day, the slides were washed with Tris buffer pH 7.6 and incubated with postantibody at room temperature for 30 minutes. After thorough washing, the slides were incubated with polymer for 30 minutes. After washing, the slides were incubated for 3 minutes with 3, 3′-diaminobenzedine tetrahydrochloride solution (0.05%) (DAB; Leica, Wetzlar, Germany) together with 0.03% hydrogen peroxide to produce a brown reaction product, counterstained with hematoxylin, washed in running tap water, then dehydrated, cleared and mounted.
7. Statistical analysis
Quantitative data are presented as means±standard deviations (SD) and inferential statistics (p-values). Statistical significance was evaluated with two-tailed t-test with significance set at p<0.05, p<0.01, or p <0.001.
RESULTS
1. Morphology and characteristics of mCherry-tagged HepG2 cells
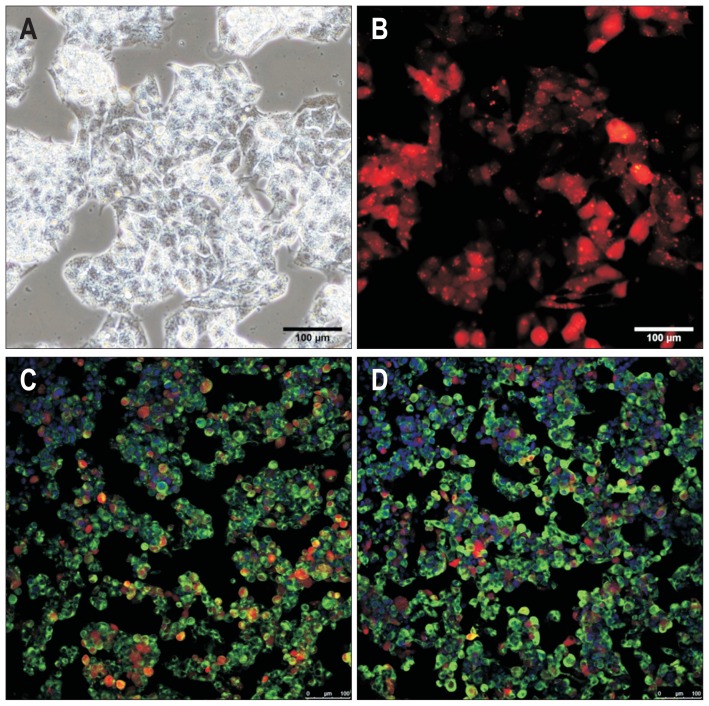
Immortalized liver-derived cells such as HepG2 cells are considered ideal for studying drug metabolism due to their unlimited availability and phenotypic stability. However, HepG2 cells exhibit relatively limited hepatic functionality compared with primary human hepatocytes in terms of genotoxic sensitivity, due to lack of drug-metabolizing enzymes, cytochromes (CYPs), and secretory functions as shown by relatively low expression of albumin, α-fetoprotein (AFP), and α-1 antitrypsin. Therefore, we mainly used HepG2 cells as a target cell source for improving their hepatic functionality in the current study. In conventional 2D monolayer culture the HepG2 cells typically displayed epithelial cell-like morphology (Fig. 1A). The HepG2 cells tagged with the red fluorescent protein, mCherry, for effectively tracing their localization and visualizing proliferative capacity were strongly positive for hepatic markers albumin and AFP (Fig. 1B–D) indicating that they had basic hepatic features under 2D monolayer culture conditions.
Fig. 1.
Expression of hepatic genes in HepG2 cells. (A) Morphology of HepG2 cells grown in two-dimensional culture. (B) HepG2 cells labeled with mCherry. (C, D) Immunofluorescence microscopy image of HepG2 cells incubated with anti-ALBUMIN and anti-AFP antibodies (green). Nuclei are stained with DAPI. Bar, 100 μm.
2. Construction of 3D printed mCherry-HepG2 hepatic structures with alginate scaffolds
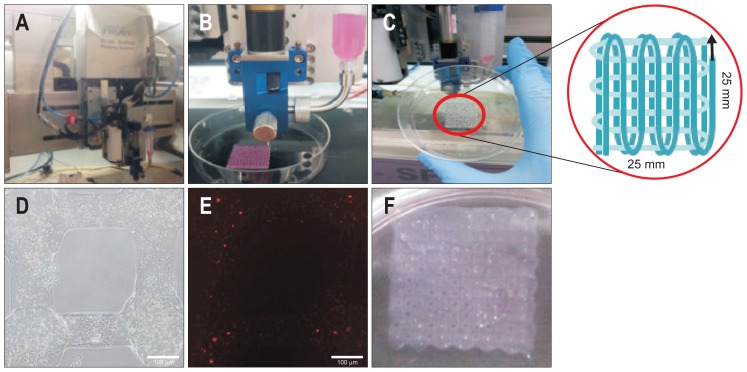
In an attempt to improve the hepatic function of the HepG2 cells, we used 3D bioprinting technology to construct 3D-printed mCherry-HepG2 hepatic structures (Fig. 2A–E). The alginate was mixed with mCherry-HepG2 cells and extruded through nozzle pressure (Fig. 2B). After crosslinking, 25 mm×25 mm structures were formed (Fig. 2C). Phase contrast microscope images revealed monolayers of compact HepG2 cells within the alginate scaffold of the designated pattern (Fig. 2D). mCherry-expressing cells were observed inside the alginate scaffold under a fluorescence microscope (Fig. 2E). Multiple layers of mCherry-HepG2 cells were constructed by repetitively adding and solidifying monolayers to be intersected at right angles (Fig. 2F). Throughout 3D bioprinting method, we can readily establish 3D-printed mCherry-HepG2 hepatic structures for mimicking liver architecture.
Fig. 2.
Construction of three-dimensional (3D) printed mCherry-HepG2 hepatic structures. (A) Instrument for 3D-bioprinting. (B) Alginate containing mCherry-HepG2 cells were extruded by nozzle pressure. (C) Cross-linked structures were deposited layer-by-layer in 25 mm×25 mm square arrays. (D) Phase contrast microscopy images of a confluent monolayer of HepG2 cells. Bar, 100 μm. (E) Fluorescence micrograph of mCherry-HepG2 cells inside the alginate scaffold. Bar, 100 μm. (F) Multilayered mCherry-HepG2 cells repetitively solidified and layered upon each other.
3. Proliferation and repopulation of mCherry-HepG2 cells in 3D-printed hepatic structure
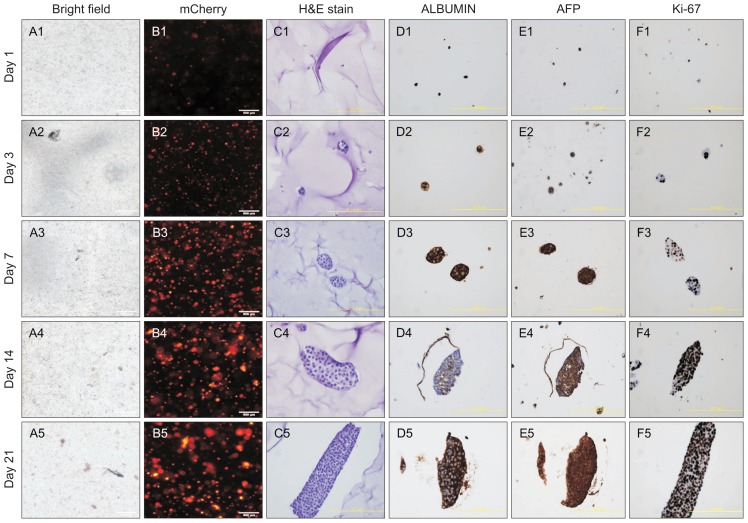
Next, we have investigated whether the 3D-printed hepatic structures functionally assisted the HepG2 cells to proliferate and populate the structures. As expected, the number of mCher-ry-HepG2 cells was stably amplified in time course manner (Fig. 3A), and we confirmed the proliferation of the cells by fluorescence microscope images (Fig. 3B). The HepG2 cells appeared to be highly compact by days 14 and 21 after seeding, indicating that they stably proliferated and repopulated to form 3D-printed hepatic structures, as revealed by hematoxylin and eosin staining analysis (Fig. 3C). Surprisingly, the cells strongly expressed ALBUMIN, AFP, and also Ki-67, demonstrating that the highly proliferating HepG2 cells hold the typical hepatic property in 3D-printed hepatic structure (Fig. 3D–F). These data suggest that the 3D-printed framework helps the HepG2 cells to proliferate without losing their hepatic identity.
Fig. 3.
Proliferation of HepG2 cells in the three-dimensional (3D) culture system. (A1–5, B1–5) Bright field image of HepG2 cells grown in 3D culture (A1–5) and immunofluorescence image of HepG2 cells labeled with mCherry (B1–5). Bar, 500 μm. (C1–5, D1–5) Images of H&E staining (C1–5) and immunohistochemistry using antibodies against ALBUMIN (D1–5), AFP (E1–5), and Ki-67 (F1–5). Bar, 400 μm. The images were captured on days 1, 3, 7, 14, and 21 after seeding the HepG2 cells on 3D-printed scaffolds.
4. Real-time PCR analysis of the 3D hepatic structures
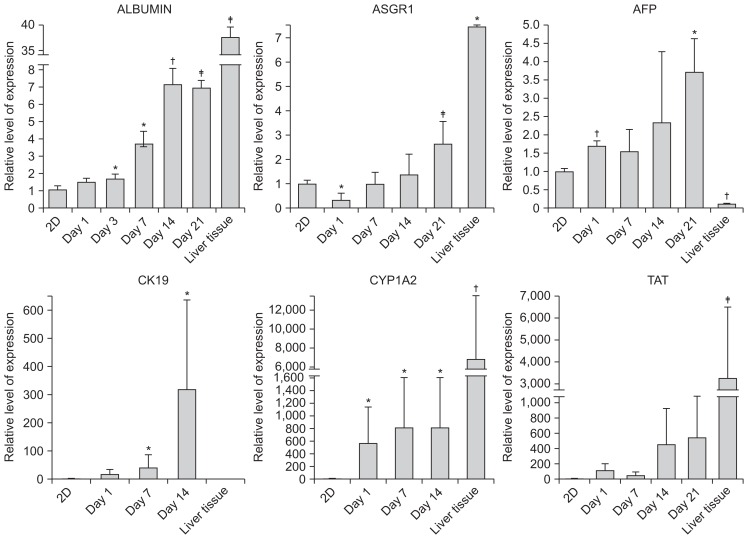
Finally, we tested if the 3D structure improved the hepatic function of HepG2 cells in terms of gene expression. To the end, we evaluated the mRNA levels of the representative hepatic markers, ALBUMIN, ASGR1, and AFP. The expression of ASGR1 and AFP had modestly increased by day 21 after seeding (Fig. 4), but the expression of ALBUMIN had already increased significantly after only 3 days of seeding (Fig. 4). Taken together, these data and those described previously show that the 3D-printed structures not only assist the HepG2 cells to multiply but also enhance their hepatic function.
Fig. 4.
Enhanced expression of hepatic markers in HepG2 cells in three-dimensional (3D) culture. Expression levels of the hepatic markers ALBUMIN, ASGPR1, AFP, CYP1A2, and TAT in HepG2 cells after culture under 3D conditions. The expression levels were normalized to those of mouse embryonic fibroblasts. *p<0.05; †p<0.01; ‡p<0.001.
DISCUSSION
End stage of liver diseases such as cirrhosis causes severe liver failure and portal hypertension. To deal with the liver failure at the end stage, liver transplantation is ultimately the only treatment at the terminal stages of disease, but the demand for liver transplantation is more than number of provision cadaveric livers or liver tissues from living donors. Therefore, human hepatocytes are considered as the most desirable source for the therapy, instead of liver organ transplantation. However, the transplantation of hepatocytes requires a number of cells in therapy, indicating that it needs an unlimited source of hepatocytes.23
Due to infinite replication capacity and differentiation potential into most of cells and tissues in our body, PSCs including embryonic stem cells or induced PSCs are regarded as alternative cell sources for therapeutic applications such as regenerative medicine and drug discovery. So far, many of strategies have been recently well developed for generating PSC-derived hepatocyte-like cells in vitro with high purity and maturity.1–4 The PSC-derived hepatocyte-like cells hold typical characters of hepatocytes with primary hepatocytes in terms of secretory functions, xenobiotic activity and detoxification functions. Moreover, they are able to repopulate after transplantation in vivo and restore the liver functions.5,7–11,24
Although the strategies for obtaining pure and high quality of hepatocyte-like cells from PSCs have been improved, however, it cannot be perfectly ruled out the risk of oncogenicity originated from residually undifferentiated cells upon differentiation. To overcome this issue, direct conversion technology of which directly convert a terminally differentiated somatic cells into another types of cells in our interest without passing through pluripotent state has been reported. In recent, several groups successfully developed the protocols of generating hepatocyte-like cells (so called as induced hepatocyte-like cells; iHeps) from somatic cells in human and mouse by ectopically expressing the defined sets of tissue-specific transcription factors.24–27 The iHeps also share hepatic properties with primary hepatocytes like as PSC-derived hepatocyte-like cells do in vitro.24–27 Similar with PSC-derived hepatocyte-like cells, the iHeps transplanted into mouse liver failure model are capable of repopulating the liver and restores normal liver structure without fusion with recipient cells.24–27
For many years, 2D culture systems have been widely used in biological research and drug screening. However, since 2D culture system cannot truly mimic the physiological environment in 3D tissues and organs including liver, the cells cultured in 2D inevitably lacks the micro-environmental influences present in natural 3D tissues.22,28 This factor has contributed to the poor predictive power of preclinical cell-based drug and toxicity screening assays.22,29,30 Furthermore, most candidate drugs passing through in vitro preclinical trials have failed to meet the desired efficacy or safety margins prior to entering subsequent clinical trials, perhaps partly for the same reason. To overcome the limitation of 2D culture system, 3D bioprinting culture models has been developed, which can reproduce true 3D pathological architecture of tissue and organs by fabricating scaffolds with a novel micro- and macro-architecture.22 Recently, Duan et al.31 have described 3D bioprinted aortic valve conduits composed of dual cell types, aortic root sinus smooth muscle cells (SMCs) and aortic valve leaflet interstitial cells (VICs), using fabricated alginate/gelatin hydrogel discs. Both SMCs and VICs were able to survive for over 7 days and displayed elevated levels of alpha-smooth muscle actin and vimentin expression, respectively. These results suggest that constructing 3D culture environment and fabricating natural structure complexity with cellular heterogeneity can be achieved by 3D bioprinting technology.
In the current study, we utilized 3D printing technology for mimicking liver tissues or organs.32–34 Multilayered-3D hepatic structures were efficiently constructed by mixing the alginate with HepG2 cells. As a consequence, we observed that mimicking the 3D hepatic structure via 3D printing technology herein not only assists the HepG2 cells to stably repopulate, but also improves their gene expression profiles. Taken together, our findings show that 3D bioprinting of liver tissue suitable for transplantation may be a realistic option for overcoming the problems of donor shortage and surgical complications and thereby offer a new paradigm in the field of liver regenerative medicine.
ACKNOWLEDGEMENTS
This work was supported by the research fund of Hanyang University (HY-2014).
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Agarwal S, Holton KL, Lanza R. Efficient differentiation of functional hepatocytes from human embryonic stem cells. Stem Cells. 2008;26:1117–1127. doi: 10.1634/stemcells.2007-1102. [DOI] [PubMed] [Google Scholar]
- 2.Sancho-Bru P, Roelandt P, Narain N, et al. Directed differentiation of murine-induced pluripotent stem cells to functional hepatocyte-like cells. J Hepatol. 2011;54:98–107. doi: 10.1016/j.jhep.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Shiraki N, Umeda K, Sakashita N, Takeya M, Kume K, Kume S. Differentiation of mouse and human embryonic stem cells into hepatic lineages. Genes Cells. 2008;13:731–746. doi: 10.1111/j.1365-2443.2008.01201.x. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan GJ, Hay DC, Park IH, et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 2010;51:329–335. doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basma H, Soto-Gutiérrez A, Yannam GR, et al. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology. 2009;136:990–999. doi: 10.1053/j.gastro.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan Y, Catana A, Meng Y, et al. Differentiation and enrichment of hepatocyte-like cells from human embryonic stem cells in vitro and in vivo. Stem Cells. 2007;25:3058–3068. doi: 10.1634/stemcells.2007-0291. [DOI] [PubMed] [Google Scholar]
- 7.Gai H, Nguyen DM, Moon YJ, et al. Generation of murine hepatic lineage cells from induced pluripotent stem cells. Differentiation. 2010;79:171–181. doi: 10.1016/j.diff.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Touboul T, Hannan NR, Corbineau S, et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–1765. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- 9.He ZY, Deng L, Li YF, et al. Murine embryonic stem cell-derived hepatocytes correct metabolic liver disease after serial liver repopulation. Int J Biochem Cell Biol. 2012;44:648–658. doi: 10.1016/j.biocel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soto-Gutiérrez A, Kobayashi N, Rivas-Carrillo JD, et al. Reversal of mouse hepatic failure using an implanted liver-assist device containing ES cell-derived hepatocytes. Nat Biotechnol. 2006;24:1412–1419. doi: 10.1038/nbt1257. [DOI] [PubMed] [Google Scholar]
- 11.Teratani T, Yamamoto H, Aoyagi K, et al. Direct hepatic fate specification from mouse embryonic stem cells. Hepatology. 2005;41:836–846. doi: 10.1002/hep.20629. [DOI] [PubMed] [Google Scholar]
- 12.Baharvand H, Hashemi SM, Kazemi Ashtiani S, Farrokhi A. Differentiation of human embryonic stem cells into hepatocytes in 2D and 3D culture systems in vitro. Int J Dev Biol. 2006;50:645–652. doi: 10.1387/ijdb.052072hb. [DOI] [PubMed] [Google Scholar]
- 13.Gieseck RL, 3rd, Hannan NR, Bort R, et al. Maturation of induced pluripotent stem cell derived hepatocytes by 3D-culture. PLoS One. 2014;9:e86372. doi: 10.1371/journal.pone.0086372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godoy P, Hewitt NJ, Albrecht U, et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol. 2013;87:1315–1530. doi: 10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mironov V, Boland T, Trusk T, Forgacs G, Markwald RR. Organ printing: computer-aided jet-based 3D tissue engineering. Trends Biotechnol. 2003;21:157–161. doi: 10.1016/S0167-7799(03)00033-7. [DOI] [PubMed] [Google Scholar]
- 16.Williams DF. On the mechanisms of biocompatibility. Biomaterials. 2008;29:2941–2953. doi: 10.1016/j.biomaterials.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 17.Bailey MJ, Schulten K, Johnson JE. The use of solid physical models for the study of macromolecular assembly. Curr Opin Struct Biol. 1998;8:202–208. doi: 10.1016/S0959-440X(98)80039-0. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu TS, Le Novère N, Levin MD, Beavil AJ, Sutton BJ, Bray D. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat Cell Biol. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- 19.Symes MD, Kitson PJ, Yan J, et al. Integrated 3D-printed reaction-ware for chemical synthesis and analysis. Nat Chem. 2012;4:349–354. doi: 10.1038/nchem.1313. [DOI] [PubMed] [Google Scholar]
- 20.Chia HN, Wu BM. Recent advances in 3D printing of biomaterials. J Biol Eng. 2015;9:4. doi: 10.1186/s13036-015-0001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones N. Science in three dimensions: the print revolution. Nature. 2012;487:22–23. doi: 10.1038/487022a. [DOI] [PubMed] [Google Scholar]
- 22.Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat Biotechnol. 2014;32:773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 23.Dhawan A, Mitry RR, Hughes RD. Hepatocyte transplantation for liver-based metabolic disorders. J Inherit Metab Dis. 2006;29:431–435. doi: 10.1007/s10545-006-0245-8. [DOI] [PubMed] [Google Scholar]
- 24.Du Y, Wang J, Jia J, et al. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell. 2014;14:394–403. doi: 10.1016/j.stem.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Huang P, He Z, Ji S, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 26.Huang P, Zhang L, Gao Y, et al. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell. 2014;14:370–384. doi: 10.1016/j.stem.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 28.Boland T, Mironov V, Gutowska A, Roth EA, Markwald RR. Cell and organ printing 2: fusion of cell aggregates in three-dimensional gels. Anat Rec A Discov Mol Cell Evol Biol. 2003;272:497–502. doi: 10.1002/ar.a.10059. [DOI] [PubMed] [Google Scholar]
- 29.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonntag F, Schilling N, Mader K, et al. Design and prototyping of a chip-based multi-micro-organoid culture system for substance testing, predictive to human (substance) exposure. J Biotechnol. 2010;148:70–75. doi: 10.1016/j.jbiotec.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Duan B, Hockaday LA, Kang KH, Butcher JT. 3D bioprinting of heterogeneous aortic valve conduits with alginate/gelatin hydrogels. J Biomed Mater Res A. 2013;101:1255–1264. doi: 10.1002/jbm.a.34420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakab K, Norotte C, Marga F, et al. Tissue engineering by self-assembly and bio-printing of living cells. Biofabrication. 2010;2:022001. doi: 10.1088/1758-5082/2/2/022001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marga F, Jakab K, Khatiwala C, et al. Toward engineering functional organ modules by additive manufacturing. Biofabrication. 2012;4:022001. doi: 10.1088/1758-5082/4/2/022001. [DOI] [PubMed] [Google Scholar]
- 34.Kwon YJ, Lee KG, Choi D. Clinical implications of advances in liver regeneration. Clin Mol Hepatol. 2015;21:7–13. doi: 10.3350/cmh.2015.21.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]