Abstract
We found the occurrence of thermophilic reversible γ-resorcylate decarboxylase (γ-RDC) in the cell extract of a bacterium isolated from natural water, Rhizobium sp. strain MTP-10005, and purified the enzyme to homogeneity. The molecular mass of the enzyme was determined to be about 151 kDa by gel filtration, and that of the subunit was 37.5 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis; in other words, the enzyme was a homotetramer. The enzyme was induced specifically by the addition of γ-resorcylate to the medium. The enzyme required no coenzyme and did not act on 2,4-dihydroxybenzoate, 2,5-dihydroxybenzoate, 3,4-dihydroxybenzoate, 3,5-dihydroxybenzoate, 2-hydroxybenzoate, or 3-hydroxybenzoate. It was relatively thermostable to heat treatment, and its half-life at 50°C was estimated to be 122 min; furthermore, it catalyzed the reverse carboxylation of resorcinol. The values of kcat/Km (mΜ−1 · s−1) for γ-resorcylate and resorcinol at 30°C and pH 7 were 13.4 and 0.098, respectively. The enzyme contains 327 amino acid residues, and sequence identities were found with those of hypothetical protein AGR C 4595p from Agrobacterium tumefaciens strain C58 (96% identity), 5-carboxyvanillate decarboxylase from Sphingomonas paucimobilis (32%), and 2-amino-3-carboxymuconate-6-semialdehyde decarboxylases from Bacillus cereus ATCC 10987 (26%), Rattus norvegicus (26%), and Homo sapiens (25%). The genes (graA [1,230 bp], graB [888 bp], and graC [1,056 bp]) that are homologous to those in the resorcinol pathway also exist upstream and downstream of the γ-RDC gene. Judging from these results, the resorcinol pathway also exists in Rhizobium sp. strain MTP-10005, and γ-RDC probably catalyzes a reaction just before the hydroxylase in it does.
Hydroxybenzoate decarboxylases (EC 4.1.1.x) are found in various microorganisms (1, 10, 12, 34, 36, 43) and catalyze the conversion of hydroxybenzoates, such as 2,3-dihydroxybenzoate (18, 30, 36), 2,5-dihydroxybenzoate (12), 3,4-dihydroxybenzoate (15), 4,5-dihydroxyphthalate (24, 29, 34), and 4-hydroxybenzoate (14, 16), to the respective phenol derivatives. Most hydroxybenzoate decarboxylases catalyze the reaction reversibly (14, 15) and require neither a cofactor nor a carbonyl group for catalytic activity (15, 36). Interestingly, the enzyme acting on 2,6-dihydroxybenzoate (γ-resorcylate) has never been reported.
Rhizobium is the typical genus of a tubercle-forming bacterium; it grows in symbiosis with the root of a plant, such as Astragalus sinicus, Trifolium repens L., or Medicago polymorpha Linn to fix nitrogen in the air (3). Therefore, much attention has been paid to the study of the Rhizobium enzymes and genes related to the regulation of symbiosis (13, 22, 39), but little information is available about the molecular structure, function, and detailed properties of the enzymes of the other metabolic pathways.
During the course of a screening experiment, we isolated a microorganism, identified as Rhizobium sp. strain MTP-10005, that shows a high level of γ-resorcylate decarboxylase (γ-RDC) activity. The enzyme also catalyzes the reversible reaction and is specifically induced by the addition of γ-resorcylate; in addition, it is thermophilic. γ-Resorcylate is an important intermediate of medicine (33) and agricultural chemicals, but it is very difficult to synthesize by traditional chemical methods, since α- and β-resorcylate are produced as by-products. Furthermore, it is practically impossible to separate the by-products from the product. Using the reverse carboxyl reaction of γ-RDC, it is expected that γ-resorcylate can be produced specifically from resorcinol. To our knowledge, there is no report on the metabolic pathway for the γ-resorcylate catabolism in bacteria and other living organisms.
We here first describe the purification and characterization of γ-RDC from Rhizobium sp. strain MTP-10005 and its gene cloning and sequencing, with emphasis on the comparison with other hydroxybenzoate decarboxylases.
MATERIALS AND METHODS
Materials.
DEAE-Toyopearl 650 M and butyl-Toyopearl 650 M were products of Tosoh, Tokyo, Japan. The plasmid purification kit was purchased from Bio-Rad Laboratories Inc., and PCR reagents were products of Takara Bio Int., Kyoto, Japan. All other chemicals were purchased from Kanto Kagaku Co. (Tokyo, Japan), Kishida Chemical Co. (Osaka, Japan), Sigma Chemical Co., Tokyo Kasei Kogyo Co. (Tokyo, Japan), and Wako Chemical Co. (Osaka, Japan) unless otherwise stated.
Screening and culture conditions.
γ-Resorcylate (GR) medium was used for the isolation of the γ-RDC-producing bacterium. It contained γ-resorcylate (3.0 g), urea (0.5 g), KH2PO4 (0.5 g), K2HPO4 (0.5 g), MgSO4 · 7H2O (0.2 g), CaCl2 · 2H2O (0.1 g), NaFe-EDTA · 3H2O (42 mg), MnSO4 · 5H2O (25 mg), ZnSO4 · 7H2O (10 mg), H3BO3 (10 mg), NiCl2 · 6H2O (25 μg), Na2MoO4 · 2H2O (250 μg), CuSO4 · 5H2O (25 μg), CoCl2 · 6H2O (25 μg), nicotinic acid (5.0 mg), glycine (2.0 mg), pyridoxine hydrochloride (500 μg), thiamine hydrochloride (500 μg), folic acid (500 μg), d-biotin (50 μg), calcium pantothenate (500 μg), and vitamin B12 (500 μg) per 1 liter of deionized water. The pH was adjusted to 7.5. The medium was solidified by the addition of agar (1.5% [wt/vol]) when necessary. A small amount of soil suspended in 0.75% NaCl solution (100 μl) or natural water (1 ml) was added to the GR medium (5 ml) and incubated at 30°C with shaking (150 rpm). After the turbidity increased, 200 μl of the culture was inoculated into new GR medium (5 ml). This operation was repeated several times. Subsequently, the culture was spread on a GR agar plate to isolate a single colony. The γ-RDC-producing bacterium, strain MTP-10005, was selected from about 100 colonies and used throughout the course of this study.
Characterization and identification of the strain.
The morphological characteristics, taxonomic characteristics, and 16S rDNA sequence were determined by NCIMB Japan Co., Ltd. (Shizuoka, Japan). The phylogenetic analysis of the 16S rDNA sequence was carried out with the ClustalW program (http://www.ddbj.nig.ac.jp/search/clustalw-j.html).
Enzyme assays.
The γ-RDC and other hydroxybenzoate decarboxylase activities were assayed with a high-performance liquid chromatographer (HPLC) (LC-10A system; Shimadzu, Kyoto, Japan) with an Inertsil ODS-2 column (0.46 by 25 cm; GL Sciences, Inc., Tokyo, Japan) and detected at 280 nm. The mobile phase was 15% acetonitrile in water containing 0.1% trifluoroacetic acid (vol/vol), with a flow rate of 0.6 ml/min. For decarboxylation, a reaction mixture (final volume, 1 ml) containing 0.9 mM γ-resorcylate or other hydroxybenzoates, 0.1 M potassium phosphate buffer (pH 7.0), and enzyme (100 μl) was used. For carboxylation, a reaction mixture (final volume, 1 ml) containing 0.12 M resorcinol or other hydroxyphenols, 0.5 M NaHCO3, 0.1 M potassium phosphate buffer (pH 7.0), and enzyme (100 μl) was used. The reaction was started by the addition of enzyme and was incubated at 30°C with shaking (70 rpm). After 5 min, 0.2 ml of 5 N hydrochloric acid was added to stop the reaction. The mixture (10 μl) was then subjected to HPLC. To find fractions containing the enzyme activity during purification, the reaction mixture (3 μl) of carboxylation (total volume, 500 μl; reaction time, 30 min) was mixed with 50 mM FeCl3 (100 μl) and deionized water (597 μl), and the absorption derived from the formation of the Fe-resorcinol complex was measured spectrophotometrically at 576 nm. One unit of the enzyme was defined as the amount of enzyme that produces 1 μmol of either resorcinol or hydroxyphenols for decarboxylation or γ-resorcylate or hydroxybenzoates for carboxylation per min.
Enzyme purification.
All procedures were done at 4°C under aerobic conditions, and column chromatographies were performed at a flow rate of 0.8 ml/min.
The cells were grown in GR medium supplemented with yeast extract (3.0 g) (GRY medium). A single colony was inoculated into a test tube (1.8 by 18 cm) containing GR medium (5 ml) and was cultivated on a reciprocal shaker (150 rpm) at 30°C. After 48 h, the culture was transferred into 2-liter Sakaguchi flasks containing GRY medium (1 liter) and cultivated at 30°C for 26 h (stationary phase). The cells were harvested by centrifugation (15,100 × g, 20 min) and washed twice with a 10 mM potassium phosphate buffer (pH 7.0) containing 0.75% NaCl. The washed cells were suspended in a 10 mM potassium phosphate buffer (pH 7.0) containing 0.02% 2-mercaptoethanol (Buffer A) and disrupted by ultrasonication (model UD-201; Tomy, Tokyo, Japan) at about 4°C for 3 min. The output was adjusted to 6, and the disruption was repeated eight times. The cell debris was removed by ultracentrifugation (183,400 × g, 90 min), and the supernatant was dialyzed against 5 liters of Buffer A and used as a crude enzyme. The crude enzyme was put on a column of DEAE-Toyopearl 650 M (2.5 by 18 cm) equilibrated with Buffer A. After the column was washed with Buffer A containing 80 mM NaCl (500 ml), absorbed proteins were eluted with a 500-ml linear gradient of 80 to 300 mM NaCl in Buffer A. Fractions containing γ-RDC activity were combined and dialyzed against Buffer A containing 1.0 M (NH4)2SO4 (Buffer B). This mixture was loaded onto a column (2.5 by 10 cm) of butyl-Toyopearl equilibrated with Buffer B. The column was washed with Buffer B (300 ml), and absorbed proteins were eluted with a 300-ml linear gradient of 1 to 0.3 M (NH4)2SO4. Fractions containing γ-RDC were combined and dialyzed against a 10 mM potassium phosphate buffer (pH 7.0) and then concentrated by ultrafiltration (Advantec ultrafilter; PO200 membrane). The enzyme solution was stored at −84°C until use.
Effect of carbon source on production of γ-RDC.
Strain MTP-10005 was cultivated in GR agar plates or modified GR agar plates that contained various other dihydroxybenzoates instead of γ-resorcylate as sole carbon sources at 30°C for 72 h. The colony formed was cultivated in the same liquid medium according to the procedure of enzyme purification, and the corresponding enzyme activity was measured with a crude extract.
Kinetics analysis.
All procedures were done at 30°C and pH 7. For decarboxylation, the maximum velocity (Vmax) and the Michaelis constant (Km) were determined by Lineweaver-Burk double reciprocal plots. For carboxylation, the initial-velocity experiment was carried out by using various concentrations of one substrate at different fixed concentrations of the other substrate. The kinetic parameters were determined from the secondary plots of intercepts versus the reciprocal concentrations of the substrate.
N-terminal and internal amino acid sequences.
The internal amino acid sequences were analyzed after the purified enzyme was digested with lysylendopeptidase (Lys-C). A reaction mixture (total volume, 202 μl) containing 18 μl of 1 M Tris-HCl (pH 9.0), 40 μl of 8 M urea, 144 μg of purified enzyme, and 4 μl of Lys-C was incubated at 37°C for 18 h. The clear solution obtained was dried by centrifugal evaporation. The Lys-C-digested peptides (1.0 nmol) were separated with a Wakosil 5C18-AR (4.6 by 250 mm; Wako Chemical Co., Kyoto, Japan) in a Shimadzu LC-10A HPLC system. A 120-min linear gradient from 0 to 60% (vol/vol) acetonitrile in 0.1% (vol/vol) trifluoroacetic acid was used to elute peptides at a flow rate of 0.7 ml/min. Peptides were monitored at 215 nm. The separated peptides (50 pmol) were transferred to a polyvinylidene fluoride membrane (Sequi-Blot membrane for protein sequencing [0.2 mm]; Bio-Rad). To determine the N-terminal amino acid sequence, 50 pmol of purified enzyme was transferred to the same membrane. The N-terminal and internal amino acid sequences were determined by Edman degradation with an automated sequencer (PPSQ-21; Shimadzu).
Sequence analysis of the gene encoding γ-RDC and putative resorcinol-metabolizing enzymes upstream and downstream of the γ-RDC gene.
On the basis of the N-terminal and internal amino acid sequences of the enzyme, two oligonucleotides, RDCN [5′-ATG CAR GGN AAR GTN GC-3′] and RDCK2 [5′-CKN GCD ATY TCD ATN GC-3′], were synthesized, and the γ-RDC gene was amplified from Rhizobium sp. strain MTP-10005 genomic DNA (506 ng) with the oligonucleotides (20 pmol) by PCR. PCR amplification was carried out with Ex Taq polymerase (Takara) in a Gene Amp PCR system 9700 (PE Applied Biosystems, Piscataway, N.J.). The resulting 0.25-kbp DNA fragment was extracted with an Ultra Clean15 DNA purification kit from gels and solutions (MO BIO Laboratories, Inc., West Carlsbad, Calif.) and ligated into a pT7 Blue T-Vector (EMD Biosciences, Inc., San Diego, Calif.) with the DNA ligation kit version 2 (Takara). The plasmid obtained was transformed into Escherichia coli (Novablue; EMD Biosciences, Inc.). White ampicillin-resistant clone cells were picked and grown in 5 ml of Luria-Bertani medium containing ampicillin (100 μg/ml). The plasmid was purified and used as the template for sequencing. Based on the partial 0.25-kb DNA sequence determined, four primers, RDCGWN1 [5′-GCA TTC AGC GAC AGG ATC ATG GT-3′], RDCGWN2 [5′-CCA TCA GCT TCA GGC GCG TAT C-3′], RDCGWC1 [5′-CTC GAA GAG CAT TTC GCA ATC CC-3′], and RDCGWC2 [5′-GTG CCC GGT GAT TAC TGG AAG GAA CT-3′], were designed, and genome-walking PCR was performed with the LA PCR in vitro cloning kit (Takara). Chromosomal DNA extracted from Rhizobium sp. strain MTP-10005 cells was digested with EcoRI or SalI and ligated to an EcoRI or SalI cassette, respectively. The DNA fragments obtained were used as the template for amplification by PCR. PCR and nested PCR were carried out successively with RDCGWN1 and C1 (primer for PCR), RDCGWC1 and C1, RDCGWN2 and C2 (primer for nested PCR), and RDCGWC2 and C2. The PCR fragments obtained were sequenced to determine the full-length DNA sequence of the γ-RDC gene and the putative resorcinol-metabolizing enzymes upstream and downstream of the γ-RDC gene by use of a DNA sequencer SQ5500E (Hitachi Electronics Engineering Co.) with the Thermo Sequenase Primer Cycle sequencing kit (Amersham Biosciences Corp.).
Analytical methods.
Polyacrylamide gel electrophoresis (PAGE) and sodium dodecyl sulfate (SDS)-PAGE were performed by the methods described by Laemmli (23) and Tulchin et al. (38), respectively. Precision Plus Protein all blue standards (Bio-Rad) were used as molecular mass markers for SDS-PAGE. The proteins in gels were stained with Coomassie brilliant blue R-250. Molecular mass was estimated by gel filtration with a column (1.6 by 60 cm) of Superdex 200 Hiload (Amersham Biosciences Corp.), with ferritin (450 kDa), catalase (240 kDa), aldolase (158 kDa), and bovine serum albumin (67 kDa) as standard proteins. The column was equilibrated with 10 mM potassium phosphate buffer (pH 7.0), and the void volume was estimated with blue dextran. Protein concentrations were measured by the method described by Bradford (4), with bovine serum albumin as the standard. Absorption spectra were recorded with a V-550 UV/VIS spectrophotometer (Jasco Corporation, Tokyo, Japan).
Nucleotide sequence accession numbers.
The DNA sequence of the gene encoding Rhizobium sp. strain MTP-10005 γ-RDC and the partial 16S rDNA sequence are available from GenBank under accession no. AB170010 and AB182998, respectively.
RESULTS
Microorganism.
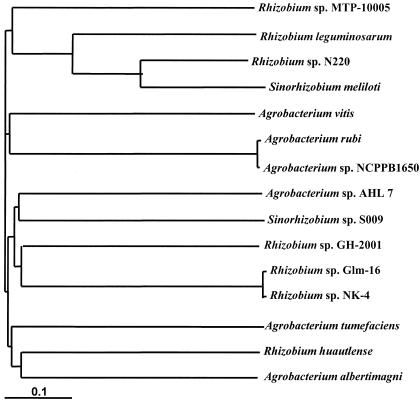
We isolated strain MTP-10005, which utilized γ-resorcylate as a sole carbon source, from a natural water sample taken from a river near Osaka, Japan. The first morphological and biochemical studies showed that this organism is a motile, gram-negative, sporulation-negative, catalase-positive, and oxidase-positive strain, and the cell form is a rod 0.7 to 0.8 by 1.5 to 2.0 μm in size. The strain formed round pale yellow colonies on nutrient agar medium and fermented glucose. These results suggested that MTP-10005 belongs to a Pseudomonas group of bacteria. Additional biochemical tests showed that strain MTP-10005 utilizes a variety of sugars and sugar alcohols, shows cytochrome oxidase activity, lacks arginine dihydrolase and gelatin hydrolase activities, and does not produce indole. These characteristics of strain MTP-10005 are similar to those of Agrobacterium tumefaciens (Rhizobium radiobacter) except that the utilization of potassium gluconate is negative. The partial 16S rDNA sequence determined is as follows: TTGAGAGTTTGATCCTGGCTCGAACGAACG CTGGCGGCAGGCTTAACACATGCAAGTCGAACGCCCCGCAAGGGGAGTGGCAGACGGGTGAGTAACGCGTGGGAATCTACCGTGCCCTGCGGAATAGCTCCGGGAAACTGGAATTAATACCGCATACGCCCTACGGGGGAAAGATTTATCGGGGTATGATGAGCCCGCGTTGGATTAGCTAGTTGGTGGGGTAAAGGCCTACCAAGGCGACGATCCATAGCTGGTCTGAGAGGATGATCAGCCACATTGGGACTGAGACACGGCCCAAACTCCTACGGGAGGCAGCAGTGGGGAATATTGGACAATGGGCGCAAGCCTGATCCAGCCATGCCGCGTGAGTGATGAAGGCCTTAGGGTTGTAAAGCTCTTTCACCGGAGAAGATAATGACGGTATCCGGAGAAGAAGCCCCGGCTAACTTCGTGCCAGCAGCCGCGGTA. The phylogenetic tree of the 16S rDNA gene sequence shows that the strain MTP-10005 locates far from A. tumefaciens and belongs to a new group alone (Fig. 1). On the basis of these results, strain MTP-10005 was identified as a novel species of Rhizobium bacterium. Rhizobium sp. strain MTP-10005 has been deposited in the collection of the Fermentation Research Institute, National Institute of Advanced Industrial Science and Technology, Tokyo, Japan, as strain FERM P-16375.
FIG. 1.
Phylogenetic tree of 16S rDNA of Rhizobium sp. strain MTP-10005.
Inducible production of γ-RDC.
In addition to γ-resorcylate, Rhizobium sp. strain MTP-10005 also utilized 2,3-dihydroxybenzoate or 3,4-dihydroxybenzoate as carbon sources but not 2,4-, 2,5-, or 3,5-dihydroxybenzoate. γ-RDC activity was not detected when 2,3-dihydroxybenzoate or 3,4-dihydroxybenzoate was used as a substrate for growth. These results suggest that the enzyme was specifically induced by the addition of γ-resorcylate and used for degradation of γ-resorcylate in the growth of Rhizobium sp. strain MTP-10005.
Purification of γ-RDC.
γ-RDC was purified about 12-fold, with a yield of 38% (Table 1). The purified enzyme was found to be homogeneous on PAGE and SDS-PAGE. The specific activity for forward reaction was 0.391 U/mg (at 30°C). The enzyme was stored at −84°C in a 10 mM potassium phosphate buffer (pH 7.0) without loss of activity for several months.
TABLE 1.
Purification of γ-RDC from Rhizobium sp. strain MTP-10005
| Step | Total activity (U) | Total protein (mg) | Specific activity (U/mg) | Yield (%) | Purification (n-fold) |
|---|---|---|---|---|---|
| Crude extract | 5.41 | 166 | 0.0325 | 100 | 1 |
| DEAE-Toyopearl 650 M | 4.80 | 14.9 | 0.321 | 88.7 | 9.88 |
| Butyl-Toyopearl 650 M | 2.05 | 5.23 | 0.391 | 37.9 | 12.0 |
Molecular mass and subunit structure.
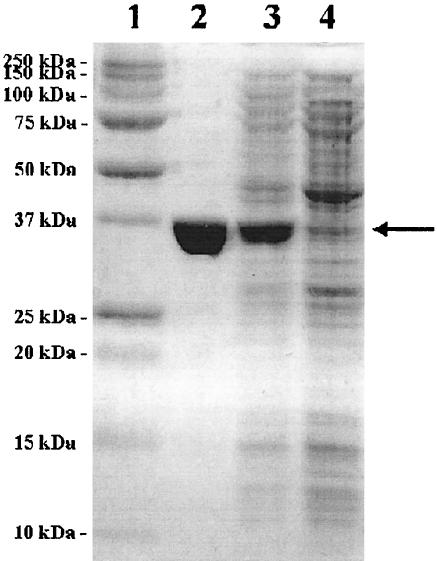
The purified enzyme migrated as a single band in SDS-PAGE with an apparent molecular mass of 37.5 kDa (Fig. 2). The molecular mass of the native enzyme on gel filtration with Superdex-200 was 151 kDa, suggesting that the enzyme is a homotetramer.
FIG. 2.
SDS-PAGE of recombinant γ-RDC. Lane 1, molecular mass markers; lane 2, wild-type γ-RDC; lane 3, crude extract of E. coli BL21 (DE3) harboring pRDC; lane 4, crude extract of E. coli BL21 (DE3) harboring pET3b.
N-terminal and internal amino acid sequences.
Twenty-three amino acid residues of the N-terminal position of the enzyme were determined to be MQGKVALEEHFAIPETLQDSAGF. The Lys-C digests of the enzyme were separated by reversed-phase HPLC, and several internal peptide sequences were determined: K1, LMDAHGIETMILSLN; and K2, AIEIARRANDVLAEE. These sequences were used for the identification of the amplified DNA fragment. The BLAST search program revealed that the N-terminal and two internal amino acid sequences show high sequence similarity with those of the hypothetical protein AGR_C_4595p from A. tumefaciens strain C58.
Effect of temperature.
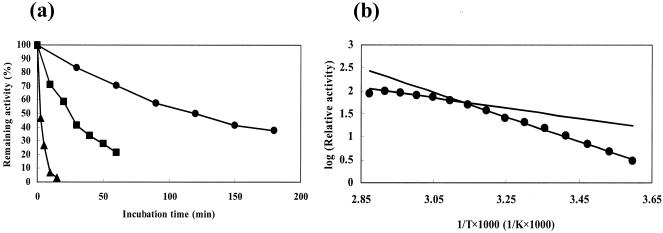
The enzyme was relatively thermostable. The thermal stability of the enzyme was examined at 50, 60, and 70°C (Fig. 3a). The half-life times at 50, 60, and 70°C were estimated to be 122, 25.5, and 2.46 min, respectively. The enzyme showed discontinuity in Arrhenius plots for decarboxylation, with a transition temperature at about 47°C (Fig. 3b). In the lower temperature range, the value of the activation energy was calculated to be 51.0 kJ/mol, whereas in the higher range, the value was about 21.5 kJ/mol. The maximum activity for decarboxylation was observed at 70°C under the standard assay conditions.
FIG. 3.
Effects of temperature on enzyme activity and stability. (a) Effects of temperature on stability. The enzyme solution was incubated in 10 mM potassium phosphate buffer, pH 7.0, at various temperatures, and the remaining activity was determined under standard assay conditions for decarboxylation. •, 50°C; ▪, 60°C; ▴, 70°C. (b) Arrhenius plot. The activation energy for the decarboxylation of γ-resorcylate to resorcinol was calculated from the Arrhenius plot.
Effect of pH.
The activity was determined under standard assay conditions at various pHs (pH 5.0 and 5.5, piperazine-HCl; pH 5.5, 6.0, and 6.5, MES-NaOH; pH 6.5, 7.0, 7.5, and 8.0, potassium phosphate; pH 8.0, 8.5, and 9.0, Tris-HCl; pH 9.0, 9.5, and 10.0, 2-[N-cyclohexylamino]ethanesulfonic acid-NaOH). The enzyme showed activity in the pH range of 5 to 10 for the decarboxylation of γ-resorcylate. The optimum pH (pH 8.0) was higher than that for the carboxylation of resorcinol (pH 7.0). The enzyme showed high activity in neutral conditions.
Substrate specificity and coenzyme requirement.
The substrate specificity for decarboxylation was studied in the presence of 0.9 mM concentrations of various substrates. In addition to 2,6-dihydroxybenzoate, the enzyme also catalyzed the decarboxylation of 2,3-dihydroxybenzoate (relative activity, 132%) but did not act on 2,4-dihydroxybenzoate, 2,5-dihydroxybenzoate, 3,4-dihydroxybenzoate, 3,5-dihydroxybenzoate, 2-hydroxybenzoate, or 3-hydroxybenzoate. Only resorcinol was carboxylated by the reverse reaction.
The coenzyme requirement for decarboxylation was also examined in the presence of various coenzymes, such as 50 μM thiamine PPi, 50 μM pyridoxal 5′-phosphate, 1 mM NAD+, and 1 mM NADP+. The enzyme did not require any coenzyme for decarboxylation. The absorption spectrum of the purified enzyme (0.362 mg/ml in a potassium phosphate buffer [pH 7.0]) also showed a lack of coenzyme, and maximum absorption was observed only at 280 nm.
Effect of inhibitors.
We examined the effect of 1 mM concentrations of various compounds on enzyme activity. The enzyme was inhibited by CuCl2 (remaining activity, 52%), monoiodoacetate (63%), and diethylpyrocarbonate (59%): the imidazole groups are probably directly or indirectly involved in the enzyme catalysis, as reported for other decarboxylases (19). No significant effect on enzyme activity was observed with CoCl2, FeCl3, MgCl2, MnCl2, NiCl2, ZnSO4, EDTA, 2,2′-bipyridyl, 1,10-phenanthroline, hydroxylamine, semicarbazide, 5,5′-dithiobis(2-nitrobenzoate), N-ethylmaleimide, 2-mercaptoethanol, or sodium borohydride.
Kinetics analysis.
For decarboxylation, the Km values for γ-resorcylate and 2,3-dihydroxybenzoate were estimated to be 70.7 and 123 μM, and the Vmax values for γ-resorcylate and 2,3-dihydroxybenzoate were determined to be 0.377 and 1.06 U/mg, respectively. For carboxylation, the Km and Vmax values for resorcinol and NaHCO3 were calculated from the secondary plots of intercepts versus the reciprocal concentrations of the other substrate. The Km values for resorcinol and NaHCO3 were estimated to be 45.7 and 22.2 mM, respectively. The Vmax values for resorcinol and NaHCO3 were determined to be 1.78 and 1.75 U/mg, respectively. The kcat/Km (mΜ−1 · s−1) values for γ-resorcylate and resorcinol were 13.4 and 0.098, respectively. These results show that the chemical equilibrium is toward decarboxylation under standard assay conditions. However, by the addition of organic solvent to the reaction mixture, the equilibrium is dramatically changed toward carboxylation, and this result suggests that this enzyme is practically applicable to the biotechnological processes for γ-resorcylate (H. Shibaboto et al., unpublished data).
Cloning and sequence analysis of the enzyme gene.
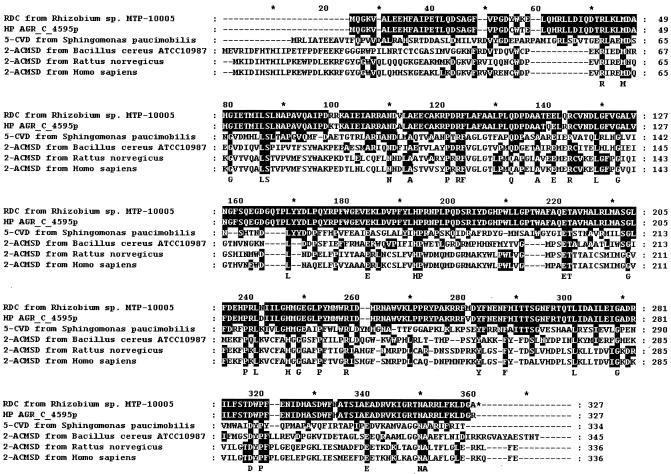
The entire sequence of the enzyme gene was determined for both strands, and the alignment of primary sequences of various decarboxylases is summarized in Fig. 4. An open reading frame of 984 bp was identified, corresponding to 327 amino acid residues with a molecular weight of 37,422. The coding region of the γ-RDC gene is preceded by the sequence of a putative bacterial Shine-Dalgarno ribosome-binding site (GGAGG) located 8 bp upstream of the starting codon, ATG. The pyrimidine-rich region was not found at the 3′ end. The G+C content of the γ-RDC gene was 60.5%. The deduced amino acid sequence was used for searching for identical sequences in the GenBank and protein databases with the BLAST program. Sequence identities were found with those of hypothetical protein AGR_C_4595p from A. tumefaciens strain C58 (96% identity), 5-carboxyvanillate decarboxylase from S. paucimobilis (32%), and 2-amino-3-carboxymuconate-6-semialdehyde decarboxylases from B. cereus ATCC 10987 (26%), R. norvegicus (26%), and H. sapiens (25%). His164 and His218 are well conserved for all six proteins, and these residues are probably essential for the catalytic activity of the decarboxylase, in consideration of the results from the inhibitory study.
FIG. 4.
Alignment of primary sequences of various decarboxylases. RDC, γ-RDC; HP AGR_C_4595p, hypothetical protein AGR_C_4595p from A. tumefaciens strain C58; 5-CVD, 5-carboxyvanillate decarboxylase; and 2-ACMSD, 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase. The shaded areas show amino acid residues identical to those of γ-RDC from Rhizobium sp. strain MTP-10005.
We also determined the DNA sequence upstream and downstream of the γ-RDC gene. The open reading frame (graC, 1,056 bp) that is homologous to maleylacetate reductase was found 3 bp downstream of the stop codon, TGA, of the γ-RDC gene (identity with maleylacetate reductase from: A. tumefaciens strain C58, 91%; Bradyrhizobium japonicum USDA 110, 57%; and Ralstonia sp. strain SJ98, 55%). The hydroxyquinol 1,2-dioxygenase homologous gene (graB, 888 bp) was also located immediately downstream of graC (identity with hydroxyquinol 1,2-dioxygenase from: A. tumefaciens strain C58, 94%; Ralstonia pickettii, 49%; and Sphingomonas sp., 44%). The phenol hydroxylase homologous gene (graA, 1,230 bp) exists immediately upstream of the γ-RDC gene. This gene is considered to be resorcinol hydroxylase.
Expression of the γ-RDC gene in E. coli.
A 986-bp NdeI-BamHI fragment containing the Rhizobium sp. strain MTP-10005 enzyme gene was ligated to pET 3b, and the pRDC obtained was used for the production of the enzyme in E. coli BL21 (DE3) cells under the control of the T7 promoter (Fig. 2). γ-RDC was detected in the soluble fractions of the cell extract (specific activity, 0.148 U/mg for decarboxylation).
DISCUSSION
We found that a bacterium, Rhizobium sp. strain MTP-10005, isolated from natural river water, abundantly produces γ-RDC, and we purified its enzyme to homogeneity for the first time. The enzyme is an inducible enzyme that occupies about 8.3% of the total soluble protein produced in the presence of 20 mM γ-resorcylate. γ-RDC was relatively thermostable to heat treatment. When the enzyme was incubated with a 10 mM potassium phosphate buffer (pH 7.0) at 50°C, more than 50% of the initial activity remained after 2 h. The activation energy for the decarboxylation of γ-resorcylate was calculated to be 51.0 kJ/mol (15 to 45°C), which is higher than those of hydroxybenzoate decarboxylases reported so far (15). The maximum temperature of γ-RDC (70°C) was higher than those of other hydroxybenzoate decarboxylases, such as Clostridium hydroxybenzoicum 4-hydroxybenzoate decarboxylase (50°C) (14), C. hydroxybenzoicum 3,4-dihydroxybenzoate decarboxylase (50°C) (15), and Aspergillus niger 2,3-dihydroxybenzoate decarboxylase (42°C) (30). Rhizobium γ-RDC consists of four identical subunits with molecular weights of about 37,500. This is not so different from the subunit structure of other hydroxybenzoate decarboxylases, which have α2, α4, α5, or α6 structures (Table 2). The differences between these enzymes in subunit structure does not reflect the difference in the thermostability of Rhizobium γ-RDC. A psychrophile, Cytophaga sp. strain KUC-1 isolated from Antarctic seawater, grows optimally at 15°C and cannot grow above 30°C. This organism produces abundantly the paradoxical thermophilic NAD(P)+-dependent aldehyde dehydrogenase and aspartase (21, 41). From an evolutionary point of view, Rhizobium sp. strain MTP-10005, Cytophaga sp. strain KUC-1, and other thermophilic-enzyme-producing nonthermophiles were probably affected genetically by a thermophile in the ancient past, and the characteristics derived from it would remain for various enzymes of nonthermophiles, not exceptionally in γ-RDC of Rhizobium sp. strain MTP-10005.
TABLE 2.
Comparison with properties of other hydroxybenzoate decarboxylases
| Decarboxylase | Source | Molecular weight | Subunit's molecular weight | Subunit structure | Km (μM) | Optimum pH | Maximum temperature (°C)a | O2 sensitivity | Other substrate | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| γ-Resorcylate decarboxylase | Rhizobium sp. strain MTP-10005 | 151,000 | 37,500 | α4 | 70.7 | 8.0 | 70 | No | 2,3-Dihydroxybenzoate | This study |
| 4-Hydroxybenzoate decarboxylase | Clostridium hydroxybenzoicum | 350,000 | 57,000 | α6 | 440 | 6.0 | 50 | Yes | 3,4-Dihydroxybenzoate | 14 |
| 3 F-4 hydroxybenzoate | ||||||||||
| Vanillate | ||||||||||
| 2,3-Dihydroxybenzoate decarboxylase | Aspergillus niger | 120,000 | 28,000 | α4 | 340 | 5.2 | 42 | No | Not found | 18,30 |
| Trichosporon cutaneum | 66,100 | 36,500 | α2 | 37.0 | 7.7 | N.D. | No | 2,3,5-Trihydroxybenzoate | 1 | |
| 2,3,6-Trihydroxybenzoate | ||||||||||
| 3,4-Dihydrozybenzoate decarboxylase | Clostridium hydroxybenzoicum | 270,000 | 57,000 | α5 | 590 | 7.0 | 50 | Yes | Not found | 15 |
| 4,5-Dihydroxyphthalate decarboxylase | Pseudomonas fluorescens | 420,000 | 66,000 | α6 | 10.0 | 6.8 | N.D. | No | N.D. | 29 |
| Pseudomonas testoseroni | 150,000 | 38,000 | α4 | 11.0 | 7.5 | N.D. | No | 4-Hydroxyphthalate | 24 |
N.D., not determined.
Similar to other hydroxybenzoate decarboxylases (15, 18, 24), γ-RDC shows high substrate specificity. In addition to γ-resorcylate, the enzyme only acts on 2,3-dihydroxybenzoate. The γ-RDC does not require any coenzyme for catalytic activity and also contains no bound carbonyl group. Therefore, the reaction mechanism is probably different from that of decarboxylases dependent on thiamine PPi (6), pyridoxal 5′-phosphate (40), NAD+ (25), NADP+ (17), or pyruvoyl (32). In cofactor-independent hydroxybenzoate decarboxylases, both the carboxyl and hydroxyl groups of a substrate are considered to be involved in binding to the active site of the enzymes (1, 19). Especially the position of the hydroxyl group(s) on the benzene ring of a substrate might be important for the determination of substrate specificity. The 2,3-dihydroxybenzoate decarboxylase from A. niger is specific for 2,3-dihydroxybenzoate and does not act on salicylate, anthranilate, 2,3-dihydroxybenzaldehyde, 2,4-dihydroxybenzoate, 3-hydroxyanthranilate, 3-hydroxybenzoate, 3,4-dihydroxybenzoate, benzoate, or 4-hydroxybenzoate (18). The hydroxyl groups in both the C-2 and C-3 positions of the benzene ring are necessary for a substrate (20). The 4-hydroxybenzoate decarboxylase from C. hydroxybenzoicum acts only on 3,4-dihydroxybenzoate, 3-F-4-hydroxybenzoate, and 3-methoxy-4-hydroxybenzoate (vanillate) (14). This result suggests that the C-4 position of the benzene ring is essential for a substrate. In addition, the γ-RDC from Rhizobium sp. strain MTP-10005 catalyzes the decarboxylation of 2,3- and 2,6-dihydroxybenzoate (γ-resorcylate) but does not act on 2,4-dihydroxybenzoate, 2,5-dihydroxybenzoate, 3,4-dihydroxybenzoate, 3,5-dihydroxybenzoate, 2-hydroxybenzoate, or 3-hydroxybenzoate. These results suggest that, for a substrate, it is necessary for two hydroxyl groups to exist on the benzene ring and for one of them to exist at the C-2 position. It is essential that all substituents on the benzene ring be adjacent to each other. Further X-ray crystallographic analysis would be needed to clarify the reaction mechanism of γ-RDC.
The primary structure of Rhizobium γ-RDC is not so similar to that of other previously reported decarboxylases, such as 5-carboxyvanillate decarboxylase from S. paucimobilis (32% identity) (28) and 2-amino-3-carboxymuconate-6-semialdehyde decarboxylases from B. cereus ATCC 10987 (26%) (31), R. norvegicus (26%) (37), and H. sapiens (25%) (8). In contrast, interestingly, the primary structure of Rhizobium γ-RDC is highly similar to that of the hypothetical protein AGR_C_4595p from A. tumefaciens strain C58 (identity 96%; accession number AAK88260). This protein would also function like γ-RDC, since the homologous genes of hydroxyquinol 1,2-dioxygenase, maleylacetate reductase, and the hypothetical resorcinol hydroxylase for the catabolism of the resorcinol exist in both Rhizobium sp. strain MTP-10005 and A. tumefaciens strain C58 (11). Recently, γ-RDC was purified from A. tumefaciens strain IAM12048, which strongly supports our hypothesis (42). His164 and His218 were conserved in all the enzymes (Fig. 4). This result agreed well with the inhibitory study.
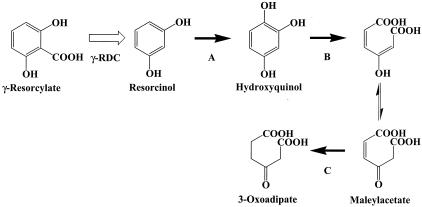
Various pathways for the biodegradation of hydroxybenzoates have been reported for several microorganisms. The 2,4-dihydroxybenzoate pathway exists in Pseudomonas sp. strain BN9 (35) and Sphingomonas sp. strain RW1 (2), while the protocatechuate (3,4-dihydroxybenzoate) pathway has been reported for Rhizobium and Agrobacterium and extensively studied for Roseobacter, and so on (5, 26, 27). In the 2,4-dihydroxybenzoate pathway, 2,4-dihydroxybenzoate is converted into 3-oxoadipate by 2,4-dihydroxybenzoate hydroxylase (1-monooxygenase) (EC 1.14.13.x), hydroxyquinol 1,2-dioxygenase (EC 1.13.11.37), and maleylacetate reductase (EC 1.2.1.32). In contrast, in the protocatechuate pathway, acetyl-coenzyme A (CoA) and succinate are produced from protocatechuate by the catalytic activity of protocatechuate 3,4-dioxygenase (EC 1.13.11.3), β-carboxy-cis-cis-muconate lactonizing enzyme (EC 5.5.1.2), γ-carboxymuconolactone decarboxylase (EC 4.1.1.44), β-ketoadipate enol-lactone hydrolase (EC 3.1.1.24), β-ketoadipate succinyl-CoA transferase (EC 2.8.3.6), and acetyl-CoA acetyltransferase (EC 2.3.1.9) and then metabolized in the citric acid cycle. In addition to these pathways, the resorcinol pathway is known in Pseudomonas putida (7) and Trichosporon cutaneum (9) but not in Rhizobium sp. bacteria. In the resorcinol pathway, resorcinol is converted into 3-oxoadipate via resorcinol hydroxylase (EC 1.14.13.x), hydroxyquinol 1,2-dioxygenase (EC 1.13.11.37), and maleylacetate reductase (EC 1.3.1.32). We found that the homologous genes of hydroxyquinol 1,2-dioxygenase and maleylacetate reductase, and the hypothetical resorcinol hydroxylase, exist immediately upstream and downstream of the γ-RDC gene. To our knowledge, there is no report on the metabolic pathway for γ-resorcylate catabolism in bacteria and other living organisms. Judging from these results, the resorcinol pathway, which is similar to that of P. putida and T. cutaneum, also exists in Rhizobium sp. strain MTP-10005, and γ-RDC probably catalyzes a reaction just before the hydroxylase in it does (Fig. 5).
FIG. 5.
Proposed metabolic pathway of γ-resorcylate in Rhizobium sp. strain MTP-10005. A, B, and C represent resorcinol hydroxylase, hydroxyquinol 1,2-dioxygenase, and maleylacetate reductase, respectively.
We are currently trying to analyze the regions upstream and downstream of the γ-RDC gene to elucidate the physiological function of γ-RDC from Rhizobium sp. strain MTP-10005 and the three-dimensional structure of the enzyme by means of X-ray crystallographic analysis to clarify the reaction mechanism of the enzyme.
Acknowledgments
This work was supported in part by a research grant from Japan Foundation of Applied Enzymology and by a research grant from Kansai University High Technology Research Center.
REFERENCES
- 1.Anderson, J. J., and S. Dagley. 1981. Catabolism of tryptophan, anthranilate, and 2,3-dihydroxybenzoate in Trichosporon cutaneum. J. Bacteriol. 146:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armengaud, J., K. N. Timmis, and R.-M. Wittich. 1999. A functional 4-hydroxysalicylate/hydroxyquinol degradative pathway gene cluster is linked to the initial dibenzo-p-dioxin pathway genes in Sphingomonas sp. strain RW1. J. Bacteriol. 181:3452-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beringer, J. E., N. Brewin, A. W. Johnston, H. M. Schulman, and D. A. Hopwood. 1979. The Rhizobium-legume symbiosis. Proc. R. Soc. Lond. Ser. B Biol. Sci. 204:219-233. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Buchan, A., E. L. Neidle, and M. A. Moran. 2004. Diverse organization of genes of the β-ketoadipate pathway in members of the marine Roseobacter lineage. Appl. Environ. Microbiol. 70:1658-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Candy, J. M., and R. G. Duggleby. 1994. Investigation of the cofactor-binding site of Zymomonas mobilis pyruvate decarboxylase by site-directed mutagenesis. Biochem. J. 300:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, P. J., and D. W. Ribbons. 1976. Metabolism of resorcinylic compounds by bacteria: alternative pathways for resorcinol catabolism in Pseudomonas putida. J. Bacteriol. 125:985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuoka, S., K. Ishiguro, K. Yanagihara, A. Tanabe, Y. Egashira, H. Sanada, and K. Shibata. 2002. Identification and expression of a cDNA encoding human alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase (ACMSD). A key enzyme for the tryptophan-niacine pathway and “quinolinate hypothesis.” J. Biol. Chem. 277:35162-35167. [DOI] [PubMed] [Google Scholar]
- 9.Gaal, A., and H. Y. Neujahr. 1979. Metabolism of phenol and resorcinol in Trichosporon cutaneum. J. Bacteriol. 137:13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallert, C., and J. Winter. 1992. Comparison of 4-hydroxybenzoate decarboxylase and phenol carboxylase activities in cell-free extracts of a defined 4-hydroxybenzoate and phenol-degrading anaerobic consortium. Appl. Microbiol. Biotechnol. 37:119-124. [Google Scholar]
- 11.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323-2328. [DOI] [PubMed] [Google Scholar]
- 12.Grant, D. J. W., and J. C. Patel. 1969. The non-oxdative decarboxylation of p-hydroxybenzoic acid, gentisic acid, protocatechuic acid and gallic acid by Klebsiella aerogenes (Aerobacter aerogenes). Antonie Leeuwenhoek 35:325-341. [DOI] [PubMed] [Google Scholar]
- 13.Harrison, J., M.-A. Pou de Crescenzo, O. Sene, and B. Hirel. 2003. Does lowering glutamine synthetase activity in nodules modify nitrogen metabolism and growth of Lotus japonicus? Plant Physiol. 133:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, Z., and J. Wiegel. 1995. Purification and characterization of an oxygen-sensitive reversible 4-hydroxybenzoate decarboxylase from Clostridium hydroxybenzoicum. Eur. J. Biochem. 229:77-82. [DOI] [PubMed] [Google Scholar]
- 15.He, Z., and J. Wiegel. 1996. Purification and characterization of an oxygen-sensitive reversible 3,4-dihydroxybenzoate decarboxylase from Clostridium hydroxybenzoicum. J. Bacteriol. 178:3539-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, J., Z. He, and J. Wiegel. 1999. Cloning, characterization, and expression of a novel gene encoding a reversible 4-hydroxybenzoate decarboxylase from Clostridium hydroxybenzoicum. J. Bacteriol. 181:5119-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwakura, M., J. Hattori, Y. Arita, M. Tokushige, and H. Katsuki. 1979. Studies on regulatory functions of malic enzymes. VI. Purification and molecular properties of NADP-linked malic enzyme from Escherichia coli W. J. Biochem. 85:1355-1365. [PubMed] [Google Scholar]
- 18.Kamath, A. V., D. Dasgupta, and C. S. Vaidyanathan. 1987. Enzyme-catalysed non-oxidative decarboxylation of aromatic acids. I. Purification and spectroscopic properties of 2,3-dihydroxybenzoic acid decarboxylase from Aspergillus niger. Biochem. Biophys. Res. Commun. 145:586-595. [DOI] [PubMed] [Google Scholar]
- 19.Kamath, A. V., N. A. Rao, and C. S. Vaidyanathan. 1989. Enzyme-catalysed non-oxidative decarboxylation of aromatic acids. II. Identification of active site residues of 2,3-dihydroxybenzoic acid decarboxylase from Aspergillus niger. Biochem. Biophys. Res. Commun. 165:20-26. [DOI] [PubMed] [Google Scholar]
- 20.Kamath, A. V., and C. S. Vaidyanathan. 1990. Structural requirements for the induction and inhibition of 2,3-dihydroxybenzoic acid decarboxylase of Aspergillus niger. Curr. Microbiol. 20:111-114. [Google Scholar]
- 21.Kazuoka, T., Y. Masuda, T. Oikawa, and K. Soda. 2003. Thermostable aspartase from a marine psychrophile, Cytophaga sp. KUC-1: molecular characterization and primary structure. J. Biochem. (Tokyo) 133:51-58. [DOI] [PubMed] [Google Scholar]
- 22.Krishnan, H. B., and S. G. Pueppke. 1991. Sequence and analysis of the nodABC region of Rhizobium fredii USDA257, a nitrogen-fixing symbiont of soybean and other legumes. Mol. Plant-Microbe Interact. 4:512-520. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Nakazawa, T., and E. Hayashi. 1978. Phthalate and 4-hydroxyphthalate metabolism in Pseudomonas testosteroni: purification and properties of 4,5-dihydroxyphthalate decarboxylase. Appl. Environ. Microbiol. 36:264-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park, S. L., and H. N. Guttman. 1973. Purification and properties of Lactobacillus plantarum inducible malic enzyme. J. Bacteriol. 116:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parke, D., and L. N. Ornston. 1986. Enzymes of the β-ketoadipate pathway are inducible in Rhizobium and Agrobacterium spp. and constitutive in Bradyrhizobium spp. J. Bacteriol. 165:288-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parke, D. 1995. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens. J. Bacteriol. 177:3808-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng, X., E. Masai, H. Kitayama, K. Harada, Y. Katayama, and M. Fukuda. 2002. Characterization of the 5-carboxyvanillate decarboxylase gene and its role in lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 68:4407-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pujar, B. G., and D. W. Ribbons. 1985. Phthalate metabolism in Pseudomonas fluorescens PHK: purification and properties of 4,5-dihydroxyphthalate decarboxylase. Appl. Environ. Microbiol. 49:374-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramachandran, A., V. Subramanian, M. Sugumaran, and C. S. Vaidyanathan. 1979. Purification and properties of pyrocatechuate decarboxylase from Aspergillus niger. FEMS Microbiol. Lett. 5:421-425. [Google Scholar]
- 31.Rasko, D. A., J. Ravel, O. A. Okstad, E. Helgason, R. Z. Cer, L. Jiang, K. A. Shores, D. E. Fouts, N. J. Tourasse, S. V. Angiuoli, J. Kolonay, W. C. Nelson, A.-B. Kolsto, C. M. Fraser, and T. D. Read. 2004. The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1. Nucleic Acids Res. 32:977-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Recsei, P. A., W. M. Moore, and E. E. Snell. 1983. Pyruvoyl-dependent histidine decarboxylases from Clostridium perfringens and Lactobacillus buchneri. Comparative structures and properties. J. Biol. Chem. 258:439-444. [PubMed] [Google Scholar]
- 33.Reid, J., R. D. Watson, J. B. Cochran, D. H. Sproull, B. E. Clayton, and F. T. Prunty. 1951. Sodium γ-resorcylate in rheumatic fever. Br. Med. J. 4727:321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribbons, D. W., and W. C. Evans. 1960. Oxidative metabolism of phthalic acid by soil pseudomonads. Biochem. J. 76:310-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stolz, A., and H.-J. Knackmuss. 1993. Degradation of 2,4-dihydroxybenzoate by Pseudomonas sp. BN9. FEMS Microbiol. Lett. 108:219-224. [DOI] [PubMed] [Google Scholar]
- 36.Subba Rao, P. V., K. Moore, and G. H. N. Towers. 1967. O-Pyrocatechuic acid carboxy-lyase from Aspergillus niger. Arch. Biochem. Biophys. 122:466-473. [DOI] [PubMed] [Google Scholar]
- 37.Tanabe, A., Y. Egashira, S. Fukuoka, K. Shibata, and H. Sanada. 2002. Purification and molecular cloning of rat 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase. Biochem. J. 361:567-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tulchin, N., L. Ornstein, and B. J. Davis. 1976. A microgel system for disc electrophoresis. Anal. Biochem. 72:485-490. [DOI] [PubMed] [Google Scholar]
- 39.Wardhan, H., M. J. McPherson, and G. R. Sastry. 1989. Identification, cloning, and sequence analysis of the nitrogen regulation gene ntrC of Agrobacterium tumefaciens C58. Mol. Plant-Microbe Interact. 2:241-248. [DOI] [PubMed] [Google Scholar]
- 40.Wu, J. Y., T. Matsuda, and E. Roberts. 1973. Purification and characterization of glutamate decarboxylase from mouse brain. J. Biol. Chem. 248:3029-3034. [PubMed] [Google Scholar]
- 41.Yamanaka, Y., T. Kazuoka, M. Yoshida, K. Yamanaka, T. Oikawa, and K. Soda. 2002. Thermostable aldehyde dehydrogenase from psychrophile, Cytophaga sp. KUC-1: enzymological characteristics and functional properties. Biochem. Biophys. Res. Commun. 298:632-637. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida, T., Y. Hayakawa, T. Matsui, and T. Nagasawa. 2004. Purification and characterization of 2,6-dihydroxybenzoate decarboxylase reversibly catalyzing nonoxidative decarboxylation. Arch. Microbiol. 181:391-397. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, X., and J. Wiegel. 1990. Isolation and partial characterization of a Clostridium species transforming para-hydroxybenzoate and 3,4-dihydroxybenzoate and producing phenols as the final transformation products. Microb. Ecol. 20:103-121. [DOI] [PubMed] [Google Scholar]