Abstract
“Candidatus Glomeribacter gigasporarum” is an endocellular β-proteobacterium present in the arbuscular mycorrhizal (AM) fungus Gigaspora margarita. We established a protocol to isolate “Ca. Glomeribacter gigasporarum” from its host which allowed us to carry out morphological, physiological, and genomic investigations on purified bacteria. They are rod shaped, with a cell wall typical of gram-negative bacteria and a cytoplasm rich in ribosomes, and they present no flagella or pili. Isolated bacteria could not be grown in any of the 19 culture media tested, but they could be kept alive for up to 4 weeks. PCR-based investigations of purified DNA from isolated bacteria did not confirm the presence of all genes previously assigned to “Ca. Glomeribacter gigasporarum.” In particular, the presence of nif genes could not be detected. Pulsed-field gel electrophoresis analyses allowed us to estimate the genome size of “Ca. Glomeribacter gigasporarum” to approximately 1.4 Mb with a ca. 750-kb chromosome and a 600- to 650-kb plasmid. This is the smallest genome known for a β-proteobacterium. Such small genome sizes are typically found in endocellular bacteria living permanently in their host. Altogether, our data suggest that “Ca. Glomeribacter gigasporarum” is an ancient obligate endocellular bacterium of the AM fungus G. margarita.
Symbiotic associations between endocellular bacteria and eukaryotic cells are common among plants (e.g., Nostoc species with Gunnera [30] and rhizobia with legumes) and animals (e.g., Buchnera species with most members of the large Hemiptera suborder Sternorrhyncha such as aphids). They are characterized as being either cyclical associations (e.g., most bacteria-plant associations), in which each partner lives independently, or permanent associations (e.g., Buchnera-insect associations), in which neither the host nor the microorganism is able to develop separately (22). In the first case, reassociation occurs by horizontal acquisition, while in the second case, bacteria are strictly vertically transmitted. In the context of permanent associations, a drastic reduction in the microbial genome size generally occurs over evolutionary time. Characterizations of various microbial genomes has revealed that many pathogenic and mutualistic obligate endobacteria have smaller genomes than their free living relatives (17, 27, 43). Genome reduction implies genetic and presumably functional loss and may reflect the dependence of the obligate endocellular bacteria on their host cell (28, 29, 37, 44). Ultimately, such obligate associations can lead to very strong integration of the endosymbionts, like in the case of mitochondria and chloroplasts (18). In the fungi, the presence of endocellular bacteria has been reported only in some Glomeromycota species (arbuscular mycorrhizal [AM] fungi and Geosiphon pyriforme) (8, 9, 35, 36) and in the ectomycorrhizal basidiomycete Laccaria bicolor (5). AM fungi are themselves obligate symbionts of a very large spectrum of plant species. They improve plant growth and resistance against diverse stresses (39). Their origin has been dated back to 353 to 462 million years (11, 31), suggesting that they were instrumental in the colonization of land by ancient plants (11, 38). AM fungi are genetically complex organisms; several genotypes, i.e., genetically different nuclei, may coexist in a single individual (21). The presence of endocellular bacteria in some of their members adds to their genetic complexity (12). The occurrence of these bacteria in AM fungi is intriguing, and their physiological role in fungal fitness as well as their potential role in mycorrhizal symbiosis are completely unknown. These bacteria have been found in several members of the Gigasporaceae (8, 9), but the bacteria present in Gigaspora margarita have been more extensively studied. On the basis of 16S ribosomal DNA (rDNA) sequence analysis, these bacteria had been initially assigned to the genus Burkholderia (9) but were recently reassigned to a new taxon named “Candidatus Glomeribacter gigasporarum” (7). They have been detected in all fungal compartments (spores, germ tube, and extra- and intraradical hyphae) except arbuscules (9). In contrast with Geosiphon pyriforme and L. bicolor, in which association with bacteria is transient, G. margarita seems to live with its endocellular bacteria in a stable association in which bacteria are transmitted vertically during fungal sporulation (6). This finding raises the question of whether the bacteria possess a free-living stage and whether their genome size is reduced as in endocellular bacteria present in insects. So far, the physiological properties of “Ca. Glomeribacter gigasporarum” are missing, while genetic and molecular data are very scarce. A few genes have been isolated from genomic spore DNA and from a G. margarita genomic library (42) and assigned to the bacterial genome, including a phosphate transporter gene (34), a vacB-like gene (33) that contributes to host cell colonization by pathogenic bacteria (Shigella flexneri and Escherichia coli), three nif genes (nifH, nifD, and nifK) (25), the mcpA (23) and cheY (24) genes which are involved in chemotaxis, a kinase gene (prkA), and a sporulation-like spoVR gene which is involved in sporulation (24).
In spite of the obligate endosymbiotic nature of the fungal host, the difficulty in accessing a large fungal biomass, and the relatively small endocellular bacterial density within the fungus, we have been able to isolate enough endocellular “Ca. Glomeribacter gigasporarum” bacteria to reveal original information on their morphology, physiology, and genome structure.
MATERIALS AND METHODS
AM fungus.
Spores of Gigaspora margarita Becker and Hall (BEG 34, deposited at the European Bank of Glomales) were produced in pot cultures with sorghum under greenhouse conditions (natural light and no cooling system) by BIORIZE (Dijon, France). Spores were collected by wet sieving and decanting in 40% (wt/vol) sucrose. The spores were then individually picked under a stereo microscope to obtain highly purified samples. In order to isolate a sufficient amount of endocellular bacteria, more than 3 × 105 spores of G. margarita were used in this study. The isolated spores were surface sterilized with 4% chloramine T three times for 10 min, rinsed with an antibiotic solution (200 mg of streptomycin liter−1 and 100 mg of gentamicin liter−1), and stored in this antibiotic solution at 4°C.
Isolation of “Ca. Glomeribacter gigasporarum” from spores.
In order to isolate the endocellular bacteria from spores, a protocol developed to isolate the endocellular bacteria of the weevil Sitophilus oryzae (Coleoptera) (19) was adapted, with slight modifications. In detail, spores were crushed in an extraction buffer (250 mM sucrose, 10 mM MES [morpholineethanesulfonic acid] [pH 6.5], 25 mM KCl, 20 mM MgCl2, 1 mM dithiothreitol) at 4°C by using a glass homogenizer. The homogenate was centrifuged at 500 × g for 2 min to eliminate major spore wall debris. The supernatant was then centrifuged at 1,000 × g for 2 min to eliminate nuclei and other debris. The newly formed supernatant was centrifuged at 10,000 × g for 10 min to pellet the bacteria. The pellet was resuspended in a minimal volume of extraction buffer. A separation buffer (250 mM sucrose, 0.5% polyethylene glycol 4000, 0.1% Ficoll, 0.1% bovine serum albumin, 30% Percoll) was added to the bacterial suspension. After centrifugation at 12,000 × g for 15 min, the bacteria were located in an opalescent band near the bottom of the tube. This band was collected from the supernatant, diluted in 5 volumes of a washing buffer (250 mM sucrose, 4 mM MES [pH 6.5], 20 mM KCl, 1 mM MgCl2), and washed three times (three successive centrifugations at 11,000 × g). The final pellet was resuspended in a minimal volume of washing buffer. All the above manipulations were conducted at 4°C.
In the first steps of the investigation, other protocols were also tested but none of them gave satisfactory results. They were developed to isolate (i) bacteroids from nodules with different centrifugations in 250 mM mannitol buffer (20), (ii) intracellular organelles from rat liver with centrifugations in 250 mM sucrose, and (iii) Buchnera from aphids by filtration (17).
Estimation of the number of isolated “Ca. Glomeribacter gigasporarum.”
Bacteria were stained for 1 min with a Live/Dead BacLight bacterial viability kit stain (dilution, 1:2,400). This kit contains two DNA staining fluorochromes (Molecular Probes, Leiden, The Netherlands), a permeant one (SYTO 9) that stains nuclei of living cells in green and a nonpermeant one (propidium iodide) that stains nuclei of dead cells in red. The number of living isolated bacteria was estimated by counting green spots by using either conventional fluorescence microscopy or confocal microscopy and image analysis. For conventional microscopy, observations were made with the 40× oil objective lens of an inverted microscope (DMIRBE; Leica, Heidelberg, Germany) equipped with epifluorescence illumination (100-W mercury lamp) at a 450- to 490-nm excitation wavelength (dichroic mirror, 510 nm; barrier, 520 nm). Ten microliters of bacterial suspension was mounted on a Thoma cell for counting, and green spots within 256 squares (corresponding to a total volume of 0.1 mm3) were counted. For confocal microscopy, observations were made with a Leica SP2 confocal microscope and an HCX PL APO 40×/numerical aperture 1.25 oil immersion lens with the 488-nm ray line of an argon laser for excitation, and the emitted light was collected between 500 and 540 nm. Z acquisitions were made throughout the total thickness of the preparation, and an image corresponding to 20 to 30 confocal planes was obtained by using the maximal projection module. Bacteria were then counted within each projection (i.e., corresponding to a volume) by using ImagePro Plus software (Media Cybernetics, Silver Spring, Md.). Twenty projections per counting experiment were used.
Morphological description of “Ca. Glomeribacter gigasporarum” by electronic microscopy. (i) Transmission and scanning electron microscopy.
For electron microscopy, bacteria isolated from 500 to 1,000 G. margarita spores were fixed in 1.5 ml of 2.5% glutaraldehyde in washing buffer (250 mM sucrose, 4 mM MES [pH 6.5], 20 mM KCl, 1 mM MgCl2) for 2 h at room temperature and dehydrated in a graduated ethanol series. Between each step, the samples were centrifuged (11,000 × g for 10 min) to change the solution.
For scanning electron microscopy, drops of bacteria in absolute ethanol were placed in the specimen holder. They were then critical point dried with CO2 as a transitional fluid and finally sputter coated with gold-palladium by using a JEOL JFC 1100 ion-sputtering device. The bacteria were observed with a Hitachi C450 scanning electron microscope at 15 kV. Photographs were taken on Illford 125 ISO film.
For transmission electron microscopy, samples were then infiltrated with a 2:1 (vol/vol) mixture of ethanol-LR White resin (Polysciences Inc., Warrington, Pa.) for 1 h, a 1:2 (vol/vol) mixture of ethanol-LR White resin for 2 h, and 100% LR White resin overnight at 4°C according to method of Balestrini et al. (3). Thin sections (0.05 μm) were stained to visualize polysaccharides with the PATAg method (32) or counterstained with uranyl acetate and lead citrate.
All the samples were observed with a CM 10 Philips transmission electron microscope.
(ii) Negative microscopy.
Bacteria were isolated from 1,000 spores, resuspended in 100 μl of sterile water, and deposited onto 200-mesh nickel grids coated with Formvar. The grids were gently blotted and negatively stained successively for 20 and 10 s on two drops of 15% (wt/vol) aqueous uranyl acetate filtered twice through a 0.2-μm membrane. Grids were blotted, left to dry, and kept in the dark until observation with a transmission electron microscope (EM 600, operating at 75 kV; Hitachi).
Cultivability and survival of “Ca. Glomeribacter gigasporarum.” (i) Cultivability tests.
Cultivability tests were carried out with purified isolated bacteria on various media at two temperatures (22 and 28°C) with or without agitation (liquid media). Five milliliters of the liquid media in 15-ml tubes and 15 ml of the solid media in petri dishes were inoculated with 10 μl of bacterial suspension (200 bacteria μl−1). The liquid media used in the tests were 40 g of tryptic soy broth (Sigma, Saint Louis, Mo.) liter−1, 5 g of soy peptone (Sigma) liter−1, 3 g of yeast extract (Difco, Detroit, Mich.) liter−1, PG medium (40), Terrific broth (40), TY medium (40), YMB medium (40), Bergersen's defined medium (40), fermentor broth (40), media 23A and 21C for Pseudomonas sp. (16), BG medium (10 g of tryptone liter −1, 1 g of yeast extract liter−1, and 1 g of Casamino Acids liter−1), Ashdown medium (2), PDB medium (Difco), yeast carbon base (Difco) with 1 g of KNO3 liter−1 or 1 g of NH4Cl liter−1, and M medium (4). Five solid media were also tested and are as follows: corn meal agar, oat meal agar, PDA, nutrient agar (all from Difco), and CGA (10 g of Casitone liter−1, 5 g of glucose liter−1, and 15 g of agar liter−1). Bacterial growth was checked after 3 weeks of incubation by looking for increasing optical density (liquid media) or colonies (solid media).
(ii) Viability tests.
The survival capacity of the isolated bacteria was investigated in a time course experiment. Living bacteria were counted weekly in bacterial suspensions (300 to 800 bacteria μl−1) incubated under various conditions. Culture tubes containing 500 μl of TY medium or 500 μl of water were inoculated with isolated “Ca. Glomeribacter gigasporarum.” Half of these bacterial suspensions were incubated at 28°C and the other half was incubated at 4°C, without agitation. For periodic counting of living bacteria in tubes, an aliquot of vortexed bacterial suspension was sampled and centrifuged at 11,000 × g for 10 min. The pellets were then washed with water and centrifuged at 11,000 × g for 10 min. The final pellets were resuspended in 5 μl of Live/Dead BacLight bacterial viability kit medium, and bacteria were counted by using conventional fluorescence microscopy as described above.
Molecular investigations: amplification of “Ca. Glomeribacter gigasporarum” genes.
DNA of “Ca. Glomeribacter gigasporarum” was extracted from bacteria isolated from 500 to 1,000 spores of G. margarita by using a WIZARD genomic DNA purification kit (Promega, Lyon, France). The final pellet was resuspended in 50 μl of sterile water. PCRs were carried out in a 25-μl volume containing 2.5 μl of 10× buffer (Promega), 0.5 μM of forward and reverse primers, 2.5 mM MgCl2, 250 μM each deoxynucleoside triphosphate, 1 U of Taq polymerase (Promega), and 1 μl of DNA solution.
For amplification of 16S rDNA with bacterium-like organism-specific primers (BLOf and BLOr) (9) and with the nonspecific primers 27f and 1495r (9), PCR cycling conditions of the PTC 200 thermocycler (MJ Research) were as follows: an initial denaturation step at 93°C for 3 min; 35 cycles at 93°C for 30 s, 55°C for 1 min, and 72°C for 1 min (2 min for amplification with 27f and 1495r primers); and a final extension step at 72°C for 10 min.
For restriction fragment length polymorphism (RFLP) analysis, 10 μl of the PCR products amplified with primers 27f and 1495r were digested in a 20-μl volume containing 2 μl of 10× buffer (Promega) and 0.5 U of restriction enzyme (AluI, SacII, or RsaI), at 37°C for 4 h.
For the pst and vacB genes, new primers were designed according to their database sequences (GenBank accession numbers AJ132617 and AJ242786, respectively). For pst product 1 (2,055 bp) and product 2 (2,184 bp) the primer used were pst1f (5′-GCAAAAATTGGTGAATGCGC-3′) and pst1r (5′-CTTCAGCGAATTCATTGGCC-3′) (product 1) and pst2f (5′-TATCAACGATATTCTGCTCAGCGC-3′) and pst2r (5′-GATTGTTGAATGTATTTACTTCGGG-3′) (product 2). For vacB product 1 (1,337 bp) and product 2 (1,851 bp) the primers used were vacB1f (5′-CTAAAGCGCGCATTCAGGGC-3′) and vacB1r (5′-ACGTTGACGATCTGACCCGC-3′) (product 1) and vacB2f (5′-GATTGAAGAGTGCATGCTGGCG-3′) and vacB2r (5′-AGAGCGGTAAAGCATCGGCC-3′) (product 2). The 23S rDNA primer pairs were designed according to the 23S rDNA conserved region of the following bacterial species which are phylogenetically close to “Ca. Glomeribacter gigasporarum”: Burkholderia gladioli Y17182, Burkholderia mallei Y17183, Burkholderia multivorans Y18704, Burkholderia pseudomallei Y17184, and Burkholderia vietnamiensis Y18705. The primers used were 23S1f (5′-CATGTGGTGGATGCCTTGGC-3′) and 23S1r (5′-ACGGTGCAGGAATATTGACC-3′) for product 1 (1,365 bp) and 23S2f (5′-TGGGGGGACGGATCGCGGAA-3′) and 23S2r 5′-TCAAGCCTTACGGGCAATTA-3′) for product 2 (1,466 bp). The primers were designed according to the oligonucleotide design program of the NTI vector advance software (InforMax). For the other genes (spoVR, mcpA, and nifH), previously described primers were used (23-25). The PCRs were performed on an Eppendorf Mastercycler apparatus, and cycle conditions were as follows: 5 min at 95°C followed by 30 cycles of 30 s at 95°C, 30 s at 57°C, and 30 s at 72°C followed by a finishing step of 10 min at 72°C.
The two vacB gene fragments were amplified by using the Expand Long Template PCR system (Roche, Basel, Switzerland) under the following conditions: 2 min at 95°C followed by 10 cycles of 30 s at 95°C, 30 s at 57°C, and 1 min 30 s at 68°C followed by 20 cycles of 30 s at 95°C, 30 s at 57°C, and 30 s at 68°C plus 20 s at each cycle, followed by a 7-min step at 68°C. The two pst and 23S fragments were amplified under the same conditions as those of the vacB amplification except that the annealing temperature was lowered to 50°C.
“Ca. Glomeribacter gigasporarum” genome size characterization. (i) Bacterial DNA preparation.
For a single bacterial plug used for one pulsed-field gel electrophoresis (PFGE) analysis, between 1 × 104 and 4 × 104 spores were used. Bacteria were extracted as described above. The quantity of living bacteria in the extract was checked under a fluorescent microscope with a Live/Dead BacLight bacterial viability kit. Approximately 108 bacteria ml−1 were embedded in agarose plugs (LM-MP agarose [Roche] at 1% in Tris-EDTA [TE, pH 7]). The plugs were treated overnight in a lysis solution (6 mM Tris-HCl [pH 7.5], 100 mM EDTA [pH 8], 1 M NaCl, 0.5% Brij, 0.2% deoxycholate, 0.5% N-laurylsarcosyl, 1 mg of lysozyme ml−1, 20 μg of RNase ml−1) at 37°C as described previously for Buchnera (14). Subsequently, the plugs were incubated in a TE (pH 7.5) solution containing a previously activated pronase (2 mg ml−1; Sigma) at 37°C during 1 day. The solution was then replaced by the same fresh solution, and plugs were incubated 2 days more at 37°C. The plugs were then rinsed twice in a TE (pH 7.5) solution for 30 min at room temperature and stored at 4°C.
(ii) Restriction enzyme assay.
Plugs were first treated in a TE solution containing a protease inhibitor, Pefabloc (200 μg per plug; Roche), for 40 min at room temperature and then treated for 40 min at 37°C in the same fresh solution. Plugs were then washed twice in a TE (pH 7.5) solution at room temperature for 30 min. The plugs were subsequently equilibrated in the restriction buffer (Invitrogen, Carlsbad, Calif.) at room temperature for 30 min. The restriction buffer was then replaced by the same fresh buffer containing the restriction enzyme (10 U of SpeI, PmeI, or CeuI; Invitrogen), and the samples were subsequently incubated for 30 min on ice. The samples were then transferred at 37°C and incubated for 3 h. After that, fresh enzyme (10 U) was added to the plug and incubated for 3 h more at 37°C. These extreme bacterial lysis conditions were used to compensate for the low amount of bacteria. Plugs were then rinsed in a TE (pH 7.5) solution and stored at 4°C. For longer digestion, a doubled quantity of enzyme was used at each step and two other steps of 3-h digestion were added.
(iii) PFGE.
PFGE was performed with a CHEF-DR DRII apparatus (Bio-Rad, Hercules, Calif.) in 1% agarose gel in 0.5× Tris-borate-EDTA. Electrophoresis conditions were as follows: 5 to 120 s at 4.5 V cm−1 for 48 h (see Fig. 5); 20 s at 4.5 V cm−1 for 12 h, followed by a switch to 5 s to 15 s at 4.5 V cm−1 for 17 h (see Fig. 6A1); 5 to 120 s at 4.5 V cm−1 for 30 h (see Fig. 6A2); 1 to 12 s at 6 V cm−1 for 15 h (see Fig. 6A3); 1 to 12 s at 6 V cm−1 for 12 h (see Fig. 6C); and 5 to 120 s at 4.5 V cm−1 for 30 h (see Fig. 6C). After separation, gels were stained with Vistra green dye (Amersham Biosciences, Buckinghamshire, United Kingdom) which was 10 times more sensitive than ethidium bromide for DNA material detection.
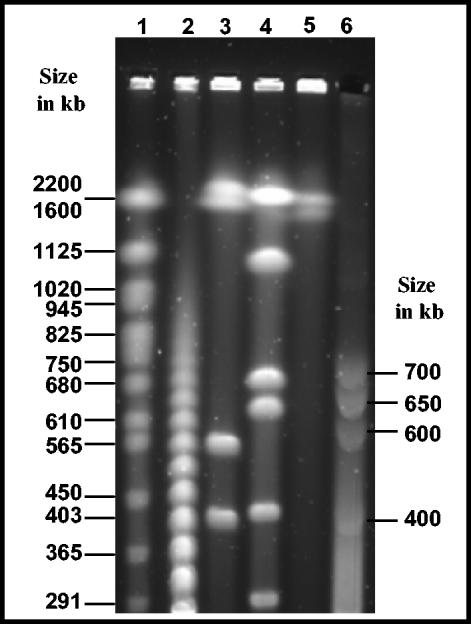
FIG. 5.
PFGE separation of intact genomic DNA of “Ca. Glomeribacter gigasporarum.” Some selected DNA sizes of either the standards or S. meliloti-digested fragments are indicated (in kb); (1) Saccharomyces cerevisiae molecular weight marker (Bio-Rad); (2) λ molecular weight marker (New England Biolabs, Beverly, Mass.); (3) CeuI/S. meliloti; (4) PmeI/S. meliloti; (5) nondigested S. meliloti; (6) nondigested “Ca. Glomeribacter gigasporarum.” All indicated sizes are in kilobases.
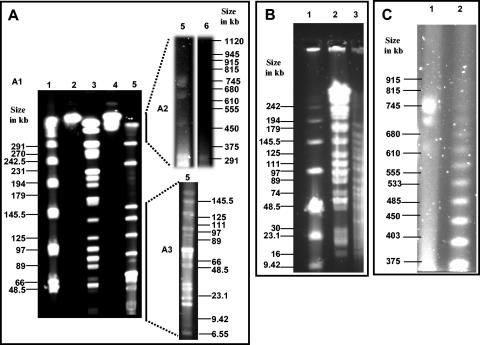
FIG. 6.
PFGE separation of digested genomic DNA of “Ca. Glomeribacter gigasporarum.” Different pulse times were used to achieve resolution in the desired size range (see Materials and Methods). Some selected DNA sizes of either the standards or S. meliloti-digested fragments are indicated (in kb). A: SpeI-digested genomic DNA of “Ca. Glomeribacter gigasporarum” under three electrophoresis conditions, A1, A2, and A3 (see Materials and Methods); (1) λ molecular weight marker (New England Biolabs), (2) nondigested S. meliloti, (3) SpeI/S. meliloti, (4) CeuI/S. meliloti, (5) SpeI/“Ca. Glomeribacter gigasporarum” under standard digestion conditions, (6) SpeI/“Ca. Glomeribacter gigasporarum” under longer digestion conditions. B: PmeI-digested genomic DNA of “Ca. Glomeribacter gigasporarum.” Plugs were all subjected to longer digestion times; (1) low-range molecular weight marker (New England Biolabs), (2) SpeI/S. meliloti, (3) PmeI/“Ca. Glomeribacter gigasporarum.” C: CeuI-digested genomic DNA. Plugs were all subjected to longer digestion; (1) CeuI/“Ca. Glomeribacter gigasporarum,” (2) λ molecular weight marker (New England Biolabs).
(iv) Southern blot hybridization.
Gels were transferred by capillarity on nylon membranes (Hybond-N; Amersham), and the membranes were subjected to Southern hybridization with the specific 32P-labeled probe generated by PCR on “Ca. Glomeribacter gigasporarum” total DNA. The labeling was performed with a Ready-To-Go DNA labeling kit (Amersham Biosciences).
RESULTS AND DISCUSSION
Due to the obligate symbiotic nature of G. margarita, conditions consisting of 6 months in 5-liter pot cultures with four sorghum plants are required to produce 15,000 fungal spores. To complete this study, more than 3 × 105 spores (100 liters of mycorrhizal sorghum cultures) were produced, collected, and manually picked to prepare clean and adequately large bacterial samples. However, the relatively small endocellular bacterial density in each fungal spore impaired the possibility to access large quantities of “Ca. Glomeribacter gigasporarum ” bacteria.
Isolation of “Ca. Glomeribacter gigasporarum” from fungal spores.
The protocol developed by Heddi et al. (19) to isolate the endocellular bacteria from the weevil Sitophilus oryzae (Coleoptera), which was slightly modified in this study (no filtration step, MES buffer instead of Pipes, and no phenylthiourea in the buffers), allowed us to consistently isolate reasonably clean endocellular bacteria. The crucial point of this method compared to the others (see Materials and Methods) was the use of a separation buffer containing Percoll, Ficoll, and polyethylene glycol that allowed satisfactory separation of bacteria from most debris.
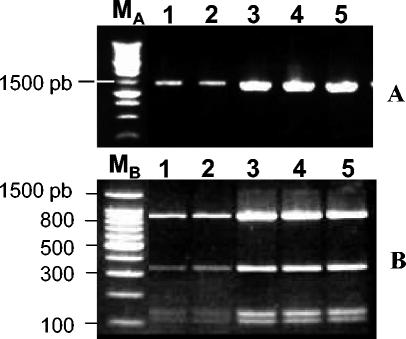
The entire 16S rDNA of isolated bacteria was amplified by PCR with universal primers 27f and 1495r (9), and RFLP analyses with several restriction enzymes (RsaI, SacII, and AluI) were carried out (Fig. 1). The results were in accordance with in silico analysis of the 16S rDNA sequence (GenBank accession number X89727) of “Ca. Glomeribacter gigasporarum,” excluding the possibility that our bacterial preparations contained a significant amount of contaminating bacteria.
FIG. 1.
RFLP analyses of the 16S rDNA of bacteria isolated from different sets of G. margarita spores (lanes 2 to 5) amplified by using the nonspecific bacterial primers 27f and 1495r (A) and digested with RsaI (B). Lane 1, same analysis with total genomic DNA extracted from G. margarita spores. MA, 1-kb ladder (Promega); MB, 100-bp ladder (Promega). pb, base pairs.
An average of ca. 13,000 bacteria per spore was obtained with a maximum of 26,000 and a minimum of 3,700 bacteria per spore (Table 1). This result is in agreement with the recent estimate (19,000 bacteria per spore) made by Bianciotto et al. (6) which is considerably smaller, but more reliable, than the initial estimate of 250,000 bacteria per spore (9). For the sets of spores 1, 9, and 12, variation coefficients of 20, 6, and 13%, respectively, were calculated, indicative of some experimental variations. The difference between sets 1 to 9 (15,500 bacteria per spore) and 10 to 12 (5,000 bacteria per spore) represents a real biological variability, a natural heterogeneity among pot cultures in the numbers of bacteria per spore. Spores from sets 10 to 12 were produced during the scorching period prevalent in Europe from June to August 2003. The temperature of the greenhouse and daily light irradiation during this period were in average 5°C and 26% above normal values, respectively (see the Meteo France website for details [http://www.meteofrance.com/FR/climat/dpt_tempsdumois.jsp?LIEUID=DEPT21]). These pots contained two to three times more spores than usual (for the same volume of substratum and time production). These conditions might have enhanced spore production without stimulating bacterial growth proportionally.
TABLE 1.
Estimation of the number of bacteria per spore of G. margaritaa
| Spore set | No. of spores used per preparation | Estimated no. of isolated bacteria | Calculated no. of bacteria/spore | Mean (SD) of No. of bacteria/sporec |
|---|---|---|---|---|
| 1 | 2,000 | 25 × 106 | 12,500 | 12,500 (2,500) |
| 2,000 | 2 × 107 | 10,000 | ||
| 1,000 | 15 × 106 | 15,000 | ||
| 2 | 1,000 | 107 | 10,000 | |
| 3 | 10,000 | 108 | 10,000 | |
| 4 | 22,000 | 35 × 107 | 16,000 | |
| 5 | 15,000 | 15 × 107 | 10,000 | |
| 6 | 15,000 | 2 × 108 | 13,500 | |
| 7 | 500 | 9 × 106b | 18,000 | 19,000 (1,414) |
| 500 | 107b | 20,000 | ||
| 8 | 500 | 13 × 106b | 26,000 | |
| 9 | 500 | 9 × 106b | 18,000 | 18,670 (1,155) |
| 500 | 107b | 20,000 | ||
| 500 | 9 × 106b | 18,000 | ||
| 10 | 40,000 | 25 × 107 | 6,300 | |
| 11 | 40,000 | 35 × 107 | 8,800 | |
| 12 | 1,000 | 4 × 106 | 4,000 | 3,690 (470) |
| 1,000 | 3 × 106 | 3,000 | ||
| 1,000 | 4 × 106 | 4,000 | ||
| 800 | 3 × 106 | 3,750 |
Bacteria were counted manually when conventional fluorescence microscopy was used or counted with ImagePro Plus software when confocal microscopy was used. The data are rounded numbers.
Bacterial counts obtained when confocal microscopy was used.
Means shown are for spore set series 1, 7, 9, and 12.
Ultrastructural morphology of isolated “Ca. Glomeribacter gigasporarum.”
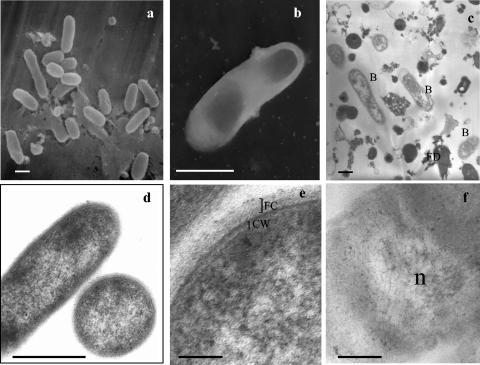
At low magnification, the samples revealed fields with a high number of clustered bacterial cells, ranging from 5 to 25 cells (Fig. 2a and c), associated with some remaining fungal debris (membrane, cell wall, and cytoplasmic materials). Fungal mitochondria comparable to those observed in G. margarita hyphae (10) were not observed in the preparations. They might have been severely damaged by osmotic shocks during bacterial isolation or they may represent an insignificant endocellular component of dormant G. margarita spores (13). Most bacteria appeared to be rod shaped (0.8 to 1.2 by 1.5 to 2.0 μm in size). When seen at higher magnifications, the bacteria appeared to be well preserved (Fig. 2d, e, and f). As for “Ca. Glomeribacter gigasporarum” living in situ in the whole spores, they presented a laminated cell wall typical of gram-negative bacteria (Fig. 2e) and a cytoplasm rich in ribosomes. An electron-transparent area probably corresponding to the chromosome was often observed (Fig. 2f). The cell surface was particularly complex, with a fibrillar coat (Fig. 2e), but flagella or pili were not visible, even upon negative staining (Fig. 2b).
FIG. 2.
Electron micrographs of isolated “Ca. Glomeribacter gigasporarum” bacteria. a: scanning electron microscopy; b: negative microscopy; c, d, e, and f: transmission electron microscopy. a, b, c, and d: bars, 0.5 μm; e and f: bars, 0.1 μm. B, bacteria; FD, fungal debris; FC, fibrillar coat; CW, cell wall; n, bacterial chromosome with fibrillar appearance.
Cultivability and survival of “Ca. Glomeribacter gigasporarum.” (i) Cultivability.
Free-living capacities of “Ca. Glomeribacter gigasporarum” were investigated. Several media suitable to sustain growth of a large spectrum of different microorganisms (rhizobia, Pseudomonas spp., Burkholderia spp., and fungi), defined or not, supplemented with various vitamins or amino acids, were tested. “Ca. Glomeribacter gigasporarum” growth was never observed in any of the tested media and chosen conditions. It is possible that growth of “Ca. Glomeribacter gigasporarum” requires more specific conditions in terms of carbon source, partial O2, partial CO2, or pH. Alternatively, this bacterium could also be an obligate symbiont, strictly dependent on its fungal host for growth.
(ii) Survival.
As no growth was observed, we tested several conditions (water and rich medium at 4 and 28°C) to determine “Ca. Glomeribacter gigasporarum” survival capacities.
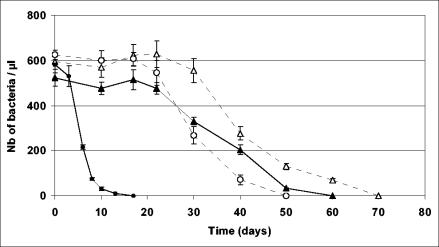
In water at 28°C (Fig. 3), the number of living “Ca. Glomeribacter gigasporarum” bacteria quickly decreased. After 2 weeks, living bacteria were no longer visible. At 4°C, the number of bacteria in the suspension was stable during 3 weeks. It then slowly decreased during the next 4 weeks. After 7 weeks, all bacteria had collapsed. In TY medium, the number of bacteria was stable for 3 weeks at both 4 and 28°C and then slowly decreased (slower than in water). After 10 weeks, no bacterium was observed. These data indicate that “Ca. Glomeribacter gigasporarum” is able to survive several weeks outside of its fungal host. The best condition to keep bacteria alive was in the TY medium at 4°C.
FIG. 3.
Survival of “Ca. Glomeribacter gigasporarum” incubated in water at (○) 4°C and (•) 28°C or in TY medium at (▵) 4°C and (▴) 28°C. Numbers (Nb) of bacteria per microliter were estimated by counting bacteria on a Thoma cell in an aliquot of the bacterial suspension stained with Live/Dead bacterial viability kit stain. Each point is the mean of three independent tubes. Vertical lines correspond to standard errors of the means (n = 3).
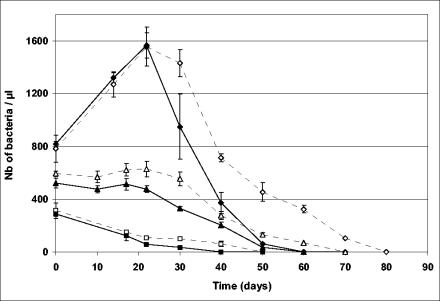
In TY medium at 4°C and at 28°C, the initial density of bacteria influenced bacterial activity and survival (Fig. 4). When the initial density was about 300 bacteria μl−1, bacteria died quickly and were no longer visible at either temperature after 1 month of incubation. When the initial density was about 800 bacteria μl−1, bacteria did not stay alive longer, but a twofold increase in bacterial population was observed during the first month of incubation at both 4 and 28°C. Bacterial population numbers grew from 800 to 1,600 bacteria μl−1 during the first 3 weeks before strongly decreasing, faster at 28°C than at 4°C. In water, even with a high initial bacterial density, such a phenomenon was not observed (data not shown). All the above experiments have been repeated at least twice (with 3 tubes per condition) and gave very similar results. This increase in bacterial number could be the result of bacterial septation with no DNA replication or the result of some actual bacterial growth (one division cycle). Additional experiments such as [3H]thymidine labeling would be necessary to conclude whether this is the case. Surprisingly, similar results were obtained with bacteria incubated at 4°C. It is possible that temperature is not the limiting factor in the physiological and rather slow processes involved.
FIG. 4.
Survival of “Ca. Glomeribacter gigasporarum” incubated in TY medium at 4°C (white symbols) and 28°C (black symbols). Three experiments were carried out with different initial bacterial densities, about 800 (diamond), 600 (triangle), and 300 (square) bacteria per μl. Numbers (Nb) of bacteria per microliter were estimated by counting bacteria on a Thoma cell in an aliquot of the bacterial suspension stained with Live/Dead bacterial viability kit stain. Each point is the mean of three independent tubes. Vertical lines correspond to standard errors of the means (n = 3).
“Ca. Glomeribacter gigasporarum” genome size estimated by PFGE analysis.
Due to the difficulty in purifying large amounts of bacteria, PFGE analyses were performed with 1 × 108 to 6 × 108 bacteria per plug isolated from 1 × 104 to 4 × 104 spores, whereas conventional PFGE analyses are usually carried out with 10 times more bacteria. This led us to use the Vistra green dye instead of ethidium bromide for nucleic acid detection (see Materials and Methods).
Analysis of nondigested DNA.
With electrophoresis conditions appropriate for separating large DNA fragments, no band larger than 1 Mb was visualized. By using ramping pulses appropriate for separation of fragments of medium size (200 kb to 1 Mb), four distinct bands with apparent sizes of 700, 650, 600, and 400 kb ± 50 kb (Fig. 5) were visualized. This four-band pattern was highly reproducible and first suggested that the genome size of “Ca. Glomeribacter gigasporarum” was around 2.35 Mb. It was unlikely that some of the observed replicons were of mitochondrial origin because no significant contamination of the bacterial preparations with mitochondria was observed by transmission electron microscopy (see above).
Analysis of digested DNA.
Subsequently, genomic DNA contained in the plugs was digested with the SpeI, PmeI, and CeuI enzymes. The restriction fragments were analyzed by PFGE at different pulse times to achieve resolution in the desired size range (Fig. 6). For the SpeI restriction analysis, two different digestion times were applied (6 h with 20 U of enzyme and 12 h with 40 U of enzyme; see Materials and Methods). The two digestion patterns were identical except for the upper band in the 6-h digestion (Fig. 6A, lane 5) which was absent from the 12-h restriction pattern (Fig. 6A, lane 6). Under appropriate electrophoresis conditions, this band was shown to correspond to the four undigested replicons (Fig. 6A2, lane 5) and was therefore shown to result from an incomplete restriction of DNA.
The sizes of all SpeI restriction fragments were summed to estimate the total size of the “Ca. Glomeribacter gigasporarum” genome to approximately 1.4 Mb (Table 2). Congruently, the PmeI digestion resulted into 16 linear fragments (Fig. 6B) whose total size was 1.35 Mb (Table 2).
TABLE 2.
Restriction fragment analysesa
| Restriction fragment (total size [kb]) | Fragment size (kb) |
|---|---|
| SpeI (1,390) | 280, 240, 150, 140, 125, 100, 80, 80, 75, 40, 30, 25, 15, 10 |
| PmeI (1,350) | 180, 150, 140, 130, 120, 110, 100, 90, 80, 75, 70, 50, 30, 25, 20, 15 |
Fragments sizes were estimated by comparison with either molecular weight standard or known S. meliloti SpeI and PmeI restriction fragments.
Hence, the estimated “Ca. Glomeribacter gigasporarum” genome size varied between 1.35 to 2.35 Mb, depending on whether it was based on intact or restricted DNA. For reasons detailed below, we believe that 1.35 Mb is more likely the right estimate.
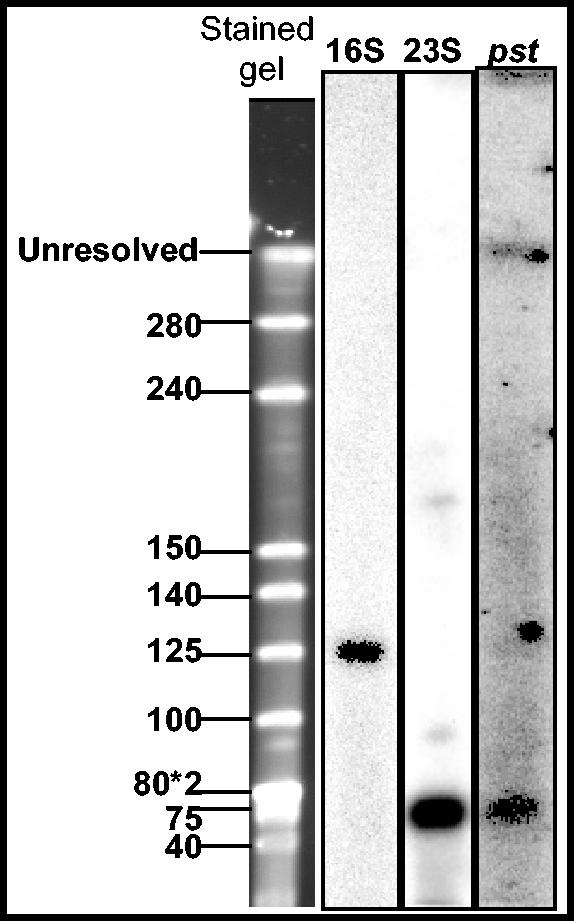
The 26-bp recognition sequence of CeuI is conserved in the 23S rRNA of many eubacteria and is therefore a useful marker for chromosomal DNA. CeuI digestion of “Ca. Glomeribacter gigasporarum” DNA gave rise to only three bands that migrated at apparent sizes of 745, 700, and 650 kb ± 50 kb (Fig. 6C). However, compared to previous experiments with undigested samples, the 745-kb band was brighter than the other bands, suggesting that this replicon had been linearized by CeuI. Independent evidence for a single copy of the rDNA operon in the genome was obtained from a Southern blot hybridization of SpeI-digested genomic DNA with 16S and 23S probes. As one SpeI site is present in the 16S rDNA gene (X89727), we expected the 16S and 23S probes to hybridize on two different SpeI fragments. Accordingly, the 16S probe hybridized to a 130-kb fragment and the 23S probe hybridized to a 70-kb fragment (Fig. 7). Altogether, these data indicated that “Ca. Glomeribacter gigasporarum” has a single chromosome (defined here as an rDNA-carrying replicon) of 745 kb ± 50 kb in size.
FIG. 7.
Southern blot hybridization with “Ca. Glomeribacter gigasporarum” markers. The transferred SpeI gel was hybridized with the 16S, 23S, and pst probes of “Ca. Glomeribacter gigasporarum.”
Examination of the CeuI-digested gel (Fig. 6, panel C) indicated the presence of only three bands instead of four in the nondigested gel (Fig. 5) (the 400-kb band was absent). We interpret this result as the existence of two different topoisomers of the chromosome that separate in native gels giving rise to two bands, one in the (apparent) 600- to 700-kb range and one in the 400-kb range. Although we have no direct evidence of this, we speculate that, similarly, the other two native replicons may actually represent two topoisomers of a single replicon. This artefactual duplication of bands in the native gels would readily explain the discrepancy in genome size mentioned above. Our current opinion is that the total size is ca. 1.4 Mb consisting of a ca. 750-kb chromosome and an additional replicon in the 650-kb size range.
Of course, we acknowledge that this characterization of the genome of “Ca. Glomeribacter gigasporarum” remains preliminary. The noncultivable nature of these bacteria and the extreme difficulty in obtaining sufficient material for laboratory studies (more than 3 × 105 spores were used for this work) prevented us from providing a definitive answer. In spite of the uncertainties indicated above, it is clear that “Ca. Glomeribacter gigasporarum” has an overall small genome size and a small (745 kb) chromosome. The smallest β-proteobacterium genome is, to our knowledge, that of Dechloromonas RCB (2 Mb) (see the U.S. Department of Energy Microbial Genome Project website [http://www.ornl.gov/sci/microbialgenomes/organisms.shtml]). The small genome size of “Ca. Glomeribacter gigasporarum” is consistent with a strict endosymbiotic nature of this bacterium. Indeed, small chromosomes such as that of “Ca. Glomeribacter gigasporarum” (745 kb) have been encountered only in Buchnera species (450 to 641 kb) (17), Wigglesworthia glossinidia (770 kb) (1), Wolbachia (950 to 1,660 kb) (41), and the primary symbiont of the sharpshooter (Cicadellinae) (680 kb) (26). All these bacteria are obligate endocellular species.
Does “Ca. Glomeribacter gigasporarum” possess nif genes?
Different “Ca. Glomeribacter gigasporarum ” genes were previously isolated from a genomic library constructed with the total DNA from G. margarita spores that included the bacterial genome (42). The nifHDK operon and cheY, mcpA, prkA, spoVR, and 16S rDNA genes were isolated by screening this library, whereas vacB and pst were first identified by using degenerated primers on genomic spore DNA (9, 23, 24, 25, 33, 34). The availability of the bacterial DNA allowed us for the first time to verify the origin of these genes. PCR experiments were first performed to detect their presence on pure genomic DNA preparations. As expected, the pst, vacB, and 16S rDNA genes could be amplified. We were not able to visualize vacB hybridization to any transferred gel for an unknown reason. pst hybridized instead to the 75-kb SpeI DNA fragment (Fig. 7).
Unexpectedly, we were repeatedly unable to amplify the nif, spoVR, and mcpA genes from the isolated “Ca. Glomeribacter gigasporarum” DNA, although the λ1NIF clone from which they had been previously isolated (23-25) gave a product of the expected size. nif primers were also tested with genomic DNA extracted from nongerminated G. margarita spores, germinated spores, and colonized roots, but we never got any PCR amplification products (data not shown). Consequently, we hypothesize that the DNA region contained in the λ1NIF clone (including the cheY and prkA genes, although these markers were not tested specifically), which presumably originated from the original “Ca. Glomeribacter gigasporarum” isolate, has been lost in the “Ca. Glomeribacter gigasporarum” isolate we have been working with in this study. Alternatively, the λ1NIF clone obtained in the G. margarita genomic library may have originated from a contaminating, unknown microorganism which possessed at least some of the genes required for nitrogen fixation. Further screening of the genomic library has indeed indicated the presence of contaminating bacterial DNA from which the nif DNA may have originated (D. Minerdi and P. Bonfante, unpublished data).
Conclusion.
The isolation protocol of “Ca. Glomeribacter gigasporarum” allowed us to obtain a reasonable amount of free bacterial cells to investigate for the first time several of their morphological, physiological, and genomic traits. They are rod shaped and heterogeneous in size, and their cell wall ultrastructure is typical of gram-negative bacteria. Free-living bacteria can stay alive for several weeks but were not culturable in any of the tested conditions. The pst, vacB, and 16S genes previously assigned to the genome of “Ca. Glomeribacter gigasporarum” (9, 33, 34) were successfully amplified, but the presence of nif, mcpA, and spoVR genes could not be confirmed.
From our knowledge of other prokaryote-eukaryote interactions (15, 37), we conclude that the small genome size of “Ca. Glomeribacter gigasporarum” bacteria and our incapacity to grow them in pure culture indicate a strict and ancient physiological dependency of the bacterial partner upon its fungal host. The reciprocal dependency of the fungus upon its bacterial partner, typical of mutualistic association, is not yet supported by any physiological or molecular evidence but is suggested by the fact that the bacteria are vertically transmitted across vegetative fungal generations (6). Only the availability of cured G. margarita spores will provide the means to experimentally identify the role of “Ca. Glomeribacter gigasporarum” in its fungal host and the significance of this bacterium-fungus association on mycorrhizal symbiosis.
Acknowledgments
We are very grateful to Bachar Blal and Marie-Line Haimet (BIORIZE) for providing a large amount of G. margarita spores. We thank Erica Lumini and Valeria Bianciotto for their great help and support, Daniela Minerdi for the screening of the genomic library, Jacques Vasse for assistance in negative microscopy, Chrisse Ngari for technical help, Thierry Liboz for access to the CHEF-DR II Bio-Rad apparatus, and A. Faccio and S. Panero for technical support in the Microscopical Laboratory (Turin).
This research was funded to G.B., J.B., and P.B. by the European Union GENOMYCA project (project no. QLK5-CT-2000-01319; http://www.dijon.inra.fr/bbceipm/genomyca/). This project provided postdoctoral fellowships to P.J. and C.C.
REFERENCES
- 1.Akman, L., and S. Aksoy. 2001. Escherichia coli gene array analysis provides insight into the biology of the obligate endosymbiont of tsetse flies. Proc. Natl. Acad. Sci. USA 98:7546-7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashdown, L. R. 1992. Rapid differentiation of Pseudomonas pseudomallei from Pseudomonas cepacia. Lett. Appl. Microbiol. 14:203-205. [DOI] [PubMed] [Google Scholar]
- 3.Balestrini, R., M. G. Hahn, A. Faccio, K. Mendgen, and P. Bonfante. 1996. Differential localization of carbohydrate epitopes in plant cell walls in the presence and absence of arbuscular mycorrhizal fungi. Plant Physiol. 111:203-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bécard, G., and J. A. Fortin. 1988. Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol. 108:211-218. [DOI] [PubMed] [Google Scholar]
- 5.Bertaux, J., M. Schmid, N. C. Prevost-Boure, J. L. Churin, A. Hartmann, J. Garbaye, and P. Frey-Klett. 2003. In situ identification of intracellular bacteria related to Paenibacillus spp. in the mycelium of the ectomycorrhizal fungus Laccaria bicolor S238N. Appl. Environ. Microbiol. 69:4243-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianciotto, V., A. Genre, P. Jargeat, E. Lumini, G. Bécard, and P. Bonfante. 2004. Vertical transmission of endobacteria in the arbuscular mycorrhizal fungus Gigaspora margarita through vegetative spore generations. Appl. Environ. Microbiol. 70:3600-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bianciotto, V., E. Lumini, P. Bonfante, and P. Vandamme. 2003. ‘Candidatus Glomeribacter gigasporarum’ gen. nov., sp. nov., an endosymbiont of arbuscular mycorrhizal fungi. Int. J. Syst. Evol. Microbiol. 53:121-124. [DOI] [PubMed] [Google Scholar]
- 8.Bianciotto, V., E. Lumini, L. Lanfranco, D. Minerdi, P. Bonfante, and S. Perotto. 2000. Detection and identification of bacterial endosymbionts in arbuscular mycorrhizal fungi belonging to the family Gigasporaceae. Appl. Environ. Microbiol. 66:4503-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianciotto, V., C. Bandi, D. Minerdi, M. Sironi, H. V. Tichy, and P. Bonfante. 1996. An obligately endosymbiotic fungus itself harbors obligately intracellular bacteria. Appl. Environ. Microbiol. 62:3005-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianciotto, V., and P. Bonfante. 1999. Presymbiotic versus symbiotic phase in arbuscular endomycorrhizal fungi, p. 229-251. In A. Varma and B. Hock (ed.), Mycorrhiza. Springer-Verlag, Berlin, Germany.
- 11.Blackwell, M. 2000. Terrestrial life—fungal from the start? Science 289:1884-1885. [DOI] [PubMed] [Google Scholar]
- 12.Bonfante, P. 2003. Plants, mycorrhizal fungi and endobacteria: a dialog among cells and genomes. Biol. Bull. 204:215-220. [DOI] [PubMed] [Google Scholar]
- 13.Bonfante, P., R. Balestrini, and K. Mendgen. 1994. Storage and secretion processes in the spore of Gigaspora margarita Becker & Hall as revealed by high-pressure freezing and freeze substitution. New Phytol. 128:93-101. [DOI] [PubMed] [Google Scholar]
- 14.Charles, H., and H. Ishikawa. 1999. Physical and genetic map of the genome of Buchnera, the primary endosymbiont of the pea aphid Acyrthosiphon pisum. J. Mol. Evol. 48:142-150. [DOI] [PubMed] [Google Scholar]
- 15.Douglas, A. E. 1998. Host benefit and the evolution of specialization in symbiosis. Heredity 80:599-603. [Google Scholar]
- 16.Gerhardt, P., R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. B. Phillips. 1981. Manual of methods of general bacteriology. American Society for Microbiology, Washington, D.C.
- 17.Gil, R., B. Sabater-Muñoz, A. Latorre, F. J. Silva, and A. Moya. 2002. Extreme genome reduction in Buchnera spp.: toward the minimal genome needed for symbiotic life. Proc. Natl. Acad. Sci. USA 99:4454-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray, M. W. 1999. Evolution of organellar genomes. Curr. Opin. Genet. Dev. 9:678-687. [DOI] [PubMed] [Google Scholar]
- 19.Heddi, A., F. Lefebvre, and P. Nardon. 1991. The influence of symbiosis on the respiratory control ratio (RCR) and the ADP/O ratio in the adult weevil Sitophilus oryzae (Coleoptera, Curculionidae). Endocyt. Cell Res. 8:61-73. [Google Scholar]
- 20.Ignatov, G., V. Vassileva, and S. Dimova-Terziivanova. 2000. The oxidative properties of mitochondria and bacteroids from root nodules of soybean treated with organic acids. Bulg. J. Plant Physiol. 26:3-14. [Google Scholar]
- 21.Kuhn, G., M. Hijri, and I. R. Sanders. 2001. Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature 414:745-748. [DOI] [PubMed] [Google Scholar]
- 22.Margulis, L., and M. J. Chapman. 1998. Endosymbioses: cyclical and permanent in evolution. Trends Microbiol. 6:342-345. [DOI] [PubMed] [Google Scholar]
- 23.Minerdi, D., R. Fani, and P. Bonfante. 2002. Identification and evolutionary analysis of putative cytoplasmic McpA-like protein in a bacterial strain living in symbiosis with a mycorrhizal fungus. J. Mol. Evol. 54:815-824. [DOI] [PubMed] [Google Scholar]
- 24.Minerdi, D., V. Bianciotto, and P. Bonfante. 2002. Endosymbiotic bacteria in mycorrhizal fungi: from their morphology to genomic sequences. Plant Soil 244:211-219. [Google Scholar]
- 25.Minerdi, D., R. Fani, R. Gallo, A. Boarino, and P. Bonfante. 2001. Nitrogen fixation genes in an endosymbiotic Burkholderia strain. Appl. Environ. Microbiol. 67:725-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moran, N. A., C. Dale, H. Dunbar, W. A. Smith, and H. Ochman. 2003. Intracellular symbionts of sharpshooters (Insecta: Hemiptera: Cicadellinae) form a distinct clade with a small genome. Environ. Microbiol. 5:116-126. [DOI] [PubMed] [Google Scholar]
- 27.Moran, N. A. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583-586. [DOI] [PubMed] [Google Scholar]
- 28.Moran, N. A., and P. Bauman. 2000. Bacterial endosymbionts in animals. Curr. Opin. Microbiol. 3:270-275. [DOI] [PubMed] [Google Scholar]
- 29.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1098. [DOI] [PubMed] [Google Scholar]
- 30.Rai, A. N., E. Söderbäck, and B. Bergman. 2000. Cyanobacterium-plant symbioses. New Phytol. 147:449-481. [DOI] [PubMed] [Google Scholar]
- 31.Redecker, D., R. Kodner, and L. E. Graham. 2000. Glomalean fungi from the Ordovician. Science 289:1920-1921. [DOI] [PubMed] [Google Scholar]
- 32.Roland, J.-C., and B. Vian. 1991. General preparation and staining of thin sections, p. 1-33. In J. L. Hall and C. Hawes (ed.), Electron microscopy of plant cells. Academic Press, London, United Kingdom.
- 33.Ruiz-Lozano, J. M., and P. Bonfante. 2000. A Burkholderia strain living inside the arbuscular mycorrhizal fungus Gigaspora margarita possesses the vacB gene, which is involved in host cell colonization by bacteria. Microb. Ecol. 39:137-144. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz-Lozano, J. M., and P. Bonfante. 1999. Identification of a putative P-transporter operon in the genome of a Burkholderia strain living inside the arbuscular mycorrhizal fungus Gigaspora margarita. J. Bacteriol. 181:4106-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scannerini, S., and P. Bonfante. 1991. Bacteria and bacteria like objects in endomycorrhizal fungi (Glomaceae), p. 273-287. In L. Margulis and R. Fester (ed.), Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. MIT Press, Cambridge, Massachusetts. [PubMed]
- 36.Schüβler, A., and M. Kluge. 2001. Geosiphon pyriforme, and endocytosymbiosis between fungus and cyanobacteria, and its meaning as a model system for arbuscular mycorrhizal research, p. 151-161. In B. Hock (ed.), The mycota IX. Springer Verlag, Berlin, Germany.
- 37.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 38.Simon, L., J. Bousquet, R. C. Lévesque, and M. Lalonde. 1993. Origin and diversification of endomycorrhizal fungi and coincidence with vascular land plants. Nature 363:67-69. [Google Scholar]
- 39.Smith, S. E., and D. J. Read. 1997. Mycorrhizal symbiosis. Academic Press, New York, N.Y.
- 40.Spaink, H. P., A. Kondorosi, and P. J. J. Hooykaas. 1998. The Rhizobiaceae. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 41.Sun, L. V., J. M. Foster, G. Tzertzinis, M. Ono, C. Bandi, B. E. Slatko, and S. L. O'Neill. 2001. Determination of Wolbachia genome size by pulsed-field gel electrophoresis. J. Bacteriol. 183:2219-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Buuren, M., L. Lanfranco, S. Longato, D. Minerdi, M. J. Harrison, and P. Bonfante. 1999. Construction and characterization of genomic libraries of two endomycorrhizal fungi: Glomus versiforme and Gigaspora margarita. Mycol. Res. 103:955-960. [Google Scholar]
- 43.Wernergreen, J. J., A. B. Lazarus, and P. H. Degnan. 2002. Small genome of Candidatus Blochmannia, the bacterial endosymbiont of Camponotus, implies irreversible specialization to an intracellular lifestyle. Microbiology 148:2551-2556. [DOI] [PubMed] [Google Scholar]
- 44.Zientz, E., F. J. Silva, and R. Gross. 2001. Genome interdependence in insect-bacterium symbioses. Genome Biol. 2:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]