Abstract
Expression of an amber suppressor tRNA should result in read-through of the 326 open reading frames (ORFs) that terminate with amber stop codons in the Escherichia coli genome, including six pseudogenes. Abnormal extension of an ORF might alter the activities of the protein and have effects on cellular physiology, while suppression of a pseudogene could lead to a gain of function. We used oligonucleotide microarrays to determine if any effects were apparent at the level of transcription in glucose minimal medium. Surprisingly, only eight genes had significantly different expression in the presence of the suppressor. Among these were the genes yaiN, adhC, and yaiM, forming a single putative operon whose likely function is the degradation of formaldehyde. Expression of wild-type yaiN was shown to result in repression of the operon, while a suppression-mimicking allele lacking the amber stop codon and extended 7 amino acids did not. The operon was shown to be induced by formaldehyde, and the genes have been renamed frmR, frmA, and frmB, respectively.
A suppressor tRNA is a tRNA with a mutation (usually) in the anticodon that allows it to recognize a stop codon and insert an amino acid in its place (reviewed in reference 6). First identified in 1965 (4, 7), they have been widely used in studies of translation, phage biology, and protein engineering. They have been critical to our understanding of the structure and function, processing, and charging of tRNAs, as well as ribosome-tRNA interaction, polarity, codon context effects, and the elucidation of the genetic code (29). Suppressors have also been used to study the impact of single amino acid substitutions and nonstandard amino acids on protein function (15, 30). Amber suppressors have been used preferentially because amber (UAG) is the least common of the three stop codons, occurring at the end of 326 out of 4,290 open reading frames (ORFs) in the Escherichia coli genome. In contrast, ochre (UAA) occurs 2,706 times and opal (UGA) occurs 1,258 times. The reasons for this dramatic skew are not known, though it is interesting that 1.5% of fecal coliform isolates (mostly E. coli) were found to contain natural amber suppressors (18). Out of 13 pseudogenes in strain K-12 (identified from the EcoGene website [26] at http://bmb.med.miami.edu/EcoGene/EcoWeb/), 6 are disrupted with amber stop codons (glvC, ybfH, yehQ, yfcU, yjiP, and b2650) and almost all are of unknown function.
For the functional characterization of the E. coli genome, we have developed methods to systematically introduce amber mutations into the chromosome and have constructed an arabinose-inducible suppressor tRNA on a high-copy-number plasmid to modulate the effects of the amber mutations (12, 13). To use this system it was important to examine the effects of the suppressor on the cell. In addition to suppressing the gene of interest, the suppressor is also expected to affect genes throughout the genome that end with amber stop codons. Reflecting the relative prevalence of amber and ochre stop codons, amber suppressors do not appear to affect the growth of their hosts, whereas isogenic ochre derivatives cause growth defects proportional to the efficiency of suppression (23, 24). It has been shown that amber suppression extends and inactivates the stringent factor RelA (3), but the full extent of extraneous effects has never before been determined. Our approach was to look for a phenotype resulting from suppressor expression. Specifically, we examined global transcription patterns to see if a stress response or other sign of physiological disruption was evident.
We have renamed the genes yaiN, adhC, and yaiM with the new designations frmR, frmA, and frmB, respectively, for reasons detailed below. For clarity, the new names will be used throughout this work.
MATERIALS AND METHODS
Microarrays.
Three independent cultures and RNA preps of two strains were labeled and hybridized. E. coli MG1655 cells carrying either pBAD/sup2 or pBAD18-Kan (10) were grown overnight in morpholinepropanesulfonic acid (MOPS) minimal medium (Teknova, Half Moon Bay, Calif.) plus 0.1% glucose and 50 μg of kanamycin/ml, then diluted 1:50 into 100 ml of MOPS minimal medium plus 0.1% arabinose and kanamycin in a 1-liter flask, and grown at 37°C in a shaking water bath to an optical density at 600 nm (OD600) of 0.2 (7 to 13 h; three to four doublings). Cells were then immediately mixed with RNAprotect bacteria reagent (QIAGEN, Valencia, Calif.). Total RNA was isolated using the Masterpure RNA purification kit (Epicentre, Madison, Wis.). The RNA pellet was resuspended in nuclease-free water, and RNA was measured by absorption at 260 nm. cDNA was synthesized and labeled using a protocol similar to that described by Rosenow et al. (25). Briefly, 10 μg of total RNA was reverse transcribed with 1,200 U of Superscript II (Invitrogen, Carlsbad, Calif.) using 500 ng of random hexamers at 42°C for 90 min. Remaining RNA was removed with 2 U of RNase H and 1 μg of RNase A for 10 min at 37°C. Synthesized cDNA was then purified with QiaQuick (QIAGEN) and fragmented to 50 to 200 bp with 0.2 U of DNase I (Epicentre) for 10 min at 37°C. The fragmented cDNA was 3′-end labeled with 25 μM biotin-N6-ddATP (Applied Biosystems, Foster City, Calif.) using 50 U of terminal transferase (New England Biolabs, Beverly, Mass.) at 37°C for 2 h. The labeled cDNA was hybridized to E. coli antisense genome arrays at 45°C for 16 h, then washed, and scanned as described in the GeneChip technical manual (Affymetrix, Santa Clara, Calif.). Signal was calculated using Microarray suite 5.0 software (Affymetrix) and normalized such that the mean ORF signal over the whole array was equal to 1,000.
Plasmid and strain construction.
All primer sequences are given in Table 1. The chromosomal deletion of frmR was made with gene gorging (13). The mutagenic plasmid insert was made by fusing two fragments consisting of about 500 nucleotides (nt) of sequence on each side of frmR. PCR was performed with a 55°C annealing temperature and Pfu Turbo polymerase (Stratagene, La Jolla, Calif.). The downstream fragment was amplified from E. coli MG1655 DNA with primers OF500 and OF501. The upstream fragment was amplified in two steps to avoid problems with the inverted repeats upstream of frmR: it was amplified first with OF498 and OF509 using five initial cycles with a 40°C annealing temperature followed by 30 cycles at 55°C and then reamplified with OF498 and OF507. The upstream and downstream fragments were then purified with QiaQuick (QIAGEN), and 4 μl of each was combined in a 45-μl reaction mixture. After five cycles, the primers OF498 and OF501 were added and it was cycled 25 more times. I-SceI sites were included in OF498 and OF501. The fusion PCR product was gel purified and cloned into pCR-BluntII-Topo (Invitrogen). After confirmation by sequencing, gene gorging was performed. One deletion was detected out of 134 colonies by colony PCR screening using primers OF502 and OF503. It was plated on Luria broth (LB), and colonies were screened for loss of the drug resistance carried by pACBSR. The plasmid-free strain, called FBSC222, was PCR amplified with OF502 and OF503 and sequenced with OF498 and OF501 to verify that frmR had been deleted without errors.
TABLE 1.
Primers used in this study
| Primer name | Sequence |
|---|---|
| OF129 | ACGTAGATCTGATATCACGGCAGAAAAGTCCAC |
| OF185 | TCTGATTTAATCTGTATCAGGCTGA |
| OF445 | CTTTCGCTAAGGATCTGCAGTG |
| OF446 | TGGGAGAGCGCTTGCATCTAAA |
| OF493 | GAGACATATGCAGATGATGAGGTGCGAAATG |
| OF494 | TCTGCATGCCTATTTAAGATAGGCACGAACCAG |
| OF495 | TCTGCATGCTCAATATGGTAATAGATTCAGCGCTTTAAGATAGGCACG |
| OF496 | GAGACATATGGAGAAACAGTAGAGAGTTGCG |
| OF497 | GGCACATAGCCTTGCTCAAATTG |
| OF498 | CACCTAGGGATAACAGGGTAATGTGATCACCAGCTACCGCTATAC |
| OF500 | TATAGTATATTGCCTGAATCTATTACCATATTGAGGAAG |
| OF501 | TAGGGATAACAGGGTAATTGCCAGAGACACTTCCGCGAC |
| OF502 | GAACGGGTATTGCCTTTGCGGA |
| OF503 | CTGAACCACTGCCAGACCAATC |
| OF507 | GTAATAGATTCAGGCAATATACTATAGGGGGGTATTCTATATGTCAATGC |
| OF509 | TATATGTCAATGCATACCCCCCTA |
| OF511 | CTAATGGGCTGATGGCAGAA |
| OF512 | GTCAACGGATTGGCTGACTT |
| OF513 | GCAAACCATGAACACGTCTG |
| OF514 | ACAGAATCACCTGGCTGGAC |
| OF515 | GATTACGACCCGGTGAGTCTT |
| OF516 | ATTTGGAGTCCGCAGCTGTT |
| OF517 | TCGTGGTGAAGCAGAACAAG |
| OF518 | CGTCGTCTTCGCTGATCTCT |
The plasmids pACB/frmR, pACB/alt and pACB/empty were constructed by cloning different inserts into a derivative of pACBSR (13). A PCR fragment containing the p15A origin, chloramphenicol resistance gene, arabinose promoter-repressor, and rrnB terminators was amplified from pACBSR with primers OF496 and OF497. Wild-type (WT) frmR was amplified from MG1655 DNA with OF493 and OF494. The alternate extended version of frmR was amplified with OF493 and OF495. The vector and inserts were digested with NdeI and SphI and ligated. The empty vector was made by filling in the restriction overhangs with the Klenow fragment of DNA polymerase and self-ligating. The plasmids were verified by amplifying and sequencing each with OF129 and OF185. Construction of the suppressor plasmids pBAD/sup1 and pBAD/sup2 has been described elsewhere (13).
Real-time PCR.
Cells were grown overnight in MOPS minimal medium plus 0.1% glucose (and 25 μg of chloramphenicol/ml for strains carrying plasmids) and then diluted 1:50 into 25 ml of MOPS minimal medium (plus chloramphenicol for plasmid strains) and either 0.1% glucose or 0.1% arabinose plus 0.01% glucose. The small amount of glucose was necessary to “jump start” the arabinose cultures since, otherwise, chloramphenicol would inhibit the protein synthesis necessary for induction of arabinose catabolism and cells could not start growing. Presumably, acetyl coenzyme A levels were too low in the saturated overnight cultures to allow chloramphenicol acetyltransferase to function. Cultures were grown to an OD600 of ∼0.2, and a sample was then immediately added to RNAprotect bacteria reagent (QIAGEN). RNA was purified using RNAeasy mini columns (QIAGEN) and eluted in 30 μl of water. Seventy microliters of 1× DNase I buffer plus 5 U of RNase-free DNase I (Epicentre) was then added and incubated at 37°C for 45 min. The RNA was then repurified using RNAeasy columns and eluted in water. A 0.5-μg aliquot of RNA was reverse transcribed with 1,200 U of Superscript III (Invitrogen) using 25 ng of random hexamers at 42°C for 90 min. The RNA was digested with 2 U of RNase H and 1 μg of RNase A for 10 min at 37°C, and then the cDNA was purified with QiaQuick (QIAGEN).
Primers were designed to amplify ∼100-bp products in the genes frmR (OF511 and OF512), frmA (OF513 and OF514), and frmB (OF515 and OF516). Thirty-microliter PCR mixtures containing 1 ng of cDNA and a 0.9 μM concentration of each primer were amplified with an ABI 7700 sequence detection system using SYBR Green PCR core reagents according to the recommended protocol (Applied Biosystems). A standard curve consisting of E. coli genomic DNA was amplified with each primer set for relative quantitation. The gene frr was used as an endogenous control for variation in the amount of cDNA in each reaction mixture (primers OF517 and OF518). frr is expressed at a fairly constant level across all our lab's collected Affymetrix array data.
RESULTS AND DISCUSSION
The plasmid pBAD/sup2 contains the Ala2 suppressor under control of the arabinose promoter (13). Ala2 was used because of its high suppression efficiency and its specificity in introducing only the correct amino acid (22). Our original construct pBAD/sup1 showed only 6% efficiency in a luciferase assay (28). Noting that the plasmid it was derived from had a different 5′ leader sequence from the transcription start site to the beginning of the tRNA, the leader sequence was modified from ACCCGTTTTTTTGGGCTAGCGAATTC to GCTGAATTC and efficiency increased to 59%. Suppression may have been low in the first construct due to poor processing by RNase P, implying that the length or sequence of the 5′ leader beyond the most-proximal 6 nt affected RNase P cleavage (5).
The growth rate of WT cells containing pBAD/sup2 was 131 ± 18 min (standard deviation) in arabinose minimal medium compared to 153 ± 29 min for cells containing empty vector. With every pUC-origin plasmid tested, different starting colonies from the same plate showed dramatically different growth rates. Slow and fast cultures when replated generated a mixture of colonies that grew both slow and fast in culture. This effect was less pronounced in glucose minimal medium and LB and did not occur with cells lacking a plasmid (73 ± 1.3 min doubling time). Colonies picked from a fresh transformation plate showed much less variation. We conclude that the slow growth rates that we observed were due to the high-copy-number plasmid and not the suppressor tRNA.
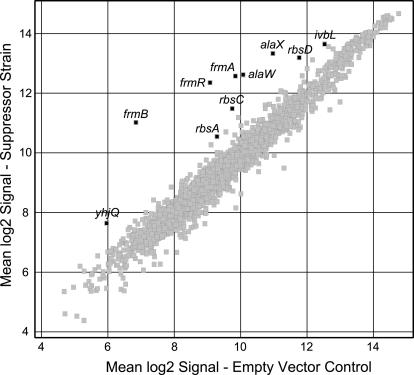
Expression patterns from strains with and without the suppressor were measured using oligonucleotide microarrays (data available at www.genome.wisc.edu/functional/microarray.htm). Cells carrying pBAD/sup2 or empty vector were grown to mid-log phase in arabinose minimal medium, and then cDNA was isolated and hybridized to Affymetrix microarrays. The physiological effects of growth in arabinose and the high-copy-number plasmids were controlled for by comparing plasmid-carrying strains grown under identical conditions. Only eight genes were significantly upregulated as a result of the suppressor, and none were downregulated (Fig. 1; Table 2). Increased signal was also detected for the tRNA genes alaX and alaW, but with a large standard error. These two genes are only 4 nt different from the suppressor tRNA sequence, and so their apparent increase is likely an artifact of cross-hybridization between the overexpressed suppressor tRNA and the alaXW probes on the array. This cross-hybridization confirms the expression of the suppressor in cells carrying pBAD/sup2. Additionally, primers were designed to detect the suppressor transcript by amplifying from the plasmid's transcriptional terminator to the anticodon of the suppressor (OF445 and OF446). These primers were used in PCR on randomly primed cDNA generated from the same RNA samples used with the microarrays. Results from this PCR agreed with the microarray results. To make sure that the plasmids had not been lost during growth, cultures were grown again under identical conditions and then plated on LB and LB-kanamycin plates. Approximately equal numbers grew on both plate types, verifying retention of the plasmids.
FIG. 1.
Scatter plot of microarray results. RNA preparations made from three independent cultures of strains carrying pBAD/sup2 or empty pBAD18 grown in arabinose minimal medium were hybridized to Affymetrix GeneChips. Each spot plots the values of the log2 signal for one gene. Only those genes that the software called either present or marginal in all three replicates of either the experimental or control samples are shown (2,297 genes). The experimental versus control log2 ratio with a 90% confidence interval was calculated from the three replicates. Those genes for which the absolute value of the log2 ratio minus the confidence interval was >0.75 are highlighted. In addition, the genes alaX and alaW are also highlighted. Details of the highlighted genes appear in Table 2.
TABLE 2.
Microarray results
| b no. | Gene | Log2 ratio | 90% conf. interval | Control log2 signal | SE | Suppressor log2 signal | SE | Gene product |
|---|---|---|---|---|---|---|---|---|
| b0355 | frmB | 4.17 | 0.69 | 6.85 | 0.46 | 11.02 | 0.22 | Putative esterase (EC 3.1.1.1) |
| b0357 | frmR | 3.27 | 0.73 | 9.08 | 0.49 | 12.35 | 0.22 | Conserved putative α-helix chain |
| b0356 | frmA | 2.72 | 0.32 | 9.85 | 0.11 | 12.57 | 0.21 | Glutathione-dependent formaldehyde dehydrogenase |
| b2397 | alaW | 2.53 | 2.96 | 10.08 | 1.02 | 12.62 | 1.93 | Alanine tRNA 2 (duplicate of alaX) |
| b2396 | alaX | 2.36 | 2.31 | 10.97 | 0.83 | 13.33 | 1.48 | Alanine tRNA 2 (duplicate of alaW) |
| b3750 | rbsC | 1.73 | 0.75 | 9.75 | 0.43 | 11.48 | 0.43 | d-Ribose high-affinity transport protein |
| b3534 | yhjQ | 1.68 | 0.63 | 5.96 | 0.30 | 7.64 | 0.42 | Conserved hypothetical protein |
| b3748 | rbsD | 1.42 | 0.44 | 11.77 | 0.14 | 13.19 | 0.30 | d-Ribose high-affinity transport system |
| b3749 | rbsA | 1.25 | 0.36 | 9.29 | 0.22 | 10.54 | 0.15 | d-Ribose high-affinity transport protein |
| b3672 | ivbL | 1.11 | 0.27 | 12.53 | 0.15 | 13.64 | 0.16 | ilvB operon leader peptide |
The stringent factor RelA was reported to be inactivated by amber suppression (3). We did not observe any change in the expression of the relA gene as a result of the suppressor, nor was there any effect related to the stringent response. This might be because the cells were not subjected to conditions that would provoke the stringent response. Presumably, amino acid biosynthesis was fully induced before the experiment began, by using minimal medium overnight cultures in which the suppressor was not induced. It seems likely that that RelA activity was irrelevant to the conditions tested, since relA is not required for growth in minimal medium (21) and steady-state levels of ppGpp are mostly determined by the activity of SpoT rather than RelA (27).
Under these conditions, few genes were affected by the suppressor. When the same criterion was used to compare data from the arabinose-grown control sample to WT cells grown in glucose, some 122 genes were significantly different (data not shown). One reason the suppressor affects so few genes may be the presence of transcriptional terminators or additional stop codons immediately downstream. Of the 326 ORFs that end with amber, 23% have another stop codon within 5 amino acids downstream, 59% have one within 20 amino acids, and 91% have another stop codon within 100 amino acids. The context in which a stop codon occurs is known to greatly affect the efficiency of protein termination (19). It is possible that context effects make extension beyond the amber stop codon very inefficient despite the suppressor. Another explanation may be that genes that end with amber have little impact on gene expression, at least under the conditions that we used. For example, microarrays show that only 31% (97 of 314) of amber-terminated ORFs are expressed at detectable levels, in contrast to 44% (1,889 of 4,337) for the whole genome (called “present” by Affymetrix software in five replicates in glucose minimal medium). Uncharacterized genes are overrepresented among amber ORFs as well, with 41% (130 of 314) having no known function, compared to 31% (1,344 of 4,337) for the whole genome.
Those few genes shown here to be upregulated by the suppressor are exceptions to this trend. Each case can be related to the presence of amber stop codons in the genes themselves or their regulators. The gene ivbL is the leader peptide for the isoleucine-valine biosynthesis operon and ends with an amber stop codon. No in-frame stop codons occur in the 105 nt downstream, so suppression presumably leads to fusion of IvbL to the downstream IlvB protein. It is important to note that suppression leads to an increase in abundance of the leader itself rather than the genes downstream (ilvB log2 ratio is only 0.87 ± 0.78), meaning that what we see is due to extension of the leader peptide affecting its own transcription or RNA half-life rather than an antitermination mechanism.
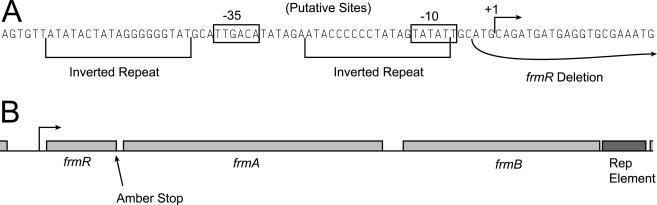
The gene yhjQ is the second in a predicted six-gene operon (2). It does not end with an amber stop codon, but yhjR immediately upstream does. Suppression should lead to a 26-amino-acid extension of YhjR. Also upregulated, the genes frmR, frmA, and frmB are part of a predicted three-gene operon (Fig. 2), and frmR ends with an amber stop codon. Suppression should lead to a 7-amino-acid extension of FrmR. The genes rbsD, rbsA, and rbsC are part of the rbsDACBK operon and do not end with amber, but the negative repressor of the operon rbsR does (17). Suppression should result in a 6-amino-acid extension or RbsR. Since suppression leads to upregulation of rbsDAC, it seems likely that the extension of RbsR decreases its ability to repress. We hypothesize that the inactivation of a repressor by amber suppression causes derepression of yhjQ and frmRAB.
FIG. 2.
Sequence and gene structure near frmR. (A) The sequence upstream of frmR reveals a potential promoter and an inverted repeat. The extent of the deletion in strain FBSC222 is shown corresponding to the ORF start codon as originally annotated. (B) Gene structure showing the putative operon and the location of the amber stop codon, drawn to scale. A downstream Rep element is of unknown significance.
To test this hypothesis, we measured the expression of frmA and frmB in strains overexpressing frmR or an extended variant. First, frmR was deleted from the chromosome by gene gorging (13). We deleted the annotated ORF (1), but four other start codons occur downstream, making the true protein start site uncertain. A putative promoter, with only one mismatch from consensus, is shown in Fig. 2. Another possible promoter with three mismatches occurs 5 nt upstream. The transcript of the latter promoter should include the annotated ORF but does not leave room for a ribosome binding site. If the promoter shown in Fig. 2 is correct, a possible ribosome binding site occurs 9 nt upstream of the fourth start codon, but then our deletion eliminates 3 nt upstream of the transcription start site. This may not affect regulation of the promoter though—a perfect 19-bp inverted repeat, possibly representing a repressor binding site or structural feature, occurs upstream in the promoter region.
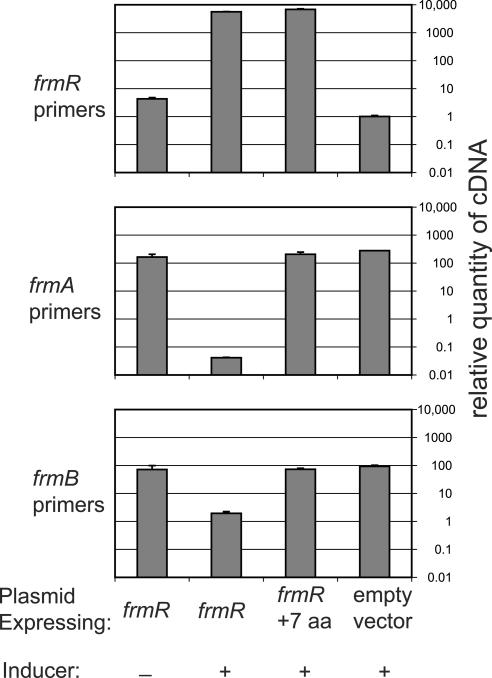
Two different versions of frmR were cloned under control of the arabinose promoter. The first was the complete annotated ORF. The second was the extended version as would result from use of the Ala2 suppressor. Alanine was substituted for the amber stop codon, and the next 6 amino acids were added to the end before an opal stop codon. These two plasmids and an empty vector control were transformed into the frmR deletion strain FBSC222. They were grown in minimal medium in either arabinose or glucose, and expression of the three genes in question was measured with real-time reverse transcription-PCR (Fig. 3). High levels of the plasmid-expressed frmR transcript were detected in both overexpressing strains, but not in uninduced and empty vector control strains. A small amount of frmR was detected in the empty vector control but was due to SYBR Green fluorescence from nonspecific PCR amplification, as observed by gel electrophoresis. Levels of the chromosomal frmA transcript were below the level of the negative control when frmR was induced and were 165 times higher than the control when frmR was repressed. The level of frmA transcript was 207 times higher than the control when the extended version was induced. A similar pattern of expression was observed for frmB. These results support our hypotheses that (i) frmR is a negative regulator of the operon and (ii) read-through of the amber stop codon and extension of the protein inactivate it. We did not test whether FrmR directly binds to the putative operator region.
FIG. 3.
Effects of frmR expression on levels of frmAB, measured with real-time PCR. Glucose overnight cultures of FBSC222 carrying the plasmids pACB/frmR, pACB/alt (denoted frmR + 7aa), or pACB/empty were inoculated into MOPS minimal medium with either arabinose (induced) or glucose (uninduced), and cDNA was isolated. SYBR Green real-time PCR was performed, and data from each sample were normalized to the housekeeping gene frr. The quantity of each transcript is expressed relative to the negative control (the empty vector strain amplified with frmR primers). Each panel shows the results from one primer pair. Error bars representing the standard deviations of three replicates were too small to be visible for some samples.
Induction of frmAB was apparently greater in the real-time PCR experiments than in the microarray experiments (38- to 207-fold versus 7- to 18-fold induction, respectively). This is probably due to the incomplete nature of suppression, indicated by suppressor efficiency. In the microarray experiments, the suppressor-extended version of FrmR may be only ∼59% of the total FrmR present in the cell, whereas 100% of FrmR was extended in the real-time PCR experiments. The effects of suppression may simply be due to a decrease in the levels of functional FrmR, or they might result from competition between the extended and nonextended versions of FrmR.
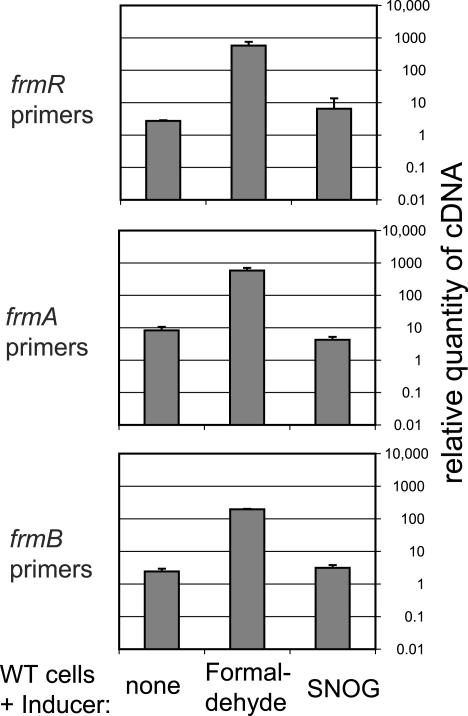
The gene frmR (previously called yaiN) is currently annotated as a putative α-helix protein, and frmB (previously yaiM) is currently annotated as a putative esterase. The gene frmA (previously adhC) encodes glutathione-dependent formaldehyde dehydrogenase, whose physiological substrate in E. coli may be either S-hydroxymethylglutathione (Km = 94 μM) (8) or S-nitrosoglutathione (SNOG; Km = 30 μM) (16). To identify the inducer of frmRAB, WT cells were subjected to either formaldehyde or SNOG (Fig. 4). The level of frmR transcript was induced by formaldehyde 215-fold over the level of uninduced cells. The operon was induced by formaldehyde but not by SNOG, in agreement with other studies (9, 20). Formaldehyde oxidation takes place in three steps: (i) it is spontaneously converted to S-hydroxymethylglutathione, (ii) then FrmA catalyzes conversion into S-formylglutathione, and then (iii) a hydrolase catalyzes conversion into formate. FrmB is 48% identical in protein sequence to the S-formylglutathione hydrolase of Paracoccus denitrificans (11). It is likely that the frmRAB operon encodes a complete pathway for degradation of formaldehyde, probably produced endogenously as a by-product of demethylation reactions. The names of the genes have been changed to reflect this probable function. frmA is conserved from bacteria to mammals, whereas frmR is conserved only among the proteobacteria. The two occur adjacent to each other in the enteric bacteria E. coli, Proteus vulgaris, Serratia marcescens, and Salmonella enterica, as well as in Xanthomonas spp. and Brucella suis.
FIG. 4.
Induction of frmRAB with formaldehyde, as measured with real-time PCR. WT strain MG1655 was grown to an OD600 of 0.15 in 0.01% glucose plus 0.1% arabinose and then split into three cultures containing no inducer, 0.25 mM formaldehyde, or 0.5 mM SNOG, and cells were harvested to make cDNA 30 min later. Relative real-time PCR and normalization were performed as for Fig. 3. The quantity is given relative to the negative control from Fig. 3, and so the data from both figures are comparable. Standard curves were generated from the same DNA.
Phenotypic analysis using BIOLOG plates was performed on a strain with a Tn5 insertion in frmB (14) (data available at www.genome.wisc.edu/functional/phenotypearray.htm for strain FB23041). Endpoint optical density was compared between plates inoculated with WT and the mutant. The largest differences in growth (those substrates with log2 ratios of <−2.0) are shown in Table 3. No log2 ratios of >+1.6 were observed, indicating that the mutation did not result in significant gain of function. No connection can be made between the observed phenotypes and formaldehyde degradation, though some of these data are questionable or have large standard deviations in WT. For instance, α-methyl-d-glucoside is known to be a nonmetabolizable glucose analog in E. coli. Regardless, some of these phenotypes may provide starting points for further investigation of the role of formaldehyde degradation in E. coli metabolism.
TABLE 3.
Phenotypic analysis
| Substrate | Plate | Testa | Well | Log2 ratio | WT ODb | FB23401 OD |
|---|---|---|---|---|---|---|
| Tween 80 | PM5 | Supplement | H12 | −4.8 | 1.0 ± 0.5 | 0.0 |
| 2′-Deoxycytidine | PM5 | Supplement | D12 | −4.7 | 1.2 ± 0.2 | 0.0 |
| α-Methyl-d-glucoside | PM2 | C source | C6 | −3.6 | 1.0 ± 0.7 | 0.1 |
| 2′-Deoxylnosine | PM5 | Supplement | C12 | −3.6 | 1.1 ± 0.4 | 0.1 |
| d-Lysine | PM3 | N source | C7 | −3.0 | 0.9 ± 0.4 | 0.1 |
| m-Inositol | PM5 | Supplement | G12 | −2.6 | 1.0 ± 0.3 | 0.2 |
| Thiophosphate | PM4 | P source | B1 | −2.4 | 1.5 ± 0.9 | 0.3 |
| N-Phthaloyl-l-glutamate | PM3 | N source | D2 | −2.2 | 0.5 ± 0.1 | 0.1 |
| Agmatine | PM3 | N source | D12 | −2.2 | 0.7 ± 0.6 | 0.1 |
| α-Keto valeric acid | PM2 | C source | E10 | −2.0 | 0.5 ± 0.3 | 0.1 |
Different BIOLOG plates were designed to test for carbon, nitrogen, or phosphorous sources or the effects of a growth supplement.
Cells were inoculated into BIOLOG plates as per the manufacturer's instructions, grown overnight at 37°C, and then the OD595 was measured in a plate reader. Data are shown from five replicates of WT (± SD) and one of FB23401 (disrupted at frmB).
Amber suppression is a powerful tool for the genetic analysis of gene function. A single mutant can be investigated with and without suppression, eliminating the complications associated with the comparison of different strains. This approach would be hindered, though, if the amber suppressor had a large number of effects on genes other than the gene of interest. In that case, it would be difficult to rule out the possibility that an observed phenotype was due to the loss of the suppressor rather than loss of gene function. In this study, the suppressor did not appear to elicit a general stress response, and the few transcriptional effects observed appeared to be inconsequential to cell function. It therefore seems feasible to study gene function by using expression analysis and amber suppression. The transcriptional changes observed here can be disregarded in such investigations as side effects of the suppressor. In future studies it may be possible to remove the problematic stop codons identified here to either ochre or opal, thereby generating an even cleaner experimental system.
Acknowledgments
We thank Guy Plunkett III for help with pseudogenes, Kai Zhao for assistance with real-time PCR, Mingzhu Liu, Yu Qiu, and Tim Durfee for assistance with hybridization of microarrays, and Tim Donohue for useful discussion.
This work was supported by NIH GM35682.
Frederick R. Blattner has financial interest in NimbleGen Systems, Inc., DNASTAR, Inc., and Scarab Genomics, Inc.
REFERENCES
- 1.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 2.Bockhorst, J., Y. Qiu, J. Glasner, M. Liu, F. Blattner, and M. Craven. 2003. Predicting bacterial transcription units using sequence and expression data. Bioinformatics 19(Suppl. 1):I34-I43. [DOI] [PubMed] [Google Scholar]
- 3.Breeden, L., and M. Yarus. 1982. Amber suppression relaxes stringent control by elongating stringent factor. Mol. Gen. Genet. 187:254-264. [Google Scholar]
- 4.Capecchi, M. R., and G. N. Gussin. 1965. Suppression in vitro: identification of a serine-sRNA as a “nonsense” suppressor. Science 149:417-422. [DOI] [PubMed] [Google Scholar]
- 5.Crary, S. M., S. Niranjanakumari, and C. A. Fierke. 1998. The protein component of Bacillus subtilis ribonuclease P increases catalytic efficiency by enhancing interactions with the 5′ leader sequence of pre-tRNAAsp. Biochemistry 37:9409-9416. [DOI] [PubMed] [Google Scholar]
- 6.Eggertsson, G., and D. Söll. 1988. Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol. Rev. 52:354-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelhardt, D. L., R. E. Webster, R. C. Wilhelm, and N. Zinder. 1965. In vitro studies on the mechanism of suppression of a nonsense mutation. Proc. Natl. Acad. Sci. USA 54:1791-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutheil, W. G., B. Holmquist, and B. L. Vallee. 1992. Purification, characterization, and partial sequence of the glutathione-dependent formaldehyde dehydrogenase from Escherichia coli: a class III alcohol dehydrogenase. Biochemistry 31:475-481. [DOI] [PubMed] [Google Scholar]
- 9.Gutheil, W. G., E. Kasimoglu, and P. C. Nicholson. 1997. Induction of glutathione-dependent formaldehyde dehydrogenase activity in Escherichia coli and Haemophilus influenzae. Biochem. Biophys. Res. Commun. 238:693-696. [DOI] [PubMed] [Google Scholar]
- 10.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harms, N., J. Ras, W. N. Reijnders, R. J. van Spanning, and A. H. Stouthamer. 1996. S-Formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway for formaldehyde detoxification? J. Bacteriol. 178:6296-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herring, C. D., and F. R. Blattner. 2004. Conditional lethal amber mutations in essential Escherichia coli genes. J. Bacteriol. 186:2673-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herring, C. D., J. D. Glasner, and F. R. Blattner. 2003. Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene 311:153-163. [DOI] [PubMed] [Google Scholar]
- 14.Kang, Y., T. Durfee, J. D. Glasner, Y. Qiu, D. Frisch, K. M. Winterberg, and F. R. Blattner. 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186:4921-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleina, L. G., and J. H. Miller. 1990. Genetic studies of the lac repressor. XIII. Extensive amino acid replacements generated by the use of natural and synthetic nonsense suppressors. J. Mol. Biol. 212:295-318. [DOI] [PubMed] [Google Scholar]
- 16.Liu, L., A. Hausladen, M. Zeng, L. Que, J. Heitman, and J. S. Stamler. 2001. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410:490-494. [DOI] [PubMed] [Google Scholar]
- 17.Lopilato, J. E., J. L. Garwin, S. D. Emr, T. J. Silhavy, and J. R. Beckwith. 1984. d-Ribose metabolism in Escherichia coli K-12: genetics, regulation, and transport. J. Bacteriol. 158:665-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall, B., and S. B. Levy. 1980. Prevalence of amber suppressor-containing coliforms in the natural environment. Nature 286:524-525. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. H., and A. M. Albertini. 1983. Effects of surrounding sequence on the suppression of nonsense codons. J. Mol. Biol. 164:59-71. [DOI] [PubMed] [Google Scholar]
- 20.Mukhopadhyay, P., M. Zheng, L. A. Bedzyk, R. A. LaRossa, and G. Storz. 2004. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl. Acad. Sci. USA 101:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neidhardt, F. C. 1963. Properties of a bacterial mutant lacking amino acid control of RNA synthesis. Biochim. Biophys. Acta 68:365-379. [DOI] [PubMed] [Google Scholar]
- 22.Normanly, J., L. G. Kleina, J. M. Masson, J. Abelson, and J. H. Miller. 1990. Construction of Escherichia coli amber suppressor tRNA genes. III. Determination of tRNA specificity. J. Mol. Biol. 213:719-726. [DOI] [PubMed] [Google Scholar]
- 23.Ohlsson, B. M., P. F. Strigini, and J. R. Beckwith. 1968. Allelic amber and ochre suppressors. J. Mol. Biol. 36:209-218. [DOI] [PubMed] [Google Scholar]
- 24.Person, S., and M. Osborn. 1968. The conversion of amber suppressors to ochre suppressors. Proc. Natl. Acad. Sci. USA 60:1030-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenow, C., R. M. Saxena, M. Durst, and T. R. Gingeras. 2001. Prokaryotic RNA preparation methods useful for high density array analysis: comparison of two approaches. Nucleic Acids Res. 29:E112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudd, K. E. 2000. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 28:60-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryals, J., R. Little, and H. Bremer. 1982. Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J. Bacteriol. 151:1261-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schultz, D. W., and M. Yarus. 1990. A simple and sensitive in vivo luciferase assay for tRNA-mediated nonsense suppression. J. Bacteriol. 172:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steege, D. A., and D. G. Söll. 1979. Suppression, p. 433-485. In R. F. Goldberger (ed.), Biological regulation and development, vol. 1. Plenum Press, New York, N.Y. [Google Scholar]
- 30.Wang, L., A. Brock, B. Herberich, and P. G. Schultz. 2001. Expanding the genetic code of Escherichia coli. Science 292:498-500. [DOI] [PubMed] [Google Scholar]