Abstract
Coupling proteins (CPs) are present in type IV secretion systems of plant, animal, and human pathogens and are essential for DNA transfer in bacterial conjugation systems. CPs connect the DNA-processing machinery to the mating pair-forming transfer apparatus. In this report we present in vitro and in vivo data that demonstrate specific binding of CP TraD of the IncFII R1 plasmid transfer system to relaxosomal protein TraM. With overlay assays and enzyme-linked immunosorbent assays we showed that a truncated version of TraD, termed TraD11 (ΔN155), interacted strongly with TraM. The apparent TraD11-TraM association constant was determined to be 2.6 × 107 liters/mol. Electrophoretic mobility shift assays showed that this variant of TraD also strongly bound to TraM when it was in complex with its target DNA. When 38 amino acids were additionally removed from the C terminus of TraD, no binding to TraM was observed. TraD15, comprising the 38 amino-acid-long C terminus of TraD, bound to TraM, indicating that the main TraM interaction domain resides in these 38 amino acids of TraD. TraD15 exerted a dominant negative effect on DNA transfer but not on phage infection by pilus-specific phage R17, indicating that TraM-TraD interaction is important for conjugative DNA transfer but not for phage infection. We also observed that TraD encoded by the closely related F factor bound to TraM encoded by the R1 plasmid. Our results thus provide evidence that substrate selection within the IncF plasmid group is based on TraM's capability to select the correct DNA molecule for transport and not on substrate selection by the CP.
In bacterial conjugation DNA is transferred unidirectionally from donor to recipient cells across two bacterial cell envelopes (15, 19, 22, 45). Conjugative DNA transfer is the main route for horizontal spread of antibiotic resistance genes (31, 44); furthermore, in Escherichia coli, biofilm formation is dependent on the presence of conjugative plasmids (16, 35). The complete set of proteins needed for DNA transfer is encoded by a conjugative plasmid. DNA transfer is mediated by a multiprotein complex that is functionally divided into two main parts: the mating pair formation (Mpf) complex for pilus assembly and disassembly, including functions required for cell-cell contact formation and maintenance between donor and recipient, and the DNA-processing (Dtr) proteins (33). Type IV secretion systems (T4SS) of pathogens such as Helicobacter pylori, Legionella pneumophila, Bordetella pertussis, or Agrobacterium tumefaciens that transport effector proteins or both DNA and proteins to eukaryotic cells are related to the Mpf systems of conjugation systems (7, 23, 25). Mpf and Dtr systems are tied together by an integral inner membrane linker protein that has been called “coupling protein” (CP) (4). CPs are essential for conjugative DNA transfer (TraD of F-like plasmids, TraG of RP4, and TrwB of R388), the VirD4 CP encoded by the Ti plasmid of A. tumefaciens is essential for T-DNA transport to plant cells (26) and has been shown to interact with the transported DNA in an early step of transport (6). CPs are actively involved in effector protein translocation mediated by protein secretion systems (8, 10, 14, 40).
The three-dimensional structure of the CP of IncW plasmid R388, TrwB, has been solved (18). On the basis of (i) the structure of a monomeric soluble protein fragment lacking the N-terminal membrane-spanning domain and (ii) the biochemical properties of the full-length protein, it has been proposed that a TrwB hexamer forms a membrane-spanning pore (27) resembling the F1 ATPase structure (18).
TraG, TraD, and TrwB bind DNA nonspecifically (30, 38), with single-stranded DNA being the preferred substrate (39), a finding that is in line with the proposed role of single-stranded DNA transport through the inner membrane. CPs can bind deoxynucleoside triphosphates; a characteristic nucleotide binding motif is present in the amino acid sequence (38). These nucleoside triphosphate binding sites have been shown to be essential for DNA transfer (2) and nucleotide binding activity (30); however, no nucleoside triphosphatase activity of purified CPs could be demonstrated (30, 38).
How do CPs interact with the transporter (the T4SS) and relaxosome parts of the conjugation machinery? An interaction between a CP and a protein with a periplasmic domain belonging to the T4SS machinery was shown recently. TraG, the CP encoded by IncH plasmid R27, was shown to interact with TrhB, which is a, VirB10-like T4SS protein anchored in the inner membrane featuring a large carboxyl domain in the periplasm (17). A similar interaction has been demonstrated in the R388 plasmid system between CP TrwB and TrwE, the homologue of TrhB in R388 (28). On the cytoplasmic side, CPs interact with relaxosomal components. In vitro experiments revealed that TraG of IncP plasmid RP4 binds TraI (38), which is the relaxase of this conjugative system. TrwB, the R388 CP, interacts with TrwA, a relaxosomal protein (28); in the F-plasmid system, interaction of F-factor CP TraD with relaxosomal protein TraM has been shown in vitro (11).
In this communication we focus on characterization of the interaction between the TraD protein and TraM encoded by IncFII plasmid R1. Since the R1 plasmid-encoded TraM protein has been extensively investigated in this laboratory (21, 34, 41-43), we wished to determine whether TraM and TraD interacts in the R1 plasmid system and whether this interaction is plasmid specific. By using affinity-tagged variants of TraD and TraM of plasmid R1 and purifying these proteins, we were able to perform in vitro experiments that clearly demonstrated that TraD binds to TraM in the IncFII R1 system. This interaction also occurred between TraDF and TraMR1. Furthermore, we could locate the main TraM binding domain on TraD and showed that it comprises only the C-terminal 38 amino acids. On the basis of our findings we propose a model for DNA substrate selection and DNA transfer in IncF plasmids.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phage.
The bacteria, plasmids, and phage used in this study are listed in Table 1. E. coli cells were grown in 2×TY medium (16 g of tryptone per liter, 10 g of yeast extract per liter, 5 g of NaCl per liter) supplemented with antibiotics when needed to the following final concentrations: ampicillin, 100 mg liter−1; kanamycin, 40 mg liter−1; chloramphenicol, 10 mg liter−1. The R17 bacteriophage was propagated on E. coli MC1061.
TABLE 1.
Strains, plasmids, and phage used in this study
| Strains, plasmid, or phage | Description | Reference or source |
|---|---|---|
| Strains | ||
| J5 | pro met λ+ | IMBMa collection |
| MC1061 | araD139 Δ(ara leu)7697 ΔlacX74 galE15 galK16 hsdR2 (rK− mK+) rpsL (Strr) mcrA mcrB1 | IMBM collection; 5 |
| BL21(DE3) | E. coli B ompT dcm hsdS gal λ− (DE3) | Stratagene |
| SCS1 | endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 | Stratagene |
| UG10 | lon::Tn10 hsdR (rK− mK+) tre thi lac rpsL (Strr) | IMBM collection |
| XL1-Blue | endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 Δ(lac) [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Plasmids | ||
| R1-16 | Derepressed, deletion-containing variant of IncFII plasmid R1 Kmr | IMBM collection |
| pCALn | ColE1 ori PT7/TT7 Amprcbp tag | Stratagene |
| pMTD11 | pCALn Δ(BamHI-HindIII) Ω(traD11) Ampr | This study |
| pMTD21 | pCALn Δ(BamHI-HindIII) Ω(traD21) Ampr | This study |
| pBTD15 | pCALn Δ(BamHI-HindIII) Ω(traD15) Ampr | This study |
| pGZ119EH | ColD replicon; Ptaq/lacIq Cmr | 24 |
| pGTD15 | pGZ119EH Δ(XbaI-HindIII) Ω(cbp-traD15) Cmr | This study |
| pMALc | pBR322ori M13ori Ptaq/Trm/lacIq Amprmbp tag | New England Biolabs |
| pMALcTraMwt | pMALc Δ(XmnI-HindIII) Ω(traM amino acids 2-127) Ampr | This study |
| pSK410ch | pMS470Δ8 Δ(NdeI-HindIII) Ω(traD-His6) Ampr | 38 |
| pSK410nh | pMS470Δ8 Δ(NdeI-HindIII) (his6-traD) Ampr | 38 |
| pBR111 | Tcr pBR322 traM R1 | 20 |
| pGK111 | Ampr pUC119 traM R1 | 20 |
| pSU2007 | Kmr derivative of IncW conjugative plasmid R388; 2.5-kb EcoRI-SalI fragment exchanged for 1.5-kb EcoRI-SalI fragment carrying Kmr-encoding gene of Tn5 | 29 |
| Phage R17 | Male-specific RNA phage | IMBM collection |
IMBM, Institut für Molekular Biologie, Biochemie und Mikrobiologie.
DNA and protein sequence analyses.
The Wisconsin Package, version 10.3 (Accelrys Inc., San Diego, Calif.), was used for analysis and in silico manipulations of DNA and protein sequences. The GenBank accession number of the traD nucleotide sequence of plasmid R1 is AY684127.
DNA techniques.
Standard molecular cloning techniques were performed in accordance with the procedures described by Sambrook et al. (36) or in accordance with the manufacturers' protocols. PCR fragments were generated with Expand high-fidelity DNA polymerase (Roche Molecular Biochemicals); all primers were purchased from MWG Biotech. The nucleotide sequences of all of the plasmid DNA constructs were verified by semiautomated DNA sequencing with an ALF-Express DNA sequencer (Amersham Biosciences). The pMTD clones were generated by using R1-16 as the template for amplification of traD sequences, the PCR fragments were subsequently cloned into pCALn vectors (Stratagene) for expression of the calmodulin binding peptide (CBP)-TraD fusion proteins. For construction of pMTD11, we used CAAAggatccCCGAAAGACGTTGCCCGGA as the forward primer (nucleotides corresponding to the traDR1 sequence are underlined, and the BamHI restriction site is in lowercase) and GTAAaagcttTCATCATCAGAAATCATCTCCC as the reverse primer (nucleotides corresponding to the traDR1 sequence are underlined, and the HindIII restriction site is in lowercase). For construction of pMTD21, the primers used for PCR amplification were CAAAggatccCCGAAAGACGTTGCCCGGA as the forward primer (nucleotides corresponding to the traDR1 sequence are underlined, and the BamHI restriction site is in lowercase) and GTAAaagcttTCATCACTCATAAGCGGCCATATCC (nucleotides corresponding to the traDR1 sequence are underlined, and the HindIII restriction site is in lowercase) as the reverse primer. pBTD15 was obtained with pMTD11 as the template, ATATATggatccGCATGGCAACAGG as the forward primer (nucleotides corresponding to the traDR1 sequence are underlined, and the BamHI restriction site is in lowercase), and GTAAaagcttTCATCATCAGAAATCATCTCCC as the reverse primer (nucleotides corresponding to the traDR1 sequence are underlined, and the HindIII restriction site is in lowercase). For maltose binding protein (MBP) fusion of traM, a PCR fragment comprising the complete traM sequence of plasmid R1 was ligated into a pMALc vector (New England Biolabs) via EcoRI and HindIII restriction sites. The template used was pBR111 containing full-length traM, and amplification was done with GTgaattcAAAACACAGTCAACAGTTGC as the forward primer (nucleotides corresponding to the traM sequence are underlined, the EcoRI restriction site is in lowercase) and GCTTaagcttTTATTATTCCTCATCATTTTCTGGAAAG as the reverse primer (nucleotides corresponding to the traM sequence are underlined, and the HindIII restriction site is in lowercase). Cloning of wild-type traM was performed as described by Verdino et al. (43). The sbmA target DNA for the electrophoretic mobility shift assay (EMSA) experiments was obtained by PCR amplification with plasmid pGK111 as the template, resulting in a 280-bp-long DNA fragment containing three TraM binding sites (forward primer, CAGGCAGATGGCTAACATCC; reverse primer, CTGATTCATCTATGAGT). With this fragment as the template, Cy5-labeled primers were used to generate the 63-bp sbmA target DNA (forward primer, GGATTCATTGGTGAAT; reverse primer, CTGATTCATCTATGAGT).
Protein purification.
Purification of His-TraDF and TraDF-His was performed as follows. E. coli SCS1 cells harboring either plasmid pSK410ch or pSK410nh were grown at 30°C to an optical density at 600 nm (OD600) of 0.5, and then the cultures were induced with 1 mM (final concentration) IPTG (isopropyl-β-d-thiogalactopyranoside). After 5 h of induction, the cells were harvested by centrifugation at 4,200 × g for 15 min at 4°C. The cell pellet was resuspended in 5 ml of spermidine mix (20 mM spermidine, 200 mM NaCl, 2 mM EDTA) per g of cell pellet and shock frozen in liquid nitrogen. The cells were lysed for 1 h at 4°C in lysis buffer (40 mM Tris-HCl [pH 7.6], 10% [wt/vol] sucrose, 50 mM NaCl, 0.25% Brij 58, 0.7 mg of lysozyme per ml) and centrifuged at 4,200 × g for 15 min at 4°C. The pellet was incubated overnight at 0°C in Triton mix (50 mM 2-[N-cyclohexylamino]ethane sulfonic acid [CHES] [pH 9.5], 1 M NaCl, 5 mM MgCl2, 1% Triton X-100) and centrifuged at 12,000 × g for 15 min at 4°C. After dialysis of the protein extract against 4 liters of buffer A (20 mM Tris-HCl [pH 8.0], 150 mM NaCl) at 4°C three times, the protein extract was centrifuged at 48,000 × g for 30 min at 4°C. The supernatant was loaded on a pre-equilibrated (with buffer A) HighTrap chelating 1-ml column (Amersham Biosciences). The fractions were eluted with an imidazole gradient ranging from 50 to 500 mM. After dialysis at 4°C against 4 liters of sodium phosphate buffer (50 mM Na2PO4 [pH 7.5], 20 mM NaCl) for 2 h, the purified proteins were stored in 20% (vol/vol) glycerol at −20°C.
Purification of MBP-TraMR1.
E. coli UG10 cells harboring recombinant plasmid pMALcTraMwt for cytoplasmic expression of an MBP-TraM fusion protein were cultivated at 30°C until the OD600 reached 0.4 to 0.7. Overexpression of fusion proteins was induced by addition of 0.3 mM (final concentration) IPTG. After 1.5 h, cells were harvested by centrifugation at 4,200 × g for 15 min at 4°C, resuspended in 2 ml of chromatography buffer (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA), and disrupted with a French pressure cell. Crude extracts were collected by low-speed centrifugation at 500 × g, diluted to 10 mg of total protein per ml, and applied to an amylose resin column with a 2-ml bed volume. The column was washed first with 20 ml of chromatography buffer and subsequently with 10 ml of chromatography buffer supplemented with complete protease inhibitor (Roche Molecular Biochemicals). Fusions proteins were eluted with 4 ml of elution buffer (chromatography buffer supplemented with 10 mM maltose). The concentration of the proteins in the obtained fractions was determined by Bio-Rad assay (Bio-Rad Laboratories).
Purification of CBP fusion proteins (TraD11, TraD21, and TraD15).
E. coli BL21(DE3) cells containing the appropriate plasmid (Table 1) were grown at 37°C to an OD600 of 0.5. Overexpression of the fusion proteins was started by supplementation with 1 mM (final concentration) IPTG. After 2 h of induction, cells were harvested by centrifugation at 4,200 × g for 15 min at 4°C and resuspended in 3 ml of CaCl2 binding buffer (50 mM Tris-HCl [pH 8.0], 10 mM NaCl, 10 mM β-mercaptoethanol, 1 mM magnesium acetate) containing the protease inhibitors pepstatin (1 μg/ml [final concentration]), PMSF (0.1 mM [final concentration]), and apoprotinin A (1 μg/ml [final concentration]). The cells were broken with a French pressure cell. After the disruption step, the broken cells were centrifuged at 27,000 × g for 30 min at 4°C and the crude extracts were applied to a calmodulin affinity resin (Stratagene) column with a 3-ml volume. The column was washed with 12 ml of CaCl2 binding buffer containing protease inhibitors as described above. The fusion proteins were eluted with 3 ml of elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM β-mercaptoethanol, 2 mM EGTA, 10 mM NaCl). The concentrations of the proteins in the eluted fractions were determined by Bio-Rad assay (Bio-Rad Laboratories). Purification of TraMR1 was performed as described previously (43).
Overlay assays.
Affinity-purified CBP fusion proteins (TraD11 and TraD21; 2 to 5 μg) were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and electrotransferred for 90 min onto Immobilon-P nitrocellulose membranes (Millipore) at 230 mA. Subsequently, the membrane was blocked with 2% milk powder in AC buffer (20 mM Tris-HCl [pH 7.6], 10% glycerol, 50 mM NaCl, 1 mM EDTA, 0.1% Tween 20) overnight. Purified TraM or MBP-TraM was used in the overlay at a concentration of 1 μg/ml diluted in AC buffer containing 2% milk powder and 1 mM dithiothreitol. After 90 min of overlay, the membrane was incubated with dilutions of anti-TraM antiserum (1:3,000 in AC buffer containing 2% milk powder) or anti-MBP antibody (1:10,000 in AC buffer containing 2% milk powder; New England Biosciences). Immunological detection was carried out with horseradish peroxidase (HRP)-coupled goat anti-rabbit immunoglobulin G (IgG; dilution of 1:15,000 in AC buffer containing 2% milk powder; Sigma-Aldrich) and photochemical detection with the ECL system (Amersham Biosciences). The membranes were washed with AC buffer after each step.
Coimmunoprecipitation.
Anti-His antibody (Clontech) was bound to protein A-Sepharose beads (Amersham Biosciences) by incubation in LB buffer (20 mM HEPES-KOH [pH 7.4], 1% Triton X-100, 10% glycerol, 250 mM NaCl) on ice for 2 h. The anti-His antibody-saturated beads were then incubated with 6.5 μg of TraM and 40 μg of His-TraD or TraD-His overnight in incubation buffer (50 mM sodium phosphate [pH 7.5], 20 mM NaCl). Proteins were eluted from the beads in 50 μl of 5× FSB buffer (200 mM Tris-HCl [pH 6.8], 15% SDS, 15% dithiothreitol, 30% glycerol, 0.1% bromophenol blue) by incubation at 37°C for 30 min, separated by SDS-PAGE, and blotted onto Immobilon-P (Millipore) nitrocellulose membrane. Immunological detection was performed as described above (overlay assays), with anti-TraM antiserum (dilution of 1:5,000) for TraM detection, anti-His IgG for TraD detection (dilution of 1:15,000 in TST buffer containing 2% milk powder), and HRP-coupled goat anti-rabbit IgG (dilution of 1:15,000 in TST buffer containing 2% milk powder). Photochemical detection was performed with the ECL system (Amersham Biosciences).
Enzyme-linked immunosorbent assay (ELISA).
Affinity-purified CBP fusion proteins were bound to the 96-well plate matrix by applying 1 μg of protein diluted in 100 μl of elution buffer (see protein purification) per well. After blocking of each well with 2% milk powder resuspended in AC buffer, MBP-TraM was added in different concentrations ranging from 10 to 5,000 ng of protein diluted in 100 μl of AC buffer each. Immunological detection was achieved with anti-MBP antibody (dilution of 1:15,000 in AC buffer containing 2% milk powder; New England Biosciences) and HRP-coupled goat anti-rabbit IgG (dilution of 1:20,000 in AC buffer containing 2% milk powder). The microplate wells were washed four times with 200 μl of AC buffer between incubation steps. The colorigenic reaction was started by addition of the substrate o-phenylenediamine (Bioclone). The A490 of the yellow complex formed after the reaction of HRP with o-phenylenediamine and H2O2 was measured with a 96-well microplate reader (model 550; Bio-Rad Laboratories). Association and dissociation constants and nonlinear regression binding curves were calculated from the data with the Prism3 program package (GraphPad Software). The algorithm used for curve fitting was y = βmax · x/(Kd + x).
EMSA.
Purified TraMR1 was incubated with Cy5-labeled target DNA containing sbmA in a mixture of BSB (12 mM HEPES-Tris, 4 mM Tris-HCl, 60 mM KCl, 10% glycerol, 1 mM EDTA) and TBE (100 mM Tris-HCl, 100 mM boric acid, 2.5 mM EDTA) for 15 min at 37°C. Affinity-purified CBP-TraD fusion proteins were added to give a final concentration of 740 nM. The samples were then incubated for an additional 15 min. Subsequently, the DNA-protein mixtures were separated on nondenaturing polyacrylamide gels. The DNA-protein complexes in the gel were visualized with a Typhoon (Amersham Biosciences) fluorescence scanner. The images were analyzed with ImageQuant 5.1 software (Amersham Biosciences).
Conjugation assays.
Overnight cultures of E. coli MC1061 donor strain cells harboring plasmids R1-16 and pGZ119EH or pGTD15 were grown in 2×TY medium supplemented with kanamycin and chloramphenicol. The overnight cultures were grown for an additional 2 h with 1 mM IPTG added for overexpression of TraD15. Forty microliters of the donor culture was diluted in 0.9 ml of fresh medium prewarmed to 37°C and incubated for 30 min without shaking. One hundred microliters of recipient E. coli J5 cells from an overnight culture was then added directly to the donor cells. Mating was allowed to proceed for 60 min at 37°C without shaking. DNA transfer was stopped by vigorous mixing for 1 min and rapid cooling on ice. Aliquots were diluted and plated on MacConkey lactose agar containing kanamycin. The conjugation frequency was determined by counting white donor and red transconjugant colonies and is expressed as the number of transconjugants per donor cell. For additional mating assays with the pSU2007 (R388 derivative, IncW) plasmid, overnight cultures of E. coli MC1061 donor strain cells harboring plasmids pSU2007 and pGZ119EH or pGTD15 were grown in 2×TY medium supplemented with kanamycin and chloramphenicol. The overnight cultures were grown for an additional 1 h with 1 mM IPTG added for overexpression of TraD15. One hundred microliters of the donor culture was mixed with 100 μl of recipient E. coli J5 cells in 0.8 ml of 2×TY. The cell suspension was filtered with 0.45-μm-pore-size filters. Mating on the filters was allowed to proceed for 60 min on 2×TY agar plates. The DNA transfer was stopped by vigorously shaking the filters in 0.9% NaCl, diluting aliquots, and plating the dilutions on MacConkey lactose agar containing kanamycin. The conjugation frequency was determined by counting white donor and red transconjugant colonies and is expressed as the number of transconjugants per donor cell.
Infection studies with RNA phage R17.
Male-specific R17 phages were propagated in R-top agar (10 g of tryptone per liter, 1 g of yeast extract per liter, 8 g of NaCl per liter, 8 g of agar-agar per liter) on the host strain, i.e., E. coli MC1061 with R1-16 and the same test plasmids as used for the conjugation assays (pGZ119EH and pGTD15), as described elsewhere (36). The overnight cultures were induced with 1 mM (final concentration) IPTG for 2 h before the phage infection assay was started. The top agar was supplemented with 1 mM IPTG and 10 mM CaCl2.
RESULTS
TraDR1 interacts in vitro with TraMR1 through the C terminus.
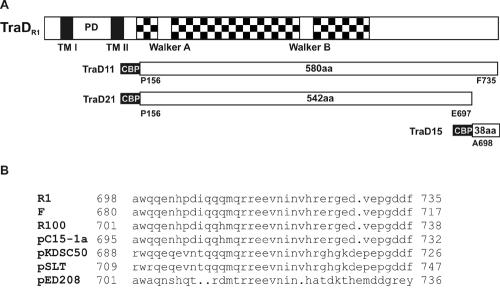
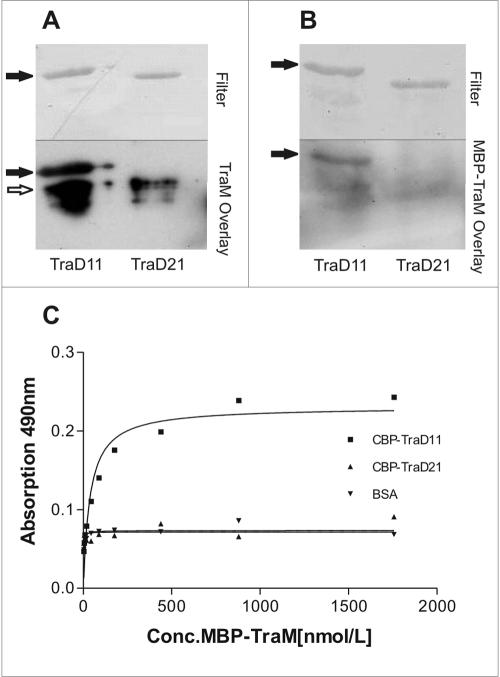
In vitro interaction between the CP TraD protein and the small DNA binding TraM protein has been described for the F-factor system (11). To investigate whether such an interaction is present in the R1 plasmid system and to determine the TraM binding domain in TraD, we constructed N-terminally truncated versions missing the transmembrane domains of TraDR1 designated TraD11 and TraD21 (Fig. 1A). Both TraD variants carry an N-terminal CBP fusion tag for purification. TraD11 contains the full C terminus, while TraD21 lacks the C-terminal 38 amino acids (Fig. 1). We carried out overlay assays in which the affinity-purified fusion proteins were first separated by SDS-PAGE and subsequently transferred to nitrocellulose. For detection of TraD-TraM interactions, purified TraM protein (Fig. 2A) or MBP-tagged TraMR1 (Fig. 2B) was added in the overlay. Bound TraM, indicating a specific interaction with TraD, was detected with anti-TraM (Fig. 2A) or anti-MBP (Fig. 2B) antiserum. As shown in Fig. 2A, there was a strong signal corresponding to the position of TraD11 (marked by filled arrows in Fig. 2) whereas no signal appeared at the position corresponding to TraD21, indicating that TraM could bind to TraD11 but not to TraD21. In this way, a possible nonspecific interaction between TraM and the CBP tag could be excluded. No signal corresponding to the position of TraD11 appeared in the control experiment without TraM as an overlay (data not shown). The signal below TraD11 and TraD21 (indicated by an open arrow) also appeared without TraM in the overlay and therefore is unspecific. To confirm these results we used an MBP-TraM fusion protein in the overlay (Fig. 2B). Again we were able to detect a strong interaction between TraD11 and MBP-TraMR1; in contrast, no interaction between TraD21 and TraMR1 could be detected (Fig. 2B). To confirm the results obtained in the overlay assays, we performed an ELISA with TraD11 or TraD21 bound to the surface of 96-well plates, incubating the wells with increasing concentrations of MBP-TraM. In Fig. 2C, binding curves derived from the quantified ELISA data are shown. A strong increase in the A490, which is indicative of MBP-TraM binding, could be seen with increasing MBP-TraM concentrations in the TraD11-coated wells only. In contrast, no increase in the absorption in TraD21-coated wells could be measured. The same was true for the bovine serum albumin (BSA)-coated wells that served as negative controls. These results confirmed data from the overlay assays and indicated that TraMR1 binds to TraDR1 and that the domain of TraDR1 binding to TraMR1 is at the very C-terminal end of the protein since a shortened version of the protein lacking the C-terminal 38 amino acids did not bind to TraMR1. The apparent association constant (kA) for TraD11 and MBP-TraM was calculated to be 2.6 × 107 liters/mol. To further characterize the TraD-TraM interaction, we performed experiments to locate the TraDR1 binding domain in TraMR1. For this purpose, two variants of MBP-TraMR1 comprising amino acids 2 to 75 and 75 to 127 were used. Neither of these variants of TraMR1 interacted with TraD11 in overlay assays or ELISAs (data not shown).
FIG. 1.
(A) CBP-TraD fusion proteins in comparison with TraDR1. In TraDR1 the checkered box represents the CP homology domain containing Walker A and B boxes, PD is the periplasmic domain, and TM I and TM II are the predicted transmembrane domains, respectively. The CBP tag is 31 amino acids long and is displayed in black with white lettering. The start and stop positions of the TraD parts of the fusion proteins are indicated. All items are drawn to scale. (B) Comparison of C-terminal sequences of TraD-like proteins. Sequences were pulled from the National Center for Biotechnology Information nonredundant protein database (posted 3 February 2004) with the NetBlast program by using 38 amino acids from the C terminus of TraDR1 as the query sequence. The sequences were aligned with the program Pileup. Note that the C termini of R1, F, and R100 are identical.
FIG. 2.
TraDR1 interacts with TraMR1 via its C terminus. Affinity-purified proteins TraD11 (69.3 kDa) and TraD21 (64.7 kDa) were separated by SDS-PAGE and transferred onto nitrocellulose. Overlays were performed with TraM (A) or MBP-TraM (B). Detection was performed with anti-TraM antiserum (A) or with anti-MBP IgG (B). The bound antibodies were visualized with the ECL system for photochemical detection. Filled arrows in the lower parts of panels A and B point to the position of TraD11, which interacted with both TraM and MBP-TraM. The open arrow indicates a nonspecific signal arising from the secondary antibody used in the experiment. In the upper part, the Coomassie-stained membrane is shown. (C) Binding curves derived from ELISA data. Purified CBP-TraD11 and CBP-TraD21 fusion proteins were bound to the surface of microplate wells and incubated with increasing concentrations (Conc.) of MBP-TraM. MBP-TraM bound to TraD11 but not to TraD21 in a concentration-dependent manner. BSA was used as a negative control. The algorithm used to calculate the nonlinear regression curve was y = βmax · x/(Kd + x). A curve calculated from one representative experiment is shown.
The C-terminal 38 amino acids of TraDR1 are sufficient to confer binding to TraMR1 in vitro.
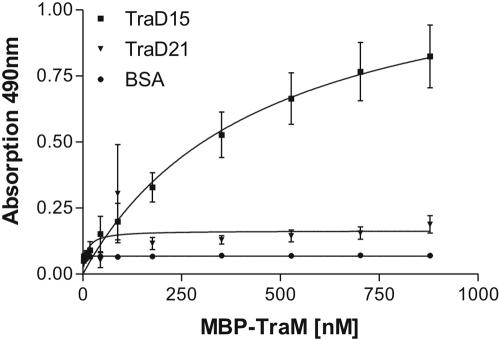
In the preceding experiments we showed that the very C-terminal part of TraDR1 is required for formation of the TraD-TraM complex. To further investigate if these 38 amino acids in TraDR1 are sufficient for binding TraMR1, we constructed a truncated version of the TraDR1 protein only retaining the 38 C-terminal amino acids with an N-terminal CBP fusion (TraD15; shown in Fig. 1A) and performed additional binding assays. In order to obtain kinetic data we carried out an ELISA in 96-well plates with TraD15 bound to the surface of the wells and probed with increasing concentrations of MBP-TraM. TraD21- and BSA-coated wells were used as negative controls, respectively. Figure 3 shows binding curves derived from the quantified ELISA data. An increase in absorption at 490 nm with increasing concentrations of MBP-TraM in the overlay on TraD15-coated wells was observed. With neither TraD21 nor BSA could such an increase in absorption be seen (Fig. 3). These results provided evidence that TraD15, comprising only 38 amino acids representing the very C terminus of TraDR1, is sufficient to mediate interaction between TraDR1 and TraMR1. From this data set the apparent association constant was calculated to be 2.2 × 106 liters/mol, which is approximately 10-fold lower than the kA determined for TraD11.
FIG. 3.
Binding curve showing the binding kinetics of the interaction of MBP-TraM with TraD15. Purified TraD15 and TraD21 fusion proteins were bound to the surface of microplate wells and incubated with increasing concentrations of MBP-TraM. BSA was used as a negative control. With increasing concentrations of MBP-TraM in the overlay, absorption increases with CBP-TraD15 but not with TraD21, indicating a specific interaction between TraM and the 38 C-terminal amino acids of TraD.
TraD binds to TraM-DNA complexes.
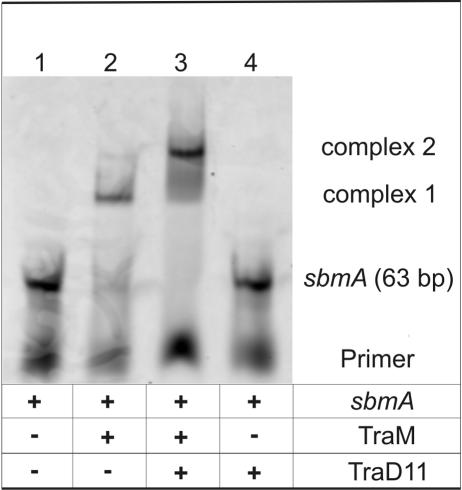
To test whether TraD can interact with TraM when TraM is bound to its target DNA, we performed EMSAs. As a DNA target sequence for TraM, a 63-bp DNA fragment comprising three binding sites (corresponding to sbmA of R1 oriT) for TraM tetramers was used. To detect interactions, TraD11 was added to the TraM-DNA complexes. As shown in Fig. 4, addition of TraD11 led to the formation of a higher-molecular-weight complex that appeared as a distinct band (lane 3). TraD11 by itself did not bind to sbmA double-stranded DNA, nor were any changes in the appearance of the probe DNA that was used in the experiment detected (Fig. 4, lanes 4). These results demonstrated that TraD bound to TraM when TraM was in a complex with its target DNA.
FIG. 4.
TraD11 interacts with TraM in a complex with its target DNA. Cy5-labeled sbmA target DNA (lane 1) was incubated with the indicated proteins. Fractions of the DNA-protein samples were separated by nondenaturing PAGE. The Cy5-labeled DNA-protein complexes in the gel were visualized with a fluorescence scanner. Free DNA is shifted to a higher-molecular-weight complex by TraM (lane 2, complex 1). The presence of TraD11 shifts this band to a slower-migrating complex (lane 3, complex 2). TraD11 alone does not interact with sbmA (lane 4). The image shown was generated by scanning the gel.
TraDF binds to TraMR1.
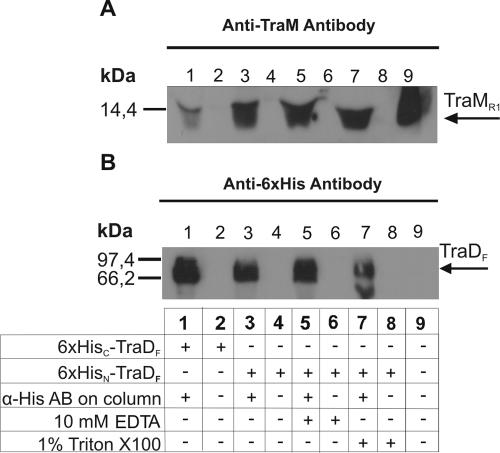
To determine whether TraD of the F plasmid is able to interact with TraM of the closely related R1 plasmid, we performed coimmunoprecipitation experiments. Affinity-purified fusion proteins of full-length TraDF, containing either a C-terminal six-His tag (TraDF-His) or an N-terminal six-His tag (His-TraDF) were incubated with TraMR1 on protein A-Sepharose beads with linked anti-His antibody. The proteins pulled down with the protein A-Sepharose beads were separated by SDS-PAGE, transferred onto nitrocellulose filters, and detected with anti-His antibody (TraDF fusions) and anti-TraM antiserum (TraMR1). As shown in Fig. 5A, we were able to detect an interaction between His-TraDF and TraMR1 (lanes 3, 5, and 7). In that case a strong TraM band could be seen when the blot was probed with anti-TraM antiserum. The presence of His-TraDF in lanes 3, 5, and 7 was verified with an anti-His antibody (Fig. 5B). Significantly less TraMR1 was pulled down by TraDF-His, as indicated by a very weak band of TraMR1 in Fig. 5A, lane 1. We interpret this to mean that TraDF and TraMR1 interacted with each other and that a C-terminal His tag in TraD negatively influenced this interaction. This result again demonstrated that the C terminus of TraD is critical for the formation of the complex. Furthermore, we observed that the TraM-TraD interaction was dependent on the salt concentration. No binding was observed with a concentration of 150 mM NaCl (data not shown), but strong binding was seen with 20 mM NaCl. Neither 10 mM EDTA nor 1% Triton X-100 had an effect on the interaction (Fig. 5, lanes 5 and 7). In an additionally performed ELISA, interaction of His-TraDF but not of TraDF-His with MBP-TraM was observed (data not shown).
FIG. 5.
TraMR1 interacts with TraDF. Coimmunoprecipitation of TraMR1 with TraDF-His (lane 1) and His-TraDF (lanes 3, 5, and 7) was performed as described in Materials and Methods. (A) Detection of TraMR1. Precipitation was strong with His-TraDF but weak with TraDF-His. Triton X-100 (lane 7) and EDTA (lane 5) had no influence on this interaction. Purified TraMR1 was used as a positive control (lane 9). (B) Detection of His-TraDF and TraDF-His. TraD could be detected in lanes 1, 3, 5, and 7, showing that TraD variants were present in approximately equal amounts in all of the experiments. Lack of a signal in lanes 2, 4, 6, and 8 shows that neither TraD nor TraM interacted with the beads used in the experiment. α-His AB, anti-His antibody.
In vivo interaction between TraDR1 and TraMR1.
Since the in vitro interaction between TraD and TraM was well established, the next step was to investigate if such an interaction occurred in vivo. For this purpose we carried out conjugation assays. We reasoned that overexpression of TraD15 in donor cells containing the R1-16 plasmid expressing wild-type TraD and TraM would inhibit DNA transfer to recipient cells. In our working hypothesis, TraD15 would act as a decoy and block TraM-TraD interaction by binding to TraM. A reduced transfer frequency would be indicative for TraM-TraD interactions in vivo. The results of these experiments are shown in Table 2. When expression of TraD15 was induced, a decrease in the frequency of plasmid R1-16 transfer from the donor strain expressing the TraD15 protein was observed. In contrast, the conjugation frequency did not change when TraD15 expression was not induced. Induction of expression per se did not affect transfer frequencies (Table 2). In order to address the question of the specificity of the interaction between TraD and TraM, we overexpressed TraD15 in donor cells harboring IncW conjugative plasmid pSU2007, which is an R388 derivative. No difference in the conjugation frequency of pSU2007 was observed in strains overexpressing TraD15 compared to the vector control (Table 2). This shows that TraD15 does not interfere with pSU2007 mating but specifically inhibits R1-16 (IncF) conjugation.
TABLE 2.
Transfer rates and R17 phage infection
| Plasmid present in donor cells | Description | Transfer frequencya of R1-16 | Phage infection efficiencyb | Transfer frequencyc of pSU2007 |
|---|---|---|---|---|
| pGZ119EH | Vector | 8.5/7.3 | 1.5 × 104/1.4 × 104 | 0.3/0.13 |
| pGTD15 | TraD15 | 8.3/0.04 | 1.9 × 104/1.2 × 104 | 0.3/0.2 |
| None | 2.3 | 2.7 × 104 | 0.17 |
E. coli MC1061 harboring R1-16 as the conjugative plasmid was used as the donor strain, and E. coli J5 was used as the recipient. Transfer frequency is expressed as the number of transconjugants per donor cell (noninduced/induced).
E. coli MC1061 harboring R1-16 was used for the phage infection assay. Phage infection efficiency is expressed as PFU per milliliter (noninduced/induced).
E. coli MC1061 harboring pSU2007 as the conjugative plasmid was used as the donor strain, and E. coli J5 was used as the recipient. Transfer frequency is expressed as the number of transconjugants per donor cell (noninduced/induced).
In addition to the conjugation experiments, we performed phage infection assays with the R17 bacteriophage. The R17 RNA phage requires a functional pilus, as well as TraD, for successful infection, and we expected here that formation of the Mpf complex would not be disturbed by a TraM protein that is blocked by TraD15. Indeed, no differences in phage infection efficiency were observed in any of the strains tested (Table 2). Taken together, these results show that overexpression of TraD15 acts on DNA transfer but not phage infection, suggesting that a specific step in DNA transfer, i.e., the TraD-TraM interaction, is blocked in that case. Importantly, the results of the RNA phage experiments also demonstrate that overexpression of TraD15 did not exert a negative effect on tra gene expression or assembly of the Mpf complex.
DISCUSSION
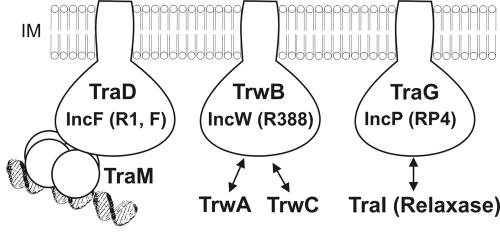
The results of in vitro experiments presented in this paper show that the major TraM interaction domain in TraD that specifically mediates interaction with TraM is constituted by 38 amino acids at the very C-terminal end of the protein. None of the TraD variants lacking these 38 residues interacted with TraM. However, we found that TraD11, comprising the complete cytoplasmic domain of TraD (amino acids 156 to 735), bound to TraM with higher affinity than did TraD15 (amino acids 698 to 735), indicating that the central part of the cytoplasmic domain contributes to the interaction with TraM. TraD11 also bound to TraM when this relaxosomal protein was bound to its target DNA sequence, sbmA from oriT of the R1 plasmid. In vivo studies corroborated the in vitro data and showed that when the 38 amino acids from the C terminus of TraD were expressed in addition to a functional conjugation system encoded by plasmid R1-16, conjugation was inhibited. Our interpretation of that inhibitory influence of TraD15 is that it acts as a molecular decoy that sequesters TraM molecules and prevents the interaction with the TraD CP, thereby reducing the possibility of successful DNA transfer. Therefore, we propose that a TraD-TraM interaction also takes place in vivo and is required for efficient cell-to-cell DNA transport. An important control here was the R17 phage infection assay. Since it is known that RNA bacteriophage R17 requires sex pili and a functional T4SS for infection of bacteria harboring plasmid R1-16 (3), we could use this assay to show that expression of TraD15 did not negatively affect the expression and assembly of the type IV secretion apparatus including sex pilus formation. It is known that RNA phages require a functional TraD protein for RNA penetration (see references cited in references 13 and 15); therefore, it is unlikely that TraD15 interferes with the function of TraD itself. Our observations are completely in line with the finding that TraD of F acts at the cell-to-cell contact stage during conjugation immediately before single-stranded DNA is transported (presumably by TraD itself) through the inner membrane of the donor cell (32). The dominant negative effect of TraD15 simply means that the physical contact between the relaxosome and the CP which is a part of the T4SS cannot be formed effectively. Such contacts have been shown to occur in other DNA transfer systems as well (Fig. 6), and these specific interactions have been proposed to be responsible for selecting the correct DNA substrate within a cell harboring a given conjugative plasmid. A role in substrate selection for the C-terminal amino acids of the TraD protein of F has been suggested earlier (37); in previously performed genetic experiments it was shown that removal of 37 amino acids from the C terminus of TraDF led to a 104-fold reduction in F-plasmid DNA transfer, whereas the ability to transfer mobilizable IncQ plasmid RSF1010 was increased 103-fold (37). Similarly, full complementation of a TraD mutant of the F plasmid was achieved with a wild-type or an N-terminally His-tagged TraD protein but not with a TraD protein carrying a His tag at the C terminus (38). Since the C-terminal amino acids that were investigated in this study are only present in IncF plasmids (Fig. 1B), our results fully support the view that this domain in TraD represents functional specialization of IncF plasmids and clearly distinguishes F-like conjugation systems from other conjugative plasmids like RP4 or R388. The observation that the 38 C-terminal amino acids of TraD did not interfere with the transfer of IncW conjugative plasmid pSU2007 supports the notion that the C-terminal 38 amino acids of TraD proteins of F-like plasmids contribute to the specificity of the interaction between the CP and the relaxosome within the F-plasmid group. The TraD-TraM interaction, which is specifically mediated by the C-terminal extension of TraD proteins of F-like plasmids, might supply F-like plasmids with a unique system for selection of its own DNA while excluding foreign DNA from the IncF plasmid T4SS (37).
FIG. 6.
CP-relaxosome interactions in conjugative plasmids. The inner membrane (IM)-anchored CPs TraD, TrwB, and TraG are depicted schematically. The interaction of each CP with a relaxosomal component is indicated.
How is the correct DNA substrate selected within the F-like group of plasmids? We observed that there is no exclusion at the level of the interaction between TraD and TraM, at least between plasmids F and R1. One piece of evidence for promiscuity at this level is the finding that TraDF interacts with TraMR1 in vitro. Furthermore, an F-plasmid mutant deficient in TraD can be fully complemented by TraDR1 (unpublished observations). Further evidence for the in vivo interaction between the transfer apparatus provided by plasmid F and TraM encoded by plasmid R1 was obtained in mobilization experiments. A plasmid carrying oriT and traM of plasmid R1 was mobilized with a high frequency by pOX38-Km, an F-plasmid derivative. The same plasmid with a TraM null mutation was not mobilized, suggesting that the cognate TraM protein was necessary for substrate recognition (21). Therefore, discrimination between different DNA substrates within closely related F-like plasmids occurs at the level of the interaction between TraM and the target DNA sequences at oriT (1, 9, 12, 41, 43). These target regions, termed sbm, are plasmid specific and show little sequence similarity (15). In the model that we propose here, substrate selection among F-like plasmids is provided by the sequence-specific DNA binding feature of the TraM protein (Fig. 6).
One important aspect of the TraM-TraD interaction that we have not been able to clarify in this work is what part of the TraM protein is the TraD binding domain. DNA binding, dimerization, and DNA sequence recognition by the 127-amino-acid-long TraM protein of R1 is conferred by the N-terminal half of the protein (21, 34), whereas the C terminus seems to be required for tetramerization (43), but neither the N-terminal nor the C-terminal half of TraMR1 alone bound to TraD. Since TraD also bound to TraM when it was in a complex with its DNA target sequence, we hypothesize that the TraD interaction domain in TraM must lie outside the N terminus. However, further experiments are required to define which part of TraM is minimally needed to confer binding to CP TraD.
Acknowledgments
We thank Erich Lanka for generously providing plasmids pSK410ch and pSK410nh and Ellen Zechner for providing the DNA sequence of R1 traD. We thank Laura Frost for critically reading the manuscript and for stimulating discussions.
This work was supported by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (grant P11844-Med) and by EU project QLK22-CT-2001-01200 within the fifth framework.
REFERENCES
- 1.Abo, T., S. Inamoto, and E. Ohtsubo. 1991. Specific DNA binding of the TraM protein to the oriT region of plasmid R100. J. Bacteriol. 173:6347-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balzer, D., W. Pansegrau, and E. Lanka. 1994. Essential motifs of relaxase (TraI) and TraG proteins involved in conjugative transfer of plasmid RP4. J. Bacteriol. 176:4285-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer, M., R. Eferl, G. Zellnig, K. Teferle, A. J. Dijkstra, G. Koraimann, and G. Högenauer. 1995. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J. Bacteriol. 177:4279-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabezon, E., J. I. Sastre, and F. de la Cruz. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254:400-406. [DOI] [PubMed] [Google Scholar]
- 5.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 6.Cascales, E., and P. J. Christie. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conover, G. M., I. Derre, J. P. Vogel, and R. R. Isberg. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305-321. [DOI] [PubMed] [Google Scholar]
- 9.Di Laurenzio, L., L. S. Frost, and W. Paranchych. 1992. The TraM protein of the conjugative plasmid F binds to the origin of transfer of the F and ColE1 plasmids. Mol. Microbiol. 6:2951-2959. [DOI] [PubMed] [Google Scholar]
- 10.Ding, Z., K. Atmakuri, and P. J. Christie. 2003. The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 11:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Disque-Kochem, C., and B. Dreiseikelmann. 1997. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J. Bacteriol. 179:6133-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fekete, R. A., and L. S. Frost. 2002. Characterizing the DNA contacts and cooperative binding of F plasmid TraM to its cognate sites at oriT. J. Biol. Chem. 277:16705-16711. [DOI] [PubMed] [Google Scholar]
- 13.Firth, N., K. Ippen-Ihler, and R. A. Skurray. 1996. Structure and function of the F factor and mechanism of conjugation, p. 2377-2401. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C. [Google Scholar]
- 14.Fischer, W., J. Puls, R. Buhrdorf, B. Gebert, S. Odenbreit, and R. Haas. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42:1337-1348. [DOI] [PubMed] [Google Scholar]
- 15.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 17.Gilmour, M. W., J. E. Gunton, T. D. Lawley, and D. E. Taylor. 2003. Interaction between the IncHI1 plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol. Microbiol. 49:105-116. [DOI] [PubMed] [Google Scholar]
- 18.Gomis-Rüth, F. X., and M. Coll. 2001. Structure of TrwB, a gatekeeper in bacterial conjugation. Int. J. Biochem. Cell Biol. 33:839-843. [DOI] [PubMed] [Google Scholar]
- 19.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koraimann, G., C. Koraimann, V. Koronakis, S. Schlager, and G. Högenauer. 1991. Repression and derepression of conjugation of plasmid R1 by wild-type and mutated finP antisense RNA. Mol. Microbiol. 5:77-87. [DOI] [PubMed] [Google Scholar]
- 21.Kupelwieser, G., M. Schwab, G. Hogenauer, G. Koraimann, and E. L. Zechner. 1998. Transfer protein TraM stimulates TraI-catalyzed cleavage of the transfer origin of plasmid R1 in vivo. J. Mol. Biol. 275:81-94. [DOI] [PubMed] [Google Scholar]
- 22.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 23.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 24.Lessl, M., D. Balzer, R. Lurz, V. L. Waters, D. G. Guiney, and E. Lanka. 1992. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J. Bacteriol. 174:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lessl, M., and E. Lanka. 1994. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell 77:321-324. [DOI] [PubMed] [Google Scholar]
- 26.Lin, T. S., and C. I. Kado. 1993. The virD4 gene is required for virulence while virD3 and orf5 are not required for virulence of Agrobacterium tumefaciens. Mol. Microbiol. 9:803-812. [DOI] [PubMed] [Google Scholar]
- 27.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 28.Llosa, M., S. Zunzunegui, and F. de la Cruz. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl. Acad. Sci. USA 100:10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez, E., and F. de la Cruz. 1988. Transposon Tn21 encodes a RecA-independent site-specific integration system. Mol. Gen. Genet. 211:320-325. [DOI] [PubMed] [Google Scholar]
- 30.Moncalian, G., E. Cabezon, I. Alkorta, M. Valle, F. Moro, J. M. Valpuesta, F. M. Goni, and F. de La Cruz. 1999. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J. Biol. Chem. 274:36117-36124. [DOI] [PubMed] [Google Scholar]
- 31.Normark, B. H., and S. Normark. 2002. Evolution and spread of antibiotic resistance. J. Intern. Med. 252:91-106. [DOI] [PubMed] [Google Scholar]
- 32.Panicker, M. M., and E. G. Minkley, Jr. 1985. DNA transfer occurs during a cell surface contact stage of F sex factor-mediated bacterial conjugation. J. Bacteriol. 162:584-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pansegrau, W., and E. Lanka. 1996. Enzymology of DNA transfer by conjugative mechanisms. Prog. Nucleic Acid Res. Mol. Biol. 54:197-251. [DOI] [PubMed] [Google Scholar]
- 34.Pölzleitner, E., E. L. Zechner, W. Renner, R. Fratte, B. Jauk, G. Högenauer, and G. Koraimann. 1997. TraM of plasmid R1 controls transfer gene expression as an integrated control element in a complex regulatory network. Mol. Microbiol. 25:495-507. [DOI] [PubMed] [Google Scholar]
- 35.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y.
- 37.Sastre, J. I., E. Cabezon, and F. de la Cruz. 1998. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J. Bacteriol. 180:6039-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schröder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schröder, G., and E. Lanka. 2003. TraG-like proteins of type IV secretion systems: functional dissection of the multiple activities of TraG (RP4) and TrwB (R388). J. Bacteriol. 185:4371-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulein, R., and C. Dehio. 2002. The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol. Microbiol. 46:1053-1067. [DOI] [PubMed] [Google Scholar]
- 41.Schwab, M., H. Gruber, and G. Högenauer. 1991. The TraM protein of plasmid R1 is a DNA-binding protein. Mol. Microbiol. 5:439-446. [DOI] [PubMed] [Google Scholar]
- 42.Stockner, T., C. Plugariu, G. Koraimann, G. Högenauer, W. Bermel, S. Prytulla, and H. Sterk. 2001. Solution structure of the DNA-binding domain of TraM. Biochemistry 40:3370-3377. [DOI] [PubMed] [Google Scholar]
- 43.Verdino, P., W. Keller, H. Strohmaier, K. Bischof, H. Lindner, and G. Koraimann. 1999. The essential transfer protein TraM binds to the DNA as a tetramer. J. Biol. Chem. 274:37421-37428. [DOI] [PubMed] [Google Scholar]
- 44.Waters, V. L. 1999. Conjugative transfer in the dissemination of beta-lactam and aminoglycoside resistance. Front. Biosci. 4:D433-D456. [DOI] [PubMed] [Google Scholar]
- 45.Zechner, E. L., F. de la Cruz, R. Eisenbrand, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 1999. Conjugative DNA transfer processes, p. 87-173. In C. M. Thomas (ed.), The horizontal gene pool: bacterial plasmids and gene spread. Harwood Academic Publishers, Amsterdam, The Netherlands.